Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel virus that belongs to the family Coronaviridae. This virus produces a respiratory illness known as coronavirus disease 2019 (COVID-19) and is to blame for the pandemic of COVID-19. Due to its massive circulation around the world and the capacity of mutation of this virus, genomic studies are much needed in to order to reveal new variants of concern (VOCs). On November 26th, 2021, the WHO announced that a new SARS-CoV-2 VOC, named Omicron, had emerged. In order to get insight into the emergence, spread and evolution of Omicron SARS-CoV-2 variants, a comprehensive phylogenetic study was performed. The results of these studies revealed significant differences in codon usage among the S genes of SARS-CoV-2 VOCs Alfa, Beta, Gamma, Delta and Omicron, which can be linked to SARS-CoV-2 genotypes. Omicron variant did not evolve out of one of the early VOCs, but instead it belongs to a complete different genetic lineage from previous ones. Strains classified as Omicron variants evolved from ancestors that existed around May 15th, 2020, suggesting that this VOC may have been circulating undetected for a period of time until its emergence was observed in South Africa. A rate of evolution of 5.61 × 10−4 substitutions/site/year was found for Omicron strains enrolled in these analyses. The results of these studies demonstrate that S genes have suitable genetic information for clear assignment of emerging VOCs to its specific genotypes.
Keywords: Coronavirus, evolution, SARS-CoV-2, Omicron, COVID-19
1. Introduction
In December, 2019, a pandemic of coronavirus disease (COVID-19) started in Wuhan, China (Li, Zai and Zhao, 2020a). This pandemic is caused by a virus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the infection by this virus leads to a severe respiratory pneumonia (Gorbalenya et al., 2020). As December 14th, 2021, there have been more than 270 million confirmed cases worldwide and the global deaths of SARS-CoV-2 disease surpasses 5 million people (World Health Organization 2021a).
SARS-CoV-2 possess a single stranded, positive-sense RNA genome of approximately 30 kilobases in length, which encodes for multiple structural and non-structural proteins. The structural proteins include the spike (S) protein, the envelope (E) protein, the membrane (M) protein, and the nucleocapsid (N) protein (Chen et al., 2020).
The replication cycle of SARS-CoV-2 starts when it infects epithelial cells using the viral S protein to bind host angiotensin-converting enzyme 2 (ACE2) and by this means fusing with cell membrane to gain cell entry (Hoffmann et al., 2020; Xia, 2021).
From the beginning of this pandemic several therapies and preventive health cares were developed, such as clinically applied monoclonal antibodies (Weinreich et al., 2021) or vaccinations (Wang et al., 2021), and both were successfully used to neutralize the virus. However, the emergence of variants of concern (VOCs) with substitutions in the S protein may make these therapies and vaccines to reduce its efficiency by escaping to anti-SARS-CoV-2 antibodies (Davies et al., 2021).
Previous studies have identified four VOCs currently circulating in the human population: VOC Alpha (B.1.1.7, first identified in the United Kingdom); VOC Beta (B.1.351, first identified in South Africa); VOC Gamma (P.1, first identified in Brazil) and VOC Delta (B.1.617.2, first isolated in India). By November 24th, 2021, a new SARS-CoV-2 VOC, now known as VOC Omicron (B.1.1.529) was first identified in South Africa (Wang & Chen, 2021).
The emergence of Omicron VOC raised a concern that this variant may reduce the efficacy of the anti-SARS-CoV-2 induced-antibodies or be more transmissible (Callaway, 2021; Zhang et al., 2021).
In order to better understand the emergence, spread and evolution of Omicron SARS-CoV-2 variants, a comprehensive phylogenetic study was performed.
2. Material and Methods
2.1. Sequences
Available and comparable complete S gene sequences of 159 Omicron SARS-CoV-2 strains isolated from November 13th to December 2nd, 2021, in South Africa, Ghana, Singapore, Portugal, Netherlands, Mexico, Malaysia, Japan, Hong Kong, Germany, Canada, Botswana, Israel, Ireland, Belgium, Austria, Australia, South Korea, USA, Sri Lanka and Switzerland, were used throughout these studies. These sequences were aligned with corresponding sequences from 97 Alfa, Beta, Gamma and Delta SARS-CoV-2 strains isolated elsewhere. Sequences were obtained from the Global Initiative on Sharing Avian Influenza Data (GISAID) database. For accession numbers, country of origin and date of isolation, see Supplementary Material Table 1.
2.2. Sequence alignment
Sequences were aligned using MAFFT version 7 program (Katoh et al., 2019).
2.3. SARS-CoV-2 genotype assignment
In order to capture local and global patterns of virus genetic diversity in a timely and coherent manner, we employed Pangolin COVID-19 genetic lineage strain assignment (Rambaut, Holmes, O'Toole et al., 2020).
2.4. Data analysis
Nucleotide frequencies and codon usage of S proteins from SARS-CoV-2 variants were calculated using the program CodonW (written by John Peden) as implemented in the Galaxy server version 1.4.4 (Afgan et al., 2018). The relationship between compositional variables and samples was obtained using Principal Component Analysis (PCA). Singular value decomposition (SVD) method was used to calculate the PCA method. The unit variance was used as the scaling method. This means that all variables are scaled so that they will be equally important (variance = 1) when finding the components. By the same approach, Heatmaps were also constructed, which is a data matrix for visualizing values in the dataset by the use of a color gradient. Rows and/or columns of the matrix are clustered so that sets of rows or columns rather than individual ones can be interpreted. PCA and Heatmaps analysis were done using the ClustVis program (Metsalu and Vilo, 2015).
To reconstruct the evolutionary history of Omicron SARS-CoV-2 strains, a Bayesian Markov Chain Monte Carlo (MCMC) approach was used as implemented in the BEAST package v2.5.2 (Bouckaert et al., 2019). First, the evolutionary model that best fit the sequence dataset was determined using the IQ-TREE program (Trifinopoulos et al., 2016). Bayesian information criterion (BIC), Akaike information criterion (AIC), and the log of the likelihood (LnL) were used to identify the best model. Both strict and relaxed molecular clock models were used to test different dynamic models (constant population size, exponential population growth, Bayesian Skyline and Birth-Death Skyline Serial). Statistical uncertainty in the data was reflected by the 95 % highest probability density (HPD) values. Results were examined using the TRACER v1.7.2 program (available from http://beast.bio.ed.ac.uk/Tracer). Convergence was assessed by effective sample sizes (ESS) above 200. Models were compared by AICM from the likelihood output of each of the models. Maximum clade credibility trees were generated by means of the use of the Tree Annotator program from the BEAST package. Visualization of the annotated trees was done using the FigTree program v1.4.4 (available at: http://tree.bio.ed.ac.uk).
3. Results
3.1. Trends in evolution across S proteins from SARS-CoV-2 VOCs variants
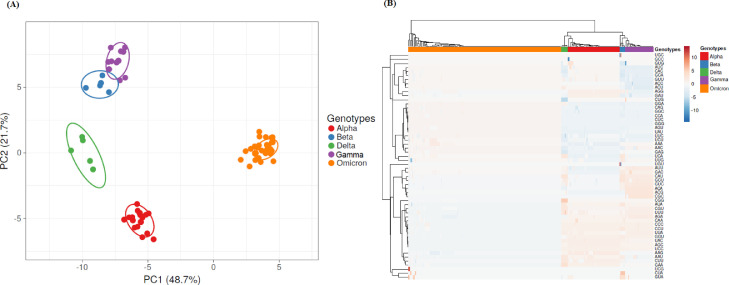
In order to gain insight into the trends of evolution of the S protein, codon usage frequencies of 256 S genes from SARS-CoV-2 strains belonging to VOCs Alpha, Beta, Gamma, Delta and Omicron were determined and PCA and Heatmap analysis were performed (for strains included in these analyses, see Supplementary Material Table 1). This analysis revealed significant differences in codon usage among the S genes of SARS-CoV-2 VOCs, which can be linked to SARS-CoV-2 genotypes (see Fig. 1 A and B). Average linkage suggest that VOCs Alfa, Beta, Gamma and Delta have a closer genetic relation among themselves and a more distant genetic relation with Omicron variants (see Fig. 1B, upper part). Moreover, average linkage also shows that Omicron isolates are not identical and heterogeneity can be observed (see Fig. 1B).
Fig. 1.
PCA of codon usage in Spike proteins from SARS-CoV-2 VOCs strains. In (A) the position of the Spike proteins in the plane conformed by the first two major components of PCA is shown. SVD was used to calculate principal components and unit variance was applied. The proportion of variance explained by each axis is shown between parentheses. Prediction ellipses are such that with probability 0.95, a new observation from the same group will fall inside the ellipse. Genotypes are indicated at the right of the figure. N = 256 data points. In (B) Heatmaps of codon usage in Spike proteins are shown. Unit variance scaling was applied. Each column corresponds to a different Spike protein from SARS-CoV-2 VOCs strains, who's genotype is shown in the upper part of the figure. Both rows and columns are clustered using correlation distance and average linkage.
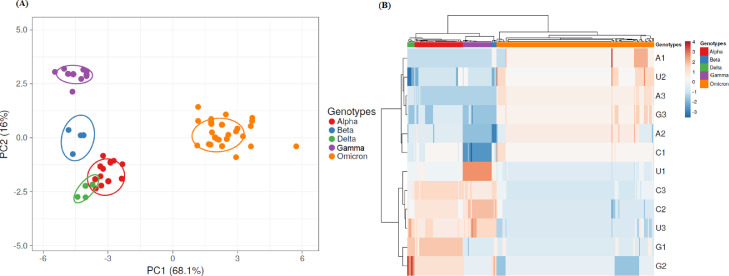
In order to study if these trends of evolution of the VOC's S genes are due to differences in nucleotide composition, an analysis of nucleotide frequencies for first, second and third codon positions were established for the complete S genes from VOCs SARS-CoV-2 variants included in these analyses and PCA and Heatmap analysis was performed (see Fig. 2 ). A significant bias in nucleotide composition frequencies was found among VOCs S proteins.
Fig. 2.
PCA of nucleotide composition in Spike proteins from SARS-CoV-2 VOCs. In (A) a PCA analysis of nucleotide frequencies for first, second and third codon positions in S protein from SARS-CoV-2 strains is shown. SVD was used to calculate principal components and unit variance was applied. The proportion of variance explained by each axis shown between parentheses. Prediction ellipses are such that with probability 0.95, a new observation from the same group will fall inside the ellipse. Genotypes are indicated at the right of the figure. N = 256 data points. In (B) Heatmaps of nucleotide frequencies in Spike proteins are shown. Frequencies for A, C, U and G at first, second and third codon positions are indicated 1 through 3. Unit variance scaling was applied. Each column corresponds to a different Spike protein from SARS-CoV-2 VOCs strains, who's genotype is shown in the upper part of the figure. Both rows and columns are clustered using correlation distance and average linkage.
3.2. Bayesian coalescent analysis of Omicron SARS-CoV-2 strains
In order to reconstruct the evolutionary history of Omicron SARS-CoV-2 population, a Bayesian MCMC approach was employed (Bouckaert et al., 2019) using 89 available and comparable full-length genomes by December 2nd, 2021, (for isolates included in these analyses see Supplementary Material Table 1). The results shown in Table 1 are the outcome of 20 million steps of the MCMC, using the HKY+I nucleotide model, a strict molecular clock and the Birth-Death Skyline Serial population model.
Table 1.
Bayesian coalescent inference of Omicron SARS-CoV-2 strains.
| Valueb | HPDc | ESSd |
|---|---|---|
| -41527.05 | -41604.75 to -41432.15 | 214.50 |
| 318.78 | 255.70 to 397.14 | 209.40 |
| -41845.83 | -41863.00 to -41830.62 | 314.10 |
| 5.613 × 10−4 | 3.051 × 10−4 to 9.014 × 10−4 | 208.10 |
| 1.539 | 0.806 to 2.251 | 210.70 |
| May 15th, 2020 | ||
| 0.310 | 0.120 to 0.570 | |
| October 10th, 2021 | ||
aSee Supplementary Material Table 1 for strains included in this analysis.
eThe rate of evolution is indicated in substitions/site/year.
ftMRCA, time of the most common recent ancestor is shown in years.
gtMRCA South Africa, time to the most recent common ancestor for all strains isolated in South Africa. The date estimated for the tMRCAs are indicated in bold.
In all cases, the mean values are shown.
HPD, high probability density values.
ESS, effective sample size.
The results of these studies suggest that strains classified as Omicron variants evolved from ancestors that existed around May 15th, 2020. A mean rate of evolution of 5.61 × 10−4 substitutions/site/year (s/s/y) was found for Omicron strains enrolled in these analyses (95 % high probability density values of 3.051 × 10−4 to 9.014 × 10−4 s/s/y). This is in agreement with previous estimations of SARS-CoV-2 rate of evolution of SARS-CoV-2 populations (6.57 × 10−4 s/s/y, Castells et al., 2020; 7.80 × 10−4 s/s/y, Lai et al., 2020; 9.90 × 10−4 s/s/y, Nie et al., 2020; 3.0 × 10−4 s/s/y, Simmonds, 2020; 1.60 × 10−3 s/s/y, Bai et al., 2020; 1.19-1.31 × 10−3s/s/y, Li, Zai and Zhao, 2020b).
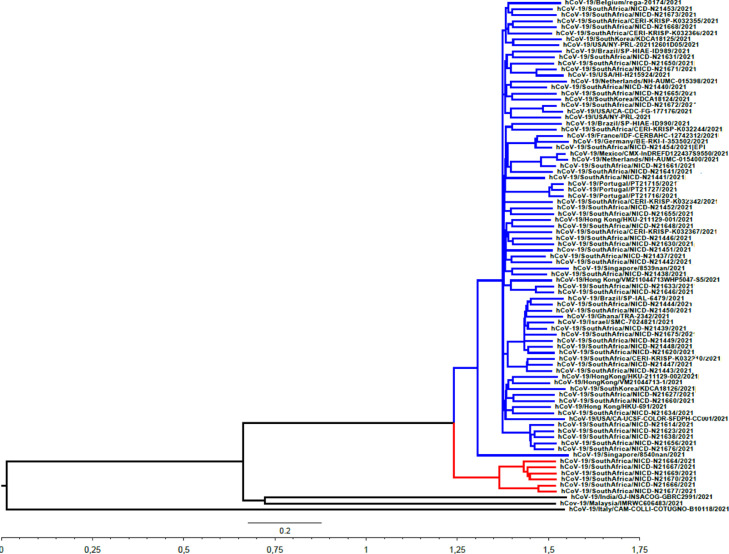
To study the phylogenetic relations among Omicron variants, maximum clade credibility trees were generated using software from the BEAST package (Bouckaert et al., 2019)(). The results of these studies are shown in Fig. 3 .
Fig. 3.
Bayesian MCMC phylogenetic tree analysis of Omicron SARS-CoV-2 strains. A maximum clade credibility tree obtained using the HKY+I model, a strict molecular clock and the Birth-Death Skyline Serial population model is shown. The tree is rooted to the Most Recent Common Ancestor (MRCA). The two main clades containing strains isolated in South Africa are shown in blue and red, respectively. Time to the MRCA is shown in years at the bottom of the figure. Bar at the bottom of the tree denotes time in years.
Strains isolated in South Africa belong to two different genetic clades, revealing that Omicron variants have diversified into two distinct genetic groups in that country. Strains isolated in Malaysia and India conform another genetic group (see Fig. 3). Inside main cluster of South African strains, strains isolated in Singapore, Hong Kong, Europe, Brazil and the USA are observed, suggesting a rapid spread of this lineage in different continents.
4. Discussion
SARS-CoV-2 S protein plays a key role in virus biology, epidemiology and adaptation of virus to its human host. Moreover, almost all vaccine candidates against SARS-CoV-2 are based on the S protein (Xia, 2021). Phylodynamic analyses can be extremely useful to study viral adaptation, particularly since new SARS-CoV-2 VOCs have emerged.
On November 26th, 2021, the WHO announced that a new SARS-CoV-2 VOC, named Omicron (initially named B.1.1.529), appeared to be increasing in almost all of South Africa's provinces, particularly Gauteng (World Health Organization, 2021b). Just three days after the announcement, cases of VOC Omicron have been detected in several other countries, including Austria, Australia, Belgium, Brazil, Canada, Czech Republic, Denmark, France, Germany, Italy, the Netherlands and the United Kingdom.
Omicron has some deletions and more than 30 substitutions in S protein, several of which (e.g., 69–70del, T95I, G142D/143–145del, K417N, T478K, N501Y, N655Y, N679K, and P681H) overlap with those in alpha, beta, gamma, or delta VOCs (GISAID, 2021). Nevertheless, Omicron also has specific S protein substitutions and insertions, like A67V, L212I, ins214EPE, G339D, S371P, S375F, N440K, N466S, S477N, Q493R, Y505H, T547K, N679K, N764K, D796Y, N856K, Q954H, N969K and L981F (GISAID, 2021).
In these studies, PCA and Heatmap analysis revealed correlation among codon usage and genotypes in the S protein from VOC's strains (see Fig. 1). These results demonstrate that S genes have suitable genetic information for clear assignment of emerging VOCs to its specific genotypes (Fig. 1). These differences are also related to biases in nucleotide composition among VOC's strains (Fig. 2). These findings highlight the latent diversity of SARS-CoV-2 that has yet to be fully explored. This is also in agreement with similar results found in other members of the family Coronvaviridae (Kumar et al., 2021).
Bayesian coalescent analysis revealed that Omicron strains evolved from ancestors that existed by May 15th, 2020. This result revealed that this VOC may have been circulating undetected for a period of time until its emergence was observed in South Africa (Petersen et al., 2021). This is in agreement with recent results suggesting that Omicron variants have been circulating much longer than previously anticipated (Kandeel et al., 2021). The earliest known case of Omicron in South Africa was a patient diagnosed with COVID-19 on Nov 9, 2021 (Karim & Karim, 2021). The results of these studies revealed that South African's Omicron strains evolved from ancestors that circulated around October 10th, 2021, just before it emerged in the South African population (see Table 1). Nevertheless, strains isolated at Singapore and Hong Kong can be traced to one of the main genetic lineages of Omicron, suggesting that although Omicron strains were first detected in South Africa, they may have also been circulating in other regions of the world undetected (see Fig. 3). Besides, Omicron strains isolated in South Africa can be assigned to two different genetic groups (see Fig. 3). This is in agreement with recent results suggesting that Omicron strains diversified in two different sub-lineages in that country (Wang & Chen, 2021).
The results of these studies revealed that Omicron variant did not evolve out of one of the early VOC's. Instead, it belongs to a complete different genetic lineage from previous VOC's. These results highlight the possibility of Omicron (or other VOCs currently unknown) may change the course of the pandemic. Whether the virus could have been circulating undetected in countries with little surveillance and sequences capacities is currently unknown (Kupferschmidk, 2021). More studies will be needed to address these important questions.
While this manuscript was in the review process, very recent studies revealed that Omicron SARS-CoV-2 viruses have diversified in three different genetic lineages: BA.1, composed by the first emerging viruses isolated in South Africa, Botswana, and elsewhere and being enrolled in the analyses shown in this work; and two more genetic lineages, BA.2, which have been remained minoritarian in several countries although its incidence became predominant in Demark (Houhamdi et al., 2022) and BA.3 (Colson et al., 2022). In order to gain insight into this recent evolutionary process, the same strains enrolled in these studies were aligned with comparable BA.2 and BA.3 strains recently isolated in different countries (for strains, accession numbers, date and country of isolation, see Supplementary Material Table 2). Then, the same Bayesian MCMC approach depicted in these studies were performed and their results are shown in Supplementary Material Fig. 1. As it can be seen in the figure, the same conclusions about the origin and phylogenetic relations of BA.1 strains were obtained. Moreover, BA.2 and BA.3 strains belong to different genetic lineages in agreement with recent results (Colson et al., 2022). More studies will be needed in order to establish the origin and evolution of BA.2 and BA.3 strains.
5. Conclusions
The results of these studies revealed significant differences in codon usage among the S genes of SARS-CoV-2 VOCs, which can be linked to SARS-CoV-2 genotypes. These results demonstrate that S genes have suitable genetic information for clear assignment of emerging VOCs to its specific genotypes. VOCs Alfa, Beta, Gamma and Delta have a closer genetic relation among themselves and a more distant genetic relation with Omicron variants. Omicron variants did not evolve out of one of the early VOCs, but instead it belongs to a complete different genetic lineage from previous VOCs. A significant bias in nucleotide composition frequencies was found among VOCs S proteins. Omicron variants evolved from ancestors that existed around May 15th, 2020 suggesting that this VOC may have been circulating undetected for a period of time until its emergence was observed in South Africa. Strains isolated in South Africa belong to two different genetic clades, suggesting that Omicron variants have diversified into two distinct genetic groups in that country.
Author individual contributions: Mercedes Paz, Fabián Aldunate, Rodrigo Arce, Irene Ferreiro were involved in the research, data curation and visualization of the results obtained. Juan Cristina was involved in conceptualization, methodology, data curation and wrote the original draft of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Agencia Nacional de Investigación e Innovación, PEDECIBA and Comisión Sectorial de Investigación Científica (Grupos I+D grant), Universidad de la República, Uruguay.
CRediT authorship contribution statement
Mercedes Paz: Data curation, Visualization, Investigation. Fabián Aldunate: Data curation, Visualization, Investigation. Rodrigo Arce: Data curation, Visualization, Investigation. Irene Ferreiro: Data curation, Visualization, Investigation. Juan Cristina: Conceptualization, Methodology, Data curation, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by Agencia Nacional de Investigación e Innovación and PEDECIBA, Uruguay. We acknowledge Comisión Sectorial de Investigación Científica, Universidad de la República, Uruguay, for support through Grupos I + D grant. We gratefully acknowledge the Originating and Submitting Laboratories for sharing newly identified coronavirus sequences through GISAID. We thank Drs. Pilar Moreno and Gonzalo Moratorio for critical reading and support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198753.
Appendix. Supplementary materials
References
- Afgan E., Baker D., Batut B., et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Jiang D., Lon J.R., et al. Comprehensive evolution and molecular characteristics of a large number of SARS-CoV-2 genomes reveal its epidemic trends. Int. J. Infect. Dis. 2020;100:164–173. doi: 10.1016/j.ijid.2020.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Beyond Omicron: what's next for COVID's viral evolution. Nature. 2021;600:204–207. doi: 10.1038/d41586-021-03619-8. [DOI] [PubMed] [Google Scholar]
- Castells M., Lopez-Tort F., Colina R., Cristina J. Evidence of increasing diversification of emerging Severe Acute Respiratory Syndrome Coronavirus 2 strains. J Med Virol. 2020;92:2165–2172. doi: 10.1002/jmv.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Delerce J., Beye M., et al. First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France. J Med Virol. 2022 doi: 10.1002/jmv.27695. 2022 doiEpub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID . 2021. Tracking of variants.https://www.gisaid.org/hcov19-variants/ (accessed December 12th, 2021) [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. bioRxiv. 2020 doi: 10.1101/2020.02.07.937862. [DOI] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houhamdi L., Gautret P., Hoang V.T., Fournier P.E., Colson P., Raoult D. Characteristics of the first 1119 SARS-CoV-2 Omicron variant cases, in Marseille, France, November-December 2021. J. Med. Virol. 2022 doi: 10.1002/jmv.27613. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M, Mohamed MEM, Abd El-Lateef HM, Venugopala KN, El-Beltagi HS. Omicron variant genome evolution and phylogenetics. J Med Virol. 2021 doi: 10.1002/jmv.27515. 2021 Dec 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Karim QA. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics. 2019;4:1160. doi: 10.1093/bib/bbx108. –116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Where did “weird” Omicron come from? Science. 2021;374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- Kumar N., Kaushik R., Tennakoon C., Uversky V.N., Mishra A., Sood R., Srivastava P., Tripathi M., Zhang K.Y.J., Bhatia S. Evolutionary Signatures Governing the Codon Usage Bias in Coronaviruses and Their Implications for Viruses Infecting Various Bat Species. Viruses. 2021;13:1847. doi: 10.3390/v13091847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;26:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Zhao Q., et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. Clustvis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research. 2015;43(W1):W566–W570. doi: 10.1093/nar/gkv468. https://doi.ogr/10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Q, Li X, Chen W, Liu D, Chen Y, Li H, Li D, Tian M, Tan W, Zai J. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020;287 doi: 10.1016/j.virusres.2020.198098. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E, Ntoumi F, Hui DS, Abubakar A, Kramer LD, et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529) - highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int J Infect Dis. 2021;114:268–272. doi: 10.1016/j.ijid.2021.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Rampant C→U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-Term Evolutionary Trajectories. mSphere. 2020;5:e00408–e00420. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cheng G. Sequence analysis of the Emerging Sars-CoV-2 Variant Omicron in South Africa. J Med Virol. 2021 Dec 12 doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Trial I. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2021. Coronavirus disease 2019 (COVID-19) Weekly epidemiological update on COVID-19 –14 December 2021.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed 12.12.2021) (accessed December 14th, 2021) [Google Scholar]
- World Health Organization . 2021. WHO 2nd Global consultation on assessing the impact of SARS-CoV-2 variants of concern on Public health interventions.https://www.who.int/publications/m/item/2nd-global-consultation-on-assessing-theimpact-of-sars-cov-2-variants-of-concern-on-public-health-interventions (accessed 26.11.2021) [Google Scholar]
- Xia X. Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design. Viruses. 2021;13:109. doi: 10.3390/v13010109/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design. Viruses. 2021;13:109. doi: 10.3390/v13010109/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Q, Liang Z, Li T, Liu S, Cui Q, Nie J, Wu Q, Qu X, Huang W, Wang Y. The significant immune escape of pseudotyped SARS-CoV-2 Variant Omicron. Emerg Microbes Infect. 2021;10:1–11. doi: 10.1080/22221751.2021.2017757.41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.