Abstract
A PCR-based reverse hybridization system (research prototype kit INNO-LiPA for H. pylori resistance) was developed and evaluated for simultaneous detection of 23S ribosomal DNA point mutations, associated with macrolide resistance in Helicobacter pylori. Fifty-seven H. pylori strains (51 natural, 6 laboratory-derived artificial, 52 resistant, and 5 susceptible strains) were tested by PCR-LiPA (detecting mutations A2115→G, G2141→A, A2142→G, A2142→C, A2143→G, A2143→C, and A2143→T), DNA sequencing, restriction fragment length polymorphism, and/or hybridization to oligonucleotide probes. Results were highly concordant, but PCR-LiPA appears to be more sensitive for the simultaneous detection of multiple mutants.
Helicobacter pylori is a gram-negative bacterium that colonizes the human stomach. Infection with H. pylori is associated with gastritis and peptic ulcer disease and may eventually result in the development of atrophic gastritis and gastric cancer (1, 7).
Infection with H. pylori can be effectively treated by a combination of proton pump inhibitors and/or H2 receptor antagonists and antibiotics. Metronidazole, amoxicillin, clarithromycin, and tetracycline are frequently included in the triple or quadruple treatment regimens used to eradicate H. pylori (5, 18). Resistance to antimicrobial agents is an important factor for the clinical outcome of anti-Helicobacter treatment. Resistance to metronidazole is observed in 10 to 50% of the cases in developed countries but can be as high as 90% in developing countries (12). Resistance to macrolides also is of importance and was found in less than 2% of the strains in The Netherlands (19) but in more than 10% of those from France as well as some other countries (2, 6, 12, 14). The prevalence of resistant strains appears to be increasing (6, 8, 11, 17).
The major cause of macrolide resistance in H. pylori is the lack of binding of the macrolides to the 23S rRNA components of the bacterial ribosome due to a modification of the target site by methylation or point mutations in the peptidyltransferase region of domain V of the 23S rRNA (21). H. pylori contains two copies of the 23S ribosomal DNA (rDNA) gene, and at least five distinct point mutations have been reported that are associated with macrolide resistance. Versalovic et al. (20) found A→G transitions at two positions (A2142→G and A2143→G). Also, an A→C transversion (A2142→C) was found to be related to resistance (15). Recently, Hultén et al. described associated mutations at two additional positions (G2115→A and G2141→A) (10). The conventional method to determine the antibiotic resistance of H. pylori is based on analysis of cultured strains by agar diffusion or the E test (12, 13), which is time-consuming and requires specific expertise. Therefore, DNA-based diagnostic methods may offer a rapid and reliable alternative approach for macrolide susceptibility testing. The present study describes a convenient method, based on PCR and reverse hybridization, for the detection of the relevant mutations in the 23S rDNA.
Bacterial strains.
A total of 57 H. pylori strains were tested for 23S rRNA mutations; 29 were obtained from Amsterdam, The Netherlands, and 28 were obtained from Bordeaux, France. All strains from Bordeaux and most strains from Amsterdam were freshly cultured from gastric biopsies. Six strains contained artificial mutations of the 23S rRNA gene (3). In addition, Helicobacter reference strains were obtained from the collection of the Laboratory of Microbiology at the University of Ghent (LMG): H. pylori (LMG 4654, 5166, 6787, 7162, 6307, 3670, 4046, 6301, and 6222), Helicobacter fennelliae (LMG 13306 and 11759), Helicobacter nemestrinae (LMG 14378), Helicobacter mustelae (LMG 8776 and 8777), Helicobacter cinaedi (LMG 9072, 7543, and 8558), Helicobacter canis (LMG 12640), Helicobacter acinonyx (LMG 12684), and Helicobacter pametensis (LMG 12678, 12637, 12681, and 12682). Also, the following non-Helicobacter reference strains were used: Arcobacter butzleri (LMG 11118), Bacillus cereus (ATCC 11778), Bacillus subtilis (ATCC 6633), Clostridium perfringens (ATCC 12916), Escherichia coli (ATCC 11775), Klebsiella pneumoniae (ATCC 13883), Listeria monocytogenes (NCTC 10527), Proteus mirabilis (ATCC 29906), Pseudomonas aeruginosa (ATCC 27853), Salmonella typhimurium (ATCC 29946), Salmonella enteritidis (ATCC 13076), and Wolinella succinogenes (LMG 7608). Finally, clinical isolates of Campylobacter jejuni, Campylobacter coli, Campylobacter lari, Campylobacter upsaliensis, Enterobacter agglomerans, Enterobacter cloacae, Haemophilus influenzae, Shigella dysenteriae, Shigella flexneri, Streptococcus faecalis, Vibrio cholerae, and Yersinia enterocolitica were tested.
Bacteria were cultured on Trypticase-soy agar plates containing 5% sheep blood (Becton Dickinson) for 3 to 5 days at 37°C under microaerobic conditions (5% O2, 10% CO2, and 80% N2). The susceptibility to clarithromycin of each strain and MICs for each strain were determined with the E test (AB Biodisk, Solwa, Sweden) (4, 9, 13) or by the standard agar dilution technique, as described previously (12, 15).
DNA isolation from cultured strains.
The H. pylori cells were harvested from the plate by suspension in 2 ml of sterile 0.9% NaCl solution and pelleted by centrifugation at 10,000 × g for 2 min. Cells were resuspended in 400 μl of a solution containing 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.1% sodium dodecyl sulfate, and 0.1 mg of proteinase K per ml and incubated for 2 to 4 h at 55°C. Proteinase K was inactivated by incubation at 95°C for 10 min. The lysates were diluted 1/100 in sterile water and directly used for PCR.
Analysis of 23S rRNA mutations by RFLP and/or direct sequencing.
Mutations A2142G and A2143G were analyzed by restriction fragment length polymorphism (RFLP) with restriction enzyme AvaII, BsaI, MboII, or BbsI as described earlier or by probe hybridization in liquid phase (15, 16). 23S rRNA sequences were determined by direct sequencing of PCR products (3, 4).
PCR-LiPA.
A fragment of the 23S rRNA gene was amplified by PCR. PCRs were performed in a volume of 50 μl, containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 200 μM concentrations of deoxynucleoside triphosphates, 0.25 U of AmpliTaq Gold, and 25 pmol of biotinylated PCR primers. Reaction mixtures were covered with mineral oil, and PCR was performed in a BioMed-60 thermocycler, under the following conditions: 9 min of preincubation at 94°C, followed by 40 cycles of 30 s at 95°C, 45 s at 62°C, and 45 s at 72°C. A final extension was performed for 5 min at 72°C.
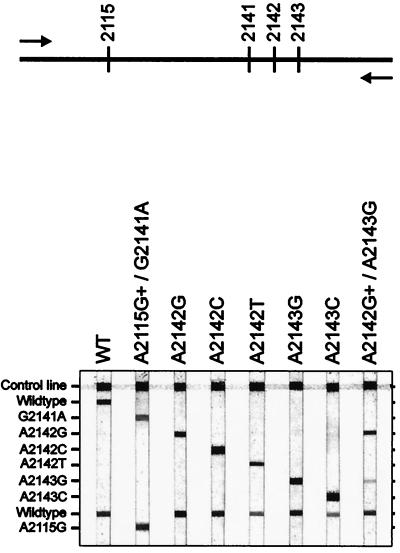
PCR products were analyzed by reverse hybridization in a reverse hybridization line probe assay (LiPA). This assay is based on hybridization to a number of oligonucleotide probes, immobilized as parallel lines on a nitrocellulose strip (Fig. 1). For each strain, 10 μl of the PCR product was analyzed on the LiPA strip, essentially as described for the LiPA detection of vacA and cagA genotypes (18a).
FIG. 1.
Outline and representative examples of the LiPA strips. The positions of probes on the strip are shown. Since we did not have access to strains that contained the A2115→G and G2141→A mutations, the specificities of the corresponding probes were determined with biotin-labelled oligonucleotides, complementary to these probes on the LiPA. WT, wild type.
Analysis of 23S rDNA mutations in H. pylori strains by PCR-LiPA.
PCR primers for the 23S rDNA of H. pylori were tested on chromosomal DNA from multiple bacterial species. PCR products of the expected size were obtained only from H. pylori, H. nemestrinae, and H. acinonyx. Specific probes were developed for reverse hybridization analysis in a line probe assay, covering the relevant positions of the 23S rDNA. The PCR fragments from H. nemestrinae and H. acinonyx showed hybridization only to the wild-type probes.
To evaluate the performance of the PCR-LiPA, a total of 57 H. pylori strains were analyzed. Of these, 51 were clinical isolates, directly derived from patients, and 6 were laboratory-derived artificial mutant H. pylori strains (3). Of the 57 strains, 52 were resistant, whereas 5 strains were susceptible (MIC < 2). Sequences of the 23S rRNA gene also were analyzed by PCR followed by a combination of direct sequencing, RFLP, and/or probe hybridization as described (3, 4, 15, 16). All results for the individual strains are shown in Table 1 and are summarized in Table 2.
TABLE 1.
Characterization of strains used in the present study
| Strain(s)a | Clarithromycin MIC (μg/ml) | 23S mutation(s) detected by:
|
|
|---|---|---|---|
| Conventional methodsb | PCR-LiPA | ||
| C3II | >256 | WTc | WT |
| 1714, 1309, 1462, 11496 | <2 | WT | WT |
| 1578–96, H2, 71, 35888, 1759, 11, 1710, 10, 1307, 1403, 1295, 2595, 2577, AB G1-2 | >256 | A2142G | A2142G |
| 1066–97, 7, 8, AB1125, CB1159, HS1146 | 4–128 | A2142G | A2142G |
| 1467, AB C1-5 | >256 | A2142C | A2142C |
| AB T1-2 | 16 | A2142T | A2142T |
| 82, 1190, 1213, 1289, 1315, 1377, 1388, 1392, 1394, 1401, 1553, MOR, FERA | >256 | A2143G | A2143G |
| 4, 15, 333, 34319, MARJE, MIS, AB G2-3 | 4–64 | A2143G | A2143G |
| AB C2-5 | 16 | A2143C | A2143C |
| 1538 | >256 | WT + A2143G | WT + A2143G |
| 1470 | >256 | A2142G | A2142G + A2143G + WT |
| LINAN 1567 | >128 | A2142C + A2143G + WT | A2142C + A2143G + WT |
| PAP | 16 | WT + A2142G | WT + A2142G |
| 140–97 | <2–>256 | WT | WT + A2142G + A2143G |
| 1264 | >256 | A2142G | A2142G + A2143G |
| 1075 | >256 | A2143G | A2142G + A2143G |
| AB T2-16 | <1 | A2143T + WT | WT |
Laboratory-derived artificial strains are underlined.
Conventional methods included direct sequence analysis, RFLP, and probe hybridization in liquid phase.
WT, wild type.
TABLE 2.
Comparative results of conventional and PCR-LiPA methods for the detection of 23S rDNA mutations in natural isolates and laboratory strains
| 23S mutation detected by conventional methods | Total no. of strains (no. of natural isolates) with 23S rDNA mutations detected by PCR-LiPA
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | A2115→G/G2141→A | A2142→G | A2142→C | A2143→G | A2142→T | A2143→C | Multiple mutations | Total | |
| Wild type | 5 (5) | 1 (1) | 6 | ||||||
| A2115→G/G2141→A | 0 | 0 | |||||||
| A2142→G | 20 (19) | 2 (2) | 22 | ||||||
| A2142→C | 2 (1) | 2 | |||||||
| A2143→G | 20 (19) | 1 (1) | 21 | ||||||
| A2142→T | 1 (0) | 1 | |||||||
| A2143→C | 1 (0) | 1 | |||||||
| A2143→T | 1 (0) | 1 | |||||||
| Multiple mutations | 3 (3) | 3 | |||||||
In 52 (91.2%) of the 57 isolates studied, PCR-LiPA yielded exactly the same results as conventional methods (direct sequencing, probe hybridization in liquid phase, and/or restriction enzyme analysis). These included the natural strains, including the wild type (n = 5) and strains with mutations A2142→G (n = 19), A2142→C (n = 1), A2143→G (n = 19), or a combination (n = 3), derived from patients, as well as five of the six artificial mutants, with mutations A2142→G, A2142→C, A2142→T, A2143→G, and A2143→C, respectively. No strains were found that contained the combined A2115→G and G2141→A mutations (10), which may indicate that such mutants are extremely rare. All strains showing discrepant results were retested by the conventional methods as well as by LiPA. In three of the clinical strains, multiple mutants were detected by conventional methods (strains 1538, LINAN 1567, and PAP), and PCR-LiPA yielded exactly the same results.
The initially seemingly discrepant results observed in five strains can all be attributed to the high sensitivity of the PCR-LiPA method. Strain 140-97, containing the wild-type 23S sequence, as determined by conventional methods, yielded a LiPA pattern indicating the presence of the wild-type sequence, as well as A2142→G and A2143→G mutations. The strain yielded variable MICs, ranging from less than 2 to more than 256, suggesting the presence of a mixture of strains. Two isolates (1264 and 1470) contained the A2142→G mutation, but PCR-LiPA also showed the presence of other mutants. Both strains were macrolide resistant, and the MICs for them were higher than 256. Similarly, the MIC for strain 1075 exceeded 256, and sequence analysis detected only the A2143→G mutation, but PCR-LiPA also detected the A2142→G mutation.
In the clinically macrolide-susceptible strain (ABT2-16), containing the artificial mutation A2143→T, the wild-type sequence as well as the mutant A2143→T sequence was detected by sequence analysis. The LiPA does not contain a probe specific for A2143→T, since it was shown earlier that the A2143→T mutation was not stable and rapidly mutated back to the wild-type sequence during subsequent passages (3). Since we tested the fourth passage of this strain, the PCR-LiPA result is consistent with these data.
Interestingly, strain C3II contained only wild-type sequences, as determined by all methods. However, this strain was clearly resistant, since the MIC for it was higher than 256. This strain is not a clinical isolate, but resistance to clarithromycin was selected in this strain in the laboratory by exposure to increased concentrations of clarithromycin (data not shown). Therefore, this strain may well have developed a different mechanism of resistance (12).
Compared to the conventional methods, the PCR-LiPA detected additional mutants and appears to provide more accurate data about the presence of bacterial variants in the culture, especially when multiple strains are present. In contrast, direct sequence analysis of PCR products will often detect only the predominant sequence. Only if different mutants are present in approximately similar concentrations are they detected by sequence analysis, whereas the LiPA specifically detects small amounts of each mutant. Also, the LiPA format permits the addition of specific probes in case additional relevant mutations are found. Thus, the LiPA provides accurate information about the presence of different 23S rDNA mutants, even if they represent only a small proportion of a bacterial population. Therefore, it could be particularly suitable to monitor the development of resistance during antibiotic therapy. The method can also be used directly in gastric biopsies, without the need for a bacterial culture (data not shown). Sometimes, the presence of multiple strains is also indicated by the presence of apparently resistant colonies within the growth inhibition zone on agar plates. This phenomenon is illustrated by the four strains in which PCR-LiPA showed the presence of multiple mutants, whereas conventional 23 rDNA analyses detected only a single wild-type or mutant sequence.
In conclusion, the PCR-LiPA offers a rapid and easy method for the detection of clinically relevant mutations in the 23S rRNA gene of H. pylori. The high sensitivity of the reverse hybridization method provides more accurate data, especially when multiple strains are present. Therefore, this method could facilitate further epidemiological and clinical studies.
REFERENCES
- 1.Blaser M J. Ecology of Helicobacter pylori in the human stomach. J Clin Investig. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broutet N, Guillon F, Sauty E, Lethuaire D, Mégraud F. Survey of the in vitro susceptibility of Helicobacter pylori to antibiotics in France—preliminary results. Gut. 1998;43:A11. [Google Scholar]
- 3.Debets-Ossenkopp Y J, Brinkman A B, Kuipers E J, Vandenbroucke-Grauls C M J E, Kusters J G. Explaining the bias in the 23S rRNA gene mutations associated with clarithromycin resistance in clinical isolates of Helicobacter pylori. Antimicrob Agents Chemother. 1998;42:2749–2751. doi: 10.1128/aac.42.10.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 5.de Boer W A, Tytgat G N J. The best therapy of Helicobacter pylori infection. Scand J Gastroenterol. 1995;30:401–407. doi: 10.3109/00365529509093298. [DOI] [PubMed] [Google Scholar]
- 6.Dore M P, Are B, Carta M, Mura I, Maida A, Realdi G. Antibiotic resistant H. pylori are extremely common in Sardinia. Gastroenterology. 1998;114:G0445. . (Abstract.) [Google Scholar]
- 7.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glupczynski Y, Goutier S, Van de Borre C, Butzler J P, Burette A. Surveillance of Helicobacter pylori resistance to antimicrobial agents in Belgium from 1989 to 1994. Gut. 1995;37:A56. . (Abstract.) [Google Scholar]
- 9.Glupczynski Y, Labbé M, Hansen W, Crokaert F, Yourassowsky E. Evaluation of the E test for quantitative antimicrobial susceptibility testing of Helicobacter pylori. J Clin Microbiol. 1991;29:2072–2075. doi: 10.1128/jcm.29.9.2072-2075.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hultén K, Gibreel A, Sköld O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Brea M, Martinez M J, Domingo D. Evolution of the resistance to several antibiotics in H. pylori over a 4-year period. Gut. 1995;37:A56. . (Abstract.) [Google Scholar]
- 12.Mégraud F. Resistance of Helicobacter pylori to antibiotics. Aliment Pharmacol Ther. 1997;11(Suppl. 1):43–53. doi: 10.1046/j.1365-2036.11.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 13.Midolo P D, Bell J M, Lambert J R, Turnidge J D, Grayson M L. Antimicrobial resistance testing of Helicobacter pylori: a comparison of E-test and disk diffusion methods. Pathology. 1997;29:411–414. doi: 10.1080/00313029700169415. [DOI] [PubMed] [Google Scholar]
- 14.Morton D, Bardhan K D. A six-year assessment of tinidazole, metronidazole, clarithromycin, tetracycline and amoxicillin resistance in Helicobacter pylori clinical isolates: a rising tide of antibiotic resistance? Gastroenterology. 1998;114:3620. . (Abstract.) [Google Scholar]
- 15.Occhialini A, Urdaci M, Doucet-Populaire F, Bébear C M, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pina M, Occhialini A, Monteiro L, Doermann H P, Mégraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi S, Itoh T, Ninomiya H, Hoshiya S, Watanabe K, Tokunaga K, Tanaka A, Nakamura N, Masubuchi N, Shingaki M, Saito S. Evolution of Helicobacter pylori antibiotic resistance in Japan (1985–1997) Gastroenterology. 1998;114:G1240. . (Abstract.) [Google Scholar]
- 18.van der Hulst R W, Keller J J, Rauws E A, Tytgat G M. Treatment of Helicobacter pylori: review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 18a.van Doorn L-J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K W, Atherton J C, Blaser M J, Quint W G V. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zwet A A, de Boer W A, Schneeberger P, Weel J F, Jansz A R, Thijs J C. Prevalence of primary Helicobacter pylori resistance to metronidazole and clarithromycin in the Netherlands. Eur J Clin Microbiol Infect Dis. 1996;15:861–864. doi: 10.1007/BF01691216. [DOI] [PubMed] [Google Scholar]
- 20.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]