Abstract
Thirty-six pyrazinamide-resistant and eight pyrazinamide-susceptible Mycobacterium tuberculosis isolates from Russia were analyzed for their pncA mutations. Thirty-one (86.1%) of the resistant isolates had a mutation either in pncA or upstream of the gene. Twenty of the 23 different mutations found in this study had not been described earlier. pncA genotype correlated well with pyrazinamidase activity and BACTEC 460 susceptibility test results.
Pyrazinamide (PZA) is the antituberculous drug of choice in modern short-course regimens with isoniazid, rifampin, and ethambutol (7). PZA appears to kill semidormant tubercle bacilli that persist in a low-pH environment and are unaffected by any other antituberculous drug (3, 9) and allows shortening of the treatment period from 12 to 18 months to 6 months when combined with isoniazid and rifampin. The exact mode of action of PZA still has to be determined, but it is thought that bacterial pyrazinamidase (PZase) converts PZA to toxic pyrazonoic acid, which mediates the direct killing effect (6, 13).
Since the global emergence of multidrug-resistant tuberculosis, the rates of resistance to PZA have also grown. PZA susceptibility testing of Mycobacterium tuberculosis is difficult because the drug is active in a relatively low-pH environment (10) and reliable susceptibility testing conditions are hard to establish for current cultivation methods (4). It has therefore been necessary to develop new methods for the detection of possible PZA resistance. In 1967, Konno et al. (6) showed a good correlation between loss of PZase activity and development of PZA resistance in M. tuberculosis strains. Although this phenotypic approach is useful in the determination of resistance, it is susceptible to errors in both execution of the test and interpretation of the results. Furthermore, the measurement of PZase requires time-consuming cultivation of M. tuberculosis.
The identification of M. tuberculosis pncA and its mutated forms in PZA-resistant isolates has been an important step in the development of genotypic methods for the rapid detection of PZA resistance (12). In some recent investigations, all of the PZA-susceptible M. tuberculosis isolates studied had a wild-type pncA sequence whereas most of the resistant isolates contained mutations in the gene itself or in the putative regulatory area upstream of it (5, 11, 12, 14).
We analyzed pncA and the upstream putative regulatory region sequences of 44 M. tuberculosis isolates from northwestern Russia, mainly St. Petersburg. The aims of the study were to compare phenotypic PZA susceptibility to the genotype and to provide more data on the geographic distribution of pncA mutations.
Forty-four M. tuberculosis clinical isolates were recovered from patients at the St. Petersburg Institute of Phthisiopulmonology from 1994 through 1997. Culture for M. tuberculosis was carried out on Löwenstein-Jensen solid medium, and susceptibility testing was performed by the BACTEC 460 radiometric method using BACTEC PZA test medium and a 100-μg/ml concentration of PZA (Becton Dickinson, Sparks, Md.). Qualitative PZase activity was assayed as described by Wayne (16).
Several loopfuls of bacterial colonies were transferred into 1.5-ml Eppendorf tubes and suspended in 200-μl aliquots of sterile 0.9% NaCl. Mycobacteria were lysed and DNA was released from them by heating in a 95°C water bath for 20 min. A 721-bp segment of the mycobacterial genome was amplified by using forward primer PP0 (5′-GCTGGTCATGTTCGCGATCG) and reverse primer PP6 (5′-GCTTTGCGGCGAGCGCTCCA), which originated 104 bp upstream and 56 bp downstream of the pncA reading frame, respectively (1). The primers were also synthesized as 5′-end biotinylated versions (Eurogentec Inc., Seraing, Belgium). The PCR conditions used have been described previously (8).
The DNA sequence was determined by using the Thermo Sequenase dye terminator cycle sequencing kit (Amersham International, plc., Little Chalfont, United Kingdom) and the ABI 373 DNA Sequencer (Applied Biosystems, Inc., Foster City, Calif.). In addition to the above-described primers, other sequencing primers (available on request) were also used for the sequencing of both strands. The data were assembled and edited by using SeqEd version 1.0.3 software (Applied Biosystems), and the sequences were compared with the published sequence of pncA in GenBank (12).
The IS6110 restriction fragment length polymorphism (RFLP) test was carried out in accordance with standard recommendations (15), and the data were analyzed by means of the GelCompar version 4.2 program (Applied Maths, Inc., Gent, Belgium) using the unweighted pair-group method of arithmetic averaging with the Dice coefficient and 1% position tolerance settings for cluster analysis.
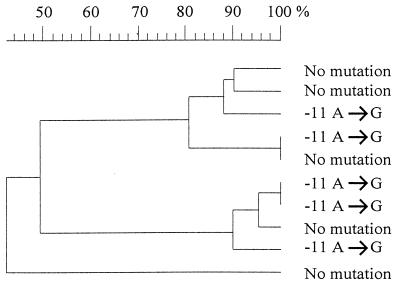
Thirty-one (86.1%) of the 36 PZA-resistant M. tuberculosis isolates had mutations in the part of the genome studied. pncA itself was mutated in 25 isolates, and 6 isolates showed a mutation in the region upstream of the gene (Table 1). Five resistant isolates and all of the susceptible isolates had wild-type pncA. A total of 23 different mutations were detected between pncA residues −12 (putative regulatory area) and 481. The most frequently encountered change was the A-to-G point mutation of residue −11 (n = 5), and this mutation has also been detected by Sreevatsan et al. (14). The RFLP test showed that these five isolates represented at least three different clones; the highest level of homology between the clones was 81% (Fig. 1). Ala102Val, Gln141Pro, and Ala161Pro amino acid shifts were found in two isolates each, and the isolates with the same mutations had identical IS6110 RFLP patterns (58 to 79% homology between the pairs). A Thr160Pro change was detected in two isolates with 44% difference between their RFLP patterns.
TABLE 1.
pncA mutations in PZA-resistant M. tuberculosis isolates from northwestern Russia
| pncA nucleotide(s) | Mutation | Amino acid change | No. of isolates |
|---|---|---|---|
| −12a | T→G | Putative regulatory area mutation | 1 |
| −11 | A→G | Putative regulatory area mutation | 5 |
| 20 | T→G | Val7Gly | 1 |
| 22a | G→T | Asp8Tyr | 1 |
| 28a | C→A | Gln10Lys | 1 |
| 28a | C→T | Gln10Stop | 1 |
| 84a | C deletion | Reading frame change | 1 |
| 139a | A→T | Thr47Ser | 1 |
| 146a | A→T | Asp49Val | 1 |
| 146a | A→G | Asp49Gly | 1 |
| 196a | T→C | Ser66Pro | 1 |
| 286a | A→G | Lys96Glu | 1 |
| 304a | G→A | Ala102Thr | 1 |
| 305a | C→T | Ala102Val | 2 |
| 309a | C→G | Tyr103Stop | 1 |
| 347a | T→G | Leu116Arg | 1 |
| 374a | T→A | Val125Asp | 1 |
| 379–389a | 11-nucleotide deletion | Reading frame change | 1 |
| 398a | T→C | Ile133Thr | 1 |
| 422 | A→C | Gln141Pro | 2 |
| 460a | A→G | Arg154Gly | 1 |
| 478a | A→C | Thr160Pro | 2 |
| 481a | G→C | Ala161Pro | 2 |
New pncA mutation identified in this study. Five PZA-resistant isolates had wild-type pncA.
FIG. 1.
GelCompar dendrogram showing the interrelationships of PZA-resistant M. tuberculosis isolates with either unmutated pncA and its putative regulatory region or a −11 A-to-G mutation in the putative pncA-regulatory region.
Twenty of the 23 pncA mutations found in this study have not been described previously (5, 11, 12, 14). The mutations were dispersed throughout pncA, except for a stretch of 80 nucleotides near the 3′ end of the gene, where no mutations were detected. Five of the PZA-resistant, PZase-negative isolates showed no mutations in the sequenced part of the genome, which is in agreement with the earlier observations that pncA is mutated in 72 to 97% of PZA-resistant M. tuberculosis strains (5, 11, 12, 14). The five isolates without pncA mutations were retested with 300- and 900-μg/ml concentrations of PZA, and all had the resistance phenotype. Subsequent IS6110 analysis showed that the isolates had different RFLP patterns (Fig. 1). The findings suggest that there are other mechanisms of PZA resistance, possibly mutations in an unidentified promoter region located further upstream of pncA or in its regulatory gene.
PZase activity was in full agreement with PZA susceptibility. All susceptible M. tuberculosis isolates produced the enzyme, whereas all resistant isolates were PZase negative. Thus, qualitative measurement of PZase activity remains a cheap and useful method for the rapid screening of PZA resistance of M. tuberculosis. The radiometric susceptibility test results were also in line with the PZase assay and sequencing results; the only discrepant findings were two clonal isolates that were PZase negative and had an Ala102Val amino acid shift in PncA but were susceptible by the BACTEC method. Nevertheless, repeated testing showed resistance to PZA, in accordance with the PZase test and the pncA genotype.
The fact that most PZA-resistant M. tuberculosis isolates have a mutated pncA gene is promising for the development of tools for the rapid detection of PZA resistance. Scorpio et al. (11) have already successfully used the PCR single-strand conformation polymorphism technique for this purpose. As new, powerful methods for the simultaneous determination of multiple drug resistance mutations will soon be available (2), continued analysis of the pncA sequences of PZA-resistant isolates is warranted. In this way, a comprehensive mutation library can be established for exploitation by these methods.
Acknowledgments
We thank M.-L. Helin, M. Kirjonen, E. Lönnblad, P. Sinkkonen, and U. Toivonen for excellent technical assistance.
The study was supported by the Finnish National Research and Development Center for Welfare and the Health/HEDEC infectious diseases project in St. Petersburg, the Finnish Anti-Tuberculosis Association Foundation, and the Sigfrid Juselius Foundation.
REFERENCES
- 1.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 3.Heifets L, Lindholm-Levy P. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am Rev Respir Dis. 1992;145:1223–1225. doi: 10.1164/ajrccm/145.5.1223. [DOI] [PubMed] [Google Scholar]
- 4.Hewlett D, Jr, Horn D L, Alfalla C. Drug-resistant tuberculosis: inconsistent results of pyrazinamide susceptibility testing. JAMA. 1995;273:916–917. [PubMed] [Google Scholar]
- 5.Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78:117–122. doi: 10.1016/s0962-8479(98)80004-x. [DOI] [PubMed] [Google Scholar]
- 6.Konno K, Feldmann F M, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 7.Maher D, Chaulet P, Spinaci S, Harries A. Treatment of tuberculosis: guidelines for national programmes. 2nd ed. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 8.Marttila H J, Soini H, Eerola E, Vyshnevskaya E, Vyshnevskiy B I, Otten T F, Vasilyef A V, Viljanen M K. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob Agents Chemother. 1998;42:2443–2445. doi: 10.1128/aac.42.9.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCune R M, Tompsett R, McDermott W. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by administration of pyrazinamide and a companion drug. J Exp Med. 1956;104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDermott W, Tompsett R. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am Rev Tuberc. 1954;70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 11.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 13.Speirs R J, Welch J T, Cynamon M H. Activity of n-propyl pyrazinoate against pyrazinamide-resistant Mycobacterium tuberculosis: investigations into mechanism of action of and mechanism of resistance to pyrazinamide. Antimicrob Agents Chemother. 1995;39:1269–1271. doi: 10.1128/aac.39.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]