Abstract
Many patients with locally advanced cervical cancer experience recurrence within the radiation field after chemoradiotherapy. Biomarkers of tumor radioresistance are required to identify patients in need of intensified treatment. Here, the biomarker potential of miR‐200 family members was investigated in this disease. Also, involvement of tumor hypoxia in the radioresistance mechanism was determined, using a previously defined 6‐gene hypoxia classifier. miR‐200 expression was measured in pretreatment tumor biopsies of an explorative cohort (n = 90) and validation cohort 1 (n = 110) by RNA sequencing. Publicly available miR‐200 data of 79 patients were included for the validation of prognostic significance. A score based on expression of the miR‐200a/b/‐429 (miR‐200a, miR‐200b, and miR‐429) cluster showed prognostic significance in all cohorts. The score was significant in multivariate analysis of central pelvic recurrence. No association with distant recurrence or hypoxia status was found. Potential miRNA target genes were identified from gene expression profiles and showed enrichment of genes in extracellular matrix organization and cell adhesion. miR‐200a/b/‐429 overexpression had a pronounced radiosensitizing effect in tumor xenografts, whereas the effect was minor in vitro. In conclusion, miR‐200a/b/‐429 downregulation is a candidate biomarker of central pelvic recurrence and seems to predict cell adhesion‐mediated tumor radioresistance independent of clinical markers and hypoxia.
Keywords: central pelvic recurrence, cervical cancer, extracellular matrix interaction, microRNA, miR‐200, radioresistance
Diagnostic biomarkers for identifying tumor radioresistance in patients with cervical cancer are highly needed. Here, we report a new candidate biomarker based on the miR‐200a/b/‐429 cluster expression in tumor biopsies. Low expression of miR‐200a/b/‐429 in patients was found to predict the risk of central pelvic recurrence after chemoradiotherapy. Additionally, miR‐200a/b/‐429 overexpression radiosensitized tumor xenografts. Moreover, the expression of the miR‐200a/b/‐429 was independent of known risk factors and was predictive of cell adhesion‐mediated radioresistance in patients.
Abbreviations
- ADC
apparent diffusion coefficients
- CI
confidence interval
- Ct
threshold cycle
- DSB
DNA double strand break
- DW‐MRI
diffusion weighted magnetic resonance imaging
- ECM
extracellular matrix
- EMT
epithelial to mesenchymal transition
- FDR
false discovery rate
- FIGO
Federation International de Gynecologie et d'Obstetrique
- GEO
Gene Expression Omnibus
- GO
gene ontology
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miRNA
microRNA
- MR
magnetic resonance
- N.S
nonsignificant
- RECIST
response evaluation criteria in solid tumors
- REMARK
reporting recommendations for tumor marker prognostic studies
- ROI
region of interest
- SEM
standard error of mean
1. Introduction
Radiotherapy, most often combined with cisplatin, is the treatment of choice for patients with locally advanced cervical cancer [1, 2, 3]. Improved treatment strategies are highly needed, since cancer recurrence after therapy is seen in about 30% of the patients, and in almost 65% at the more advanced stages. Many patients develop a highly radioresistant tumor, and more than 25% of all recurrences are located within the radiation field [1, 2, 3]. These patients have limited further treatment options [4, 5]. A better understanding of the molecular mechanisms behind tumor radioresistance in cervical cancer can lead to new therapeutic approaches [6]. Moreover, such knowledge would help development of biomarkers for identifying patients at risk of recurrence in the radiation field, as current clinical markers like tumor stage, tumor size and lymph node status are insufficient for this purpose [7].
microRNAs (miRNAs) are important regulators of biological processes affecting the response of tumor cells to radiation, including DNA damage repair, hypoxia tolerance, cell survival, and proliferation [8]. The small, noncoding RNAs downregulate expression of their target genes by degrading mRNAs and inhibiting translation. They have a large potential as biomarkers due to high stability in tissue specimens and biofluids [9], but no candidates for radioresistance have yet been evaluated in a decent number of cervical cancer patients. A comprehensive molecular characterization of 228 tumors of patients mainly treated with surgery identified downregulation of miR‐200a and miR‐200b and upregulation of some of their target genes as frequent events in cervical cancer [10]. These miRNAs therefore seem to be functionally active and promising candidates for further exploration in patients receiving chemoradiotherapy.
The miR‐200 family consists of nine mature miRNAs in two clusters located on chromosome 1 (miR‐200a‐5p, miR‐200a‐3p, miR‐200b‐5p, miR‐200b‐3p, and miR‐429) and chromosome 12 (miR‐200c‐5p, miR‐200c‐3p, and miR‐141‐5p miR‐141‐3p) [11]. miR‐200a has been included in two prognostic miRNA‐signatures in cervical cancer [12, 13], but the role of the miR‐200 family in tumor radioresistance has not been addressed in this disease. The members regulate large networks of genes that combined suppress various biological processes including epithelial to mesenchymal transition (EMT) and are often found to be downregulated in cancer [11]. EMT involves interaction between the extracellular matrix (ECM) components collagens and glycoproteins, and cell surface molecules like integrins and growth factor receptors. Such interactions regulate several downstream pathways that have been implicated in the radiation response of tumor cells [14, 15]. Moreover, members of the miR‐200 family have been found to be downregulated by hypoxia [16, 17, 18], which is a tumor feature associated with failure of chemoradiotherapy in cervical cancer [19, 20, 21]. It is therefore likely that the miR‐200 family plays a role in the radioresistance of this disease.
The recurrence pattern of cervical cancer patients receiving chemoradiotherapy includes distant metastases and locoregional recurrence within the pelvic radiation field [4]. A higher radiation dose is given centrally to the tumor region than to the lateral part of the pelvis. A distinction between central and lateral pelvic recurrence is therefore needed to identify radioresistant tumors. In this work, we assessed a unique set of site‐specific recurrence data of 200 patients, as well as expression data of the miR‐200 family and their predicted target genes for all tumors. Our aim was to investigate whether members of the miR‐200 family play a role in the radioresistance of cervical tumors and may be potential biomarkers of central pelvic recurrence after chemoradiotherapy. We further aimed to clarify whether hypoxia, as assessed by a 6‐gene hypoxia classifier from previous work [19], was involved in this resistance mechanism. Proof‐of‐principle radiation experiments in a cervical cancer model system with overexpressed miRNAs were performed to validate our conclusion from the clinical analyses. Our results provide novel insight into a role of the miR‐200 family in cervical cancer radioresistance that is independent of tumor hypoxia and may be exploited for a more precise identification of patients with risk of central pelvic recurrence.
2. Materials and methods
2.1. Patient cohorts
The study included 200 patients with locally advanced squamous cell carcinoma of the uterine cervix enrolled in our prospective chemoradiotherapy protocol at the Norwegian Radium Hospital from 2001 to 2012 (Table S1). Totally 1–4 tumor biopsies were taken at diagnosis, snap‐frozen, stored at −80 °C, and used for miRNA and gene expression analysis. The patients were assigned to two different cohorts based on the Illumina bead array version that was used for gene expression profiling, that is, HT‐12 v4 (explorative cohort, 90 patients) or WG‐6 v3 (validation cohort 1, 110 patients) [19]. Tumor volume, pathologic lymph nodes, and Federation International de Gynecologie et d'Obstetrique (FIGO) stage were determined by magnetic resonance (MR) imaging or computed tomography at diagnosis, according to the REsponse evaluation Criteria In Solid Tumors (RECIST) v1.1guidelines [22].
All patients received external radiotherapy, followed by intracavitary brachytherapy. Concurrent cisplatin (40 mg·m−2 weekly) was given according to tolerance. Follow‐up was performed as described [19]. Site‐specific recurrence data were collected and classified as central (pelvic recurrence within the center of the radiation field), lateral (pelvic recurrence within the radiation field outside the central region), and/or distant (metastases outside the radiation field). The research was conducted in accordance with the Declaration of Helsinki. The study was approved by The Regional Committee for Medical and Health Research Ethics in South East of Norway (REC 2016/2179), and written informed consent was obtained from all patients. The study adheres to the REporting recommendations for tumor MARKer prognostic studies (REMARK) guidelines [23].
2.2. miRNA profiling and calculation of miR‐200a/b/‐429 score
miRNA expression profiles of the 200 patients in explorative cohort and validation cohort 1 were generated from biopsies with a tumor cell fraction above 50%. The tumor cell fraction was estimated by a person experienced in cervical cancer pathology. This estimation was based on visual inspection of a hematoxylin‐ and eosin‐stained histological section from the biopsy. Total RNA from samples in the explorative cohort was isolated from frozen sections, using miRNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Total RNA from samples of validation cohort 1 was isolated from frozen sections, using TRIzol® Reagent (Thermo Fisher Scientific, Waltman, MA, USA) followed by double precipitation with isopropanol, according to the manufacturer's recommendation. RNA was further purified by precipitation with 5 m lithium chloride and washed with 70% ethanol. Following either RNA isolation protocols, the RNA pellet was dissolved in RNase‐free water and stored at −80 °C until labeling. RNA concentration was measured by a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), and samples with RNA integrity number above 6.5 were used. The stability of miRNAs in stored tissue samples was assessed in a previous study and showed no degradation during storage time [24]. RNA of different samples from the same tumor was pooled.
Small RNA preparation for sequencing was performed according to Illumina TruSeq Small RNA library protocols (Illumina Inc., San Diego, CA, USA). In brief, indexed 3′‐ and 5′‐end RNA adapters were ligated to isolated total RNA (1 μg) per sample followed by reverse transcription and library amplification. The cDNA was purified by cutting out bands corresponding to 140–160 bp (length of miRNA + adapter and index sequences) on a PAGE gel, pooling, and precipitation with ethanol. Single‐end sequencing of the cDNA molecules was performed on the Illumina HiSeq 2500 platform. Real‐time analysis, base calling, and filtering of low‐quality reads were performed by Illumina's software packages (Illumina Inc.). fastq data were quality checked, and reads were aligned, quantified, and annotated using the miRDeep2 algorithm with miRBase v21 for annotation of mature miRNAs [25]. Reads per million annotated mature miRNAs were used for normalization of read counts. Log2‐transformed data were used in the analyses. Data have been deposited in the Gene Expression Omnibus (GEO), accession number GSE178629.
A miR‐200a/b/‐429 score was calculated for each patient by averaging the median centered, log2‐transformed expression levels of miR‐200a‐5p, miR‐200a‐3p, 200b‐5p, miR‐200b‐3p, and miR‐429.
2.3. External miRNA data set
Publicly available miRNA data of a third cohort (n = 79 tumor samples, validation cohort 2) were included for the validation of prognostic significance. The cohort included patients treated with radiation and concurrent cisplatin at Princess Margaret Cancer Centre in Toronto, Canada, between 2000 and 2007. Progression‐free survival and miRNA data were available for the ‘training cohort’ (‘TLDA data, Training cohort’) in the publication by How et al. [26].
miRNA data were derived from snap‐frozen biopsies taken at diagnosis, as described [26]. Total RNA was isolated from frozen biopsies with a tumor cell fraction above 70%, using Norgen Total Purification kit (Norgen Biotek, Thorold, ON, Canada). miRNA expression profiles were measured with RT‐qPCR array, using the TaqMan Low Density Array (TLDA) Human MicroRNA A Array v2.0 and the 7900HT Real‐Time PCR System (Applied Biosystems, Carlsbad, CA, USA). miRNA expression levels were calculated using the mean threshold cycle (Ct) of the three control RNAs RNU44, RNU48 and U6 for normalization. None of the miRNAs had any Ct‐values above 37. A miR‐200a/b/‐429 score was calculated as described in Section 2.2, but based on only miR‐200a‐3p, miR‐200b‐3p, and miR‐429, since data of miR‐200a‐5p and miR‐200‐5p were not available.
2.4. Gene expression profiling and hypoxia status
The gene expression data and hypoxia status of all patients in the explorative cohort and validation cohort 1 are parts of a data set used in previous work [19] and are available in GEO (GSE72723). In brief, gene expression profiling of fresh‐frozen tumor biopsies was performed using Illumina bead array versions HT‐12 v4 (explorative cohort) and WG‐6 v3 (validation cohort 1) with 47 231 and 48 803 transcripts, respectively (Illumina Inc.). The same RNA isolation protocols that were used for miRNA profiling were applied for each sample. Signal extraction and quantile normalization were performed using the software provided by the manufacturer (Illumina Inc.), and log2‐transformed gene expression data were used in the analyses [19]. A 6‐gene hypoxia signature was derived from the gene expression data of ERO1A, DDIT3, KCTD11, P4HA2, STC2, and UPK1A, as described in previous work [19]. The signature has a predefined cutoff of zero, which was used for dichotomous classification of tumors according to their hypoxia status as more or less hypoxic.
2.5. Target gene prediction
Candidate target genes for miR‐200a‐5p/‐3p, 200b‐5p/‐3p, and miR‐429 were collected from the miRGate database, including both validated and computationally predicted targets from several databases [27]. The expression values of the candidates were correlated with the corresponding miRNA expressions of the explorative cohort and validation cohort 1 separately, using Spearman correlation analysis (two‐sided) with the Benjamini–Hochberg procedure [28] to control the false discovery rate (FDR). Genes with a significant negative correlation (nominal P < 0.05 in both cohorts and FDR q‐value < 0.1 in at least one cohort) were considered as potential target genes. These requirements were used to compromise between the risk of losing true discoveries and the risk of keeping noisy, irrelevant data. The result was visualized in a regulatory network with miR‐200a‐5p/‐3p, 200b‐5p/‐3p, and miR‐429 as nodes and their potential target genes as interaction partners. Interactions with an FDR q‐value < 0.1 were included in the network. The network was created by the cytoscape software v3.6.1 [29].
2.6. Generation of SiHa cell line with stable miR‐200a/b/‐429 overexpression
miRNA overexpression rather than repression was chosen due to low endogenous expression of the miR‐200 family in cervical cancer cell lines. In accordance with the histology of the clinical tumor samples (Table S1), the cervical squamous cell carcinoma cell line SiHa (ATCC® HTB‐35™ from ATCC, LGC standards, Wesel, Germany) was used. Cell line authentication was performed with Powerplex 16 (Promega, Madison, WI, USA) to identify DNA STR profiles. Cells were cultured at 37 °C in DMEM medium containing Glutamax (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with FBS (10%) and PenStrep (1%) in 5% CO2 and 95% room air. All cell line experiments were performed between passage two and 14. Mycoplasma testing was conducted at a regular basis with MycoAlert™ Mycoplasma Detection Kit (Lonza, Cologne, Germany).
A stably expressed miR‐200a/b/‐429 SiHa cell line and a control SiHa cell line (empty vector), both with co‐expressed GFP, were generated by lentiviral transduction. pLenti 4.1 Ex miR200b‐200a‐429 GFP (Addgene plasmid #35533; Addgene, Cambridge, MA, USA) was a gift from G. Goodall [30]. Empty vector control plasmid, pLenti 4.1 Ex GFP was derived from pLenti 4.1 Ex miR200b‐200a‐429 GFP by removal of the miRNA precursor sequence with the restriction enzymes Nhe1 (R3131S; New England Biolabs, Ipswich, MA, USA) and AvrII (R0174S; New England Biolabs). Lentivirus was generated by co‐transfection of packaging plasmids and the pLenti 4.1 Ex miR200b‐200a‐429 GFP or pLenti 4.1 Ex GFP (control) construct into HEK293T cells. The 293T cells produced lentiviral particles for 3 days before isolation from the culture medium. SiHa cells were infected with the generated lentivirus. Stably transduced cells (SiHa mir‐200b‐200a‐429‐GFP or SiHa control‐GFP) were selected by culturing in puromycin (1 μg·mL−1) for 20 days. Subsequent cell culturing conditions were according to common practice, using DMEM medium containing Glutamax (Gibco) supplemented with FBS (10%) and PenStrep (1%) in 5% CO2 and 95% air.
Stably transduced cells were further selected based on GFP intensity by cell sorting, using a SH800 Cell Sorter (Sony, Tokyo, Japan), and based on expression level of miR‐200a‐5p/‐3p, miR‐200b‐5p/‐3p and miR‐429 measured by RT‐qPCR. For RT‐qPCR analysis, total RNA was isolated with the miRNeasy kit (Qiagen), dissolved in nuclease‐free water, and stored at −80 °C prior to use. RNA concentration was measured on a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). cDNA was synthesized from 10 ng of total RNA using the miRCURY LNA™ microRNA PCR, Polyadenylation, and cDNA synthesis kit II (Qiagen). Predeveloped microRNA LNA™ PCR primer sets (Qiagen) were used for amplification of the selected miRNAs: hsa‐miR‐200a‐3p; (prod. no. 204707), hsa‐miR‐200a‐5p (prod. no. 206063), hsa‐miR‐200b‐3p (prod. no. 2060071), hsa‐miR‐200b‐5p (prod. no. 204144), and hsa‐miR‐429; (prod.no. 205901). Hsa‐miR‐151a‐5p (prod. nr 204007) and hsa‐miR‐152‐3p (prod. nr 204294) were used as reference miRNAs for normalization of miRNA expression in the RT‐qPCR analysis, based on our previous work [24]. The miRCURY Universal RT miRNA PCR system (Qiagen) and Bio‐Rad Real‐Time PCR thermocycler (Bio‐Rad, Hercules, CA, USA) were used for amplification and detection of miRNAs according to manufacturer's protocols. Amplification was analyzed using the sds Software v2.3 (Applied Biosystems). Reverse transcriptase negative controls and nontemplate controls were included and showed no amplification. Relative miRNA expression level was calculated using the comparative ΔCt (cycle threshold) method by normalizing the Ct value of each miRNA to the mean Ct value of the reference miRNAs: ΔCtmiRNA = CtmiRNA − (CtmiR‐151a‐5p + CtmiR‐152‐3p/2). A Z2 Coulter particles counter and size analyzer (Beckman Coulter, Brea, CA, USA) with a 100‐μm capillary was used to measure cell proliferation and cell size, respectively.
2.7. Western blotting
Protein extracts from cell lines were prepared by lysing the samples in RIPA‐buffer (Thermo Fisher Scientific) containing Halt™ Phosphatase Inhibitor Cocktail and Halt™ Protease Inhibitor Cocktail EDTA‐Free (Thermo Fisher Scientific). Lysates were incubated for 30 min on ice followed by centrifugation for 15 min at 16 000 g . The supernatants were added Pierce Lane Marker Reducing Sample Buffer (Thermo Fisher Scientific) and boiled at 95 °C for 5 min. The protein extracts (20 μg) were electrophoresed in a Mini‐PROTEAN® TGX Stain‐Free™ Protein Gel (Bio‐Rad) and transferred to a Trans‐Blot Turbo RTA Mini 0.2 μm PVDF membrane (Bio‐Rad). Mouse anti‐E‐cadherin monoclonal antibody (#610181; BD Biosciences, San Jose, CA, USA) was used as primary antibody and diluted 1 : 5000 in PBS with 5% dry milk. HRP‐conjugated polyclonal donkey anti‐mouse‐IgG was used as secondary antibody (715‐035‐150; Jackson Immunoresearch, West Grove, PA, USA). The blot was imaged in a Chemidoc MP (Bio‐Rad) using LumiGlo (KPL, Gaitersburg, MD, USA) substrate for detection. Total protein was used as loading control and detected with Stain‐Free technology imaging (Bio‐Rad) of the membrane after transfer.
2.8. Clonogenic survival assay of irradiated cells
miR‐200a/b/‐429‐overexpressing cells or control cells were grown to 60–70% confluence and exposed to 4 or 8 Gy of ionizing radiation in single fractions at a dose rate of 1.0 Gy·min−1 with a 160 kV X‐ray generator (Faxitron Cabinet X‐ray; Faxitron, Tucson, AZ, USA). Cells were immediately trypsinized (0.05% trypsin; Sigma‐Aldrich, St. Louis, MO, USA), diluted, and seeded into 60‐mm culture dishes for clonogenic survival assays. After 14 days in culture, cells were fixed in ethanol (70%) and stained with methylene blue (0.5%). Colonies with > 50 cells were counted, and survival fraction was calculated. Plating efficiencies were calculated from nonirradiated cells.
2.9. Cell cycle distribution and DNA double strand break (DSB) detection by flow cytometry
miR‐200a/b/‐429‐overexpressing cells or control cells were cultured until they reached 60–70% confluence and irradiated with 8 Gy at a dose rate of 1.0 Gy·min−1. Samples were collected at the timepoints 0, 0.5, 1, 2, 6, 24, and 48 h after radiation, incubated with LIVE/DEAD™ Fixable Near‐IR Dead Cell Stain (L34975; Life Technologies AS, Carlsbad, CA, USA) on ice for 30 min, fixed in ice cold ethanol (70%), and stored at −20 °C before analyses.
About 1 × 106 cells of each sample were added 2.5 × 105 ethanol fixed lymphocyte leukemia REH cells. The REH cells were applied in a 1 : 1 mixture of irradiated (4 Gy) and nonirradiated cells and were used for normalization to reduce effect of intersample staining variations. The samples were rehydrated in PBS and incubated for 30 min in room temperature with the DSB marker mouse anti‐γH2Ax (#05‐636; Millipore, Burlington, MA, USA) and rabbit anti‐phospho‐histone H3 (Ser10) mitosis marker (#06‐570; Millipore) diluted 1 : 500 in PBS with 5% nonfat milk. The samples were further washed in PBS, incubated for 30 min in room temperature with the secondary antibodies goat anti‐mouse IgG Alexa Fluor 594 (ab150116; Abcam, Cambridge, MA, USA) and donkey anti‐rabbit IgG AlexaFluor 647 (A31573; Thermo Fisher Scientific), washed in PBS, and resuspended in PBS containing 1.5 μg·mL−1 Hoechst 33258 (Sigma‐Aldrich) for DNA staining. They were thereafter run on a LSR II yellow laser flow cytometer (BD Biosciences) and analyzed with flowjo software (v10; TreeStar Inc., San Jose, CA, USA).
Gating of cell populations is illustrated in Fig. S1. The whole population of SiHa cells stained with LIVE/DEAD Near‐IR was distinguishable from unstained REH cells with the red laser (640 nm). The median γH2Ax intensity of nonmitotic, live miR‐200a/b/‐429‐overexpressing cells or control cells was normalized to the median γH2Ax intensity of the REH population in the same sample.
2.10. Generation of SiHa xenograft tumors with stable miR‐200a/b/‐429 overexpression
Tumor xenografts with stable miR‐200a/b/‐429 overexpression were established in female athymic nude mice (Hsd:Athymic Nude‐Foxn1nu; originally supplied by Envigo, Indianapolis, IN, USA). The mice were bred in‐house at Department of Comparative Medicine at our institution and kept in specific pathogen‐free environment with food and water supplied ad libitum, 12‐h light‐dark cycle, temperature of 22 ± 1 °C and 65 ± 5% humidity. Mice were monitored at daily base. All procedures were conducted according to the European Laboratory Animal Science Association (FELASA) regulations, and studies were approved by the Norwegian Food Safety Authority (FOTS #8344).
At an average age of 6–7 weeks, the mice were injected with miR‐200a/b/‐429‐overexpressing cells or control cells (2 × 106) intramuscularly in both hind legs for characterization studies of unirradiated tumors. Tumor growth was monitored by anatomical T2‐weighted (T2W) MR imaging (MRI) at regular timepoints. Tumor doubling time (Td) was calculated for each tumor in the exponential growth phase and fitted to an exponential growth regression model. The curves were used to calculate mean Td for each group of tumors. At the day of tumor resection, diffusion weighted (DW)‐MRI was used to assess tumor apparent diffusion coefficients (ADC) as a measure of cell density. The median tumor volume was 252 mm3. The hypoxia marker pimonidazole (60 mg·kg−1; Hypoxyprobe Inc., Burlington, MA, USA) was administered intraperitoneally 90–120 min before the mice were euthanized by cervical dislocation and tumors were resected, paraformaldehyde‐fixed (4%), and paraffin‐embedded for digital histopathology.
2.11. Radiation experiments on xenograft tumors
For radiation experiments on xenografts, mice at an age of 6–7 weeks were injected with miR‐200a/b/‐429‐overexpressing cells or control cells intramuscularly in the right hind leg. At a tumor volume of 200–250 mm3, the mice were anesthetized by subcutaneous injection with a mixture of 10–15 mg·kg−1 xylasin (Rompun®; Bayer AG, Leverkusen, Germany), 5–10 mg·kg−1 butorphanol (Torbugesic®; Zoetis, Berlin, Germany), and 15–20 mg·kg−1 zolazepam and tiletamine (Zoletil®; Virbac, Carros, France) before irradiation with a single dose of 8 Gy (1.6 Gy·min−1) from an X‐ray source (Faxitron MultiRad 225; Faxitron). Postirradiation tumor growth and ADC values were assessed once every 3 or 4 days by T2W‐ and DW‐MRI, respectively. Mice were sacrificed by cervical dislocation when the tumor volume had reached at least two times the volume at baseline (day 0), or at latest 82 days after inoculation.
2.12. Magnetic resonance imaging
Magnetic resonance imaging of xenograft tumors was conducted with a Bruker Biospec 7.05 T bore magnet (Bruker Biospin AG, Fallanden, Switzerland) with a mouse quadrature volume coil. Animals were anesthetized with a continuous supply of sevoflurane (3–4% in O2; Baxter, Guyancourt, France). The body core temperature was monitored and kept at 37 °C during scanning.
To assess tumor volume, anatomical T2W images were acquired with a fast spin echo pulse sequence, using an echo time of 31.05 ms, field of view of 3 × 3 cm, matrix of 256 × 256, slice thickness of 0.8 mm, and repetition time of 3000 ms. DW‐MR images for ADC measurement in the characterization studies were acquired with DW single‐shot fast spin echo sequence (field of view of 3 × 3 cm; matrix of 64 × 64; slice thickness of 0.8 mm; repetition time of 1100 ms; echo time of 26 ms; diffusion weightings or b‐values of 200, 400, 700 and 1000 s·mm−2). DW‐MRI for assessing effect of radiation was acquired with DW echo planar imaging (field of view of 3 × 3 cm; matrix of 128 × 128; slice thickness of 0.8 mm; repetition time of 1500 ms; echo time of 17.89 ms; diffusion weightings or b‐values of 200, 300, 500, 700 and 800 s·mm−2). ADC values were extracted voxel‐wise from the indicated ranges of b‐values, and calculated using in‐house developed scripts in matlab (MathWorks, Natick, MA, USA). This was done by fitting the log‐transformed signal intensities (S) to the linear equation ln(S(b)/S0) = −bADC + c, using a linear least square fit algorithm. Tumor regions of interest (ROIs) were defined in the T2W images and used to construct ADC maps. Areas were defined as necrotic if ADC > 0.0011 mm2·s−1. The median ADC value of each tumor was used for subsequent analyses.
2.13. Digital histopathology
Serial sections (4 μm) were made through the central region of paraffin‐embedded xenograft tumors and prepared for histological staining with standardized protocols using the Dako EnVision™ Flex+ System (K8012; Dako, Carpinteria, CA, USA). Staining with rabbit anti‐GFP (1 : 5000, ab290; Abcam) for detection of transduced tumor cells, rabbit anti‐Ki‐67 (1 : 1000, ab15580; Abcam) for assessment of proliferating cells and necrosis, and rabbit anti‐pimonidazole (1 : 3500, PAb27HAP; Hypoxyprobe Inc.) to quantify hypoxic fraction was performed and visualized using 3,3′‐diamenobendizine (DAB) as chromogen. Hematoxylin was used for counterstaining. Stained tumor sections were digitized (0.46 μm·pixel−1, 20× mode) using a whole slide scanner (Nanozoomer‐NZ Digital Slide scanner; Hamamatzu Photonics, Hamamatsu, Japan). Comparable ROI‐boundaries were drawn for adjacent sections stained for different markers and used in the subsequent analyses (Fig. S2).
In‐house developed software programs in matlab (MathWorks) were used for image analyses. Color deconvolution was performed to separate the brown DAB color information in the image, as described [3]. Necrotic regions were defined in the Ki‐67 stained sections by manual outlining of decellularized areas in RGB images (Fig. S3A). Necrotic fraction was calculated as total necrotic area relative to ROI. The necrotic areas were excluded from other analyses. Hypoxia was defined in pimonidazole stained sections. Segmentation and thresholding of pimonidazole‐positive regions were performed as described [31]. Briefly, the hypoxic regions were defined as the regions with an intensity value larger than a manually decided intensity threshold in the DAB‐image (Fig. S3B). Hypoxic fraction was calculated as total hypoxic area relative to ROI after exclusion of necrosis. To quantify cell density, segmentation of GFP‐positive cells was first performed by intensity thresholding of the color deconvoluted DAB image to define tumor parenchyma (Fig. S4). The area of GFP‐positive cells was quantified as total GFP‐positive area relative to ROI after exclusion of necrosis. Cell nuclei were thereafter identified in binary Ki‐67 maps by applying segmentation thresholding on the color deconvoluted DAB images (Fig. S4). All tumor cells were Ki‐67 positive. Binary structures with size > 200 pixels were assumed to be clusters of cells and further divided by applying the Watershed function on the Euclidean distance transformed binary image (matlab). Cells segmented in necrotic areas were removed from the binary map. A binary Ki67‐positive cell mask was further created by including only the segmented cells within the tumor parenchyma (Fig. S4), as defined in the adjacent GFP‐stained section. Cell density was calculated as number of Ki‐67‐positive cell nuclei relative to total GFP‐positive area.
2.14. Statistical analysis
All statistical analyses were performed using r [32] v3.5.1, brb‐arraytools v4.5.1 [33], ibm spss v25 (IBM Corp., Chicago, IL, USA), sigmaplot statistical software package v14 (Systat Software Inc., San Jose, CA, USA), x‐tile software v3.6.1 [34], g*power software v3.1.9.7 [35], and david v6.8 web tool [36]. A significance level of P < 0.05 was considered statistically significant, if not otherwise stated. Cox uni‐ and multivariate proportional hazards (PH) survival analyses were performed with endpoints central pelvic control, lateral pelvic control, distal control, and progression‐free survival (survival without central, lateral, and/or distal recurrence). Assumptions of PHs in the Cox regression models were evaluated graphically using log‐minus‐log plots. The multivariate analyses were performed by forward and backward stepwise Cox PH regression to test the prognostic and predictive value of the miR‐200a/b/‐429 score, using conventional clinical markers and hypoxia status as covariates. Only the covariates with P < 0.1 in univariate analysis were included. Kaplan–Meier survival curves were compared using log‐rank test. Stratification of patients in low or high score groups for these curves was based on the optimal cut‐off for the miR‐200a/b/‐429 score when associated with the different endpoints, as determined by using the x‐tile software.
The functional annotation tool david v6.8 was used to identify enriched gene ontology (GO)‐terms, that is, biological processes, cellular components and molecular functions, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the list of potential target genes. The global human transcriptome or the total miR‐200a/b/‐429 target gene list collected from the miRGate analysis was selected as background in two separate GO‐ and KEGG analyses. The default GO category ‘GO Direct’ was used for each GO term. The Benjamini–Hochberg procedure [28] was used to control FDR, and FDR q‐value < 0.01 was considered statistically significant.
Pearson product moment or Spearman rho two‐sided correlation test was used to search for correlation between parameters. Kruskal–Wallis, chi‐square, Fisher exact, Mann–Whitney U‐ or Student's T‐test was applied for comparison of groups. The choice between parametric and nonparametric test was based on conditions of normality and type of data (continuous or categorical). Variability of miRNAs over the range of measured values (heteroscedasticity) was checked by visual inspection of plots showing residuals versus predicted values and statistically with the White test.
Characterization and radiation response studies on xenografts were performed with 23–30 tumors and 8–10 tumors in each group, respectively. The sample sizes were above the estimated number of tumors needed to detect a significant difference between two groups in a two‐sided Student's t‐test with P < 0.05, power > 0.8, and 95% confidence interval (CI). These calculations were based on doubling time data of SiHa xenograft tumors from previous work [31]. Endpoint in xenograft tumor growth delay analyses and Kaplan–Meier survival curves was fraction of tumors with a volume below 1.5 times the baseline volume (day 0). An event was defined as the time point when the tumor reached this size (T 1.5×). Matched tumor growth data of nonirradiated xenografts from the characterization experiments were used for comparison. Kaplan–Meier survival curves were compared by Holm–Sidak method for multiple comparisons.
3. Results
3.1. Expression of miR‐200a/b/‐429 has prognostic impact in three independent cohorts
Expression of the nine miR‐200 members was analyzed in relation to progression‐free survival in the explorative cohort to identify candidates for further analysis. All miRNAs were detected and their expression level varied considerably across the patients (Fig. S5A). Univariate Cox regression analysis revealed a significant association between miRNA expression and progression‐free survival for each of the miR‐200 members on chromosome 1 (miR‐200a‐3p, P = 1.3 × 10−2; miR‐200a‐5p, P = 4.3 × 10−2; miR‐200b‐3p, P = 4.1 × 10−2; miR‐200b‐5p, P = 2.4 × 10−2 and miR‐429, P = 6.7× 10−3), where patients with a low expression had poor survival probability compared to the others (Table 1). No prognostic significance was seen for any miR‐200 members on chromosome 12 (miR‐200c‐3p/‐5p and miR‐141‐3p/‐5p).
Table 1.
Cox regression analyses of progression‐free survival in the explorative cohort (90 patients).
| Chromosome 1 | Chromosome 12 | ||||||
|---|---|---|---|---|---|---|---|
| miRNA | P | HR | 95% CI | miRNA | P | HR | 95% CI |
| miR‐429 | 6.7 × 10−3 | 0.40 | 0.20–0.78 | miR‐141‐3p | 1.4 × 10−1 | 0.67 | 0.40–1.14 |
| miR‐200a‐3p | 1.3 × 10−2 | 0.44 | 0.23–0.84 | miR‐141‐5p | 4.9 × 10−1 | 0.79 | 0.40–1.56 |
| miR‐200a‐5p | 4.3 × 10−2 | 0.52 | 0.27–0.98 | miR‐200c‐3p | 3.7 × 10−1 | 0.71 | 0.33–1.50 |
| miR‐200b‐3p | 4.1 × 10−2 | 0.49 | 0.25–0.97 | miR‐200c‐5p | 1.5 × 10−1 | 0.65 | 0.36–1.17 |
| miR‐200b‐5p | 2.4 × 10−2 | 0.43 | 0.21–0.90 | ||||
The miR‐200 members on chromosome 1 were highly co‐expressed, with Pearson correlation coefficients (r) in the range of 0.74–0.93 (P < 1.0 × 10−3 for all; Fig. S5B). Plots of residuals against predicted values showed no patterns of nonconstant variance over the range of measured values for any of the miRNAs, and no significant heteroscedasticity was found (Fig. S6). Since all five mature miRNAs showed prognostic significance and were highly correlated, their expression level was used to calculate a score, termed miR‐200a/b‐429 score for each patient, as outlined in Section 2.2. The score showed prognostic significance when used as a continuous variable [P = 1.3 × 10−2; hazard ratio (HR), 0.40; 95% CI, 0.19–0.82], consistent with results for the individual miRNAs (Table 1). A cut‐off value that allocated about 30% of the patients to a group with low score, in line with the expected recurrence rate when using progression‐free survival as endpoint, showed the strongest association to outcome in a log‐rank test (Fig. 1A).
Fig. 1.
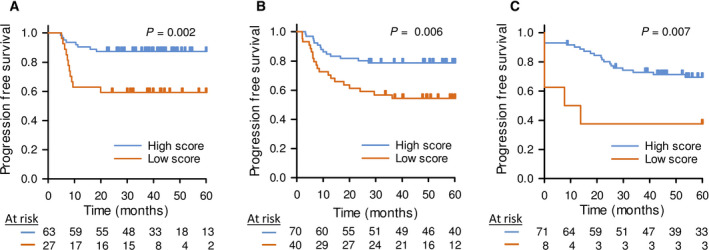
Prognostic significance of the miR‐200a/b/‐429 score in independent cohorts. Kaplan–Meier curves showing progression‐free survival of patients stratified with high‐ and low miR‐200a/b/‐429 score in the (A) explorative cohort (n = 90), (B) validation cohort 1 (n = 110) and (C) validation cohort 2 (n = 79). (A–C) 60 months follow up data were used. P‐values from log‐rank test and number of patients at risk are indicated. Patients were divided into a high and low score group based on the strongest association to progression‐free survival.
The prognostic impact of the miR‐200a/b/‐429 score was further evaluated in validation cohorts 1 and 2. For validation cohort 1, with miRNA sequencing data of patients treated at our institution, a strong association to outcome was found when comparing about 30% of the patients with the lowest miR‐200a/b‐429 score with the others (Fig. 1B). Significant association to outcome was also found in validation cohort 2, with RT‐qPCR miRNA data of patients treated at Princess Margaret Cancer Centre in Toronto, when allocating about 10% of the patients to the high‐risk group (Fig. 1C). These findings validated the prognostic impact of the miR‐200a/b/‐429 score and demonstrated its robustness across technologies and cohorts treated at different institutions.
3.2. miR‐200a/b/‐429 is an independent predictive marker of central pelvic recurrence
To identify the site of recurrence that was most strongly associated with the miR‐200a/b/‐429 score, the explorative cohort and validation cohort 1 were merged into a cohort of 200 patients to increase the statistical power. The recurrence sites included central pelvic in 21% of the recurrences, lateral pelvic in 20% and distant metastasis in 59% (Fig. 2A,B). There was a significant correlation between the miR‐200a/b/‐429 score and central recurrence in univariate Cox regression analysis, using continuous score data (P = 6.3 × 10−3; HR = 0.45; CI 95%, 0.25–0.80; Table 2). Moreover, in log‐rank test, a group of 13% of the patients with the lowest score had a significantly higher risk of central recurrence compared to the others (Fig. 2C). No association with lateral pelvic or distal recurrence was found, neither in univariate Cox analyses on continuous score data (P = 3.7 × 10−1 and P = 3.4 × 10−1, respectively; Table 2) or in log‐rank test on patients groups (Fig. 2D,E).
Fig. 2.
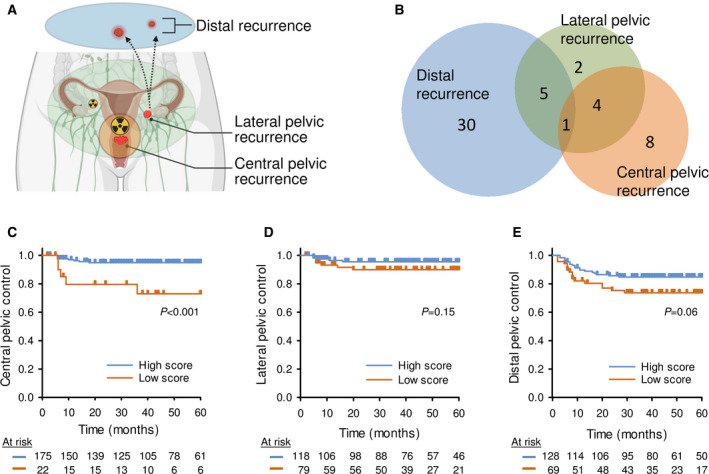
Association between the miR‐200a/b/‐429 score and tumor recurrence at different sites. (A) Schematic illustration of recurrence sites in cervical cancer. Created with BioRender.com. (B) Venn diagram showing site‐specific numbers of recurrences (n = 50) in patients of the explorative cohort and validation cohort 1 combined (n = 200). Kaplan–Meier curves of patients with high or low miR‐200a/b/‐429 score, using (C) central pelvic control, (D) lateral pelvic control, and (E) distal control as endpoint (n = 200). (C–E) 60 months follow up data were used; P‐values from log‐rank test and number of patients at risk are indicated; censored data adjusted for recurrence site were used in each analysis; patients were divided into a high and low score group based on the strongest association to the endpoint in each panel.
Table 2.
Cox regression analysis of tumor recurrence at different sites for 200 patients. N.S, nonsignificant.
| Factor | Univariate analysis a | Multivariate analysis b | ||||
|---|---|---|---|---|---|---|
| P | HR | 95% CI | P | HR | 95% CI | |
| Central pelvic control c | ||||||
| Lymph node status | 1.3 × 10−2 | 5.15 | 1.42–18.73 | 2.8 × 10−2 | 4.67 | 1.18–18.54 |
| Tumor volume d | (6.3 × 10−2) | 3.06 | 0.94–9.97 | N.S | – | – |
| FIGO stage e | (1.0 × 10−1) | 2.56 | 0.83–7.85 | N.S | – | – |
| Hypoxia status f | 3.4 × 10−3 | 6.87 | 1.89–25.00 | 1.9 × 10−2 | 4.78 | 1.29–17.74 |
| miR‐200a/b/‐429 score | 6.3 × 10−3 | 0.45 | 0.25–0.80 | 1.1 × 10−2 | 0.43 | 0.23–0.83 |
| Lateral pelvic control c | ||||||
| Lymph node status | 9.9 × 10−3 | 7.37 | 1.61–33.67 | 4.8 × 10−2 | 4.81 | 1.01–22.85 |
| Tumor volume d | 1.5 × 10−2 | 6.56 | 1.40–30.00 | N.S | – | – |
| FIGO stage e | 8.6 × 10−3 | 7.74 | 2.23–25.84 | 8.3 × 10−3 | 5.27 | 1.53–18.15 |
| Hypoxia status f | (7.6 × 10−2) | 2.81 | 0.89–8.87 | N.S | – | – |
| miR‐200a/b/‐429 score | N.S | – | – | – | – | – |
| Distal control c | ||||||
| Lymph node status | 1.1 × 10−2 | 2.39 | 1.22–4.68 | N.S. | – | – |
| Tumor volume d | 7.2 × 10−4 | 3.53 | 1.70–7.32 | 3.0 × 10−2 | 2.35 | 1.09–5.08 |
| FIGO stage e | 2.0 × 10−6 | 5.05 | 2.61–9.77 | 1.6 × 10−4 | 3.82 | 1.90–7.68 |
| Hypoxia status f | (6.5 × 10−2) | 1.85 | 0.96–3.57 | N.S. | – | – |
| miR‐200a/b/‐429 score | N.S | – | – | – | – | – |
Results for the miR‐200a/b/‐429 score are highlighted in bold.
Variables with P > 0.05 and ≤ 0.1 in the univariate analysis are shown in parentheses.
Variables with P ≤ 0.1 in the univariate analysis were included in multivariate analysis. The same results were obtained for forward and backward selection.
Site of recurrence was unknown for three patients.
Patients were divided into two groups based on the median tumor volume of 36.6 cm3.
Patients were divided into two groups based on FIGO stage IB–IIB or IIIA–IVA.
Patients were divided into two groups based on a less hypoxic or more hypoxic tumor.
The miR‐200a/b/‐429 score was further related to conventional clinical markers, including FIGO stage, tumor size, and lymph node status, as well as hypoxia status. There was no association between the score and any of these markers (Table S2). In unitivariate Cox regression analysis, lymph node and hypoxia status, in addition to the miR‐200a/b/‐429 score, showed significant association to central pelvic recurrence. Moreover, the miR‐200a/b/‐429 score retained its statistical significance together with lymph node and hypoxia status in multivariate analysis (Table 2). These results suggested a potential of miR‐200a/b/‐429 expression as a biomarker of central pelvic recurrence.
3.3. miR‐200a/b/‐429 overexpression increases radiosensitivity in tumor models in vivo
To determine whether the miR‐200a/b/‐429 cluster had a direct role in regulation of radiation response, radiation experiments were conducted in cell lines and xenograft tumors with stable overexpression of these candidates. The expression level of each of the five mature miRNA species was successfully increased in the stable mir‐200b‐200a‐429‐GFP transduced SiHa cell line compared to control‐GFP SiHa cell line (Fig. S7A). Protein level of E‐cadherin was higher (P = 7.4 × 10−3) and cell size smaller (P = 1.8 × 10−3) in miRNA‐overexpressing cells than in control cells (Fig. S7B–D), in line with the known function of these miRNAs in EMT suppression [37]. miRNA overexpression had no significant effect on cell survival or proliferation rate (Fig. S7E,F), but a minor decrease in fraction of cells in G1‐phase of the cell cycle and an increase in S‐phase fraction were seen (Fig. S7G,H). miRNA‐overexpressing tumors (n = 23) showed a higher cell density both by histopathology and MRI‐based ADC values compared to control tumors (n = 30) (Fig. S8A,B), consistent with the smaller cell size observed in vitro. These tumors also had a higher fraction of necrosis, but there was no difference in hypoxic fraction or growth rate (Fig. S8C–F). Our model system therefore seemed to express an EMT‐suppressive phenotype as expected from published work [38], without showing enrichment of radiosensitive features like fraction of cells in G2/M phase in vitro or oxygenation in vivo.
Radiation treatment (RT) of cell lines with 8 Gy led to a larger decrease in clonogenic survival fraction in miRNA‐overexpressing cells than in control cells (P = 1.4 × 10−2) (Fig. 3A). The difference was small, but was consistent with a higher fraction of mitotic cells for the miRNA‐overexpressing cells at 48 h after irradiation, and therefore presumably a larger extent of mitotic catastrophe in these cells (Fig. 3B). Both miRNA‐overexpressing cells and controls showed, however, the same elevated G2/M‐fraction at 48 h and a considerable arrest in these cell cycle phases (Fig. S9A,B). Moreover, no difference was found in the kinetics of DSB repair between miRNA‐overexpressing cells and controls, as judged from the γH2Ax levels (Fig. 3C). Thus, miR‐200a/b/‐429 overexpression led to only a small increase in cellular radiosensitivity and no detectable effect on DSB repair capacity in vitro.
Fig. 3.
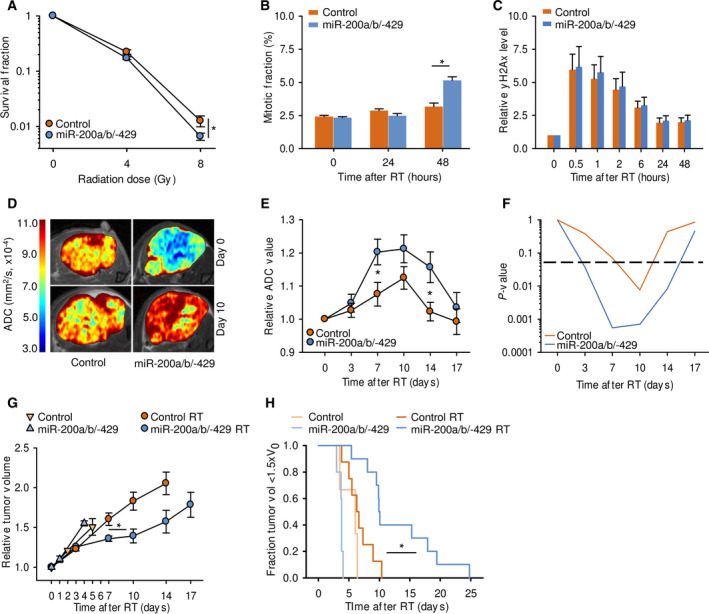
Radiation response of tumor models with stable miR‐200a/b/‐429 overexpression. (A) Clonogenic survival fraction of miRNA‐overexpressing cells and controls after exposure to 4 or 8 Gy, three independent experiments (mean ± SEM; *P < 0.01 from Students T‐test). (B) Mitotic fraction in miRNA‐overexpressing cells and controls after exposure to 8 Gy, four independent experiments (mean ± SEM; *P < 0.05 from Student's T‐test). (C) Relative γH2Ax level of miRNA‐overexpressing cells and controls after exposure to 8 Gy. Median γH2Ax intensity of each sample are presented relative to nonirradiated cells, four independent experiments (mean ± SEM). No significant difference between groups was observed with Student's T‐test. (D) ADC maps of representative tumors before (day 0) and 10 days after exposure to 8 Gy. ADC‐level scale bar is included. (E) ADC in miRNA‐overexpressing tumors (n = 10) and controls (n = 8) after exposure to 8 Gy. Data relative to ADC at day 0 are shown (mean ± SEM; *P < 0.05 from Students T‐test). (F) Significance level (P‐values) of the relative change in ADC from day 0 in miRNA‐overexpressing tumors and in controls based on the data in (E) (one sample Student's T‐test; stapled line, significance level P = 0.05). (G) Tumor volume of miRNA‐overexpressing xenograft tumors (4 nonirradiated, 10 irradiated) and controls (3 nonirradiated, 8 irradiated) after exposure of the irradiated groups to 8 Gy. Data relative to volume at day 0 are shown (mean ± SEM; *P = 0.01 from Student's T‐test on T 1.5×, irradiated miRNA‐overexpressing tumors versus controls). (H) Kaplan–Meier curves of the tumors shown in (G), using fraction of tumors with a volume below 1.5 times the baseline volume (day 0). The Holm–Sidak method was used for all pairwise multiple comparisons (*P < 0.05).
In xenograft tumors, the radiosensitizing effect of miR‐200a/b/‐429 overexpression appeared to be more prominent. The miRNA‐overexpressing tumors showed a significantly higher and more persistent increase in ADC after RT with 8 Gy compared to control tumors (P < 5.0 × 10−2; Fig. 3D–F), reflecting a greater loss of cellularity and development of necrosis [39, 40]. Moreover, RT led to a significant larger growth delay in these tumors than in irradiated controls (P = 1.3 × 10−2; Fig. 3G, Table S3). The radiation response enhancement factor (EF) by miR‐200a/b/‐429 overexpression was found to be as high as 6.6 (Table S3). RT caused a marginal, nonsignificant growth delay in control tumors (Fig. 3G), showing that a dose of 8 Gy had minor effect in these tumors with originally low miR‐200a/b/‐429 expression. The increased radiation response in miRNA‐overexpressing tumors was also evident when Kaplan–Meier curves were compared between the experimental groups (P = 1.9 × 10−2; Fig. 3H). Altogether, although miR‐200a/b/‐429 overexpression had no large effect on radiosensitivity in vitro, it seemed to significantly increase radiosensitivity in vivo. This supported our hypothesis that low miR‐200a/b/‐429 expression in tumors is associated with increased radioresistance.
3.4. miR‐200a/b/‐429 target genes show enrichment of processes involving interactions with ECM
To better understand the role of miR‐200a/b/‐429 as regulators of radiation response in patients, we evaluated potential target genes of the miRNAs. In total 7613 unique, validated or predicted target genes of miR‐200a‐5p/‐3p, 200b‐5p/‐3p, and/or miR‐429 were collected from the miRGate database. Out of these, 225 genes showed significant inverse correlation between gene and miRNA expression in both the explorative cohort and validation cohort 1 (Table S4). They were therefore considered as potential target genes in our patient cohorts. The strong inverse correlation implied that these genes were upregulated in tumors with low miRNA expression. The correlation data in Table S4 were used to construct a regulatory network with the miRNAs as nodes and their target genes as interaction partners. We used a cutoff FDR q‐value of 0.1 to identify interactions; that is pairs of miRNA and target gene, for the network. The network visualized pronounced overlap in the target genes of the five miRNAs, where 98 out of 225 (43%) genes had an interaction with more than one miRNA (Fig. 4A). Moreover, each miRNA had more than 30 potential target genes (range 31–118).
Fig. 4.
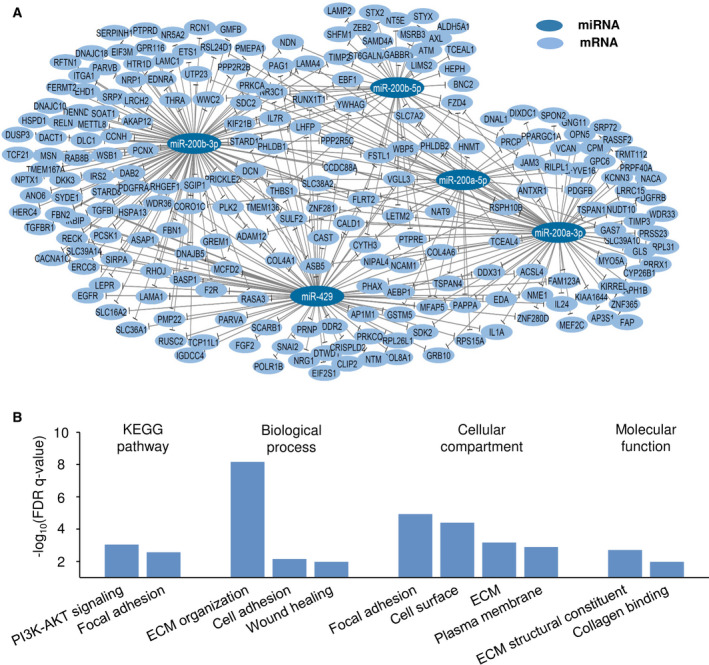
Regulatory network and biological processes associated with miR‐200a/b/‐429. (A) miR‐200a/‐b/‐429 regulatory network. Each miRNA in the miR‐200a/‐b/‐429 cluster is used as node. The interaction partners are their potential target genes, identified from correlation analysis of miRNA and gene expression in the explorative cohort (n = 90) and validation cohort 1 (n = 110). The network was visualized using cytoscape software v3.6.1. (B) Enriched KEGG pathways and GO terms in the list of 225 potential miR‐200a/b/‐429 target genes. Y‐axis denotes significance level from the functional annotation tool david v6.8.
Gene ontology and KEGG pathway analysis of the list of all 225 potential target genes was further performed to identify biological processes, cellular compartments, and molecular functions that were enriched, using the global human transcriptome as background. Several interconnected pathways and processes appeared significant, including focal adhesion, ECM organization and the PI3K‐AKT pathway (Fig. 4B; Table S5A). In line with this result, cellular compartments and molecular functions related to ECM were enriched. The list of all 7613 miRGate registered, candidate target genes for the five miRNAs contained genes involved in a large variety of biological processes. A GO and KEGG pathway analysis of the 225 potential target genes was also performed by using these 7613 genes as background. The analysis led to similar results as those presented in Fig. 4B (Table S5B). Downregulation of miR‐200a/b/‐429 therefore seemed to be associated with cell‐ECM interactions and activation of PI3K‐AKT signaling. Moreover, these biological processes represented a specific part of the repertoire of processes controlled by miR‐200a/b/‐429.
4. Discussion
Our study identifies a role of the miR‐200a/b/‐429 cluster in tumor radioresistance of cervical cancer. By assessing one of the largest cohorts of patients to date with paired miRNA and site‐specific recurrence data, we showed that miR‐200a/b/‐429 had prognostic impact that was mainly caused by an association with central pelvic recurrence. A unique data set of hypoxia status was also available for all patients and showed that this radioresistance mechanism was not involved. Tumor models with stable miR‐200a/b/‐429 expression were developed for proof‐of‐principle studies, which validated a role of the miRNAs in radiation response. Further, to verify functional activity of miRNAs and understand which pathways they regulate, analysis of their target genes in patient tumors is essential. By assessing global gene expression data of all patients, we addressed this in an explorative manner that helped to propose the resistance mechanisms involved. Our results encourage development of miR‐200a/b/‐429 as a diagnostic biomarker, identifying patients in need for radiosensitizing therapy.
Expression of mature miRNAs derived from both the 5′ and 3′ arm of the miR‐200a and miR‐200b precursor duplex, in addition to miR‐429, was associated with cancer progression in our survival analyses. Moreover, potential target genes were identified for all five members. It is therefore likely that all of them, including the 5p/3p pairs, are functionally active in cervical cancer and regulate different genes, in accordance with reports on 5p/3p pairs of other miRNAs in cancer cells [41, 42]. In addition, all members showed considerable variation in expression level across patients, which is a requirement for further biomarker development [43]. Although highly co‐expressed, all five members were therefore considered in our work. In contrast, expression of miR‐141 or miR‐200c seemed to be less important for chemoradiotherapy resistance in cervical cancer.
Our data suggested that miR‐200a/b/‐429 is associated with treatment resistance locally in the tumor, and not with development of metastases. This hypothesis was further supported by radiation experiments in tumor models. It should be noted, however, that downregulation of miR‐200a/b/‐429 also might increase resistance to the concurrent cisplatin, as indicated for miR‐200b in experimental studies of ovarian cancer [44]. The miR‐200a/b/‐429 target genes were shown to be involved in ECM organization and cell adhesion processes. Such EMT processes are known to be regulated by these miRNAs [11], and are key mechanisms for maintaining a mesenchymal phenotype in cancer cells [45]. Moreover, cancer cell's interaction with ECM has been associated with radioresistance in several cancer types, a mechanism referred to as cell adhesion‐mediated radioresistance [14, 15, 46]. Involvement of this resistance mechanism in our patients is likely. Hence, it was supported by experimental studies, showing a larger radiosensitizing effect of miR‐200a/b/‐429 overexpression in tumors compared to cell lines. Thus, the effect in cell lines was minor and much less than the radiosensitizing effect of for example oxygen in SiHa cells, where the cell death fraction after 8 Gy of irradiation is 10‐fold higher under normoxic than hypoxic conditions [47]. EMT has been implicated in chemotherapy resistance in previous work on cervical cancer, but only few studies have addressed its importance for radioresistance [48]. Our work therefore revealed a novel radioresistance mechanism in this disease that appears to involve interactions between cancer cells and ECM and is mediated by miR‐200a/b/‐429 downregulation.
A limitation of our experimental studies was the use of a single model system. However, the experiments were guided by findings in our patient material, and aimed to provide a proof‐of‐principle regarding radiation response. Moreover, the model was tested both in vitro and in vivo. Our conclusions are therefore most likely sound and robust with the use of a single model system.
Several downstream pathways mediated by EMT and cell adhesion can be involved in the development of radioresistance. We found enrichment of genes in the PI3K‐AKT pathway among the potential miR‐200a/b/‐429 target genes in patients, indicating that this pathway is important. Downregulation of the miR‐200 family has been associated with activation of this pathway in other cancer types, promoting cell proliferation and survival and regulating intracellular pathways in tumor invasion [49]. Moreover, mutation of pathway member PI3KCA and phosphorylation of AKT have been associated with poor prognosis in cervical cancer [50, 51, 52, 53, 54], showing that PI3K‐AKT is important in progression of this disease. Other pathways controlled by miR‐200a/b/‐429 may, however, also influence radiosensitivity, like focal adhesion‐mediated chromatin modulation via the cytoskeleton [55].
Tumor hypoxia was explored in our work as a potential mechanism underlying downregulation of miR‐200a/b/‐429. This hypothesis was based on studies in cell lines from breast cancer, colorectal cancer and in human endothelial cells [16, 17, 18]. However, our studies on both patient material and model system indicated that this regulation mechanism is not important in cervical cancer. Other mechanisms like epigenetic modulation of the miR‐200a/b/‐429 cluster have been demonstrated in some cancer types [56], and should be investigated. Analysis of tumor hypoxia against our site‐specific recurrence data further showed that its correlation with locoregional control, as reported in previous work [19] mainly reflected an effect on central pelvic recurrence. Hence, we here propose that at least two independent tumor radioresistence mechanisms are important in cervical cancer, one involving downregulation of miR‐200a/b/‐429 and another hypoxia.
The association between the miR‐200a/b/‐429 score and central pelvic recurrence was independent of conventional clinical markers. The expression level of this miRNA cluster may therefore contain information about tumor radioresistance that is not covered by current diagnostics. This encourages development of a diagnostic test for patients referred to chemoradiotherapy, measuring miR‐200a/b/‐429 expression. Decision about the optimal test assay, definition of a cutoff score for classifying patients to the low expression group, and establishment of an internal control would be important aspects of such development. An RT‐qPCR kit, which includes reference miRNAs with stable expression in tumor biopsies [24], could be an appropriate low‐cost approach. It should also be considered whether a refined set of miRNAs would improve the specificity and sensitivity of the test. Moreover, investigations of whether the miRNAs are detectable in blood or plasma and their potential as a blood based test would be of considerable interest. Successful establishment of a test would help to select patients for clinical trials on strategies to overcome tumor resistance, and to exclude patients with no expected benefit. Several exciting strategies are upcoming, like combination therapy with radiation and targeted radiosensitizers, including inhibitors of the PI3K‐AKT pathway and of cell–matrix interactions [57], or escalation of the radiation dose within normal‐tissue tolerance by proton therapy [58].
5. Conclusion
We have identified miR‐200a/b/‐429 as an independent predictor of central pelvic recurrence in cervical cancer and a promising candidate biomarker for tumor radioresistance. Downregulation of miR‐200a/b/‐429 represents a new radioresistance mechanism that operates independently of hypoxia and seems to involve interaction between cancer cells and ECM.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
AN and HL conceived and designed the project. AN, THi, VES, E‐KA, TSE, and BG acquired the data and performed the experiments. AN, THi, THo, VES, and TSE analyzed the data and interpreted the results. CSF, THo, GBK, and TS provided resources and data curation. AN and HL wrote and edited the paper with input from all other authors.
Supporting information
Fig. S1. Representative example showing gating of cell subpopulations.
Fig. S2. IHC‐staining of xenograft tumor sections.
Fig. S3. Definition of necrotic and hypoxic areas in xenograft tumor sections.
Fig. S4. Cell segmentation.
Fig. S5. Expression of the miR‐200 family in cervical cancer.
Fig. S6. Heteroscedasticity analysis.
Fig. S7. In vitro characterization of miR‐200a/b/‐429‐overexpressing SiHa cells.
Fig. S8. In vivo characterization of miR‐200a/b/‐429‐overexpressing SiHa tumors.
Fig. S9. Radiation effect on cell cycle distribution in miR‐200a/b/‐429‐overexpressing SiHa cells.
Table S1. Patient characteristics.
Table S2. miR‐200a/b/‐429 expression score versus clinical markers and hypoxia status for 200 patients.
Table S3. Tumor growth delay (TDG) in miR‐200a/b/‐429‐overexpressing xenografts after radiation treatment (RT).
Table S4. Spearman correlation between miRNA and gene expression for the potential target genes of miR‐200a/b/‐429.
Table S5. GO and KEGG enrichment analyses.
Acknowledgements
Financial support was received from The Norwegian Cancer Society (grant numbers 107438 and 182451) and The South‐Eastern Norway Regional Health Authority (grant numbers 2015020 and 2020044). Technical assistance from I. D. Rein, Flow Cytometry Core Facility at Oslo University Hospital and D. Trinh, Dept. of Pathology at Oslo University Hospital, is highly appreciated.
Contributor Information
Anja Nilsen, Email: anja.nilsen@rr-research.no.
Heidi Lyng, Email: heidi.lyng@rr-research.no.
References
- 1. Kobayashi R, Yamashita H, Okuma K, Ohtomo K, Nakagawa K. Details of recurrence sites after definitive radiation therapy for cervical cancer. J Gynecol Oncol. 2016;27:e16. 10.3802/jgo.2016.27.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Legge F, Chiantera V, Macchia G, Fagotti A, Fanfani F, Ercoli A, et al. Clinical outcome of recurrent locally advanced cervical cancer (LACC) submitted to primary multimodality therapies. Gynecol Oncol. 2015;138:83–8. 10.1016/j.ygyno.2015.04.035 [DOI] [PubMed] [Google Scholar]
- 3. Teh J, Yap SP, Tham I, Sethi VK, Chua EJ, Yeo R, et al. Concurrent chemoradiotherapy incorporating high‐dose rate brachytherapy for locally advanced cervical carcinoma: survival outcomes, patterns of failure, and prognostic factors. Int J Gynecol Cancer. 2010;20:428–33. 10.1111/IGC.0b013e3181d4a0d1 [DOI] [PubMed] [Google Scholar]
- 4. Gadducci A, Tana R, Cosio S, Cionini L. Treatment options in recurrent cervical cancer (Review). Oncol Lett. 2010;1:3–11. 10.3892/ol_00000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshida K, Kajiyama H, Utsumi F, Niimi K, Sakata J, Suzuki S, et al. A post‐recurrence survival‐predicting indicator for cervical cancer from the analysis of 165 patients who developed recurrence. Mol Clin Oncol. 2018;8:281–5. 10.3892/mco.2017.1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adiga D, Eswaran S, Pandey D, Sharan K, Kabekkodu SP. Molecular landscape of recurrent cervical cancer. Crit Rev Oncol Hematol. 2021;157:103178. 10.1016/j.critrevonc.2020.103178 [DOI] [PubMed] [Google Scholar]
- 7. Lin LL, Lakomy DS, Ning MS, Simpkins F, Jhingran A. Combining novel agents with radiotherapy for gynecologic malignancies: beyond the era of cisplatin. Int J Gynecol Cancer. 2020;30:409–23. 10.1136/ijgc-2020-001227 [DOI] [PubMed] [Google Scholar]
- 8. Korpela E, Vesprini D, Liu SK. MicroRNA in radiotherapy: miRage or miRador? Br J Cancer. 2015;112:777–82. 10.1038/bjc.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. 10.1016/j.molonc.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network , Albert Einstein College of Medicine , Analytical Biological Services , Barretos Cancer Hospital , Baylor College of Medicine , Beckman Research Institute of City of Hope , et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–84. 10.1038/nature21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Humphries B, Yang C. The microRNA‐200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–98. 10.18632/oncotarget.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, et al. A microRNA expression signature for cervical cancer prognosis. Can Res. 2010;70:1441–8. 10.1158/0008-5472.CAN-09-3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pedroza‐Torres A, Fernandez‐Retana J, Peralta‐Zaragoza O, Jacobo‐Herrera N, Cantu de Leon D, Cerna‐Cortes JF, et al. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 2016;142:557–65. 10.1016/j.ygyno.2016.07.093 [DOI] [PubMed] [Google Scholar]
- 14. Henke E, Nandigama R, Ergün S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front Mol Biosci. 2019;6:160. 10.3389/fmolb.2019.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu T‐S, Lin B‐R, Chang H‐H. Radio resistance mechanisms of cancers: an overview and future perspectives. Biol Med. 2015;s2:002. https://pdfs.semanticscholar.org/fa05/d46f6a33e63cd2060de72f459a4fae1dded6.pdf [Google Scholar]
- 16. Bartoszewska S, Kochan K, Piotrowski A, Kamysz W, Ochocka RJ, Collawn JF, et al. The hypoxia‐inducible miR‐429 regulates hypoxia‐inducible factor‐1α expression in human endothelial cells through a negative feedback loop. FASEB J. 2015;29:1467–79. 10.1096/fj.14-267054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shang Y, Chen H, Ye J, Wei X, Liu S, Wang R. HIF‐1alpha/Ascl2/miR‐200b regulatory feedback circuit modulated the epithelial‐mesenchymal transition (EMT) in colorectal cancer cells. Exp Cell Res. 2017;360:243–56. 10.1016/j.yexcr.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 18. van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. 10.1038/ncomms6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fjeldbo CS, Julin CH, Lando M, Forsberg MF, Aarnes EK, Alsner J, et al. Integrative analysis of DCE‐MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia‐related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22:4067–76. 10.1158/1078-0432.CCR-15-2322 [DOI] [PubMed] [Google Scholar]
- 20. Fyles A, Milosevic M, Hedley D, Pintilie M, Levin W, Manchul L, et al. Tumor hypoxia has independent predictor impact only in patients with node‐negative cervix cancer. J Clin Oncol. 2002;20:680–7. 10.1200/JCO.2002.20.3.680 [DOI] [PubMed] [Google Scholar]
- 21. Hockel M, Schlenger K, Aral B, Mitze M, Schaffer U, Vaupel P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Can Res. 1996;56:4509–15. [PubMed] [Google Scholar]
- 22. van Persijn van Meerten EL, Gelderblom H, Bloem JL. RECIST revised: implications for the radiologist. A review article on the modified RECIST guideline. Eur Radiol. 2010;20:1456–67. 10.1007/s00330-009-1685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72. 10.1200/JCO.2004.01.0454 [DOI] [PubMed] [Google Scholar]
- 24. Nilsen A, Jonsson M, Aarnes EK, Kristensen GB, Lyng H. Reference microRNAs for RT‐qPCR assays in cervical cancer patients and their application to studies of HPV16 and hypoxia biomarkers. Transl Oncol. 2019;12:576–84. 10.1016/j.tranon.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. How C, Pintilie M, Bruce JP, Hui AB, Clarke BA, Wong P, et al. Developing a prognostic micro‐RNA signature for human cervical carcinoma. PLoS One. 2015;10:e0123946. 10.1371/journal.pone.0123946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andres‐Leon E, Gonzalez Pena D, Gomez‐Lopez G, Pisano DG. miRGate: a curated database of human, mouse and rat miRNA‐mRNA targets. Database. 2015;2015:bav035. 10.1093/database/bav035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25:60–83. 10.3102/10769986025001060 [DOI] [Google Scholar]
- 29. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR‐200 family and miR‐205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 31. Hillestad T, Hompland T, Fjeldbo CS, Skingen VE, Salberg UB, Aarnes EK, et al. MRI distinguishes tumor hypoxia levels of different prognostic and biological significance in cervical cancer. Can Res. 2020;80:3993–4003. 10.1158/0008-5472.CAN-20-0950 [DOI] [PubMed] [Google Scholar]
- 32. R Core Team . A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. https://www.R‐project.org/ [Google Scholar]
- 33. Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB‐ArrayTools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 34. Camp RL, Dolled‐Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res. 2004;10:7252–9. 10.1158/1078-0432.CCR-04-0713 [DOI] [PubMed] [Google Scholar]
- 35. Faul F, Erdfelder E, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–60. 10.3758/BRM.41.4.1149 [DOI] [PubMed] [Google Scholar]
- 36. Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module‐centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial‐mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial‐to‐mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Afaq A, Andreou A, Koh DM. Diffusion‐weighted magnetic resonance imaging for tumour response assessment: why, when and how? Cancer Imaging. 10(Spec no A):S179–88. 10.1102/1470-7330.2010.9032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koh DM, Scurr E, Collins D, Kanber B, Norman A, Leach MO, et al. Predicting response of colorectal hepatic metastasis: value of pretreatment apparent diffusion coefficients. AJR Am J Roentgenol. 2007;188:1001–8. 10.2214/AJR.06.0601 [DOI] [PubMed] [Google Scholar]
- 41. Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA‐5p and ‐3p co‐expression and cross‐targeting in colon cancer cells. J Biomed Sci. 2014;21:95. 10.1186/s12929-014-0095-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, et al. miR‐126 and miR‐126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–94. 10.1038/ncb2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hristova VA, Chan DW. Cancer biomarker discovery and translation: proteomics and beyond. Expert Rev Proteomics. 2019;16:93–103. 10.1080/14789450.2019.1559062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu J, Zhang X, Huang Y, Zhang Q, Zhou J, Zhang X, et al. miR‐200b and miR‐200c co‐contribute to the cisplatin sensitivity of ovarian cancer cells by targeting DNA methyltransferases. Oncol Lett. 2019;17:1453–60. 10.3892/ol.2018.9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial‐mesenchymal transition. Cell Adh Migr. 2015;9:317–24. 10.1080/19336918.2015.1016686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sandfort V, Koch U, Cordes N. Cell adhesion‐mediated radioresistance revisited. Int J Radiat Biol. 2007;83:727–32. 10.1080/09553000701694335 [DOI] [PubMed] [Google Scholar]
- 47. Pajonk F, Grumann T, McBride WH. The proteasome inhibitor MG‐132 protects hypoxic SiHa cervical carcinoma cells after cyclic hypoxia/reoxygenation from ionizing radiation. Neoplasia. 2006;8:1037–41. 10.1593/neo.06634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett. 2015;356:321–31. 10.1016/j.canlet.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 49. Guo L, Wang J, Yang P, Lu Q, Zhang T, Yang Y. MicroRNA‐200 promotes lung cancer cell growth through FOG2‐independent AKT activation. IUBMB Life. 2015;67:720–5. 10.1002/iub.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim TJ, Lee JW, Song SY, Choi JJ, Choi CH, Kim BG, et al. Increased expression of pAKT is associated with radiation resistance in cervical cancer. Br J Cancer. 2006;94:1678–82. 10.1038/sj.bjc.6603180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lachkar B, Minaguchi T, Akiyama A, Liu S, Zhang S, Xu C, et al. Prognostic significance of PIK3CA mutation in stage IIB to IVA cervical cancers treated by concurrent chemoradiotherapy with weekly cisplatin. Medicine (Baltimore). 2018;97:e11392. 10.1097/MD.0000000000011392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McIntyre JB, Wu JS, Craighead PS, Phan T, Köbel M, Lees‐Miller SP, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. 2013;128:409–14. 10.1016/j.ygyno.2012.12.019 [DOI] [PubMed] [Google Scholar]
- 53. Schwarz JK, Payton JE, Rashmi R, Xiang T, Jia Y, Huettner P, et al. Pathway‐specific analysis of gene expression data identifies the PI3K/Akt pathway as a novel therapeutic target in cervical cancer. Clin Cancer Res. 2012;18:1464–71. 10.1158/1078-0432.CCR-11-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013;119:3776–83. 10.1002/cncr.28288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deville SS, Cordes N. The extracellular, cellular, and nuclear stiffness, a trinity in the cancer resistome – a review. Front Oncol. 2019;9:1376. 10.3389/fonc.2019.01376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic modulation of the miR‐200 family is associated with transition to a breast cancer stem‐cell‐like state. J Cell Sci. 2013;126:2256–66. 10.1242/jcs.122275 [DOI] [PubMed] [Google Scholar]
- 57. Hintelmann K, Kriegs M, Rothkamm K, Rieckmann T. Improving the efficacy of tumor radiosensitization through combined molecular targeting. Front Oncol. 2020;10:1260. 10.3389/fonc.2020.01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vanderwaeren L, Dok R, Verstrepen K, Nuyts S. Clinical progress in proton radiotherapy: biological unknowns. Cancers. 2021;13:604. 10.3390/cancers13040604 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative example showing gating of cell subpopulations.
Fig. S2. IHC‐staining of xenograft tumor sections.
Fig. S3. Definition of necrotic and hypoxic areas in xenograft tumor sections.
Fig. S4. Cell segmentation.
Fig. S5. Expression of the miR‐200 family in cervical cancer.
Fig. S6. Heteroscedasticity analysis.
Fig. S7. In vitro characterization of miR‐200a/b/‐429‐overexpressing SiHa cells.
Fig. S8. In vivo characterization of miR‐200a/b/‐429‐overexpressing SiHa tumors.
Fig. S9. Radiation effect on cell cycle distribution in miR‐200a/b/‐429‐overexpressing SiHa cells.
Table S1. Patient characteristics.
Table S2. miR‐200a/b/‐429 expression score versus clinical markers and hypoxia status for 200 patients.
Table S3. Tumor growth delay (TDG) in miR‐200a/b/‐429‐overexpressing xenografts after radiation treatment (RT).
Table S4. Spearman correlation between miRNA and gene expression for the potential target genes of miR‐200a/b/‐429.
Table S5. GO and KEGG enrichment analyses.


