Abstract
The aac(2′)-Ia gene in Providencia stuartii encodes a 2′-N-acetyltransferase capable of acetylating both peptidoglycan and certain aminoglycoside antibiotics. Regulation of the aac(2′)-Ia gene is influenced in a positive manner by the product of the aarP gene, which encodes a small transcriptional activator of the AraC (XylS) family. In this study, we demonstrate the sequence requirements at the aac(2′)-Ia promoter for AarP binding and activation.
Providencia stuartii is a gram-negative pathogen that is a leading cause of urinary-tract infections in patients undergoing chronic catheterization (16, 22, 28, 29). Eradication of P. stuartii infections can be extremely difficult due to the multiple antibiotic resistances that are often present in this bacterium (11, 15). An intrinsic, chromosomal acetyltransferase [AAC(2′)-Ia] is present in P. stuartii and confers high-level aminoglycoside resistance when overexpressed (4, 24, 25, 31). In addition, this enzyme serves a housekeeping role in the O acetylation of peptidoglycan (21). Intrinsic chromosomal aminoglycoside acetyltransferases in other bacteria have been described previously, and a summary of these enzymes has been reported (26).
A central activator of aac(2′)-Ia gene is AarP, a transcriptional activator similar to the MarA, SoxS, and Rob proteins (14). The MarA, SoxS, and Rob proteins have been shown to activate an intrinsic multiple-antibiotic resistance phenotype (Mar) and a defense regulon against superoxide-generating agents in Escherichia coli (1, 2, 5, 9, 10, 12, 13, 18–20, 30). In addition to activating the aac(2′)-Ia gene, increased expression of AarP activates a Mar phenotype in both P. stuartii and E. coli (14). Presumably each of these proteins is capable of activating a common set of genes due to the extensive homology in their helix-turn-helix DNA binding motif (9).
Prior to this study, it was not clear whether AarP influenced expression of aac(2′)-Ia by a direct or indirect mechanism. In this report, the aac(2′)-Ia sequences required for the in vivo activation by AarP are identified. DNA binding studies using purified AarP protein suggest that activation of aac(2′)-Ia by AarP is mediated by an interaction with the aac(2′)-Ia promoter region.
Bacterial strains and growth conditions.
Escherichia coli DH5α (Gibco/BRL, Gaithersberg, Md.) and XL1-Blue (Stratagene, La Jolla, Calif.) were used as hosts for plasmids. P. stuartii PR50 has been described previously (24). Luria-Bertani broth was used for the growth of cells, and the following antibiotics were used at the indicated concentrations for selections: ampicillin, 150 μg/ml for E. coli and 300 μg/ml for P. stuartii; chloramphenicol, 100 μg/ml for P. stuartii.
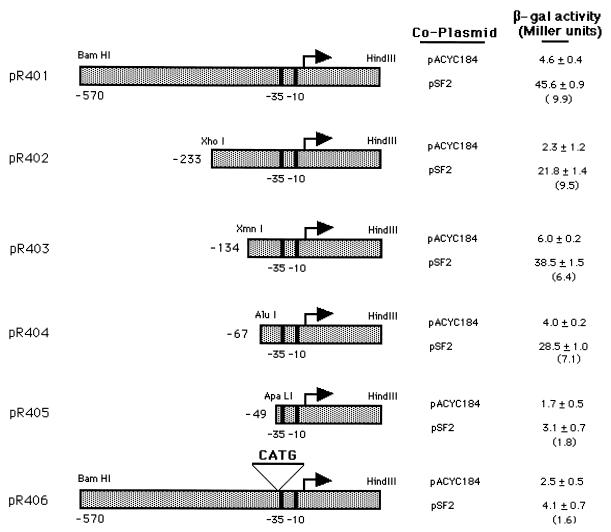
Upstream aac(2′)-Ia sequences required for activation by AarP.
We previously reported that the presence of aarP on a multicopy pACYC184 derivative (pSF2) was capable of 10-fold activation of an aac(2′)-lacZ fusion contained on a separate compatible plasmid, pR401 (14). To determine the sequences required for this activation, a series of progressive 5′ deletions of the full-length aac(2′)-Ia promoter were constructed. As shown in Fig. 1, plasmid pR402 contains a 456-bp XhoI-HindIII fragment inserted into pQF50 digested with SalI and HindIII. pR403 contains a 357-bp XmnI-HindIII fragment cloned into pQF50 digested with SmaI and HindIII. pR404 contains a 292-bp AluI-HindIII fragment cloned into pQF50 digested with SmaI and HindIII. To construct pR405, plasmid pBC.KS-aac(2′) was first digested with ApaLI followed by Klenow treatment to create blunt ends. This was followed by digestion with HindIII, which released a 273-bp fragment that was cloned into pQF50 digested with SmaI and HindIII. Plasmid pR406 was constructed in two steps. First the aac(2′)-Ia promoter region from pR401 was cloned into pACYC184 as a BamHI-HindIII fragment. This plasmid was then linearized with ApaLI, which cuts at position −49 in the aac(2′)-Ia promoter region end filled with Klenow treatment in the presence of deoxynucleoside triphosphates. Religation then created a 4-bp insertion of CATG at position −47. This promoter fragment was then recloned into pQF50 as a BamHI-HindIII fragment to create pR406. Each of these derivatives was electroporated into PR50, and then either pSF2 or pACYC184 was introduced into each strain. The ability of AarP to activate each promoter derivative was determined by analysis of β-galactosidase accumulation in sodium dodecyl sulfate (SDS)-chloroform-treated cells by the method of Miller (17). Plasmid pR401 was activated 9- to 10-fold in the presence of pSF2 (Fig. 1). Deletion derivatives extending to −233 (pR402), −134 (pR403), or −67 (pR404) relative to the start of transcription still supported a seven- to ninefold activation by pSF2. However, pR405 containing a deletion extending to an ApaI site at −49 failed to support activation by AarP. This ApaI site was centered within a region of dyad symmetry that represented a potential sequence required for activation by AarP. To examine this possibility, pR406 containing a 4-base insertion of CATG at −47 was assayed for activation. The presence of this insertion in the context of the full-length promoter resulted in a severe reduction in the activation conferred by pSF2, 1.5-fold compared to the 10-fold increase seen in pR401.
FIG. 1.
Deletion analysis of aac(2′)-Ia sequences required for AarP-mediated activation. A series of 5′ deletion derivatives of the aac(2′)-Ia promoter were constructed and fused to the lacZ gene in pQF50 (7). P. stuartii PR50 containing each deletion derivative fused to lacZ was then transformed with a compatible plasmid containing aarP (pSF2) or with a pACYC184 control. The ability of each aac(2′)-Ia promoter derivative to support activation by AarP was determined by analysis of β-galactosidase (β-gal) accumulation according to the method of Miller (17). A representative experiment is shown, with the values in parentheses representing the fold induction conferred by the presence of pSF2 containing the aarP gene, relative to pACYC184. Standard deviations are also given, and repeat experiments gave similar induction values to those shown.
Purification of AarP.
To determine if the observed activation by AarP was mediated by a direct interaction with the aac(2′)-Ia promoter region, a His6-AarP protein was constructed by PCR amplification with primers to introduce the His6 tag at the amino terminus. DNA sequence analysis confirmed that no mutations were introduced from the PCR amplification. The His6-AarP fusion protein was shown to function in vivo as indicated by the 48-fold activation of an aac(2′)-lacZ fusion observed in P. stuartii when His6-AarP was introduced (data not shown). To express the fusion protein via the T7 RNA polymerase, an EcoRI-SalI fragment containing the His6-aarP construct and the ribosomal binding site from pDS56/RBSII (2a) was cloned into pET24(+) (Novagen) digested with the same enzymes to create pET24.His(6)-aarP.
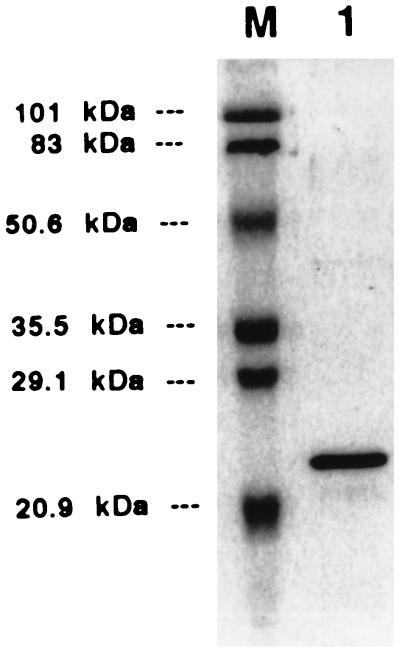
The His6-AarP protein was purified by affinity chromatography using Talon resin (Clontech, Palo Alto, Calif.). Initial attempts to overexpress and purify AarP from E. coli BL21 (DE3) gave reproducibly poor yields. Examination of codon usage for AarP revealed a number of rare codons for E. coli; therefore, purification of AarP was subsequently done in P. stuartii PR50 harboring pET24.His(6)-aarP. To introduce the T7 RNA polymerase gene into P. stuartii, a 4.4-kb BamHI fragment from pAR1219 (6) containing both gene I from phage T7 driven by the lac UV5 promoter and the E. coli lacI gene was cloned into pACYC184 (3) linearized with BamHI to create p184.T7. Cells were grown at 37°C in 400 ml of Luria-Bertani broth to an optical density at 600 nm of ∼0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 2 mM and cultures were returned to 37°C for an additional 3 h. Cells were disrupted by passing twice through a French pressure cell at 10,000 lb/in2, and cellular debris was removed by centrifuging twice at 35,000 × g for 30 min at 4°C. The His6-AarP protein was purified with Talon affinity resin by a batch purification method as described by the manufacturer (Clontech). Protein concentration was determined by the method of Bradford. The purified His6-AarP fusion protein was visualized on SDS–12% polyacrylamide gels after Coomassie blue staining and was greater than 90% pure as shown in Fig. 2.
FIG. 2.
Purification of AarP protein. A His6-AarP fusion protein was purified from an induced culture of P. stuartii PR50 as described in Materials and Methods. The purity of the final preparation was examined on an SDS–12% polyacrylamide gel stained with Coomassie blue. Prestained molecular mass standards (Bio-Rad) are in lane M, and the estimated molecular masses are indicated.
Binding of AarP to the aac(2′)-Ia promoter.
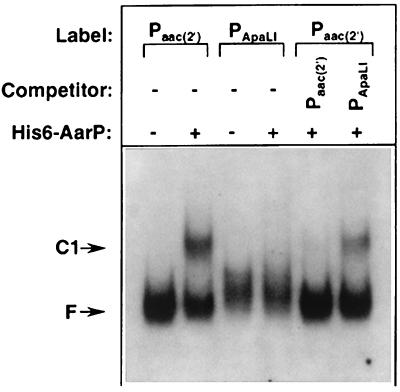
Mobility gel shift analysis (8) was performed with the purified His6-AarP protein and fragments of the aac(2′)-Ia promoter region generated by PCR amplification and end labeled with digoxigenin-11-2′-deoxy-uridine-5′-triphosphate followed by gel purification.
DNA binding reactions contained 50 mM KCl, 10 mM Tris (pH 8.0), 2 mM dithiothreitol, bovine serum albumin (0.4 mg/ml), 10% (vol/vol) glycerol, and 4 fmol of labeled DNA fragment and purified His6-AarP protein as described in the text. Binding reaction mixtures were incubated for 20 min at room temperature and then loaded onto 5% polyacrylamide gels. Gels were electrophoresed at 200 V, washed twice for 20 min in 1× Tris-borate-EDTA buffer, and transferred to nylon membranes with a Bio-Rad (Hercules, Calif.) Semi-Dry blotting system. Filters were developed with LumiPhos 530 (Boehringer Mannheim, Indianapolis, Ind.) and exposed to X-ray film.
The ability of AarP to bind the aac(2′)-Ia promoter is demonstrated by the presence of a DNA fragment with decreased mobility in the presence of AarP (Fig. 3) relative to that observed when no AarP protein is present (Fig. 3). In the presence of a DNA fragment containing the 4-base insertion centered at position −47, there was no observable binding by AarP, a result in agreement with the in vivo analysis (Fig. 1). Control experiments using an excess of unlabeled specific and nonspecific competitor DNAs indicated that the observed binding was specific for the aac(2′)-Ia promoter (Fig. 3).
FIG. 3.
Binding of purified His6-AarP to aac(2′)-Ia promoter sequences. Binding reactions were set up, subjected to electrophoresis on polyacrylamide gels, and visualized as described in the text. Approximately 2.5 fmol of labeled wild-type aac(2′)-Ia promoter fragment [Paac(2′)] or the mutant promoter with a 4-bp insertion at −47 (PApaLI) was included in the reaction mixtures. Purified His6-AarP (20 pmol) and unlabeled competitor DNA fragments (1.25 pmol) were added to the binding reactions where indicated. The designation F denotes free unbound fragment, and C1 indicates an AarP complex with the aac(2′)-Ia promoter.
Concluding remarks.
The results of this study indicate that the AarP protein directly interacts with the aac(2′)-Ia promoter region to mediate transcriptional activation. The in vivo activation of deletion derivatives of the aac(2′)-Ia promoter and a 4-bp insertional mutation have indicated that bases at position −47 are important for activation. The purified AarP protein has been shown to bind the wild-type aac(2′)-Ia promoter region but was unable to bind a mutant derivative containing a 4-bp insertion at position −47. The sequences at position −47 include a GCA motif centered at position −47, which may be equivalent to the GCAY motif proposed to be important in SoxS binding (13a).
The AarP protein is a member of the AraC (XylS) family (23) and like the SoxS and MarA proteins is missing the portion involved in ligand binding (5, 9, 23, 30). Therefore, like that of SoxS and MarA, the regulation of gene expression mediated by AarP appears to be the result of changes in expression (5, 9, 23, 30). Studies will now focus on the mechanisms that regulate aarP expression and result in the subsequent activation of aac(2′)-Ia and genes involved in the multiple-antibiotic resistance phenotype in P. stuartii.
Acknowledgments
We are grateful to S. F. J. LeGrice for plasmid pDS56.
This work was supported by a Merit Review award from the Department of Veterans Affairs.
REFERENCES
- 1.Amabile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Certa U, Bannwarth W, Stüber D, Gentz R, Lanzer M, LeGrice S, Guillot F, Wendler I, Hunsmann G, Bujard H, Mous J. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 1986;5:3051–3056. doi: 10.1002/j.1460-2075.1986.tb04605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A C Y, Cohen S N. Construction and characterization of amplifiable DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevereau M, Daniels P J L, Davies J, LeGoffic F. Aminoglycoside resistance in bacteria mediated by gentamicin acetyltransferase II, an enzyme modifying the 2′ amino group of aminoglycoside antibiotics. Biochemistry. 1974;13:598–603. doi: 10.1021/bi00700a030. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S P, Hächler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davanloo P, Rosenberg A H, Dunn J J, Studier F W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farinha M, Kropinski A M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambino L, Gracheck S J, Miller P F. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1993;175:2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hächler H, Cohen S P, Levy S B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkey P M. Providencia stuartii: a review of a multiply antibiotic resistant organism. J Antimicrob Chemother. 1984;13:209–226. doi: 10.1093/jac/13.3.209. [DOI] [PubMed] [Google Scholar]
- 12.Jair K-W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E., Jr Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic resistance and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jair K-W, Yu X, Skarstad K, Thöny B, Fujita N, Ishihama A, Wolf R E., Jr Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Li Z, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 14.Macinga D R, Parojcic M M, Rather P N. Identification and analysis of aarP, a transcriptional activator of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1995;177:3407–3413. doi: 10.1128/jb.177.12.3407-3413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHale P J, Keane C, Dougan G. Antibiotic resistance in Providencia stuartii isolated in hospitals. J Clin Microbiol. 1981;13:1099–1104. doi: 10.1128/jcm.13.6.1099-1104.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHale P J, Walker F, Scully B, English L, Keane C T. Providencia stuartii infections: a review of 117 cases over an eight year period. J Hosp Infect. 1981;2:155–165. doi: 10.1016/0195-6701(81)90024-4. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Miller P F, Gambino L, Sulavik M C, Gracheck S J. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994;38:1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 20.Nunoshiba T, Hidalgo E, Amábile Cuevas C F, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payie K G, Rather P N, Clarke A J. Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptidoglycan in Providencia stuartii. J Bacteriol. 1995;177:4303–4310. doi: 10.1128/jb.177.15.4303-4310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penner J L, Hinton N A, Hamilton L J, Hennessey J N. Three episodes of nocosomial urinary tract infections caused by one O-serotype of Providencia stuartii. J Urol. 1981;125:668–671. doi: 10.1016/s0022-5347(17)55157-5. [DOI] [PubMed] [Google Scholar]
- 23.Ramos J L, Rojo F, Zhou L, Timmis K N. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 1990;18:2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rather P N, Orosz E, Hare R, Miller G, Shaw K J. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J Bacteriol. 1993;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rather P N, Orosz E. Characterization of aarA, a pleiotrophic negative regulator of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1994;176:5140–5144. doi: 10.1128/jb.176.16.5140-5144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rather P N. Origins of the aminoglycoside modifying enzymes. Drug Resist Updates. 1998;1:258–291. doi: 10.1016/s1368-7646(98)80044-7. [DOI] [PubMed] [Google Scholar]
- 27.Skarstad, K., B. Thöny, D. Hwang, and A. Kornberg. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268:5365–5370. [PubMed]
- 28.Warren J W. A prospective microbiological study of antibiotic resistant bacteriuria in patients with chronic indwelling urethral catheters. J Infect Dis. 1982;146:719–723. doi: 10.1093/infdis/146.6.719. [DOI] [PubMed] [Google Scholar]
- 29.Warren J W. Providencia stuartii: a common cause of antibiotic-resistant bacteriuria in patients with long-term indwelling catheters. Rev Infect Dis. 1986;8:61–67. doi: 10.1093/clinids/8.1.61. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Wiess B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi M, Mitsuhashi S, Kobayashi F, Zenda H. A 2′-N-acetylating enzyme of aminoglycosides. J Antibiot. 1974;27:507–515. doi: 10.7164/antibiotics.27.507. [DOI] [PubMed] [Google Scholar]