Abstract
The COVID-19 pandemic has caused tremendous damage to the world. In order to quickly and accurately diagnose the virus and contain the spread, there is a need for rapid, sensitive, accurate, and cost-effective SARS-CoV-2 biosensors. In this paper, we report on a novel biosensor based on angiotensin converting enzyme 2 (ACE-2)-conjugated vertically-oriented silicon nanowire (vSiNW) arrays that can detect the SARS-CoV-2 spike protein with high sensitivity and selectivity relative to negative controls. First, we demonstrate the efficacy of using ACE-2 receptor to detect the SARS-CoV-2 spike protein via a capture assay test, which confirms high specificity of ACE-2 against the mock protein, and high affinity between the spike and ACE-2. We then report on results for ACE-2-conjugated vSiNW arrays where the biosensor device architecture is based on a p-n junction transducer. We confirm via analytical modeling that the transduction mechanism of the biosensor involves induced surface charge depletion of the vSiNWs due to negative electrostatic surface potential induced by the spike protein after binding with ACE-2. This vSiNW surface charge modulation is measured via current-voltage characteristics of the functionalized biosensor. Calibrated concentration dependent electrical response of the vSiNW sensor confirms the limit-of-detection for virus spike concentration of 100 ng/ml (or 575 pM). The vSiNW sensor also exhibits highly specific response to the spike protein with respect to negative controls, offering a promising point-of-care detection method for SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Metal-assisted chemical etching (MACE), Silicon nanowire biosensor, Point-of-care device, Label-free detection
1. Introduction
Coronavirus disease 2019 (COVID-19) is a human infectious disease emerged in late 2019 that is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Based on the rapid increase in human infection, the World Health Organization has classified the COVID-19 outbreak as a pandemic. Among people without vaccination or immunity, early diagnosis and containment are critical for limiting the spread of the virus [1]. One approach to detect SARS-CoV-2 infection is the reverse transcription polymerase chain reaction (RT-PCR) using nasopharyngeal (NP) or throat swab samples [2]. It involves incubation, RNA extraction, reverse transcriptase, PCR amplification, and spectrophotometry [3]. This approach has good specificity but intermediate sensitivity, with false-negative rates ranging between 4% and 29% [4,5]. In particular, the sensitivity and false-negative rates can aggravate 5 days after symptom onset [6,7]. According to the Center for Disease Control [8], while RT-PCR based tests are highly specific and sensitive, they are costly (~$75–$100/test) with moderate to high complexity sample processing requiring trained professional staff, and the turnaround time for results could range from several hours to 1–3 days. The considerable lag between sample collection and the time individuals are informed of the results is a major concern and can lead to preventable spread of SARS-CoV-2. RT-PCR based tests also require Clinical Laboratory Improvement Amendments (CLIA) certification which can be costly to obtain and maintain for resource constrained rural clinics [9].
Several inexpensive point-of-care (POC) antigen-detection rapid diagnostic tests (Ag-RDTs) are currently commercially available. World Health Organization (WHO) recommends minimum sensitivity of 80% and specificity of 97% for Ag-RDTs [10]. Peeling et al. [11] reported in an extensive global study of Ag-RDTs that almost all of the single antigen tests for SARS-CoV-2 meet the WHO criteria. WHO recognizes that despite lower sensitivity than RT-PCR tests, Ag-RDTs offer the possibility of rapid, inexpensive detection of SARS-CoV-2 in individuals who have high viral loads and hence are at high risk of transmitting the infection to others. However, some key shortcomings are that Ag-RDTs are typically non-modular i.e., are designed to detect only one virus, and cannot differentiate between SARS-CoV-2 and other common respiratory viruses. Ag-RDTs tests also only provide qualitative results with limited ability to identify the stage of infection.
Alternatively, recently there has been increasing efforts in the development of angiotensin-converting enzyme 2 (ACE2) receptor-based biosensors by multiple research groups around the world [[12], [13], [14], [15], [16]]. This is because ACE2 is the main receptor used by SARS-CoV-2 for cellular entry, and infection of SARS-CoV-2 begins when the spike protein binds to ACE2 [17]. In a recent study, Ozono et al. [18] reported the binding affinity of ACE2 with five variants having global spread and mutations in the spike protein, and noted that four out of five variants showed significantly increased binding affinity to ACE2. From these results, one can infer that variants with a higher binding affinity to ACE2 are more contagious. Accordingly, implementing ACE2 in a biosensor can be an effective strategy for screening variants with high transmissibility.
Recently Park et al. [13] demonstrated a high sensitivity (~165 virus copies/ml) dual-gate field-effect transistor (FET) using ACE2 as the receptor. However, the dual-gate FET architecture was rather complex to manufacture and based on non-CMOS compatible materials, such as tin oxide, titanium nitride, among others; all of which can make it challenging to scale up this biosensor architecture for global monitoring of the pandemic. The work also did not confirm selective detection of the spike with respect to negative controls. Another biosensor architecture was reported by Pinals et al. [15] based on ACE2-single-walled carbon nanotube (SWCNT) optical sensing. The team demonstrated that the ACE2-SWCNT nanosensors exhibit a 73% fluorescence turn-on response within 5 s of exposure to 35 mg/L SARS-CoV-2 virus-like particles. SWCNTs are expensive and spectroscopic measurements needed for the optical characterization require bulky equipment that renders this sensor architecture unsuitable for inexpensive portable biosensors.
In this manuscript, we present results on a scalable bioelectronic sensor platform, which converts the binding of target biomolecules to electrical signals in a rapid and compact formfactor, enabling POC testing of viruses, such as COVID-19, by minimally trained individuals qualifying it for eventual CLIA waiver certification. Our biosensor architecture is based on silicon nanowires (SiNWs) that various teams [19,20] including ours [21] have demonstrated to have higher sensitivity than planar Si biosensors because of the high surface-to-volume ratio of the SiNWs. However, not all SiNWs-based biosensors are suited for COVID-19 detection, because their manufacturing steps are not complementary metal oxide semiconductor (CMOS) compatible while also being low-yield, slow, and require high-cost equipment [[22], [23], [24], [25]]. To address these challenges of SiNWs-based biosensors, we employ the metal-assisted chemical etching (MACE) process [26,27] to fabricate large area arrays of vertically-oriented SiNWs (vSiNWs) for our biosensor. The MACE process is CMOS-compatible and does not require any vacuum-based equipment and can be scaled up cost-effectively. Fig. S1 shows the scanning electron microscope images of the resulting vSiNWs from the MACE process. After the vSiNW MACE etching, the Si substrate is turned into an electrically active p-n junction diode via a number of nanofabrication steps shown in Fig. S1. Our simple device architecture allows us to electrically probe millions of NWs concurrently via a top and bottom contact scheme (Fig. 1(c)), and since each NW acts as an anchor for the target molecule, the sensor architecture results in highly sensitive detection of the biomolecules.
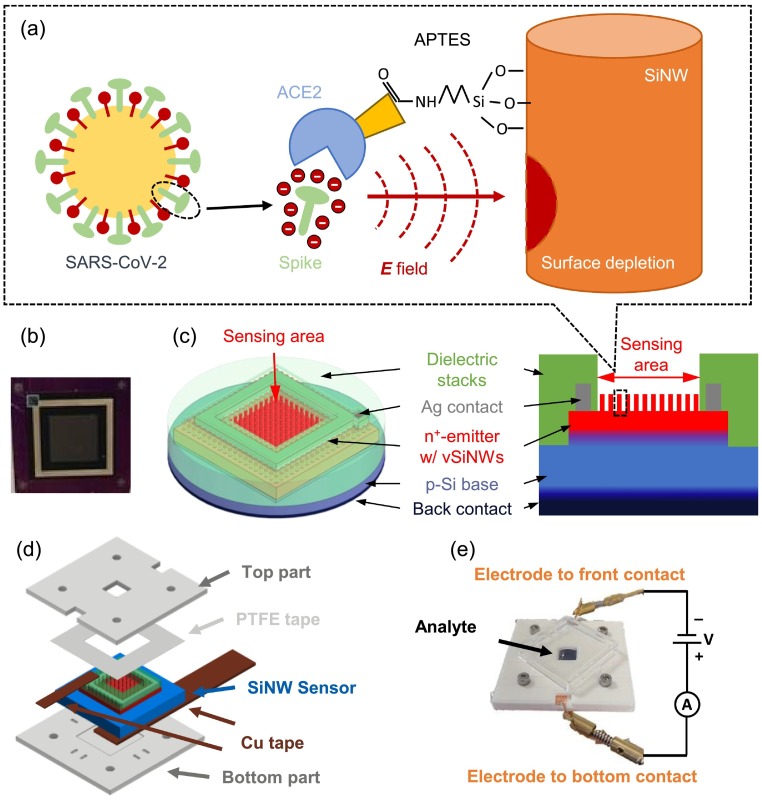
Fig. 1.
vSiNW biosensor and the encapsulation for SARS-CoV-2 detection. (a) Schematics of the detection mechanism of the vSiNW biosensor. Spike proteins exist on the surface of SARS-CoV-2, and the spike can specifically bind to the ACE-2 protein that is immobilized on the surface of vSiNWs. Negative electrostatic surface potential is induced by the spike protein after binding, which enhances the surface recombination effect if the vSiNWs are n+-doped. (b) Photo of a fabricated vSiNW biosensor. (c) Titled top and cross-sectional view schematic diagrams of the vSiNW biosensor. (d) Structural schematic of the ABS encapsulation of the vSiNW sensor, the assembly starts from the bottom. (e) I(V) testing system of the vSiNW biosensor with the sensor encapsulation, an external source meter, the analyte on the sensing area, and an ABS cover on top.
We functionalized the vSiNWs-based biosensors for SARS-CoV-2 detection by first treating the sensor surface with (3-Aminopropyl) triethoxysilane (APTES) and then immobilizing human angiotensin converting enzyme 2 (ACE-2) on APTES. Since ACE-2 interacts with the spike protein of SARS-CoV-2 with high affinity [28], we decided to utilize this interaction to detect presence of the virus. The spike protein is on the surface of SARS-CoV-2 molecule, and the specific binding of spike and ACE-2 proteins is found to be critical to the immobilization of SARS-CoV-2 during the infection of human cells [29]. A similar biochemical process was simulated on the surface of vSiNWs when the ACE-2 functionalized sensor was immersed in the solution of spike protein. We expect the polar spike protein to induce an electrostatic surface potential that can modulate the carrier density of vSiNWs substantially [30], as shown in Fig. 1(a). The detection of the coronavirus was thereby enabled by the conversion of this charge density modulation in the n-doped vSiNWs, into an electrical signal. Relative to RT-PCR, advantages of the vSiNW sensor platform are manifold, including: (i) results are obtained within 1 h, allowing rapid containment of infected individuals, (ii) due to the low expense of fabricating Si devices, the cost of each sensor is expected to be $10 to $15 (at scale production of 100,000 sensors/year), (iii) the sensor is small and portable, and can be used in a wide range of public settings for on-the-spot results, and (iv) it does not require scientifically qualified personnel for operation reducing labor costs included for virus testing.
2. Material and methods
2.1. Design of probes to detect SARS-CoV-2 spike protein
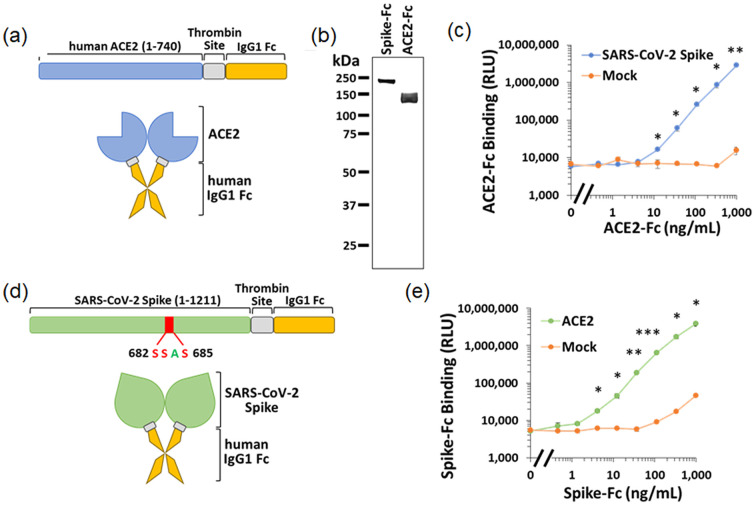
The ACE-2 protein is bound to the surface of cells and serves as an entry receptor for SARS-CoV-2 [31]. To this end, we first generated a soluble form of ACE-2, by introducing a stop codon at the C-terminal position of the extracellular domain (at amino acid position 741). This domain was linked to the Fc region of human IgG1 (Fig. 2 (a)), which allows purification of the chimeric protein using Protein A-coated beads and renders the protein dimeric. A thrombin cleavage site was introduced between the ACE-2 and the Fc segment for removal of the latter after purification [32]. Molecular weight and purity of the chimeric protein was validated by silver staining of samples analyzed by polyacrylamide gel electrophoresis (Fig. 2(b)). To determine the ability of the ACE-2-Fc probe to detect spike, we used a cell-based enzyme-linked immunosorbent assay (ELISA) [33,34]. For this purpose, we transfected human osteosarcoma (HOS) cells with a plasmid that expresses the SARS-CoV-2 spike protein. Cells were then incubated with soluble ACE-2 and its binding was detected using a horseradish peroxidase-conjugated secondary antibody and measured by luminescence. The limit of detection for binding of ACE-2-Fc to the spike-expressing cells was 12.5 ng/ml (96.2 pM) (Fig. 2(c)).
Fig. 2.
Design and validation of the ACE-2-Fc and Spike-Fc soluble probes. (a) Composition of the ACE-2-Fc chimeric protein. The extracellular domain of human ACE-2 was fused to the Fc region of human IgG1. A thrombin cleavage site was introduced to allow removal of the Fc region. (b) Analysis of the purity and size of the ACE-2-Fc and Spike-Fc probes by polyacrylamide gel electrophoreses, followed by silver staining. (c) Detection of the ACE-2-Fc probe by cell-based ELISA. HOS cells were transfected to express on their surface the SARS-CoV-2 spike protein. As controls, some samples were transfected with an empty (mock) vector. Binding of ACE-2-Fc to the cells was detected using an HRP-conjugated goat anti-human IgG preparation. Significance of the differences between binding to cells that express spike and mock-transfected cells was calculated using an unpaired t-test: *, p < 0.05; **; p ≤ 0.005; ***, p ≤ 0.0005. (d) Composition of the soluble Spike-Fc probe. To generate a probe that contains both S1 and S2 subunits, the furin cleavage site was disrupted by substitution of the Arg-Arg-Ala-Arg motif to Ser-Ser-Ala-Ser. (e) Measurements of Spike-Fc binding to HOS cells transfected by a plasmid that expresses the ACE-2 protein or a negative control (mock) plasmid.
To quantify the ability of the ACE-2-activated vSiNWs to detect spike, we also generated a soluble form of the spike protein. It is composed of the ectodomain of spike (amino acids 1–1211), a Thrombin cleavage site, and the Fc region of human IgG1 (Fig. 2(d)). To maintain subunits S1 and S2 of spike associated, we abrogated the furin cleavage site at positions 682–685 by substituting the existing Arg-Arg-Ala-Arg cleavage motif to Ser-Ser-Ala-Ser. Binding efficiency of the chimeric Spike-Fc construct to ACE-2 was measured by cell-based ELISA using HOS cells transfected with ACE-2. As shown in Fig. 2(e), binding of Spike-Fc to the ACE-2-expressing cells was detected at a concentration as low as 0.4 ng/ml (2.3 pM). We note that viral loads in the upper airways during early stages of infection are ~106–109 RNA copies per ml [35]. Assuming 30 spikes per virion [36], this corresponds to 0.05–50 pM of spike protein. Thus, the detection limit of the Spike-Fc probe using the cell-based ELISA system is within this range.
2.2. Preparation of testing solutions for the vSiNW biosensor
For testing the vSiNW sensor, two types of proteins were prepared. Spike proteins were prepared and used for the specific binding with the ACE-2 on the functionalized sensor surface. Bovine serum albumin (BSA) was used as a disturber for the specificity test, because its structure is analogous to that of the human serum albumin (HSA) [37]. Both BSA and spike were dissolved in the phosphate-buffered saline (PBS) at a serum-like pH of 7.4, and the concentration of all PBS solutions was 0.1× giving a sufficiently high Debye length of around 2.4 nm [38]. An isoelectric point (pI) study of the spike protein [39] showed that it has a pI of around 5, which means it applied a substantial negative potential in PBS.
2.3. vSiNW biosensor fabrication and characterization approaches
The photo and schematic of the vSiNW sensor are shown in Fig. 1(b) and (c), respectively. For further details on the biosensor fabrication process, please refer to Sections S1–S4 of the Supporting Information document. Briefly, a n+-p junction was formed on the front side of a lightly doped p-type Si substrate. The vSiNWs were turned into a highly-doped n+-emitter using ammonium dihydrogen phosphate (NH4H2PO4, ADP), a phosphorous (P) dopant of Si [40]. The back side of the Si substrate was highly p-doped via an aluminum (Al) based back surface field. After biosensor fabrication, the resulting vSiNWs were ~ 350 nm in length with a density of around 1010 SiNWs per cm2 area (see Fig. S1). The sensing area was 6.2 mm by 6.2 mm, and the top contact was silver (Ag) based and the back contact of the sensor is Ag-coated Al. On the front side, the Ag-contact and Si area outside the sensing area was coated with sputtered dielectric materials, namely silicon dioxide (SiOx) and silicon nitride (SiNx) to minimize degradation of the Ag-contact during biosensor functionalization steps.
We designed a 3D-printed mount for testing the biosensors such that sufficient volume of the spike solution can be maintained on the front surface of the biosensor. The 3D-printed mount was made of two acrylonitrile butadiene styrene (ABS) plastic accessories and attached to the front and back sides of the vSiNW biosensor to form a cuvette that could contain a maximum of 250 μL of liquid on the sensing area, as shown in Fig. 1(d). Polytetrafluoroethylene (PTFE) tapes were inserted between the top plastic part and the vSiNW sensor for fluid leak proofing. Biosensor characterization was performed on a benchtop Keithley 2400 source meter unit (SMU) without illumination, as in Fig. 1(e). Front and back contacts of the sensor were extended by copper (Cu) tape for contacting with the probes connected to the SMU inputs and outputs. The analyte testing solution was added in the cuvette to cover the biosensor sensing area, and a plastic cover was capped on top to prevent evaporation and contamination of the solution during experiments. When the voltage source was turned on, the vSiNW biosensor operated as an n+-p diode, and the measured dark current I as a function of voltage V, I(V), was measured six times during each experiment. After assembly of the ABS parts with the sensor, the sensing area was washed by adding and removing 250 μl of PBS twice using a micropipette, then 150 μl of PBS was added, and the first I(V) curve was measured with the plastic cap on, denoted by I PBS(V). After that, the PBS was extracted, and 150 μl of testing solution was added, followed by an immediate measurement of the I(V) curve, denoted by I 0min(V). Then after each 20 min, an I(V) curve was measured until 1 h after the addition of the testing solution, which generated three I(V) curves, I 20min(V), I 40min(V), and I 60min(V). The electrical response of the sensor was then determined by the normalized I(V) change before and 1 h after the addition of the testing solution, denoted by ΔI% and defined by:
| (1) |
Equation (1) shows that the first and last of the six I(V) curves were used to determine ΔI%, while the other four were plotted to illustrate the gradual change of the I(V) curves from 0 min to 60 min.
2.4. Biosensor testing protocols
First, the specificity of the vSiNW sensor against BSA was demonstrated, for which four combinations of functionalization and incubation were evaluated. For the functional sensors, ACE-2 was attached to the sensor surface, whereas for the control sensors, the ACE-2 immobilization was skipped, which means only BSA was used to terminate all the APTES molecules on the sensor. Then two types of incubation solutions were involved, each containing spike protein or BSA in the PBS. Combining the two types of functionalization and two types of incubation solutions, it resulted in four types of experiments. The positive test was conducted by testing the ACE-2-functionalized sensor with the spike protein (denoted by AS test or AS sensor), and three negative control tests included the ACE-2-functionalized sensor with the BSA (AB), the BSA-functionalized sensor with the spike (BS), and the BSA-functionalized sensor with the BSA (BB). The concentration of the spike was 7.5 μg/ml, and the BSA concentration was determined by maintaining the same density of molecules (in count/ml) as in the spike solution, which was approximately 2.5 μg/ml. In total, three AS sensors, three AB sensors, two BS sensors, and two BB sensors were tested. Then the concentration response of the sensor was demonstrated. All the vSiNW biosensors were ACE-2-functionalized, and the AS and AB tests for the specificity experiment corresponded to the spike concentrations of 7.5 μg/ml and 0 μg/ml, respectively. In addition, two other spike concentrations were tested, 0.06 μg/ml and 1.5 μg/ml, with one sensor at each concentration, and BSA was also added in those solutions to maintain the same density of molecules as the 7.5 μg/ml solution.
3. Results and discussion
3.1. Modeling of the vSiNW biosensor
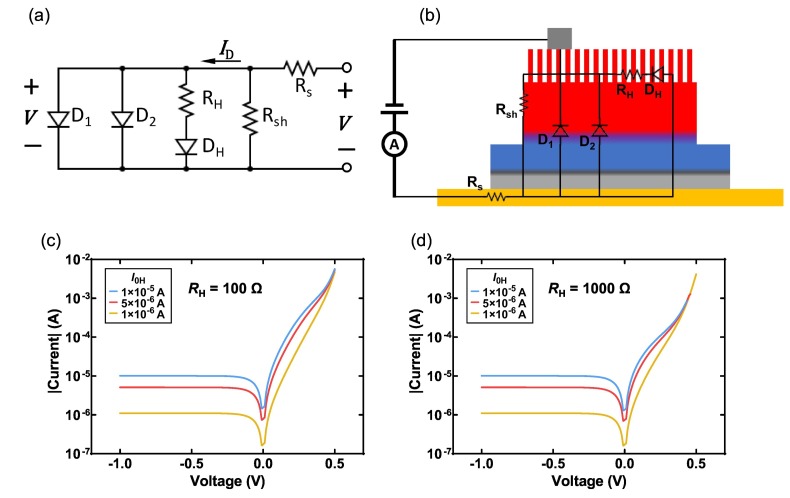
Devices with a p-n junction can be described using the Shockley equation for the diode model [41], from which the effective ideality factor n eff can be approximated. However, using the measured I(V) curve of a vSiNW biosensor, it was found that the effective ideality factor n eff as a function of V exhibits a “hump” that is much greater than two, under forward bias (Fig. S3). This effect indicates an enhanced front surface recombination that has been previously documented on nanotextured Si solar cells [42]. As a result, a “hump” diode, in addition to the traditional diffusion- and recombination-related diodes due to the p-n junction is induced, and the dark current model of device can be described using three diodes, referred as the three-diode model (Fig. 3(a)) [43,44]. A possible source of each component of the three-diode model can be found in Fig. 3(b). The two diodes D1 and D2 are caused by the p-n junction-related current, and the R s and R sh are the equivalent series and shunt resistances. The third diode DH, the “hump” diode, is due to the surface recombination-related saturation current I 0H.The diode is also connected in series with an equivalent resistance R H because it is away from the contact. Given the I(V) curve under high reverse and forward biases, the R s and R sh can be found from the derivative of I(V) (see Section S6 of Supporting Information), and the effects of R s and R sh can then be eliminated from the I(V) curve by
| (2) |
| (3) |
where V D and I D are the voltage across D1 and the sum of the current through all the diodes, respectively. The calibrated I(V) curve, I D(V D), is defined as the I D-V D curve, then ΔI% can also be calibrated by substituting I and V in Eq. (1) with I D and V D in Eq.s (2) and (3), which is denoted by ΔI D%,
| (4) |
Fig. 3.
Study of the three-diode model for the vSiNW biosensor. (a) Equivalent circuit diagram of the proposed three-diode model for the vSiNW biosensor. (b) Example schematic diagram of the possible location of each component of the circuit in (a). Variations of simulated ID-VD curve for different values of I0H and RH = 100 Ω (c) and 1000 Ω (d). Other parameters: I01 = 10−11 A, I02 = 1 × 10−7 A, n1 = 1, n2 = 2, nH = 2.5, Rs = 0 Ω, Rsh → ∞.
Using ΔI D%, the variations in R s and R sh during experiment can be eliminated. However, there are still seven coupled variables in the model: the saturation current (I 01, I 02, or I 0H) and ideality factor (n 1, n 2, or n H) for each diode (D1, D2, or DH), and R H. Regarding those variables, the following assumptions are made that due to the binding of ACE-2 and spike on the surface of the vSiNW sensor: (i) junction-related diodes D1 and D2 are not affected since the n+-emitter thickness is much greater than the Debye length of heavily doped Si [45], (ii) the ideality factor of DH is fixed at 2.5 since it is dependent on the source of DH [43,44]. Then the only variables that may change the I(V) curve, are I 0H and R H, whose effects on the simulated I D-V D curve are illustrated in Fig. 3(c) and (d). From simulation, the characteristic effect of I 0H on the I D-V D curve is found to be such that as I 0H increases, induced by enhanced surface recombination, the I D(V D) increases, and the increase is significant only at V D lower than a cutoff voltage V cutoff. Besides, the V cutoff is determined by R H since the higher the R H, lower the V cutoff. Furthermore, the effect of R H is found only significant at V > V cutoff, therefore we can determine the cause of the change in I D-V D by checking the V range where the change is significant: the I D(V D) change at V D < 0 is from I 0H, whereas the I D(V D) change at V D ⩾ 0.5 V is from R H.
In order to further demonstrate and find the major source of variation in the vSiNW sensor caused by a change in surface potential, a water-gate experiment was conducted by simulating the electrostatic potential of the spike protein using an external voltage source (see Section S7 of Supporting Information), from which it has been found that the change in I 0H is dominating, and for negative (positive) surface potential, the I 0H increases (decreases) due to surface depletion (passivation) effect, and it is also roughly found that V cutoff ≈ 300 mV.
3.2. Testing of the vSiNW biosensor and calibration of I(V) curves
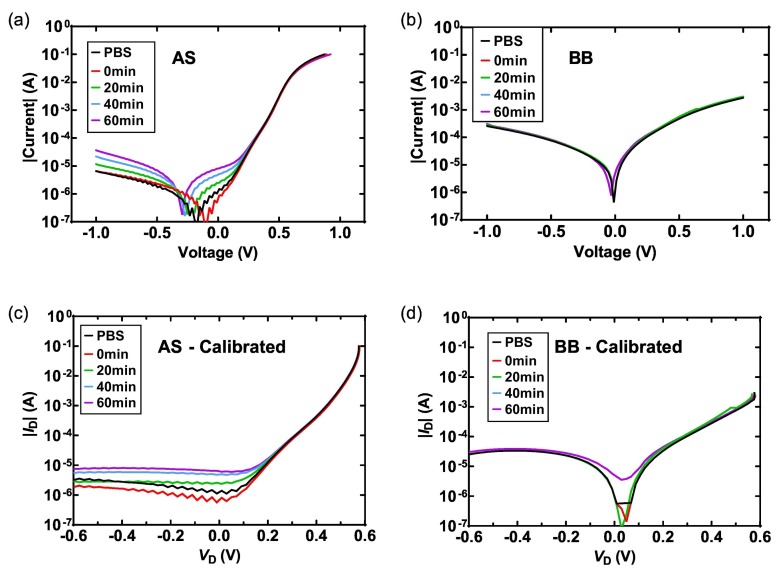
Given the analysis above, the vSiNW biosensor can then be tested with spike and BSA testing solutions. Theoretically, if the ACE-2 or (and) the spike is missing in the experiment, the non-specific binding of ACE-2-BSA, BSA-spike, or BSA-BSA will not induce randomly oriented electrostatic potential on the surface, hence the change of I(V) curves will be negligible. As for the specific binding of ACE-2 and spike proteins, net negative surface potential will be applied on the surface, which enhances the surface recombination effect of the n+-type vSiNW and emitter and also increases I 0H. This effect has been found on the AS sensors, as the I(V) change of an AS sensor from I PBS to I 60min is presented in Fig. 4(a). When the incubation time increases, the sensor shows a clear increase in I, and after 1 h, an increase in I(V) of an order of magnitude is observed at V < 0.3 V, indicating a substantial increase in I 0H induced by the negative surface potential. As a comparison, a BB sensor shows only has a small change in its I(V) curve after 1 h in the BSA solution (Fig. 4(b)). The I(V) curves are then calibrated to eliminate the effects of R s and R sh, as in Fig. 4(c) and (d), from which the conclusion can be drawn that the I 0H has increased on the AS sensor whereas the increase is less significant on the BB sensor.
Fig. 4.
Calibration of I(V) curves. Measured I(V) curves of an AS (a) and a BB sensor (b). Calibrated I(V) (ID-VD) curves of the AS (c) and the BB sensor (d).
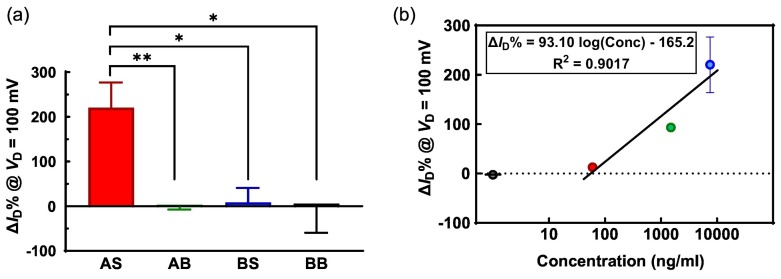
For each sensor in the experiment, ΔI D% is calculated using Eq. (4). Although from Fig. 4 and the water-gate experiment (Fig. S4), all the values of ΔI D% at V D < 300 mV can be used for demonstration, in this work, we choose to use ΔI D% at V D = 100 mV. The main reason is that due to the limitation of the source meter, the low current values of sensors at −0.4 V < V D < 0.1 V are noisy (such as in Fig. 4(c)). Besides, the estimation of R s and R sh could have uncertainties due to measurement errors, hence the ΔI D% values at low biases are still more reliable than those at high biases. ΔI D% at 100 mV of all the sensors for specificity experiment are presented in Fig. 5(a), where the high specificity of the vSiNW biosensor is demonstrated clearly. The significantly low ΔI D% for the negative controls with respect to the positive control indicates that non-specific binding cannot induce strong modulation of the biosensor's electronic response. The statistics of these controlled experiments confirm the high specificity response of the vSiNW biosensor for the spike protein when ACE2 is conjugated on the vSiNWs surface.
Fig. 5.
Specificity and concentration response results of the vSiNW biosensor for spike protein detection. (a) Statistics of ΔID% at VD = 100 mV for the positive test AS and three negative control tests AB, BS, and BB. (b) ΔID% at VD = 100 mV of the ACE-2-functionalized vSiNW biosensors at different concentrations of spike testing solutions, where the black circle represents the ΔID% for 0 ng/ml. (*: p < 0.05, **: p < 0.01, unpaired t-test between AS and each control group).
Furthermore, by testing at two more concentrations of the spike solution, ΔI D% at 100 mV at those concentrations are plotted in Fig. 5(b). The data for 0 ng/ml corresponds to the AB functionalization scheme in Fig. 5(a), which is a good negative control for the sensor. By analyzing the other groups with spike concentrations higher than zero, a semi-logarithmic relation between the concentration and ΔI D% is found, which has been theoretically demonstrated by Nair et al. [46] The regression equation indicates a strong ΔI D% response of 93.10% per decade increase of concentration, hence the sensitivity or the resolution of concentration is high in the semi-log region. The ΔI D% is close to zero at 100 ng/ml or 575 pM (ng/ml to pM conversion is explained above in Section 2.1) of spike protein, which defines the limit-of-detection (LOD) for our biosensor. This LOD is significantly higher than some other reported works [15] for ACE2 receptor-based biosensors and lower than others [13]. Therefore, we have successfully demonstrated a proof-of-principle for a vSiNW based biosensor platform for sensitive and selective detection of SARS-CoV-2. Future efforts will involve utilizing antibodies instead of ACE2 as the receptor for the spike protein given antibodies have many fold higher binding affinity to the protein than ACE2 [47], which would result in significantly improved sensitivity and LOD of our biosensor platform.
4. Conclusion
In this work, a vSiNWs-based biosensor for SARS-CoV-2 detection is demonstrated. The diode-type vSiNW biosensor with an area density of 1010 vSiNWs per cm2 and average vSiNW length of ~350 nm is fabricated using a scalable MACE process. ACE-2 is chosen as the functionalization protein, with successful assay tests showing specific binding of ACE-2 and SARS-CoV-2 spike proteins. A three-diode model of the sensor is proposed, which indicates that the I(V) characteristics of the sensor can be used to detect biomolecules that exhibit electrostatic polarity. Sensitive and specific detection of the SARS-CoV-2 spike protein utilizing the vSiNW biosensor is presented and confirmed. This work demonstrates promising capabilities of the vSiNW biosensor platform for use in POC SARS-CoV-2 detection.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sbsr.2022.100487.
Appendix A. Supplementary data
Supplementary material
References
- 1.Guglielmi G. The explosion of new coronavirus tests that could help to end the pandemic. Nature. 2020;583(7817):506–509. doi: 10.1038/d41586-020-02140-8. [DOI] [PubMed] [Google Scholar]
- 2.Pokhrel P., Hu C.P., Mao H.B. Detecting the coronavirus (COVID-19) ACS Sensors. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin S.A., Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005;109(4):365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- 4.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., del Campo R., Ciapponi A., Sued O., Martinez-Garcia L., Rutjes A., Low N., Bossuyt P.M., Perez-Molina J.A., Zamora J. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One. 2020;15(12):1–19. doi: 10.1371/journal.pone.0242958. e0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection – challenges and implications. N. Engl. J. Med. 2020;383(6) doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 6.Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369 doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 7.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 2020;173(4):262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Interim Guidance for Antigen Testing for SARS-CoV-2. 2022. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html#table1
- 9.Ambulos N.P., Jr. The good, the bad and the ugly: getting CLIA-certification for a basic research lab. J. Biomol. Tech. 2013;24(Suppl):S21. [Google Scholar]
- 10.Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection. 2021. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays in: W.H. Organization (Ed.)
- 11.Peeling R.W., Olliaro P.L., Boeras D.I., Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect. Dis. 2021;21(9):e290–e295. doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y., Peng Y., Lin C., Long L., Hu J., He J., Zeng H., Huang Z., Li Z.-Y., Tanemura M., Shi J., Lombardi J.R., Luo X. Human ACE2-functionalized gold “virus-trap” nanostructures for accurate capture of SARS-CoV-2 and single-virus SERS detection. Nano-Micro Letters. 2021;13(1):109. doi: 10.1007/s40820-021-00620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S., Kim H., Woo K., Kim J.-M., Jo H.-J., Jeong Y., Lee K.H. SARS-CoV-2 variant screening using a virus-receptor-based electrical biosensor. Nano Lett. 2022;22(1):50–57. doi: 10.1021/acs.nanolett.1c03108. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinals R.L., Ledesma F., Yang D., Navarro N., Jeong S., Pak J.E., Kuo L., Chuang Y.-C., Cheng Y.-W., Sun H.-Y., Landry M.P. Rapid SARS-CoV-2 spike protein detection by carbon nanotube-based near-infrared nanosensors. Nano Lett. 2021;21(5):2272–2280. doi: 10.1021/acs.nanolett.1c00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nascimento E.D., Fonseca W.T., de Oliveira T.R., de Correia C.R.S.T.B., Faça V.M., de Morais B.P., Silvestrini V.C., Pott-Junior H., Teixeira F.R., Faria R.C. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sensors Actuators B Chem. 2022;353 doi: 10.1016/j.snb.2021.131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11(1):4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12(1):848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K., Park C., Kwon D., Kim D., Meyyappan M., Jeon S., Lee J.S. Silicon nanowire biosensors for detection of cardiac troponin I (cTnI) with high sensitivity. Biosens. Bioelectron. 2016;77:695–701. doi: 10.1016/j.bios.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G.-J., Ning Y. Silicon nanowire biosensor and its applications in disease diagnostics: a review. Anal. Chim. Acta. 2012;749:1–15. doi: 10.1016/j.aca.2012.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Smith R., Duan W., Quarterman J., Morris A., Collie C., Black M., Toor F., Salem A.K. Surface modifying doped silicon nanowire based solar cells for applications in biosensing. Adv. Mater. Technol. 2019;4(2):1800349. [Google Scholar]
- 22.Yu G.H., Lieber C.M. Assembly and integration of semiconductor nanowires for functional nanosystems. Pure Appl. Chem. 2010;82(12):2295–2314. [Google Scholar]
- 23.Patolsky F., Zheng G.F., Lieber C.M. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat. Protoc. 2006;1(4):1711–1724. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 24.Hochbaum A.I., Fan R., He R.R., Yang P.D. Controlled growth of Si nanowire arrays for device integration. Nano Lett. 2005;5(3):457–460. doi: 10.1021/nl047990x. [DOI] [PubMed] [Google Scholar]
- 25.Wagner R.S., Ellis W.C. Vapor-liquid-solid mechanism of single crystal growth (new method growth catalysis from impurity whisker epitaxial + large crystals Si E) Appl. Phys. Lett. 1964;4(5):89. [Google Scholar]
- 26.Toor F., Miller J.B., Davidson L.M., Duan W.Q., Jura M.P., Yim J., Forziati J., Black M.R. Metal assisted catalyzed etched (MACE) black Si: optics and device physics. Nanoscale. 2016;8(34):15448–15466. doi: 10.1039/c6nr04506e. [DOI] [PubMed] [Google Scholar]
- 27.Toor F., Miller J.B., Davidson L.M., Nichols L., Duan W.Q., Jura M.P., Yim J., Forziati J., Black M.R. Nanostructured silicon via metal assisted catalyzed etch (MACE): chemistry fundamentals and pattern engineering. Nanotechnology. 2016;27(41) doi: 10.1088/0957-4484/27/41/412003. [DOI] [PubMed] [Google Scholar]
- 28.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair P.R., Alam M.A. Design considerations of silicon nanowire biosensors. IEEE T Electron. Dev. 2007;54(12):3400–3408. [Google Scholar]
- 31.Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020;43(7):648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waugh D.S. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 2011;80(2):283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson J., Zhai Y., Salimi H., Espy N., Eichelberger N., DeLeon O., O’Malley Y., Courter J., Smith A.B., 3rd, Madani N., Sodroski J., Haim H. Induction of a Tier-1-like phenotype in diverse Tier-2 isolates by agents that guide HIV-1 Env to perturbation-sensitive, nonnative states. J. Virol. 2017;91(15) doi: 10.1128/JVI.00174-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLeon O., Hodis H., O’Malley Y., Johnson J., Salimi H., Zhai Y., Winter E., Remec C., Eichelberger N., Van Cleave B., Puliadi R., Harrington R.D., Stapleton J.T., Haim H. Accurate predictions of population-level changes in sequence and structural properties of HIV-1 Env using a volatility-controlled diffusion model. PLoS Biol. 2017;15(4) doi: 10.1371/journal.pbio.2001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 36.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Lu J.M., Peukes Julia, Xiong Xiaoli, Kräusslich H.G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. Structures, conformations and distributions of SARS-CoV-2 spike protein trimers on intact virions. Nature. 2020;588(7838):498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandeville J.S., Tajmir-Riahi H.A. Complexes of dendrimers with bovine serum albumin. Biomacromolecules. 2010;11(2):465–472. doi: 10.1021/bm9011979. [DOI] [PubMed] [Google Scholar]
- 38.Lloret N., Frederiksen R.S., Møller T.C., Rieben N.I., Upadhyay S., De Vico L., Jensen J.H., Nygård J., Martinez K.L. Effects of buffer composition and dilution on nanowire field-effect biosensors. Nanotechnology. 2013;24(3) doi: 10.1088/0957-4484/24/3/035501. [DOI] [PubMed] [Google Scholar]
- 39.Krebs F., Scheller C., Grove-Heike K., Pohl L., Wätzig H. Isoelectric point determination by imaged CIEF of commercially available SARS-CoV-2 proteins and the hACE2 receptor. Electrophoresis. 2021;42(6):687–692. doi: 10.1002/elps.202100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan W., Gao B., Haque K.A.S.M.E., Toor F. Performance enhancement techniques for the front and back of nanostructured “black silicon” solar cells. J. Photon. Energy. 2018;8(3) [Google Scholar]
- 41.Shockley W. The theory of p-n junctions in semiconductors and p-n junction transistors. Bell Syst. Tech. J. 1949;28(3):435–489. [Google Scholar]
- 42.Hernando F., Gutierrez R., Bueno G., Recart F., Rodriguez V. 2nd World Conference and Exhibition on Photovoltaic Solar Energy Conversion, Vienna, Austria. 1998. Humps, a surface damage explanation. [Google Scholar]
- 43.McIntosh K.R. 2001. Lumps, Humps and Bumps: Three Detrimental Effects in the Current-Voltage Curve of Silicon Solar Cells. [Google Scholar]
- 44.McIntosh K.R., Black L.E. On effective surface recombination parameters. J. Appl. Phys. 2014;116(1) [Google Scholar]
- 45.Sze S.M., Ng K.K. Third edition ed. Wiley-Interscience; 2007. Physics of Semiconductor Devices. [Google Scholar]
- 46.Nair P.R., Alam M.A. Screening-limited response of NanoBiosensors. Nano Lett. 2008;8(5):1281–1285. doi: 10.1021/nl072593i. [DOI] [PubMed] [Google Scholar]
- 47.Loo Y.M., McTamney P.M., Arends R.H., Abram M.E., Aksyuk A.A., Diallo S., Flores D.J., Kelly E.J., Ren K., Roque R., Rosenthal K., Streicher K., Tuffy K.M., Bond N.J., Cornwell O., Bouquet J., Cheng L.I., Dunyak J., Huang Y., Rosenbaum A.I., Reddy V.P., Andersen H., Carnahan R.H., Crowe J.E., Jr., Kuehne A.I., Herbert A.S., Dye J.M., Bright H., Kallewaard N.L., Pangalos M.N., Esser M.T. The SARS-CoV-2 monoclonal antibody combination, AZD7442, is protective in non-human primates and has an extended half-life in humans. Sci. Transl. Med. 2022;14(635) doi: 10.1126/scitranslmed.abl8124. eabl8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material