Streptogramin antibiotics are mixtures of two chemically unrelated A and B compounds that act synergistically in vivo against gram-positive pathogens, such as staphylococci, streptococci, and enterococci (8, 11). Resistance against B compounds is very widespread among enterococci and is mediated via the ermB gene cluster (e.g., on Tn917) that confers macrolide-lincosamide-streptogramin B resistance (7). The synergistic mixture of streptogramins A and B overcomes resistance to B compounds but is inactive in resistance to A compounds. The only known resistance mechanism against streptogramin A compounds in enterococci is mediated by the streptogramin acetyltransferase SatA (9). Enterococcus faecium isolates with satA-mediated resistance have been found in samples of human and animal origins, indicating a possible spread of resistance genes or resistant bacteria among different ecosystems (10).
We isolated a quinupristin-dalfopristin-resistant E. faecium UW1965 from a sewage treatment plant in Germany. The resistance determinant was transferred to a susceptible recipient, producing the transconjugant UW1965K1. UW1965K1 is resistant to quinupristin-dalfopristin (MIC ≥ 16 μg/ml) and virginiamycin M (A compound; MIC, 16 μg/ml), whereas the MIC of each antibiotic for the recipient was 1 μg/ml. PCR amplification for the satA gene was negative.
In staphylococci, resistance to streptogramin A compounds is mediated by two mechanisms: (i) acetylation of the streptogramin A via acetyltransferases (Vat, VatB, and VatC [1–3]) and (ii) efflux due to an ABC transporter (Vga and VgaB [4, 5]). PCR amplification for the vat, vatB, vatC, and vga genes failed to produce any product. The putative protein sequences of the known streptogramin acetyltransferases in staphylococci and enterococci contain three conserved motifs (2). Corresponding primers, satI and satJ, have been made, producing a 144- to 147-bp fragment for vat, satA, and vatB (2). PCR performed with these primers resulted in a ca. 150-bp fragment for UW1965K1. A digoxigenin-labelled probe of the amplified fragment was prepared, hybridizing with a 5.5-kbp fragment of EcoRI-digested plasmid DNA from the transconjugant. The corresponding plasmid fragment was cloned into pUC18 and sequenced.
The resulting DNA sequence (Fig. 1) did not show significant identity with other gene sequences from GenBank on the DNA level (6). One suitable open reading frame (ORF) was found, giving rise to a putative 214-amino-acid (214-aa) protein. A comparison of amino acid similarities indicated rather significant homology between streptogramin acetyltransferases and the new putative acetyltransferase, designated SatG (Fig. 2). Based on the sequence for satG, two primers, satG1 and satG2, have been designed. Preliminary results of a search for streptogramin-resistant enterococci (E. faecium, E. hirae, and E. durans) revealed the existence of the satG gene in 9 of 23 isolates from sewage, 6 of 24 isolates from broiler samples, and all 17 isolates from poultry manure. Of 62 quinupristin-dalfopristin-resistant E. faecium (QDREF) isolates from hospitals in Germany, 9 were positive for satG. The high number of satG QDREF isolates from poultry meat and manure may be due to selection of these bacteria by use of virginiamycin as a feed additive, and spread of the resistance via the food chain to humans is very likely. This hypothesis is being investigated.
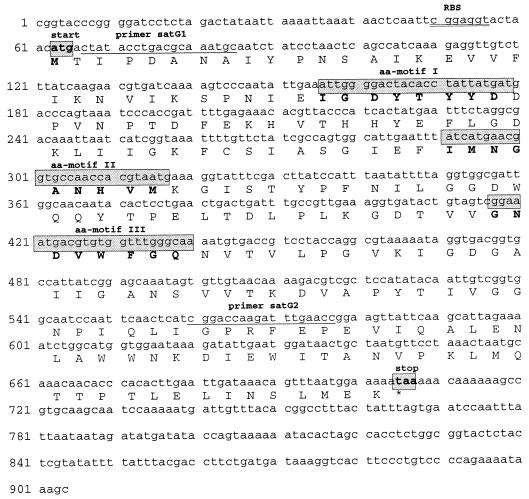
FIG. 1.
A 904-bp sequence located on the 5.5-kbp cloned fragment in pUC18 (GenBank accession no. AF139725). The ORF begins at nucleotide 63 with an ATG start codon preceding a putative ribosomal binding site (RBS) (double-underlined) at positions 50 to 57. The predicted gene sequence encodes a protein of 214 aa which shows significant homology with other streptogramin acetyltransferases (aa motifs I, II, III; see also Fig. 2). The locations of the primers satG1 and satG2, specific only for the satG sequence, are underlined (plus strand).
FIG. 2.
Alignment of amino acid sequences of acetyltransferases from staphylococci and enterococci (1–3, 9) conferring resistance to streptogramin A antibiotics. Identical residues are indicated by asterisks. Highly conserved regions in different streptogramin A acetyltransferases—motifs I, II, and III—are boldfaced. Primers satI and satJ have been designed on the basis of the corresponding nucleotide sequences in motifs II and III (2).
REFERENCES
- 1.Allignet J, Loncle V, Simmenel C, Delepierre M, El Solh N. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91–98. doi: 10.1016/0378-1119(93)90350-c. [DOI] [PubMed] [Google Scholar]
- 2.Allignet J, El Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2029. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allignet J, Liassine N, El Solh N. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother. 1998;42:1794–1798. doi: 10.1128/aac.42.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allignet J, Loncle V, El Solh N. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene. 1992;117:45–51. doi: 10.1016/0378-1119(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 5.Allignet J, El Solh N. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene. 1997;202:133–138. doi: 10.1016/s0378-1119(97)00464-2. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur M, Brisson-Noël A, Courvalin P. Origin and evolution of genes specifying resistance to macrolide, lincosamide, and streptogramin antibiotics: data and hypothesis. J Antimicrob Chemother. 1987;20:783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- 8.Bouilla H J, Perri M B, Kauffman C A, Zervos M J. Comparative in-vitro activity of quinupristin/dalfopristin against multidrug-resistant Enterococcus faecium. Diagn Microbiol Infect Dis. 1996;25:127–131. doi: 10.1016/s0732-8893(96)00123-x. [DOI] [PubMed] [Google Scholar]
- 9.Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother. 1993;37:2119–2125. doi: 10.1128/aac.37.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner G, Klare I, Witte W. Association between quinupristin/dalfopristin resistance in glycopeptide-resistant Enterococcus faecium and the use of additives in animal feed. Eur J Clin Microbiol Infect Dis. 1998;17:401–402. doi: 10.1007/BF01691571. [DOI] [PubMed] [Google Scholar]
- 11.Zervos M J. Vancomycin-resistant Enterococcus faecium infections in the ICU and quinupristin/dalfopristin. New Horizons. 1996;4:385–392. [PubMed] [Google Scholar]