Abstract
About 1.2 to 33% of high-altitude populations suffer from Monge’s disease or chronic mountain sickness (CMS). Number of factors such as age, sex, and population of origin (older, male, Andean) contribute to the percentage reported from a variety of samples. It is estimated that there are around 83 million people who live at altitudes > 2500 m worldwide and are at risk for CMS. In this review, we focus on a human “experiment in nature” in various high-altitude loca-tions in the world—namely, Andean, Tibetan, and Ethiopian populations that have lived under chronic hypoxia conditions for thousands of years. We discuss the adaptive as well as mal-adaptive changes at the genomic and physiological levels. Although different genes seem to be involved in adaptation in the three populations, we can observe convergence at genetic and signaling, as well as physiological levels. What is important is that we and others have shown that lessons learned from the genes mined at high altitude can be helpful in better understanding and treating diseases that occur at sea level. We discuss two such examples: EDNRB and SENP1 and their role in cardiac tolerance and in the polycythemic response, respectively.
Keywords: High-altitude adaptation, Chronic mountain sickness, Genomics, Polycythemic response, Cardiovascular response
Introduction
Stable and continuous oxygen supply to tissues is essential for the functional integrity of cells and survival of mammals, include-ing humans. To ensure sufficient oxygen delivery to all cells in the body, animals have developed a sophisticated physiological system through millions of years of evolution. This system includes an oxygen uptake site (the lungs), an O2 carrier (eryth-rocytes), circulating and dispensing channels (the vasculature), and a muscular contracting pump (the heart). The precise regulation of the various components of such a system is critical for keeping O2 homeostasis; a malfunction of any component of this O2 delivery system might limit O2 supply, resulting in cellular hypoxia. Such hypoxia is a rather common pathophysiologic feature in many human diseases, including cardiovascular, respi-ratory, and neurologic diseases [1,2]. Hence, understanding the mechanisms that regulate O2 homeostasis is critical for develop-ing novel diagnostic markers and therapeutic strategies.
A number of strategies have been used to learn about and dissect such mechanisms. For example, (a) some investigators have studied the mechanisms that lead to injury as a result of hypoxia in the brain or heart of mammals [3, 4]; (b) others have studied the mechanisms that are responsible for susceptibility and tolerance in vertebrates or invertebrates [5–7], re-spectively; and (c) still others have used an interesting and attractive approach of studying animals and humans that have lived at high altitude for thousands of years to determine how these high-altitude dwellers confronted the severe challenge of hypoxic environments [8]. About 1.2 to 33% of high-altitude populations suffer from Monge’s disease or chronic mountain sickness (CMS) [9–15] and the prevalence of CMS depends on a number of factors such as age, sex, altitude, and population of origin. A number of these populations have been under natural selection ~ 5000 to possibly 70,000 years [16–20] for which multiple hypoxia-adaptive strategies have been selected [8, 21–23]. It is estimated that there are around 83 million people who live at altitudes > 2500 m worldwide and are at risk for CMS [24]. Among human highlander populations, the Ethiopians in Africa, the Tibetans in Asia, and the Andeans in South America have been the most studied using a variety of tools to dissect the adaptive mechanism(s) at the level of ge-nomics, molecular biology, and physiological responses [8, 21, 24–26].
The fact that makes highlander populations even more interesting is that some sub-populations (e.g., the Andean high-landers) have not adapted as well as other sub-populations. In fact, about 15.4% of the Andean highlanders (from Cerro de Pasco (4330 m), age group 30–39; and around 33% by age group 50–59), mostly males, are maladapted to low levels of inspired O2 and suffer from CMS, also known as Monge’s disease [9, 10, 13, 27]. These populations in the Andes offer the opportunity to study the differences between adaptation and mal-adaptation and hence help in dissecting the mechanisms underlying adaptation or the lack thereof. From a phenotypic point of view, subjects with CMS suffer mostly from neurologic, cardiovascular, and hematologic ailments. Signs and symptoms of CMS include headache, dizziness, breath-lessness, palpitations, sleep disturbance, mental fatigue, con-fusion, pulmonary and systemic hypertension, heart failure, and polycythemia [9, 10, 28–35]. Polycythemia (hematocrit > 65%) and hypoxemia (O2—percent of oxygen saturation of hemoglobin < 85%) render CMS subjects very vulnerable to stroke and myocardial infarction mainly due to an increase in blood viscosity resulting in a sluggish circulation and obstruct-tion of the blood supply to susceptible organs such as brain and heart [9, 10, 30, 33, 34].
Genomic signature of hypoxia adaptation
Under the selection pressure of hypoxia, specific mutations that confer an evolutionary advantage increase in frequency in the population under selection, relative to other control populations. For a Mendelian genotype, selection would lead to a hard sweep, or to a soft selective sweep with standing variation. However, the adaptation to chronic hypoxia is polygenic and involves many biological pathways. Individuals carrying different adaptive mutations are competing, slowing the fixation process and the strength of the signal. Moreover, geographically isolated populations could have evolved different adaptation mechanisms. Researchers have applied different genomic strategies to samples to uncover these mechanisms. One approach is to look for an association between allele frequency difference in highlanders (case) and lowlanders (control) or adapted (non-CMS) and non-adapted (CMS) individuals. These association tests may not have high power due to the polygenic nature of adaptation, but have been useful to identify genes with strong frequency differential.
An alternative strategy exploits the fact that hypoxia adaptation in humans is relatively recent (< 1000 generations), with a strong selection. Regions under selection have long haplotypes with low diversity, which can be identified with smaller sample sizes. Studies have used exome sequencing [36] and genotyping [23, 37], but whole genome studies with a dense sampling of variants and no ascertainment bias offer the best results [38, 39].
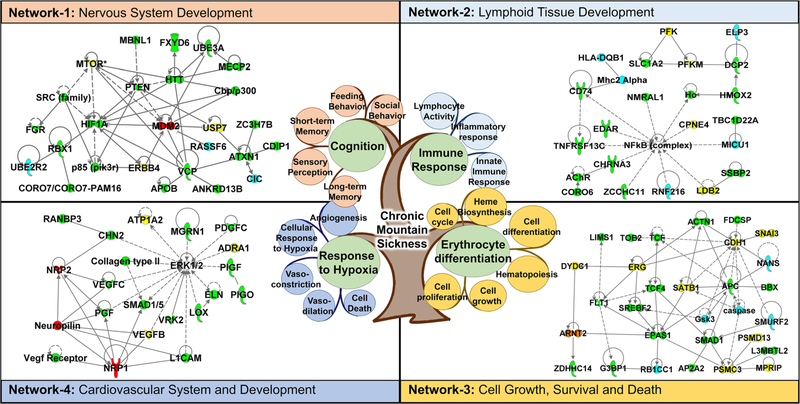
Genomic studies to date have yielded a wealth of valuable information. Importantly, they have increased our apprecia-tion of the complexity of the molecular response to high-altitude adaptation. A survey of the literature has suggested that > 1000 genes are potentially involved in high-altitude adaptation to hypoxia in different human populations across the world (supplementary information Table S1) [23, 29, 37, 39–58]. (For the selection of genes, we picked only the prior-itized genes from each referenced article. The prioritization is usually based on significance levels reported in the respective article). Using a knowledge-based bioinformatics tool, i.e., Ingenuity Pathway Analysis (IPA) analysis tools (QIAGEN Bioinformatics, Redwood City, CA), we found that many cell signaling pathways were enriched in this dataset (see Table S2 for the top signaling pathways), suggesting that physiological adaptation to high altitude requires coordinated cellular signaling at a molecular level. Furthermore, many of the candidate genes also formed various networks that are predicted to regulate biological processes and physiological functions during development. For example, interactions between the candidate genes that were obtained by analyzing Andean, Ethiopian, and Tibetan highlander populations suggested that there is a coordination of functions and developmental processes in various organs/tissues, such as the function and development of the neuronal, hematological, and cardiovascular systems as well as the gene-gene interactions regulating cell death and survival (Fig. 1). Therefore, it is interesting to note that although different genetic networks involved genes that were identified in different human populations, we hypothesized that these genes may nonetheless regulate similar biological processes and physiological functions.
Fig. 1.
Representative genetic interactions (as deciphered by IPA analysis) regulating biological processes and physiological functions that are involved in human adaption to high altitude. The central figure shows the complexity of the pathology linked to chronic mountain sickness. It involves various physiological responses such as erythrocyte differentiation, immune response, and response to hypoxia. Network 1 shows the gene interactions regulating post-translational modification, cell-to-cell signaling and interaction, and nervous system development and function. Network 2 shows the genetic network regulating hematological system development and function, immunological disease, and lymphoid tissue structure and development. Network 3 depicts the genetic interactions regulating cell death and survival, gene expression, and cell cycle particularly during hematopoiesis. Network 4 shows the genetic network regulating cardiovascular system development and function, tissue development, and organismal development. Color code: yellow—genes identified in Andean population, blue—genes identified in Ethiopian population, green—genes identified in Tibetan population, and red—genes identified in both Andean and Tibetan populations
Human high-altitude studies have shed light on the function of genes involved in adaptation
Most studies that aim to understand the genetic basis for adaptation to high altitude rely on statistical association between the gene(s) and phenotype. While this is an important first step towards understanding the complexity of phenotypes such as CMS, ultimate tests (functional tests) must uncover causal relationships. Functional and mech-anistic studies are currently limited in the field. Table 1 shows studies that have functionally tested the role of specific genes and uncovered mechanism(s) explaining certain traits. Model organisms such as Drosophila and mice provide good in vivo model systems for studying the mechanism(s) related to hypoxia tolerance— endothelin receptor type B (EDNRB) is a good example for such a study (Table 1). Besides the examples mentioned in Table 1, hypoxia-inducible factor (HIF) and its pathway components have been analyzed extensively using mouse models (as reviewed in [37]). However, there are limitations to using model systems. First, there are genetic differences between the multiple species and hence, a true ortholog for a particular gene may not be available. Second, in certain instances, the loss of function of a gene can cause embryonic lethality (e.g., sentrin-specific protease 1 (SENP1) KO are embryonic lethal due to severe anemia) and make it difficult to study the function in vivo. In vitro human or murine cell lines provide another useful alternative resource for testing the functional role of genes and have been an invaluable asset over decades. Nevertheless, some-times the lack of specific cell lines or phenotype can restrict the studies particularly in complex disorders such as CMS. Recently, some studies (Table 1) (HMOX2, EPAS1) have used cell lines to validate the molecular mechanisms linked with these genes [60, 61]. With the advent of newer technology, molecular and genetic tools have further facilitated these studies. For example, using hematopoietic stem cells (HSCs) from peripheral blood and colony proliferation assays (BFU), the study by Lorenzo et al. elegantly shows how a missense mutation in the EGLN1 gene is functionally linked to erythropoiesis in the Tibetan population [43]. Similarly, our recent study (Azad et al. [58]) has built a model of excessive erythropoiesis using induced pluripotent stem cells (iPSCs) generated from the Andean population (CMS and non-CMS) and their differentiation into RBCs under normoxia and hypoxia. Indeed, we were able to replicate hypoxia-induced poly-cythemia, in the dish, as well as functionally link SENP1 to the phenotype of excessive erythropoiesis. In spite of the fact that the iPSC technology has certain limitations, such as maturation of the somatic cells being differentiated, this system has tremendous potential and the field is progressing and refining the techniques rapidly. With the development of more efficient differentiation methods (such as 3D organoids, expansion techniques of HSCs), not only do we generate the right tissue type, but we can also improve on tissue maturation [63–66]. Such functional studies will tremens-dously increase our understanding of the mechanism(s) and will become a key factor in personalized medicine.
Table 1.
Human high-altitude studies that have shed light on the mechanism or function of the genes involved in adaptation
| Gene(s) | Population | Functional assay | Species used to assess function | Phenotype measured | Study |
|---|---|---|---|---|---|
| EGLN1 | Tibetan | In vitro BFU assays | Human | Excessive erythrocytosis | Lorenzo et al. 2014 [43] |
| PHD2 | Tibetan | IP and WB assays using in vitro cell lines | Human cell | Decreased ability of the mutant PHD2 to associate with the HSP90 cochaperone p23 | Song et al. 2014 [57] |
| SENP1 | Andean | In vitro iPSC-derived assays, BFU assays | Human | Excessive erythrocytosis | Azad et al. 2016 [58] |
| EDNRB | Ethiopian | In vivo assays | Mouse | Cardiac tolerance under acute hypoxia | Stobdan et al. 2015 [59] |
| EPAS1 | Tibetan | In vitro cell lines | Human | Repression and activation of EPAS1 targets | Xu et al. 2014 [60] |
| ANDP32D, SENP1 | Andean | Hypoxia tolerance | Drosophila | Eclosion rate under hypoxia | Zhou et al. 2013 [39] |
| CIC, LIPE, PAFAH1B3 | Ethiopian | Hypoxia tolerance | Drosophila | Eclosion rate under hypoxia | Udpa et al. 2014 [38] |
| HMOX2 | Tibetan | EMSA using in vitro cell lines | Human | Binding of transcriptional factor Sp1 | Yang et al. 2016 [61] |
| GUCY1A3 | Kyrgyz | In vitro reporter cell lines | Chinese hamster and Human | NO sensitivity | Wilkins et al. 2014 [62] |
Founder effects in the Andean, Tibetan, and Ethiopian populations
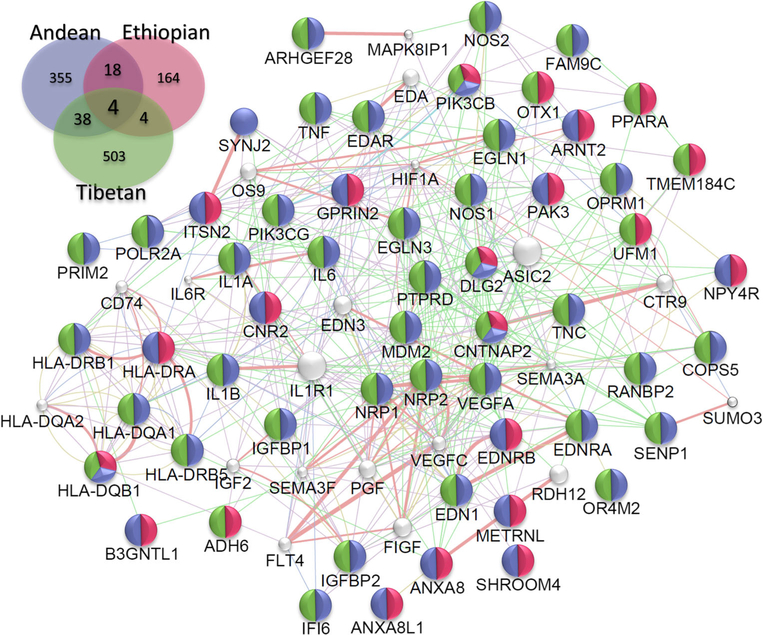
Indigenous human populations in the Andean, Tibetan, and Ethiopian regions are descendants of colonizers who came to these regions at different time periods [8, 21, 22, 24, 25, 67]. These populations are therefore great examples of three inde-pendent selection experiments-in-nature under similar selection pressure (in terms of extreme conditions, such as hypobaric hypoxia). Physiologically, it appears that different adaptation mechanisms have emerged. For example, in terms of the hematological response, a sub-population of Andeans, living at the same altitude as others in the Andes, represents a classical model with a phenotype of “excessive erythrocytosis and arterial hypoxemia.” Tibetans show “normal hemoglobin concentration with arterial hypoxemia” and Ethiopian highlanders maintain hemoglobin concentrations and arterial oxygen saturation within the ranges of sea-level populations [8, 21,22, 67]. Indeed, the Andean pattern of adaptation is characterized by higher hemoglobin concentrations, low oxygen saturation, and arterial oxygen levels [24, 68]. The Tibetan pattern is characterized by sea-level hemoglobin levels (unless at a very high levels of elevation), low oxygen saturation, and around 10% lower arterial O2 content. Ethiopians (particularly the Amhara ethnic group) have similar levels of hemoglobin concentration, oxygen saturation, and arterial O2 content to healthy sea-level individuals [24, 52, 68]. It seems that evolutionary selection related to oxygen utilization, transport, and homeostasis as well as founder effects have helped in the shaping of different patterns of adaptation in the three high-altitude populations. It is very likely, also, that natural selection is still operating and we are therefore observing different stages of the effects of selection on these three populations. These studies suggest that there are genetic differences that can be responsible for differences in the phenotypes we observe in these populations. Based on the data available, the three different populations must have adapted to the same selection pressure through different routes (i.e., genes, pathways, or mechanism(s)) to achieve a common goal of adaptation to a strong hypoxic stress and selection pres-sure. For Tibetans, EGLN1 and EPAS1 have been under positive selection, as demonstrated in multiple studies [43, 69–71]. In the Andean population, numerous studies, including ours, have pointed to different candidate genes, including ANP32D, SENP1, G allele NOS3, and VEGF loci that play a role in adaptation [39, 72–74]. In Ethiopians, CBARA1, VAV3, ARNT2, THRB, CIC, LIPE, and PAFAHIB3 have been associated to adaptation [19, 38, 52, 54]. Although we have observed different sets of genes in the three populations that are most likely related to founder effects, they converge on similar pathways to mitigate or counteract the negative effects of low oxy-gen. Figure 2 shows the genes that are common between the three populations and Fig. 3 shows the gene ontology (GO) processes that are significant in them. Besides convergence at the genetic level, what is also fascinating is that there is convergence at the level of physiological processes, such as erythropoietic and cardiovascular regulation and function.
Fig. 2.
Convergence at the genetic level. Insert is the Venn diagram depicting the common genes shared between three HA populations. We used Genemania v3.4.1 app on Cytospace v3.4.0. The network is based on the common gene (n = 64) that are shared at least in two populations. We restrict our analysis to Homo sapiens and options restricted to finding top 20 related genes with automatic weighting process. The color in each node depict the HA population from which the particular gene is reportedly found associated
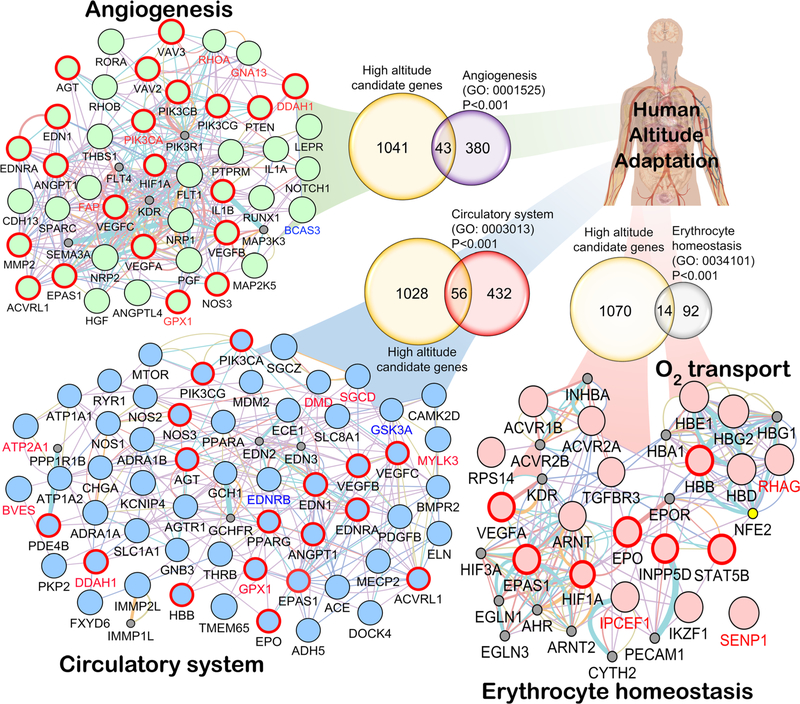
Fig. 3.
Convergence in the major biological processes. Anticipating the predominant role of biological processes involved in hypoxia response, e.g., circulatory system (GO: 0003013), angiogenesis (GO: 0001525), erythrocyte homeostasis (GO: 0034101), and O2 transport (GO: 0015671), the HA selected genes involved in these specific GO classes were picked for network analysis from Gene Ontology Consortium (geneontology.org). The Venn diagram (except “oxygen transport” (GO: 0005344) because there were only 13 genes in this GO category and 5 were common) depicts the common gene(s) shared between HA candidate genes and the respective GO category’s gene list. The probability of gene enrichment at HA for these GO categories was significant (Fisher’s exact test P < 0.001). We used Genemania v3.4.1 app on Cytospace v3.4.0. Each network is restricted to Homo sapiens with weighting process only related to GO biological process-based
Convergence of common genes at a genetic level
The three major high-altitude (HA) human populations that have been greatly studied seem to have successfully adapted or are in the process of adaptation. Keeping in mind the spatial and temporal differences in their HA res-idence, it would be intriguing to see whether this adaptation in these populations has followed similar or different genetic pathways. Since the selection pressure on these three populations is shared, one could hypothesize that the genomic signature detected in these populations could teach us about their genetic evolution and the genetic path taken for adaptation. Interestingly, there are > 1000 genes, reported in different studies, that are associated to HA adaptation/mal-adaptation in one or more of these populations (Table S1). In order to determine whether there was any genetic convergence between these three populations, we used two different approaches. First, using genetic network analysis tools, we investigated whether there is a cross-talk between genes shared by at least two populations. Second, we studied the major biological processes (GO class) that emerged from the genes mined in all three populations. We found 64 genes that were common between at least two populations of which four genes (PIK3CB, HLA-DQB1, CNTNAP2, and DLG2) were common in all the three populations (Fig. 2). Remarkably, there was cross-talk between 62 out the 64 genes mentioned above (Fig. 2). Functionally, these genes are related mostly with the circulatory system, angiogenesis, and immunity. It is interesting to note in Fig. 2 that out of the 64 genes shared between at least two HA populations, 10 belonged to the “immune response.” Foll et al. 2014 have also studied convergent evolution in high-altitude populations [75] but they used a more restrictive definition of convergence, namely, they used shared single-nucleotide polymorphisms (SNPs) being under selection in different populations and identified 362 shared SNPs encompassing multiple genes, including EGLN1 and EPAS1. Furthermore, in the enrichment analysis, they reported two big clusters of overlap-ping gene sets: one of them was linked to immune response.
Convergence at physiological systems
In order to study the convergence at physiological level, using a total of 19,000 human genes [76, 79], we tested if any of the GO process terms had a significant overlap with the 1084 genes suggested as being under selection as found in the literature (Table S1, Fig. 3). We found that 43 genes belonging to Angiogenesis (423 genes), 56 genes belonging to the Circulatory system (488 genes), 14 genes belonging to Erythrocyte homeostasis (106 genes), and 5 genes belonging to Oxygen transport (13 genes) all had significant overlap with the genes under selection (1084 genes, Fisher’s exact test; P < 0.001; Fig. 3). Regarding immune response genes and pathways, we did not observe any statistical significance in terms of an overlap with genes under selection. Our analysis points to three major pathways that contain genes responding to selection pressure at high altitude as shown in Fig. 3.
At physiological levels, heart, lungs, the vasculature, and red blood cells regulate oxygen homeostasis. If this is the case, by investigating the genes responsible for this adaptation to hypoxia, we will be able to determine whether similar or different genetic pathways led to the same phenotype. For example, both Tibetans and Ethiopians have similar Hb levels but they achieved this adaptation through different genes. There are various studies in the Tibetan population that have shown a strong association of Hb levels with EPAS1 and EGLN1. However, in Ethiopians, several studies have not observed any association between Hb levels and these loci [37, 53, 54] but instead with the following genes: THRB, ARNT2, VAV2, and VAV3.
Another interesting aspect is that specific genes can have pleiotropic effects on different physiological responses. Erythropoietin (EPO) is a classic example: it not only induces the maturation of red blood cells from erythroid progenitors and mediates erythropoiesis but also mediates non-erythroid processes such as angiogenesis, and immune regulation [77]. Similarly, VEGF, besides its angiogenic function, also activates pathways associated with nitric oxide (NO) synthesis and thus induces vasodilation and improves blood supply to cardiac cells [78]. It also plays a role in maturation and proliferation of erythroid cells [79–81]. In order, to understand the convergence of genes in terms of their physiological response (based on signaling pathways and the genes involved in these pathways), we analyzed the erythropoietic and circulatory systems (based on the literature) in detail as discussed in the sections below.
Convergence of genes associated with erythropoiesis
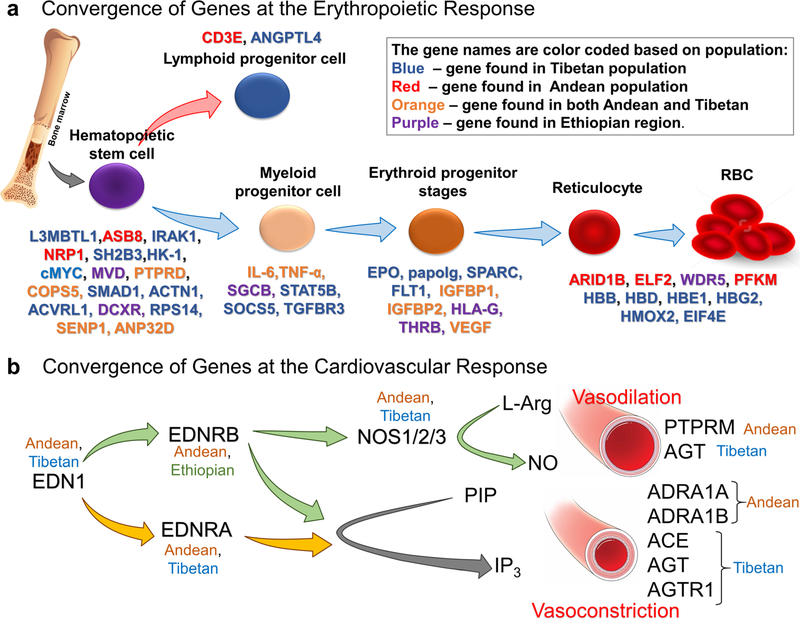
Erythropoiesis is a dynamic and tightly regulated process. The regulation of erythropoiesis occurs at multiple levels through multiple proteins. What is surprising is that there is a convergence of genes in all the three populations at various stages of erythropoiesis. Figure 4a shows genes mined from all three populations (Andean, Tibetan, or Ethiopian) which are involved at various stages of erythroid development (Fig. 4a). HIFs (HIF1 and HIF2) are key regulators particularly under hypoxia [1, 80–83]. They play a critical role in EPO synthesis as well as iron metabolism and their regulation under hypoxic conditions [81, 82]. Besides the regulation of HIF1 by hydroxylation (PHDs/VHL), it is also regulated by post-translational sumoylation (SUMO1) and hence, through desumoylase such as SENP1 [84]. Hematopoietic stem cells continuously differ-entiate into all blood lineages and therefore are crucial for red blood cell regulation. The expansion and regulation of these HSCs are regulated by a number of signaling pathways involving BMP, JAK/STAT, and TNFa as well as transcriptional regulators such as GATA1, SOCS5, and PU.1 [85]. Genes obtained from high-altitude studies such as SMAD1, TNFa, TGFBR3, HLA, and IGFBP1 have been associated with the abovementioned signaling pathways [40, 55, 56]. Erythroid progenitor stages, such as BFU-e, respond and interact with a number of cytokines including EPO, SCF, IGF-1, corticosteroids, IL-3, and IL-6 [85]. Certain genes such as ARID1B, HBB, HBD, HB1, and HBG2 are involved in the maturation of reticulocytes and production of hemoglobin by modulating globin switching from embryonic, to fetal, to adult hemoglobin [86]. It is interesting to observe that genes such as SPARK, HK-1, SH2B3, and L3MBTL1, which have been linked to erythropoietic defects at sea level, are found in these high-landers, possibly reinforcing our idea that the information obtained from high-altitude studies can have a major impact on the understanding of diseases that afflict us at sea level [87, 88].
Fig. 4.
Convergence at erythropoietic response and cardiovascular system. a Genes that were mined from high-altitude studies that have been involved in erythropoiesis. There is regulation at multiple time points of erythroid maturation and differentiation by various genes shown in the figure. The genes found in various populations are color-coded. Blue—Tibetan, red—Andean, orange—genes found in both Andean and Tibetan, and purple—Ethiopian population. b Cardiovascular homeostasis at HA primarily depends on the vasoconstriction and vasodilation response to hypoxia. Important genes from RAAS (ACE, AGT, and AGTR1), endothelin pathway (EDN1, EDNRA, and EDNRB), and the three NOS isoforms (NOS1, NOS2, and NOS3) involved in NO signaling pathway are all reported in different HA population studies
Convergence of genes associated with the circulatory system
The body’s responses to altitude hypoxia are extremely robust under both acute challenge (e.g., increased cardiac output with tachycardia) and chronic conditions (e.g., right ventricular hypertrophy). Researchers have long focused on physiological systems, such as the renin-angiotensin-aldosterone system (RAAS) [89–91], the endothelin system [89], and the NO signaling system [91, 92], just to name a few that are believed to be important in cardiovascular homeostasis in human adaptation to HA. Accordingly, the pharmacological interventions are also designed to target these systems. Drugs like angiotensin-converting enzyme (ACE) inhibitor and Bosentan are well-known pharmacological agents used to treat HA illnesses [93, 94]. NO itself is used to treat pulmonary edema at HA [95]. Remarkably, most of the genes from the aforemen-tioned pathways are also found to be associated with altitude adaptation (Figs. 3 and 4b). Specifically, these are ACE, AGT, and AGTR1 from RAAS, all the candidate genes from the endothelin pathway and the three NOS isoforms, i.e., NOS1, NOS2, and NOS3 of the NO signaling pathway. Important genes from the adrenergic pathway involved in vasoconstriction, ADRA1A and ADRA1B (both α1 receptors), have also been mined from genomics. A coincidental prominence of a1 receptors here might align with its peripheral abundance which override the vasodilation mediated by β-adrenoreceptors [96]. In addition to these well-known target genes, there are additional genes identified from these populations which are equally important in cardiovascular homeostasis. For example, transcript-tion factors PPARα and PPARγ, which are also reported to be involved in hypoxia responses, VEGFB and VEGFC, reportedly involved in angiogenesis and protein serine/ threonine kinase activity genes, PIK3CB and PIK3CG, are a few important genes in this class of gene ontology. In addition, ATP1A1, involved in maintaining the electro-chemical gradients of Na and K ions across the plasma membrane, and ATP2A1, involved in translocation of cal-cium from the cytosol to the sarcoplasmic reticulum lumen and therefore, in muscular excitation and contraction, are also found to be selected for in humans living at HA.
Translating high-altitude lessons into low-altitude medicine: the EDNRB story
Since we have been interested in hypoxia adaptation, inju-ry, and survival of tissues during hypoxic stress, in the past, we have argued that learning about HA physiology and biology in human dwellers might be one way of learning about mechanisms that can actually occur in disease states in humans at sea level. Therefore, we hypothesized that understanding high-altitude physiology and biology will allow us to better understand the molecular mechanisms of human diseases at sea level, especially those involving hypoxia and ischemia. In one such endeavor, we analyzed the whole genome for genetic variation in HA Ethiopians, an East-African HA population [38]. Using cross-population tests of selection and searching for genomic regions indicative of selective sweeps, we discovered regions that were significantly associated with HA adaptation. The gene EDNRB was present in one of these selected regions [38]. More precisely, there was a large block of 52 differential SNPs (single-nucleotide polymorphisms that were significantly divergent in allele frequency between CMS and non-CMS individuals) in the regulatory region of EDNRB. These SNPs (spanning approximately 170 kb) are in the regulatory region (upstream of the promoter re-gion) and also contain several transcription factor-binding sites [38, 59]. Although we could not test EDNRB messenger RNA (mRNA) levels in the human subjects, a relatively lower level of this gene seems beneficial when tested in heterozygote knockout (KO) mice [59]. Interestingly, a recent study in the Tibetans showed that the EDNRB levels were significantly lower in the healthy controls [97] suggesting that it could be protective and have a beneficial effect. It is interesting to note that Bosentan, an EDNRA/ B antagonist, is given to HA residents to reduce the high-altitude increase in pulmonary artery pressure [93]. In order to elucidate the potential role of this gene in hypoxia, we generated a knockout mouse of this particular gene, which also correlated with the antagonist Bosentan, and tested for hypoxia tolerance. We discovered that the het-erozygous KO mice (EdnrB−/+) were resistant not only to moderate but also to extreme hypoxia by maintaining higher cardiac output and peripheral perfusion and better O2 delivery to vital organs [59]. This was also indicated by lower serum lactate levels in the EdnrB−/+ mice under extreme hypoxia. The gene, EDNRB, encodes a G proteincoupled receptor which activates a phosphatidylinositol-calcium second messenger system [98]. The ligand endothelin (ET) that activates this receptor also binds and activates the other receptor subtype, endothelin receptor type A (EDNRA). Both receptors are known to have critical roles in regulating cardiovascular function largely with opposing activities, EDNRA in vasoconstriction and EDNRB in vasodilatation [99]. Of the two receptors, EDNRB has some intriguing functions. For example, it has a role as a vasoconstrictor [99] in some tissues, in neural crest cell migration [8], and different human cancers [100–102]. Unlike EDNRA, which are only present on the cell membrane, EDNRB are present on both plasma lemma and nuclear membrane and are reportedly involved in regulating nuclear Ca2+ signaling [103].
With respect to cardiac tolerance to hypoxia in our EdnrB−/+ mice, there is ample evidence that indicates its possible role in cardiac pathological conditions. For example, the relative density of EDNRB in the heart is about 1:4 that of EDNRA [104]. However, in cardiac-specific EdnrA KO mice, the study of EDNRA did not elicit much of a function during baseline or stressful conditions for this type of receptors [105]. This would indicate an important role for EDNRB in cardiac tissues. In fact, in patients with ischemic heart disease, the expression of vascular EDNRB increases considerably [106, 107]. The downregulation of EDNRB, as we saw in the EdnrB−/+ mice, might then be postulated as an appropriate therapeutic strategy for these patients. Other evidence that supports our notion has come from the cardiac failure seen during septic shock and, as stated above, Bosentan improves cardiac performance and microcirculatory blood flow [108, 109].
The fact that this gene was found as a one of the “candidate gene” in our whole genome sequence analysis of a HA human population [38] and was subsequently con-firmed to have a role in hypoxia tolerance using a mammalian model organism (i.e., mice) [59] was remarkable. As discussed by Prchal J. [110] in his commentary, this phenotype would have “escaped the prediction of its impact” had the experiments not been appropriately de-signed. As a result, we now believe that our findings and approach can be a model example of “translational medicine.” Also, the fact that this gene was mined from a HA human population gives us additional confidence that such an approach can be fruitful.
SENP1: another story underlying polycythemia in high-altitude dwellers
Since polycythemia is a predominant trait in some high-altitude dwellers (CMS or Monge’s disease) in the Andes, we took advantage of this “experiment in nature” using iPSC technology in order to understand its molecular basis. Although an increase in hemoglobin increases O2-carrying capacity, this adaptive trait can have deleterious effects since blood viscosity increases which in turn can induce serious morbidities, such as myocardial infarction and stroke [29, 30]. We have replicated the phenotype of “hypoxia-induced excessive erythropoiesis in the dish” in CMS subjects and were successful in transforming the human-derived iPS cell lines from both CMS and non-CMS subjects into rather mature RBCs [58]. Furthermore, in order to mimic high-altitude hypoxia as well as the bone marrow’s hypoxic niche [111–114], we exposed these iPS cells to 5% O2 early on during erythroid differentiation. Interestingly, CMS cells responded to hypoxia by having an exaggerated erythropoietic response (measured quantitatively by FACS) and non-CMS cells showed a blunted response with the sea-level controls having a moderate response. By modeling the phenotype into an in vitro platform, we can probe the system further to uncover mechanisms. Using molecular tools such as short hairpin RNA (shRNA) and fused overexpression con-structs, we were able to delineate the critical role of SENP1 and the regulation of its downstream targets, such as GATA1 and BclxL [58]. SENP1 is a major regulator of erythropoiesis in CMS subjects and has many target genes one of which is HIF1α [84, 115]. From our whole genome sequence analysis of the CMS and non-CMS subjects from the Andean region, we found 66 SNPs that were signify-cantly divergent in allele frequency between CMS and non-CMS individuals (differential SNPs) [39, 58]. Three of these differential SNPs are transcription factor-binding sites, and they overlap with different regulatory regions such as promoter as well as enhancer sites [58]. Indeed, we observed a significant upregulation of SENP1 in CMS subjects under hypoxia at the mRNA as well as protein level [58].
Besides erythropoiesis, SENP1 plays an important role in resistance to senescence, in regulating inflammatory response in diseases such as diabetes, and in regulating androgen receptor and prostate cancer development, to name a few examples [116, 117]. This suggests that by understanding the mechanism(s) of action of this gene (targets, effectors, or desumoylase), we not only gain insight of one disease or pathology but we can also potentially shed light on other conditions.
Role of epigenetics
While the previously described studies focused primarily on elucidating genomic adaptation to hypoxia, the role of epigenetics in hypoxic response and adaptation has thus far been poorly characterized in a functional manner in any organism. However, recently, there are studies that focus on the various epigenetic marks and their impact on the response to hypoxia stress (e.g., survival, tolerance, growth, and development) and the long-term heritability of these modifications.
To date, some of the most promising studies from a clinical perspective involve histone deacetylase enzymes (HDACs). It has been shown recently that by inhibiting the zinc-dependent classes of HDACs (I, Ila, Ilb, and IV) or stimulating the NAD+-dependent class (sirtuins), infarct volume may be decreased and behavioral outcomes improved following an ischemic insult [118]. Also, several enzymes in the lysine demethylase (KDM) family have also been implicated in hypoxia response, though the implications have yet to be fully studied. A number of KDM genes are activated in hypoxia, including KDMla, KDM2a, KDM2b, KDM3a, KDM4c, and KDM5b, while KDM6b is inactivated [119–123]. Additionally, it has been shown that environmental factors present during development, including hypoxia, may have a profound impact on phenotype, and this adaption may be encoded via DNA methylation and other epigenetic marks [124]. In our own work, we have observed significant and substantial changes to the global methylation landscape in primary hippocampal neurons [125]. DNA methylation has been strongly implicated as a mediator of this developmental response in a study of rats exposed to neonatal intermittent hypoxia for 10 days, showing that hypoxic stress during development can cause a predisposition to exaggerated hypoxic response in the carotid body as an adult 30 days later [126]. In addition, a number of anti-oxidant enzyme genes were observed to have been differentially expressed [126]. A follow-up study published earlier this year by the same group showed a strong link between long-term hypoxic stress in adult rats exposed to 30 days of intermittent hypoxia and a similar phenotype that exhibited hypertension [62, 127]. Another study that investigated the genetic and epigenetic differences between Ethiopian highlanders and lowlanders found significant methylation changes nearby to a number of known hypoxia response genes including glutathione S-transferase (GSTP1), protein regulator of cytokinesis 1 (PRC1), protein tyrosine phosphatase receptor type O (PTPRO), ring finger protein 146 (RNF146), and Ras-related GTP-binding D (RRAGD) [52]. Clearly, there remain many unanswered questions regarding the role epigenetics play in functional adaptation to hypoxic stress. Indeed, now that we have more tools in our kit of techniques, additional questions can be asked and answered.
Summary and future directions
Integration of omics (genomics, transcriptomics, or epigenomics) will be the important next step towards understanding the molecular basis and the mechanism(s) of complex disorders such as CMS. With the development of in vitro and in vivo technologies, such as CRISPR technology and induced pluripotent stem cells and differentiation protocols, it has become increasingly possible to validate the function of adaptive variants in humans and con-nect the phenotype to the genotype. While advances in technology have facilitated our ability to ask questions and mine further the genetic pathways and possibly the epigenetic role in the adaptation of organisms to natural challenges including extremes of environments, increasing efforts in the precise description of the phenotype has also become essential.
Supplementary Material
Acknowledgements
Our study is funded by NIH grant 1P01HL098053 and 5P01HD32573 to GGH, VB, and AA who were supported in part by grants from the NSF (DBI-1458557, IIS-1318386) and NIH (1R01GM114362). Dr. Vineet Bafna is a co-founder, has an equity interest, and receives income from Digital Proteomics, LLC. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. DP was not involved in the research presented here. The authors declare no competing financial interests.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00109-017-1584-7) contains supplementary material, which is available to authorized users.
References
- 1.Semenza GL (2000) HIF-1 and human disease: one highly involved factor. Genes Dev 14:1983–1991 [PubMed] [Google Scholar]
- 2.Semenza GL (2014) Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol 9:47–71 [DOI] [PubMed] [Google Scholar]
- 3.Nathaniel TI, Williams-Hernandez A, Hunter AL, Liddy C, Peffley DM, Umesiri FE, Imeh-Nathaniel A (2015) Tissue hypoxia during ischemic stroke: adaptive clues from hypoxia-tolerant animal models. Brain Res Bull 114:1–12 [DOI] [PubMed] [Google Scholar]
- 4.Drew KL, Harris MB, LaManna JC, Smith MA, Zhu XW, Ma YL (2004) Hypoxia tolerance in mammalian heterotherms. J Exp Biol 207:3155–3162 [DOI] [PubMed] [Google Scholar]
- 5.Haddad GG (2006) Tolerance to low O2: lessons from inverte-brate genetic models. Exp Physiol 91:277–282 [DOI] [PubMed] [Google Scholar]
- 6.Boutilier RG (2001) Mechanisms of cell survival in hypoxia and hypothermia. J Exp Biol 204:3171–3181 [DOI] [PubMed] [Google Scholar]
- 7.Larson J, Drew KL, Folkow LP, Milton SL, Park TJ (2014) No oxygen? No problem! Intrinsic brain tolerance to hypoxia in vertebrates. J Exp Biol 217:1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beall CM (2006) Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr Comp Biol 46:18–24 [DOI] [PubMed] [Google Scholar]
- 9.Villafuerte FC, Corante N (2016) Chronic mountain sickness: clinical aspects, etiology, management, and treatment. High Altitude Med Biol 17:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao H, Wang D, Zhao X, Wu Y, Yin G, Meng L, Wang F, Ma L, Hackett P, Ge RL (2017) Cerebral edema in chronic mountain sickness: anew finding. Sci Rep 7:43224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richalet JP, Rivera M, Bouchet P, Chirinos E, Onnen I, Petitjean O, Bienvenu A, Lasne F, Moutereau S, Leon-Velarde F (2005) Acetazolamide—a treatment for chronic mountain sickness. Am J Respir Crit Care Med 172:1427–1433 [DOI] [PubMed] [Google Scholar]
- 12.Sahota IS, Panwar NS (2013) Prevalence of chronic mountain sickness in high altitude districts of Himachal Pradesh. Indian J Occup Environ Med 17:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monge C, Leonvelarde F, Arregui A (1989) Increasing prevalence of excessive erythrocytosis with age among healthy high-altitude miners. N Engl J Med 321:1271–1271 [DOI] [PubMed] [Google Scholar]
- 14.Leon-Velarde F, Maggiorini M, Reeves JT, Aldashev A, Asmus I, Bernardi L, Ge RL, Hackett P, Kobayashi T, Moore LG et al. (2004) Consensus statement on chronic and subacute high altitude diseases. High Alt Med Biol 6:147–157 [DOI] [PubMed] [Google Scholar]
- 15.Penaloza D, Arias-Stella J (2007) The heart and pulmonary circulation at high altitudes—healthy highlanders and chronic mountain sickness. Circulation 115:1132–1146 [DOI] [PubMed] [Google Scholar]
- 16.Aldenderfer M (2011) Peopling the Tibetan plateau: insights from archaeology. High Alt Med Biol 12:141–147 [DOI] [PubMed] [Google Scholar]
- 17.Simonson TS (2015) Altitude adaptation: a glimpse through various lenses. High Altitude Med Biol 16:125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M, Kong QP, Wang HW, Peng MS, Xie XD, Wang WZ, Jiayang DJG, Cai MC, Zhao SN et al. (2009) Mitochondrial genome evidence reveals successful late Paleolithic settlement on the Tibetan plateau. Proc Natl Acad Sci USA 106:21230–21235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abir M (1992) The Oromo of Ethiopia—a history 1570–1860 - Hassen, M. Int J Middle East Stud 24: 344–346 [Google Scholar]
- 20.Rademaker K, Hodgins G, Moore K, Zarrillo S, Miller C, Bromley GR, Leach P, Reid DA, Alvarez WY, Sandweiss DH (2014) Paleoindian settlement of the high-altitude Peruvian Andes. Science 346:466–469 [DOI] [PubMed] [Google Scholar]
- 21.Beall CM (2004) Andean, Tibetan and Ethiopian patterns of human adaptation to high-altitude hypoxia. Integr Comp Biol 44: 522–522 [DOI] [PubMed] [Google Scholar]
- 22.Beall CM, Decker MJ, Brittenham GM, Kushner I, Gebremedhin A, Strohl KP (2002) An Ethiopian pattern of human adaptation to high-altitude hypoxia. Proc Natl Acad Sci U S A 99:17215–17218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bigham A, Bauchet M, Pinto D, Mao XY, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Herraez DL et al. (2010) Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet 6: e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall CM (2014) Adaptation to high altitude: phenotypes and genotypes. Annu Rev Anthropol 43(43):251–272 [Google Scholar]
- 25.Beall CM (2007) Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A 104: 8655–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronen R, Zhou D, Bafna V, Haddad GG (2014) The genetic basis of chronic mountain sickness. Physiology 29:403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monge CC, Whittembury J (1976) Chronic mountain sickness. Johns Hopkins Med J 139:87–89 [PubMed] [Google Scholar]
- 28.Dainiak N, Spielvogel H, Sorba S, Cudkowicz L (1989) Erythropoietin and the polycythemia of high-altitude dwellers. Mol Biol Erythropoiesis 271:17–21 [DOI] [PubMed] [Google Scholar]
- 29.Mejia OM, Prchal JT, Leon-Velarde F, Hurtado A, Stockton DW (2005) Genetic association analysis of chronic mountain sickness in an Andean high-altitude population. Haematologica 90:13–18 [PubMed] [Google Scholar]
- 30.Monge C, Arregui CA, Leonvelarde F (1992) Pathophysiology and epidemiology of chronic mountain sickness. Int J Sports Med 13:S79–S81 [DOI] [PubMed] [Google Scholar]
- 31.Moore LG (2001) Human genetic adaptation to high altitude. High Altitude Med Biol 2:257–279 [DOI] [PubMed] [Google Scholar]
- 32.Monge C, Lozano R, Whittembury J (1965) Effect of blood-letting on chronic mountain sickness. Nature 207:770. [DOI] [PubMed] [Google Scholar]
- 33.Naeije R (2010) Physiological adaptation of the cardiovascular system to high altitude. Prog Cardiovasc Dis 52:456–466 [DOI] [PubMed] [Google Scholar]
- 34.Naeije R, Vanderpool R (2013) Pulmonary hypertension and chronic mountain sickness. High Alt Med Biol 14:117–125 [DOI] [PubMed] [Google Scholar]
- 35.Wright AD, Birmingham Medical Research Expeditionary S (2004) Medicine at high altitude. Clin Med 6:604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS et al. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bigham AW, Lee FS (2014) Human high-altitude adaptation: for-ward genetics meets the HIF pathway. Genes Dev 28:2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udpa N, Ronen R, Zhou D, Liang JB, Stobdan T, Appenzeller O, Yin Y, Du YP, Guo LX, Cao R et al. (2014) Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol 15:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou D, Udpa N, Ronen R, Stobdan T, Liang J, Appenzeller O, Zhao HW, Yin Y, Du Y, Guo L et al. (2013) Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am J Hum Genet 93:452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, Parra EJ, Akey JM, Moore LG, Shriver MD (2009) Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics 4:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eichstaedt CA, Antao T, Pagani L, Cardona A, Kivisild T, Mormina M (2014) The Andean adaptive toolkit to counteract high altitude maladaptation: genome-wide and phenotypic analysis of the Collas. PLoS One 9:e93314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valverde G, Zhou H, Lippold S, de Filippo C, Tang K, Lopez Herraez D, Li J, Stoneking M (2015) A novel candidate region for genetic adaptation to high altitude in Andean populations. PLoS One 10:e0125444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenzo FR, Huff C, Myllymaki M, Olenchock B, Swierczek S, Tashi T, Gordeuk V, Wuren T, Ri-Li G, McClain DA et al. (2014) A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet 46:951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang BB, Zhang YB, Zhang F, Lin HB, Wang XM, Wan N, Ye ZQ, Weng HY, Zhang LL, Li X et al. (2011) On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One 6:e17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang GD, Fan RX, Zhai WW, Liu F, Wang L, Zhong L, Wu H, Yang HC, Wu SF, Zhu CL et al. (2014) Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the Tibetan plateau. Genome Biol Evol 6:2122–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuren T, Simonson TS, Qin G, Xing JC, Huff CD, Witherspoon DJ, Jorde LB, Ge RL (2014) Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLoS One 9:e88252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu SH, Li SL, Yang YJ, Tan JZ, Lou HY, Jin WF, Yang L, Pan XD, Wang JC, Shen YP et al. (2011) A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol 28: 1003–1011 [DOI] [PubMed] [Google Scholar]
- 48.Jeong C, Ozga AT, Witonsky DB, Malmstrom H, Edlund H, Hofman CA, Hagan RW, Jakobsson M, Lewis CM, Aldenderfer MS et al. (2016) Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc Natl Acad Sci U S A 113:7485–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji FY, Sharpley MS, Derbeneva O, Alves LS, Qian P, Wang YL, Chalkia D, Lvova M, Xu JC, Yao W et al. (2012) Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci U S A 109:7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Jiang CH, Luo YJ, Liu FY, Gao YQ (2016) Interaction of CARD14, SENP1 and VEGFA polymorphisms on susceptibility to high altitude polycythemia in the Han Chinese population at the Qinghai-Tibetan plateau. Blood Cells Molecules and Diseases 57: 13–22 [DOI] [PubMed] [Google Scholar]
- 51.Zhang YB, Li X, Zhang F, Wang DM, Yu J (2012) A preliminary study of copy number variation in Tibetans. PLoS One 7:e41768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A (2012) The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet 8:e1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huerta-Sanchez E, Degiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, Cardona A, Montgomery HE, Cavalleri GL, Robbins PA et al. (2013) Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol Biol Evol 30:1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G et al. (2012) Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol 13:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tekola-Ayele F, Adeyemo A, Chen GJ, Hailu E, Aseffa A, Davey G, Newport MJ, Rotimi CN (2015) Novel genomic signals of recent selection in an Ethiopian population. Eur J Hum Genet 23:1085–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng Y, Yang ZH, Zhang H, Cui CY, Qi XB, Luo XJ, Tao XA, Wu TY, Ouzhuluobu B et al. (2011) Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol 28:1075–1081 [DOI] [PubMed] [Google Scholar]
- 57.Song D, Li LS, Arsenault PR, Tan Q, Bigham AW, Heaton-Johnson KJ, Master SR, Lee FS (2014) Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem 289(21):14656–14665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azad P, Zhao HW, Cabrales PJ, Ronen R, Zhou D, Poulsen O, Appenzeller O, Hsiao YH, Bafna V, Haddad GG (2016) Senp1 drives hypoxia-induced polycythemia via GATA1 and Bcl-xL in subjects with Monge’s disease. J Exp Med 213:2729–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stobdan T, Zhou D, Ao-Ieong E, Ortiz D, Ronen R, Hartley I, Gan Z, McCulloch AD, Bafna V, Cabrales P et al. (2015) Endothelin receptor B, a candidate gene from human studies at high altitude, improves cardiac tolerance to hypoxia in genetically engineered heterozygote mice. Proc Natl Acad Sci U S A 112:10425–10430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu XH, Huang XW, Qun L, Li YN, Wang Y, Liu C, Ma YY, Liu QM, Sun K, Qian F et al. (2014) Two functional loci in the promoter of EPAS1 gene involved in high-altitude adaptation of Tibetans. Sci Rep 4:7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang DY, Peng Y, Ouzhuluobu B, Cui CY, Bianba WLB, Xiang K, He YX, Zhang H et al. (2016) HMOX2 functions as a modifier gene for high-altitude adaptation in Tibetans. Hum Mutat 37:216–223 [DOI] [PubMed] [Google Scholar]
- 62.Wilkins MR, Aldashev AA, Wharton J, Rhodes CJ, Vandrovcova J, Kasperaviciute D, Bhosle SG, Mueller M, Geschka S, Rison S, Kojonazarov B, Morrell NW, Neidhardt I, Surmeli NB, Aitman TJ, Stasch JP, Behrends S, Marletta MA (2014) Alpha 1-A680T variant in GUCY1A3 as a candidate conferring protection from pulmonary hypertension among Kyrgyz highlanders. Circ-Cardiovasc Gene 7(6):920–U505 [DOI] [PubMed] [Google Scholar]
- 63.Cao L, Tan L, Jiang T, ZhuX C, Yu JT (2015) Induced pluripotent stem cells for disease modeling and drug discovery in neurode-generative diseases. Mol Neurobiol 52:244–255 [DOI] [PubMed] [Google Scholar]
- 64.Hossain MK, Dayem AA, Han J, Saha SK, Yang GM, Choi HY, Cho SG (2016) Recent advances in disease modeling and drug discovery for diabetes mellitus using induced pluripotent stem cells. Int J Mol Sci 17:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooi L, Sidhu K, Poljak A, Sutherland G, O’Connor MD, Sachdev P, Munch G (2013) Induced pluripotent stem cells as tools for disease modelling and drug discovery in Alzheimer’s disease. J Neural Transm 120:103–111 [DOI] [PubMed] [Google Scholar]
- 66.Stemeckert JL, Reinhardt P, Scholer HR (2014) Investigating human disease using stem cell models. Nat Rev Genet 15:625–639 [DOI] [PubMed] [Google Scholar]
- 67.Beall CM, Brittenham GM, Strohl KP, Blangero J, Williams-Blangero S, Goldstein MC, Decker MJ, Vargas E, Villena M, Soria R et al. (1998) Hemoglobin concentration of high-altitude Tibetans and Bolivian Aymara. Am J Phys Anthropol 106:385–400 [DOI] [PubMed] [Google Scholar]
- 68.Beall CM, Decker MJ, Brittenham GM, Kushner I, Gebremedhin A, Strohl KP (2002) An Ethiopian pattern of human adaptation to high-altitude hypoxia. ProcNatlAcad Sci U S A 99:17215–17218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB et al. (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329:72–75 [DOI] [PubMed] [Google Scholar]
- 70.Xiang K, Ouzhuluobu PY, Yang Z, Zhang X, Cui C, Zhang H, Li M, Zhang Y, Bianba et al. (2013) Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol 30:1889–1898 [DOI] [PubMed] [Google Scholar]
- 71.Yang J, Jin ZB, Chen J, Huang XF, Li XM, Liang YB, Mao JY, Chen X, Zheng Z, Bakshi A et al. (2017) Genetic signatures of high-altitude adaptation in Tibetans. Proc Natl Acad Sci USA 114:4189–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Appenzeller O, Minko T, Qualls C, Pozharov V, Gamboa J, Gamboa A, Wang Y (2006) Gene expression, autonomic function and chronic hypoxia: lessons from the Andes. Clin Auton Res 16: 217–222 [DOI] [PubMed] [Google Scholar]
- 73.Leon-Velarde F, Mejia O (2008) Gene expression in chronic high altitude diseases. High Alt Med Biol 9:130–139 [DOI] [PubMed] [Google Scholar]
- 74.Gonzales GF, Chaupis D (2014) Higher androgen bioactivity is associated with excessive erythrocytosis and chronic mountain sickness in Andean highlanders: a review. Andrologia. 10.1111/and.12359 [DOI] [PubMed] [Google Scholar]
- 75.Foll M, Gaggiotti OE, Daub JT, Vatsiou A, Excoffier L (2014) Widespread signals of convergent adaptation to high altitude in Asia and America. Am J Hum Genet 95:394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML (2014) Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum Mol Genet 23:5866–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghezzi P, Brines M (2004) Erythropoietin as an antiapoptotic, tissue-protective cytokine. Cell Death Differ 11:S37–S44 [DOI] [PubMed] [Google Scholar]
- 78.Senger DR(2010) Vascular endothelial growth factor: much more than an angiogenesis factor. Mol Biol Cell 21:377–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Achaz G (2009) Frequency Spectrum Neutrality Tests: one for all and all for one. Genetics 183:249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Semenza GL (2000) HIF-1: mediator of physiological and patho-physiological responses to hypoxia. J Appl Physiol 88:1474–1480 [DOI] [PubMed] [Google Scholar]
- 81.Haase VH (2010) Hypoxic regulation of erythropoiesis and iron metabolism. Am J Physiol-Renal Physiol 299:F1–F13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Semenza GL (2009) Involvement of oxygen-sensing pathways in physiologic and pathologic erythropoiesis. Blood 114:2015–2019 [DOI] [PubMed] [Google Scholar]
- 83.Rankin EB, Biju MP, Liu QD, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH (2007) Hypoxia-inducible fac-tor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Investig 117:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng J, Kang X, Zhang S, Yeh ET (2007) SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131:584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hattangadi SM, Wong P, Zhang LB, Flygare J, Lodish HF (2011) From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 118:6258–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schechter AN (2008) Hemoglobin research and the origins of molecular medicine. Blood 112:3927–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giani FC, Fiorini C, Wakabayashi A, Ludwig LS, Salem RM, Jobaliya CD, Regan SN, Ulirsch JC, Liang G, Steinberg-Shemer O et al. (2016) Targeted application of human genetic variation can improve red blood cell production from stem cells. Cell Stem Cell 18:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perna F, Gurvich N, Hoya-Arias R, Abdel-Wahab O, Levine RL, Asai T, Voza F, Menendez S, Wang L, Liu F et al. (2010) Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q-polycythemia vera. Blood 116:2812–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Charu R, Stobdan T, Ram RB, Khan AP, Qadar Pasha MA, Norboo T, Afrin F (2006) Susceptibility to high altitude pulmonary oedema: role of ACE and ET-1 polymorphisms. Thorax 61: 1011–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stobdan T, Ali Z, Khan AP, Nejatizadeh A, Ram R, Thinlas T, Mohammad G, Norboo T, Himashree G, Qadar Pasha M (2011) Polymorphisms of renin—angiotensin system genes as a risk factor for high-altitude pulmonary oedema. J Renin-Angiotensin-Aldosterone Syst 12:93–101 [DOI] [PubMed] [Google Scholar]
- 91.Stobdan T, Karar J, Pasha MA (2008) High altitude adaptation: genetic perspectives. High Alt Med Biol 9:140–147 [DOI] [PubMed] [Google Scholar]
- 92.Beall CM, Laskowski D, Strohl KP, Soria R, Villena M, Vargas E, Alarcon AM, Gonzales C, Erzurum SC (2001) Pulmonary nitric oxide in mountain dwellers. Nature 414:411–412 [DOI] [PubMed] [Google Scholar]
- 93.Kojonazarov B, Isakova J, Imanov B, Sovkhozova N, Sooronbaev T, Ishizaki T, Aldashev AA (2012) Bosentan reduces pulmonary artery pressure in high altitude residents. High Alt Med Biol 13:217–223 [DOI] [PubMed] [Google Scholar]
- 94.Plata R, Cornejo A, Arratia C, Anabaya A, Perna A, Dimitrov BD, Remuzzi G, Ruggenenti P, Commission on Global Advancement of Nephrology RSotISoN (2002) Angiotensin-converting-enzyme inhibition therapy in altitude polycythaemia: a prospective randomised trial. Lancet 359:663–666 [DOI] [PubMed] [Google Scholar]
- 95.Scherrer U, Vollenweider L, Delabays A, Savcic M, Eichenberger U, Kleger GR, Fikrle A, Ballmer PE, Nicod P, Bartsch P (1996) Inhaled nitric oxide for high-altitude pulmonary edema. N Engl J Med 334:624–629 [DOI] [PubMed] [Google Scholar]
- 96.Lundvall J, Hillman J, Gustafsson D (1982) Beta-adrenergic dila-tor effects in consecutive vascular sections of skeletal muscle. Am J Physiol 243:H819–H829 [DOI] [PubMed] [Google Scholar]
- 97.Wu S, Hao G, Zhang S, Jiang D, Wuren T, Luo J (2016) Cerebral vasoconstriction reactions and plasma levels of ETBR, ET-1, and eNOS in patients with chronic high altitude disease. Mol Med Rep 14:2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tykocki NR, Watts SW (2010) The interdependence of endothelin-1 and calcium: areview. Clin Sci (Lond) 119:361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneider MP, Boesen EI, Pollock DM (2007) Contrasting actions of endothelin ET(A) and ET(B) receptors in cardiovascular disease. Annu Rev Pharmacol Toxicol 47:731–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen C, Wang L, Liao Q, Huang Y, Ye H, Chen F, Xu L, Ye M, Duan S (2013) Hypermethylation of EDNRB promoter contrib-utes to the risk of colorectal cancer. Diagn Pathol 8:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cruz-Munoz W, Jaramillo ML, Man S, Xu P, Banville M, Collins C, Nantel A, Francia G, Morgan SS, Cranmer LD et al. (2012) Roles for endothelin receptor B and BCL2A1 in spontaneous CNS metastasis of melanoma. Cancer Res 72:4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zuiverloon TC, Beukers W, van der Keur KA, Munoz JR, Bangma CH, Lingsma HF, Eijkemans MJ, Schouten JP, Zwarthoff EC (2012) A methylation assay for the detection of non-muscle-invasive bladder cancer (NMIBC) recurrences in voided urine. BJU Int 109:941–948 [DOI] [PubMed] [Google Scholar]
- 103.Merlen C, Farhat N, Luo X, Chatenet D, Tadevosyan A, Villeneuve LR, Gillis MA, Nattel S, Thorin E, Fournier A et al. (2013) Intracrine endothelin signaling evokes IP3-dependent increases in nucleoplasmic Ca(2)(+) in adult cardiac myocytes. J Mol Cell Cardiol 62:189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuc RE, Maguire JJ, Davenport AP (2006) Quantification of endothelin receptor subtypes in peripheral tissues reveals downregulation of ET(A) receptors in ET(B)-deficient mice. Exp Biol Med (Maywood) 231:741–745 [PubMed] [Google Scholar]
- 105.Kedzierski RM, Grayburn PA, Kisanuki YY, Williams CS, Hammer RE, Richardson JA, Schneider MD, Yanagisawa M (2003) Cardiomyocyte-specific endothelin A receptor knockout mice have normal cardiac function and an unaltered hypertrophic response to angiotensin II and isoproterenol. Mol Cell Biol 23: 8226–8232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dagassan PH, Breu V, Clozel M, Kunzli A, Vogt P, Turina M, Kiowski W, Clozel JP (1996) Up-regulation of endothelin-B receptors in atherosclerotic human coronary arteries. J Cardiovasc Pharmacol 27:147–153 [DOI] [PubMed] [Google Scholar]
- 107.Dimitrijevic I, Edvinsson ML, Chen Q, Malmsjo M, Kimblad PO, Edvinsson L (2009) Increased expression of vascular endothelin type B and angiotensin type 1 receptors in patients with ischemic heart disease. BMC Cardiovasc Disord 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krejci V, Hiltebrand LB, Erni D, Sigurdsson GH (2003) Endothelin receptor antagonist bosentan improves microcirculatory blood flow in splanchnic organs in septic shock. Crit Care Med 31:203–210 [DOI] [PubMed] [Google Scholar]
- 109.Wanecek M, Weitzberg E, Alving K, Rudehill A, Oldner A (2001) Effects of the endothelin receptor antagonist bosentan on cardiac performance during porcine endotoxin shock. Acta Anaesthesiol Scand 45:1262–1270 [DOI] [PubMed] [Google Scholar]
- 110.Prchal JT (2015) Genetic selection by high altitude: beware of experiments at ambient conditions. Proc Natl Acad Sci U S A 112:10080–10081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A (2010) Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 7:150–161 [DOI] [PubMed] [Google Scholar]
- 112.Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, Zaher W, Mortensen LJ, Alt C, Turcotte R et al. (2014) Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Suda T, Takubo K, Semenza GL (2011) Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9: 298–310 [DOI] [PubMed] [Google Scholar]
- 114.Tiwari A, Wong CS, Nekkanti LP, Deane JA, McDonald C, Jenkin G, Kirkland MA (2016) Impact of oxygen levels on human hematopoietic stem and progenitor cell expansion. Stem Cells Dev. 10.1089/scd.2016.0153 [DOI] [PubMed] [Google Scholar]
- 115.Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng EC, Chen H, Krause DS, Min W (2010) SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J Exp Med 207:1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bawa-Khalfe T, Cheng J, Lin SH, Ittmann MM, Yeh ETH (2010) SENP1 induces prostatic intraepithelial neoplasia through multiple mechanisms. J Biol Chem 285:25859–25866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shao L, Zhou HJ, Zhang HF, Qin LF, Hwa J, Yun Z, Ji WD, Min W (2015) SENP1-mediated NEMO deSUMOylation in adipo-cytes limits inflammatory responses and type-1 diabetes progress-sion. Nat Commun 6:8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Langley B, Sauve A (2013) Sirtuin deacetylases as therapeutic targets in the nervous system. Neurotherapeutics 10:605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hala D, Huggett DB, Burggren WW (2014) Environmental stressors and the epigenome. Drug Discov Today Technol 12:e3–e8 [DOI] [PubMed] [Google Scholar]
- 120.Luo W, Chang R, Zhong J, Pandey A, Semenza GL (2012) Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci U S A 109:E3367–E3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prickaerts P, Adriaens ME, Beucken TV, Koch E, Dubois L, Dahlmans VE, Gits C, Evelo CT, Chan-Seng-Yue M, Wouters BG et al. (2016) Hypoxia increases genome-wide bivalent epigenetic marking by specific gain of H3K27me3. Epigenetics Chromatin 9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salminen A, Kaarniranta K, Kauppinen A (2016) Hypoxia-inducible histone lysine demethylases: impact on the aging process and age-related diseases. Aging Dis 7:180–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ueda J, Ho JC, Lee KL, Kitajima S, Yang H, Sun W, Fukuhara N, Zaiden N, Chan SL, Tachibana M et al. (2014) The hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a mecha-nistic link between angiogenesis and tumor growth. Mol Cell Biol 34:3702–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Frisancho AR (2009) Developmental adaptation: where we go from here. Am J Hum Biol 21:694–703 [DOI] [PubMed] [Google Scholar]
- 125.Hartley I, Elkhoury FF, Heon Shin J, Xie B, Gu X, Gao Y, Zhou D, Haddad GG (2013) Long-lasting changes in DNA méthylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS One 8:e77859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nanduri J, Makarenko V, Reddy VD, Yuan G, Pawar A, Wang N, Khan SA, Zhang X, Kinsman B, Peng YJ et al. (2012) Epigenetic regulation of hypoxic sensing disrupts cardiorespiratory homeostasis. Proc Natl Acad Sci U S A 109:2515–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nanduri J, Peng YJ, Wang N, Khan SA, Semenza GL, Kumar GK, Prabhakar NR (2017) Epigenetic regulation of redox state mediates persistent cardiorespiratory abnormalities after long-term intermittent hypoxia. J Physiol 595:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.