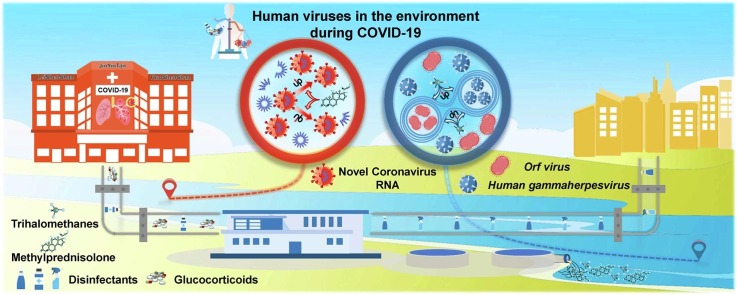
Graphical abstract
Keywords: COVID-19, Virome, Glucocorticoids, Trihalomethanes, Human Virus, SARS-CoV-2
Abstract
Due to extensive COVID-19 prevention measures, millions of tons of chemicals penetrated into natural environment. Alterations of human viruses in the environment, the neglected perceiver of environmental fluctuations, remain obscure. To decipher the interaction between human viruses and COVID-19 related chemicals, environmental samples were collected on March 2020 from surroundings of designated hospitals and receivers of wastewater treatment plant effluent in Wuhan. The virus community and chemical concentration were respectively unveiled in virtue of virome and ultra-high-performance liquid chromatography-tandem mass spectrometry. The complex relationship between virus and chemical was ulteriorly elaborated by random forest model. As an indicator, environmental viruses were corroborated to sensitively reflect the ecological disturbance originated from pandemic prevention supplies. Chemicals especially trihalomethanes restrained the virus community diversity. Confronting this adverse scenario, Human gammaherpesvirus 4 and Orf virus with resistance to trihalomethanes flourished while replication potential of Macacine alphaherpesvirus 1 ascended under glucocorticoids stress. Consequently, human viruses lurking in the environment were actuated by COVID-19 prevention chemicals, which was a constant burden to public health in this ongoing pandemic. Besides, segments of SARS-CoV-2 RNA were detected near designated hospitals, suggesting environment as a missing link in the transmission route. This research innovatively underlined the human health risk of pandemic prevention supplies from the virus - environment interaction, appealing for monitoring of environmental viruses in long term.
1. Introduction
Human viruses in the environment pose considerable effects to public health and microbial ecology. Environment matrices were gradually recognized as huge reservoirs of human viruses (Labadie et al., 2020) with the assistance of rapidly developed non-culture methods such as metagenomics and virome (Paez-Espino et al., 2016, Schulz et al., 2020). In aquatic environments, gastroenteritis-related Adenovirus, Astrovirus and Rotaviruses were detected in rivers, reservoirs (Wang et al., 2020), and even drinking water (Mehle et al., 2018). In terrestrial ecosystems, the prevalence of Influenza A virus was over 80% in soil matrix associated with poultry (Lau et al., 2019). Apart from causing disease, Influenza A virus threatened public health for indirectly initiating the expression of antibiotic resistance in human microbes (Zhang et al., 2020). With the raging of COVID-19 pandemic, the footprint of waterborne SARS-CoV-2 was tracked in waste water treatment plant (WWTP) systems all over the world (Carducci et al., 2020). Besides, the ubiquitous human viruses in the environment affect the microbial ecology through antagonistic or mutualistic interaction. As a consequence of antagonistic relationship (Paul et al., 2021), bacteria that grazed on Echovirus via proteolytic enzymes could outcompete their counterparts (Olive et al., 2020). Norovirus in the environment attached to pathogenic bacteria via histo-blood group antigens (Amarasiri and Sano, 2019) and heightened the bacterial adherence as well as their co-infection to eukaryotic cells (Neu and Mainou, 2020). In addition, human viruses, as the proxy of human-originated contaminants (Farkas et al., 2020), would be sensitive perceivers of environmental turbulence. During COVID-19 when anthropogenic activities to prevent the spread of pandemic tremendously altered environment, human viruses lying in the intersection of “One Health” perspective (Shaheen et al., 2022) should be elaborated to suppress the potential side effect.
Human viruses in the environment were regulated by various parameters such as temperature, relative humidity and pH (Carratalà et al., 2020). Nevertheless, the interaction between human viruses and chemicals originated from pandemic prevention supplies that were excessively used during COVID-19 remain misty. For the sake of disinfection, chlorine and quaternary ammonia surfactant containing biocidal agents were exorbitantly used in hospitals and wastewater treatment plants (WWTPs) (Elsaid et al., 2021). In Wuhan, the average usage of disinfectants in 26 WWTPs was roughly 40 t day−1 normally (Text S1) but it ascended to 84.64 t day−1 during pandemic (Wuhan Water Affairs Bureau, 2020). As a recommended adjuvant therapy for treating COVID-19 patients (Sterne et al., 2020), glucocorticoids were clinically used with a proportion of 44.5% (Guan et al., 2020). The consumption rate of glucocorticoids in Wuhan was reckoned at 44,168 mg day−1 on March 2020 (Text S2). Environment, the reservoir of human viruses, was inevitably confronted with the lash from residues and by-products derived in disinfectants and glucocorticoids. To what extent chemicals related with COVID-19 prevention i.e., chlorine, trihalomethanes, quaternary ammonium surfactants, and glucocorticoids shaped human viruses in the environment was obscure.
To delve into the interplay between chemicals from COVID-19 prevention supplies and human virus community in the environment, soil and water samples were collected from designated hospital surroundings and downstream of WWTPs in Wuhan on March 2020. Human virus community in this special period was manifested by means of metagenomics and virome. Random forest models on various taxonomic levels were executed to investigate the environment - virus interaction with total chlorine, trihalomethanes, quaternary ammonium surfactants, and glucocorticoids taken into consideration. Furthermore, mechanism underlying the bilateral relationship was explored from the dimension of virus viability represented by replication, recombination and repair genes.
2. Materials and methods
This study aimed to unravel the effect of excessively used pandemic prevention supplies on human viruses in the environment and the response of human viruses to the lash from pandemic prevention supplies. To this end, environmental samples were collected from surroundings of hotspots in Wuhan city i.e., designated hospitals and WWTPs on March 2020. Consumptions of glucocorticoids and disinfectants were calculated to measure the environmental burden from overused pandemic prevention supplies. Virome was conducted to illustrate the virus community; and UPLC-MS/MS was adopted to detect chemicals generated by pandemic prevention supplies. Random forest model was constructed to identify the interaction between human viruses and chemicals. Genes related with viability were picked out to clarify the mechanism of human viruses - pandemic prevention supplies interplay.
2.1. Sample collection
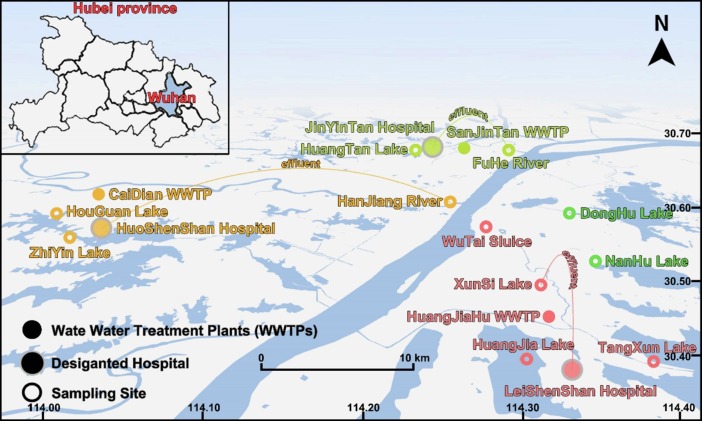
As one of the first cities that took measures to suppress COVID-19 (Zhou et al., 2020), Wuhan was typical in evaluating the ecological impact of pandemic prevention supplies. Designated hospitals for novel coronavirus pneumonia, the biggest consumer of drugs, were considered as source of glucocorticoids in the environment during COVID-19. WWTPs, the major consumer of disinfectants, linked chemicals and environment with effluent (Larsen and Wigginton, 2020). Medical wastewater from JinYinTan Hospital, HuoShenShan Hospital, and LeiShenShan Hospital was ultimately discharged to FuHe River, HanJiang River, and XunSi River after disposal of SanJinTan WWTP, CaiDian WWTP, and HuangJiaHu WWTP (Fig. 1 ). Hence, water and soil samples were collected at 11 sites from surroundings of designated hospitals and receivers of WWTPs (Table S1). To meet the demand of following virome analysis, 500 g of topsoil (0–10 cm) and 1.0 L surface water (∼0.5 m) were simultaneously gathered (Liu et al., 2013, Hu et al., 2021). The concentration of Fe3+ was adjusted to 1 mg L-1 through adding FeCl3 solution for flocculating virus particles (John et al., 2011). Virus particles were then enriched by ultracentrifugation and 0.22 μm filtration. Soil samples and 0.22 μm membranes were stored at −80 °C for subsequent treatment.
Fig. 1.
Distribution of environmental samples on March 2020 in Wuhan. Surroundings of designated hospitals (JinYinTan hospital, HuoShenShan hospital, LeiShenShan hospital), receivers of effluent from WWTPs that disposed of medical sewage (FuHe River, HanJiang River, XunSi River), along with inland lakes (DongHu Lake, NanHu Lake) were thoroughly taken into account.
2.2. Virome sequencing and processing
DNA and RNA were respectively extracted by PowerSoil DNA Isolation Kit (Qiagen, German) and RNeasy PowerSoil Total RNA Kit (Qiagen, German) (Wang et al., 2019). Reverse transcription reaction was conducted with PrimeScript RT Master Mix (Takara, Japan). Library construction and sequencing were carried out on Illumina HiSeq 2500 platform. To diminish ribosomal and host sequences, raw data were first processed by Trimmomatic (Bolger et al., 2014) and BWA alignment (Li and Durbin, 2009). For downstream virome analyses, clean data were assembled with Megahit v1.1.2 (Li et al., 2016) and clustered to unique contigs through CDHIT v4.7 (Fu et al., 2012). Two strategies i.e., database alignment (blast v2.9.0 + ) and hidden Markov models (Paez-Espino et al., 2016) were concurrently adopted to identify virus contigs. The RPKM (Reads Per Kilobase per Million mapped reads) value of each contig (Liu et al., 2020) was calculated to determine virus abundance. Information about virus functional genes was acquired through blastp (v2.9.0 + ) with UniProtKB/Swiss-Prot database as blueprint. Especially, zoonotic viruses and viruses that exclusively host human were categorized as human viruses in this research. A catalogue comprising sequences annotated as human viruses was constructed as human viral genomes.
2.3. Determination of chemicals from COVID-19 prevention supplies
Four types of chemicals derived from pandemic prevention supplies i.e., total chlorine, trihalomethanes represented by trichloromethane, quaternary ammonium surfactants represented by benzalkyl dimethylammonium compounds, and glucocorticoids represented by methylprednisolone were taken into consideration in this study. Prior to chemical detection, water samples and soil samples treated with ultrasonic were enriched by solid-phase extraction (SPE) (Xu et al., 2019). Subsequently, concentrations of 4 trihalomethanes, 5 quaternary ammonium surfactants, and 40 glucocorticoids were determined by ultra-high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) (Chen et al., 2021). Detailed information referring to concentration of above chemicals were exhibited in Table S2.
2.4 Estimation of COVID-19 prevention supply.
Consumptions of glucocorticoids and disinfectants were estimated in designated hospitals and WWTPs during COVID-19. According to the COVID-19 designated hospitals in Wuhan, the whole city could be divided to three regions (Fig. 1). In every region, designated hospital and WWTP would respectively be the major consumer of glucocorticoids and disinfectants. Thus, the estimated amount of glucocorticoids and disinfectants could stand for consumption of pandemic prevention supplies in each region.
Mglucocorticoids: amount of glucocorticoids used in COVID-19 designated hospital
Ndiagnosed: number of COVID-19 cases
p: usage rate of glucocorticoids, 18.6% (Guan et al., 2020)
W: average weight of adult in China, 65 kg
Cglucocorticoids: clinical dose of glucocorticoids recommended by Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7), 1 mg kg−1 day−1
Mdisinfectant: amount of disinfectant used in WWTPs during COVID-19
Vwater: volume of sewage in WWTP
Ccl: the concentration of disinfectant that would meet the standard for effluent on non-pandemic period, 10 mg L-1
f: the exceeding degree of disinfection during COVID-19, 2.12 (Wuhan Water Affairs Bureau, 2020)
To quantify the overuse degree of pandemic prevention supplies, the conventional usage of glucocorticoids before COVID-19 was estimated.
Mglucocorticoids_0: conventional value of glucocorticoids usage
Npatient: number of patients before pandemic, 66441 day−1 (Wuhan Municipal Health Commission, 2020)
Uglucocorticoids: average usage of glucocorticoids per patient, 2.04 mg day−1 (Zhang et al., 2020)
2.4. Data analyses
Relationships between chemicals and viruses were expatiated based on random forest model. Random forest regression models were constructed between chemicals and virus composition on order, family, genus, and contig levels. To a considerable extent, chemicals with high model accuracy were supposed to robustly affect virus community. Response of viruses to chemicals was calibrated as importance i.e., the descent of model accuracy after eliminating this virus. An aggregate of viruses with high importance (increase in mean squared error, IncMSE > 1%) in models (Wright et al., 2020) was prescribed as group that sensitively responded to chemicals. Phylogenetic relationship of viruses was demonstrated by Cytoscape v3.8.0. Principal components analysis was performed to distinguish geological distribution of virus community regulated by chemicals. Mantel test and Pearson correlation was used to examine the relationship between chemical and virus community (Hu et al., 2020).
3. Results
3.1. Amount of pandemic prevention supplies used during COVID-19
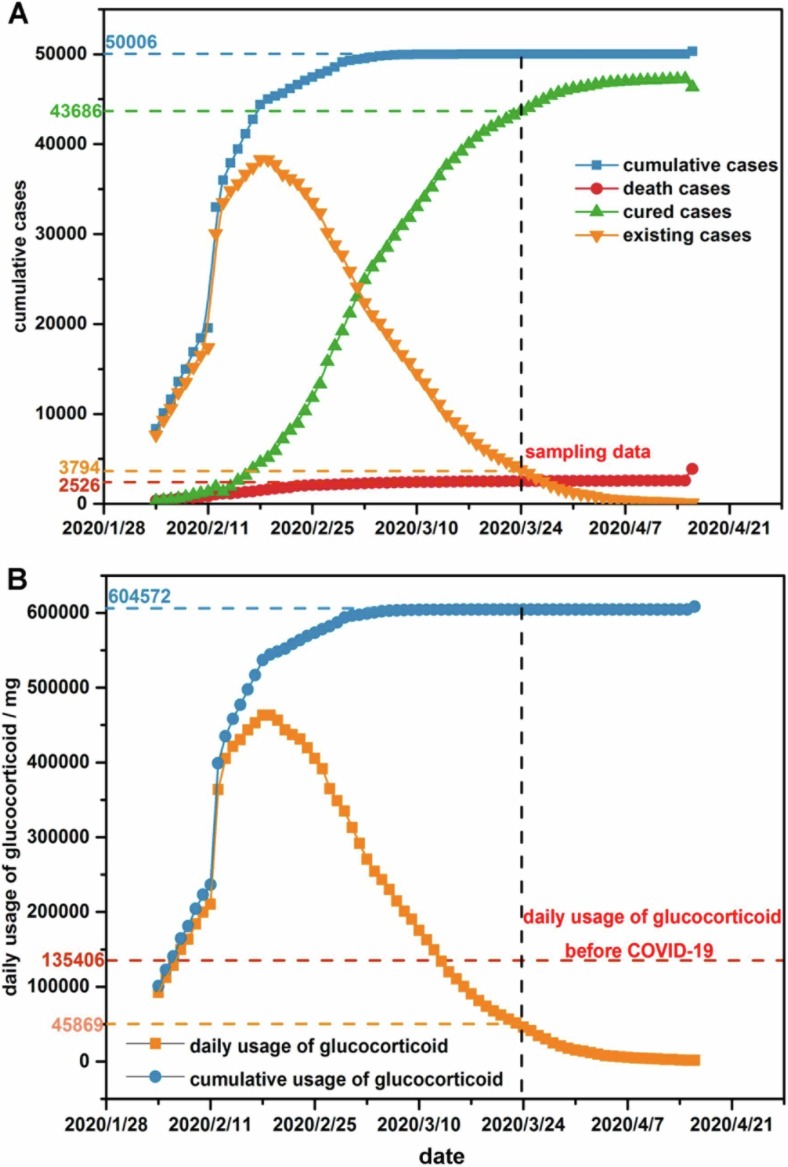
To clarify the environmental pressure imposed by excessively used COVID-19 prevention supplies, a rough calculation was drawn to specify the amount of glucocorticoids and disinfectants. The sampling date (2020/3/24) was between the inflection point and zero point of the case curve, representing scenario under impact from prevention supplies during COVID-19 (Fig. 2 A). The usage of glucocorticoids form 2020/2/7 to 2020/3/12 exceeded the conventional value before COVID-19, leading to a 1.83 times burden from glucocorticoids (Fig. 2B). The consumption of glucocorticoids in JinYinTan Hospital, HuoShenShan, and LeiShenShan Hospital on sampling date reached up to 6673.68 mg day−1, 11654.76 mg day−1, and 12597.78 mg day−1. To cater to the strict sterilizing demand during COVID-19, the daily amount of disinfectant consumed in Wuhan city rose by 2.12 times. Specifically, the usage of disinfectants in SanJinTan WWTP, CaiDian WWTP, and HuangJiaHu WWTP respectively climbed to 6.36 t day−1, 4.24 t day−1, and 2.12 t day−1.
Fig. 2.
Estimation of COVID-19 prevention supplies usage. (A) COVID-19 case curve in Wuhan from 2020/2/4 (reception day of HuoShenShan Hospital) to 2020/3/24 (sampling date) and (B) daily usage of glucocorticoids during COVID-19.
3.2. Virus community during COVID-19
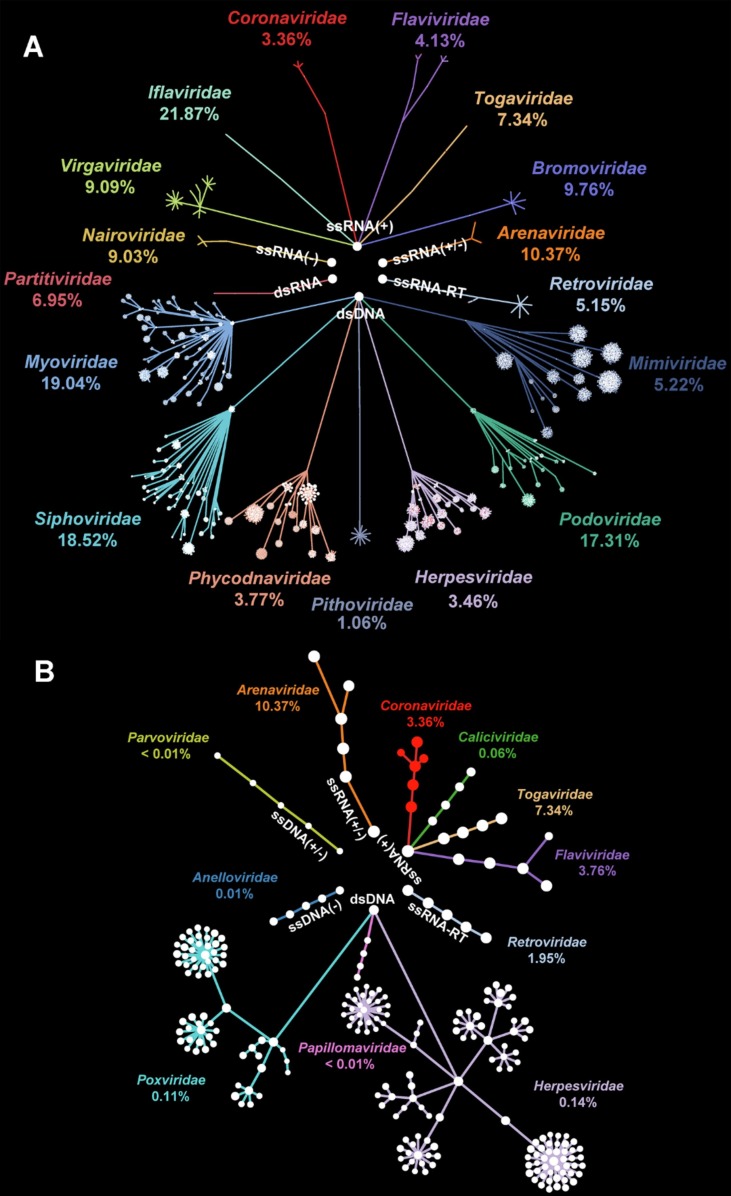
To comprehensively illustrate the composition of viruses during COVID-19, read-based and denovo methods were simultaneously adopted to identify viral genomes. A final catalog comprising 62,356 DNA contigs and 4,530 RNA contigs were screened under sequencing depth at 30 G per sample. Compared to aquatic viruses (bray-curtis dissimilarity: 0.90), viruses in soil samples were significantly more homogenized (bray-curtis dissimilarity: 0.82; p < 0.01) with minor families taking up > 90% sequences (Fig. S1). Consequently, aquatic samples were assumed to be more dynamic during COVID-19 and were targeted in following analyses. DNA virus community in water was dominantly occupied by phages i.e., Myoviridae (19.04%), Siphoviridae (18.52%), and, Podoviridae (17.31%) affiliated to Caudovirales (Fig. 3 A). Besides, dsDNA Herpesviridae (3.46%) whose members could infect human was ubiquitous in all samples. Aquatic RNA viruses were represented by invertebrate ssRNA(+) Iflaviridae (21.87%) and animal ssRNA(+/-) Arenaviridae (10.37%) (Fig. 3A). Notably, vertebrate ssRNA(+) Coronaviridae was detected in water samples with average abundance at 3.36%.
Fig. 3.
Virus community in the environment of Wuhan during COVID-19. Phylogenetic relationship of (A) total viruses with family abundance > 1% and (B) human viruses in water samples. Every dot represented a taxa annotated as the same family. The connection of dots represented the affiliation between taxon.
Focusing on human viral genomes in the environment, an aggregate consisted of 189 contigs subject to 26 species were extracted (Fig. 3B). An overwhelming proportion of human viruses was taken up by 20 species of DNA viruses that incorporated 179 contigs. Among DNA human viruses, Herpesviridae and Poxviridae that embraced 17 species stood out as representatives (Fig. 3B). Specifically, Human gammaherpesvirus 4 (0.06%) related to various neurological diseases and Molluscum contagiosum virus (0.04%) connected with infectious papules ranked top 2 by percentage. By contrast, Only 6 species with average abundance at 4.47% were recognized in human RNA virus community (Fig. 3B). Guanarito mammarenavirus (10.37%), the hemorrhagic pathogen, preponderated in human disease related RNA virome. To be vigilant, gene segments of SARS-CoV-2 were assembled in samples nearby designated hospitals i.e., HouGuan Lake (0.51%) and HuangJia Lake (36.38%) (Fig. S2) (Table S3).
3.3. Effect of pandemic prevention chemicals on virus community
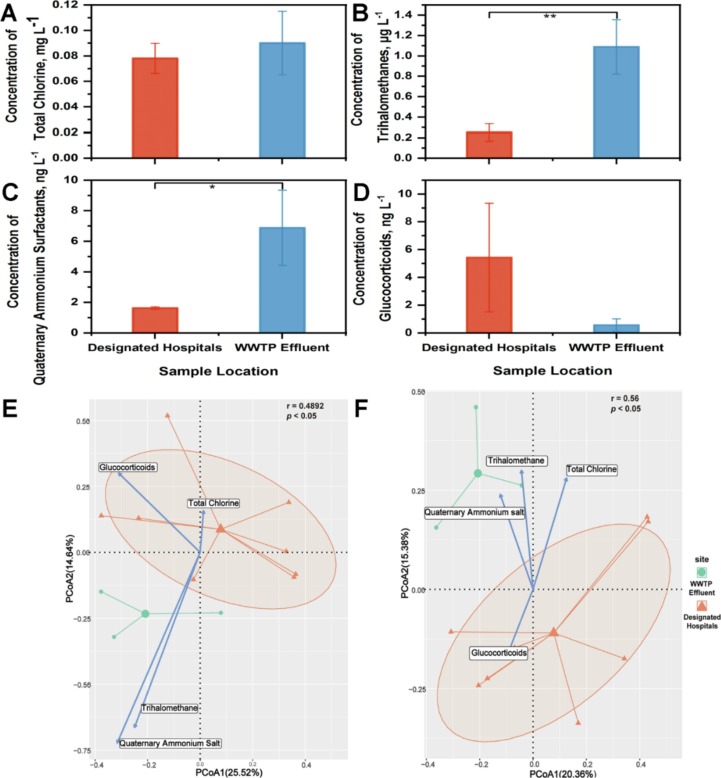
The variation of virus community was demonstrated under the massive input of COVID-19 related chemicals i.e., total chlorine, trihalomethanes, quaternary ammonium surfactants, and glucocorticoids. Samples from receivers of WWTP effluent and surroundings of designated hospitals were respectively featured by high concentration of trihalomethanes and glucocorticoids (Fig. 4 A–D). Correspondingly, the virus community was significantly divergent (adonis: r = 0.49, p < 0.05) with trihalomethanes as one of the vital influencing factors (Fig. 4E). Similar pattern was observed when it comes to human virus community. The geological variation of human viruses was critically shaped by trihalomethanes (adonis: r = 0.56, p < 0.05) (Fig. 4F). Moreover, significant negative relationships were seen between community diversity and trihalomethanes (richness: r = -0.70, p < 0.05; Shannon index: r = -0.87, p < 0.001). Human virus community also became homogeneous with concentration of trihalomethanes elevating (richness: r = -0.73, p < 0.05; Shannon index: r = -0.85, p < 0.001).
Fig. 4.
The geological discrepancy of virus community and chemicals generated from pandemic prevention supplies and. Comparison of (A) total chlorine, (B) trihalomethanes, (C) quaternary ammonium surfactants, and (D) glucocorticoids between samples in nearby water of designated hospitals and samples in receivers of WWTP effluent. Principal Components Analysis of (E) total viruses and (F) human viruses. * indicated p < 0.05 in t test; ** indicated p < 0.01 in t test.
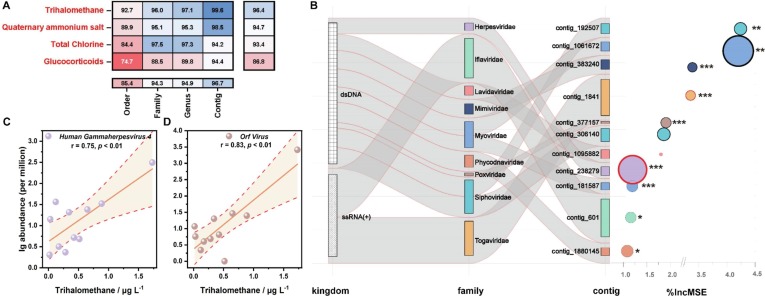
To further verify the impact of chemicals generated from pandemic prevention supplies on virus community, random forest model was conducted on order, family, genus, and contig levels. Concentrations of chemicals were determined by virus composition via regression model in which the accuracy indicated the extent of influence. Models built on contig and phylum levels averagely held the highest (96.7%) and lowest (86.8%) accuracy (Fig. 5 A). Regression was most successful when ascertaining the concentration of trihalomethanes (96.4%) (Fig. 5A).
Fig. 5.
Interactions between viruses and chemicals based on random forest regression analyses. (A) accuracies of random forest models when determining concentrations of chemicals with virus composition on order, family, genus and contig levels. (B) viruses with %IncMSE (increase in mean squared error) > 1% in models for trihalomethanes. (C) correlation between trihalomethanes and Human Gammaherpesvirus 4. (D) correlation between trihalomethanes and Orf Virus.
3.4. Response of viruses to pandemic prevention chemicals
To delineate the bidirectional associations, viruses that sensitively responded to chemicals were evaluated. As the most decisive factor (Fig. 5A), trihalomethanes were identified as the major environmental stress to virus community. Thence, 11 contigs affiliated to 9 families were picked out as sensitive viruses to trihalomethanes (Fig. 5B). Among the 11 contigs, 4 types were phages from dsDNA viurs Myoviridae and Siphoviridae. Confronting the disturbance from trihalomethanes, contig_1880145 (%IncMSE = 4.24), a member of algal dsDNA virus Phycodnaviridae, was the first to bear the brunt, followed by contig_601 (%IncMSE = 4.20) belonged to invertebrate ssRNA(+) virus Iflaviridae. Notably, 3 human viruses including Human gammaherpesvirus 4 (p < 0.001), Orf virus, and Semliki Forest virus (p < 0.001) entered the list.
To further elucidate the response of viruses in the environment to COVID-19 related chemicals, relationships between sensitive taxa and trihalomethanes were dissected. Sharp declines of contig_383240 (Mimiviridae) (pearson correlation: r = -0.79, p < 0.01) and contig_1061672 (Myoviridae) (r = -0.67, p < 0.05) were seen under high concentration of trihalomethanes. Contrarily, a boost in contig_192507 (Siphoviridae) corresponded with the increase of trihalomethanes (r = 0.67, p < 0.05). It was noteworthy that trihalomethanes were considerably favorable for the existence of sensitive human viruses i.e., Human gammaherpesvirus 4 (r = 0.75, p < 0.01) (Fig. 5C) and Orf virus (r = 0.83, p < 0.01) (Fig. 5D).
3.5. Potential human health risk resulted from viruses - pandemic prevention chemicals interactions
The latent hazard to human health from pandemic prevention supplies was distinguished by human viruses - trihalomethanes interaction. Samples in receiving water of WWTP effluent were marked by high concentration of trihalomethanes and relatively barren virus diversity (Fig. 4A–D) (Fig. S3). Whereas, human pathogens tremendously governed virus community in these sites. In HanJiang River that accepted treated medical wastewater from HuoShenShan Hospital, Crimean-Congo hemorrhagic fever virus, a member of Orthonairovirus causing human febrile illness, surprisingly reached the proportion of 98.65%. Receiving wastewater form JinYinTan hospital, FuHe River was notable with the prevalence of a human arbovirus - Aroa virus (41.39%) (Table S3). At XunSi River in which LeiShenShan hospital discharged wastewater, the abundance of Orf virus, a zoonotic virus causing contagious ecthyma, increased by at least 30 times compared to other sites (Table S3). Additionally, human viruses i.e., Human gammaherpesvirus 4 (pearson correlation: r = 0.75, p < 0.01) (Fig. 5C) and Orf virus (r = 0.83, p < 0.01) (Fig. 5D), that sensitively responded to trihalomethanes displayed a significant ascend in abundance. The abundance of Human Gammaherpesvirus 4 and Orf Virus multiplicatively increased by 9.90 and 32.06 times per 1 μg L-1 rise in trihalomethanes.
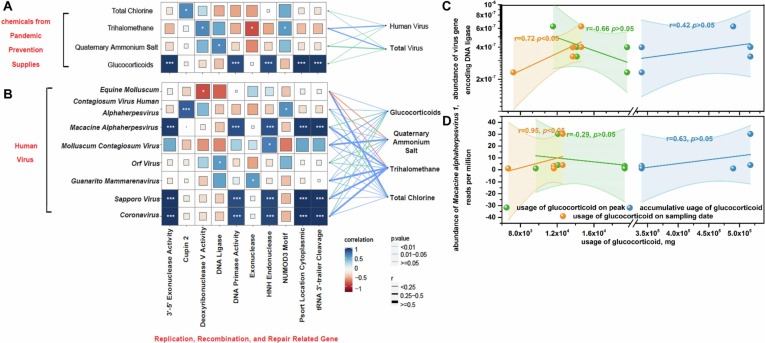
To take a deeper insight into the health threat from human viruses - pandemic prevention supplies connection, genes concerned with replication, recombination and repair were analyzed. Among the chemicals, glucocorticoids were conspicuous for links with several genetic material related genes (Fig. 6 A) although their influence on virus abundance was not as strong as other chemicals (Fig. 5A). As a marker gene of virus viability, RNA polymerase gene positively correlated with glucocorticoids (r = 0.71, p < 0.005). The abundance of genes regulating DNA primase activity (essential genes in replication) (r = 0.91, p < 0.001), 3′–5′ exonuclease activity (ensuring the high-fidelity in replication) (r = 0.98, p < 0.001), and HNH endonuclease (required for genome packaging reactions) (r = 0.91, p < 0.001) was raised under glucocorticoids. The viability of human viruses represented by the abundance of these genes ascended by 13.34 times with 1 ng L-1 rise in glucocorticoids. Additionally, positive correlation between virus gene that encoded DNA lipase, the most fundamental element in duplication, and usage of glucocorticoids (r = 0.72, p < 0.05) was observed (Fig. 6C). Human viruses i.e., Macacine alphaherpesvirus 1, Sapporo virus, and SARS-CoV-2 were found to be objects of effect from glucocorticoids because they simultaneously related with above genes as well as glucocorticoids (Fig. 6B). Not coincidently, the concentration of glucocorticoids in Huangjia Lake (500 m from LeiShenShan Hospital) was at least 9.8 times higher than that in other samples while the proportion of SARS-CoV-2 reached peak at 36.38% in this site. Especially, the abundance of Macacine alphaherpesvirus 1 escalated with the expanded usage of glucocorticoids (r = 0.95, p < 0.05) (Fig. 6D).
Fig. 6.
Interactions between chemicals and virus potential viability. Correlations between virus replication, recombination and repair genes and (A) chemicals and (B) human viruses. Correlations between usage of glucocorticoids and (C) gene encoding DNA lipase and (D) human virus Macacine alphaherpesvirus 1. Given the high concentration of glucocorticoids in surroundings of designated hospitals, 5 samples from nearby lakes including HuangTan Lake, ZhiYin Lake, HouGuan Lake, HuangJia Lake, and TangXun Lake were taken into consideration for analyzing relationship between usage of glucocorticoids and human viruses. Usage of glucocorticoids on peak, accumulative usage of glucocorticoids, and usage of glucocorticoids on sampling date were the estimated consumption of glucocorticoids based on the maximum admitted patients, total admitted patients, and existing patients in designated hospitals.
4. Discussion
4.1. Trace of SARS-CoV-2 RNA in the environment
RNA segments of novel coronavirus in the water implied the involving of environmental mediums on the transmission route. Unpredictably, RNA fragments of novel coronaviruses distinguished to SARS-CoV-2 were discovered in water near designated hospitals. Distinct from researches that quantified SARS-CoV-2 in raw sewage (Randazzo et al., 2020), wastewater (Trottier et al., 2020), or receiving water of WWTP effluent (Guerrero-Latorre et al., 2020) through RT-qPCR, this work was one of the first researches determining that gene segments of SARS-CoV-2 existed in natural water irrelevant with wastewater discharging. To be vigilant, RNA fragments of SARS-CoV-2 in natural water implied transmission pathway in which aquatic environment served as intermediate host. Presumably, the genetic material of novel coronavirus in water nearby designated hospital might originate from COVID-19 patients through direct contact or bioaerosol (Setti et al., 2020). Considered to be a fecal transmitted virus (Ding and Liang, 2020), SARS-CoV-2 was supposed to persist in water environment (Kumar et al., 2020). Although SARS-CoV-2 could not survive the reinforced disinfection process in WWTPs (WHO, 2020), integrated enveloped particles of SARS-CoV-2 were found to be stable in natural water for 12 days (Wurtzer et al., 2021). Despite the existence, whether SARS-CoV-2 was viable in genuine aquatic condition was vital to evaluate the possibility of dissemination through environment. Attempts were made to assess the infectivity of SARS-CoV-2 in water from aspect of cytopathic effect (Rimoldi et al., 2020). Further endeavor were imperative to evaluate the menace of SARS-CoV-2 in natural environment.
4.2. Succession of virus community initiated by COVID-19 prevention supplies
Chemicals generated from pandemic prevention supplies led to ecological impact through re-constructing virus community. It was deemed that the environment pollution level descended sharply due to the sweeping and strict lockdown (Muhammad et al., 2020), but the secondary pollution from medical treatment and disinfection substances imperiled ecological balance (Bhat et al., 2021). In particular, glucocorticoids from treatment and total chlorine, trihalomethanes, quaternary ammonium surfactants from disinfectants burst into environment through medical and household waste. Microbes in the environment undoubtedly were confronted with the interference of chemicals above, which has been ignored so far. This research shed light on the short-term dynamic of viruses, underlining the ecological damage of pandemic prevention supplies from dimension of environment microbial especially viral community. Generally, the succession of microbial community followed the cycle of collapse, reorganization, exploitation, and conservation (Shabarova et al., 2021). Pathogens i.e., human viruses tended to dominate the community during succession, which was similar in other researches (Revetta et al., 2013). Apart from the above chemicals, antibiotics were consumed in an inordinate scale with the clinical prevalence at 74.6% (Langford et al., 2021). The residue of antibiotics would inevitably induce antimicrobial resistance (AMR) in the environment even at a trace concentration (Sun et al., 2019). Viruses that mediated horizon gene transfer were proved to participate in the acquisition and dissemination of AMR (Debroas and Siguret, 2019). Therefore, excessively used antibiotics during COVID-19 were likely to interfere with the succession of human viruses via bacteria-virus interaction. Distinguished with the chemical pollution, biological contaminants including human viruses to some extent were more persistent for their ability to proliferate. Although the consumption of non-antiviral drugs i.e., glucocorticoids gradually returned to normal with the clearance of COVID-19 cases, disinfectants were still massively used in the ongoing pandemic (Chu et al., 2021). As a strategy to resist the trihalomethanes that continuously entered environment, it was assumed that the virus community would be dominated by Human gammaherpesvirus 4 (Fig. 5C) and Orf virus (Fig. 5D) in the long time scale. To comprehensively depict the succession process, subsequent monitoring of human viruses in the environment should be attached great importance.
4.3. Excessive disinfection in favor of human virus resistance
Viruses especially specific human viruses whose abundance soared with trihalomethanes withstood side effect of exaggerated disinfection through resistance. High concentration of trihalomethanes in receiving water of WWTP effluent corresponded with excessive usage of disinfectants during COVID-19 (Fig. 4A). Nevertheless, extravagant disinfection was found to be the villain of microbial resistance (Lu and Guo, 2021). Under disinfection, transforms of physiological state, e.g., increasing membrane permeability, SOS response and stress response offered an easier access for microorganisms to resistance (Cai et al., 2021). Equipped with horizontal transfer, viruses served as antibiotic resistance gene pool (Debroas and Siguret, 2019). Gene that specifically methylated the adenine in position 37 of tRNA (1) favored resistance under oxidative stress (Golovina et al., 2009). In this research, this gene was detected in virus genome and enriched in receiving water of WWTP effluent. Also, the long-term exposure of disinfection byproduct was in favor of gene mutation (Kurasam et al., 2018). Under the induction of trihalomethanes during COVID-19, it was possible for Human gammaherpesvirus 4 and Orf virus that strongly correlated with trihalomethanes (Fig. 5 C-D) in this study to obtain resistance through gene mutation or horizontal transfer. Moreover, herpesvirus (Bello-Morales et al., 2020) as well as Orf virus (Lan et al., 2016) were discovered to unite as vesicles. Depending on this morphology, viruses could efficiently combated with adversities e.g., disinfection (Zhang et al., 2021). Despite the prevalence of Human gammaherpesvirus 4 (Young and Rickinson, 2004) and Orf virus (Zhang et al., 2014), few researches focused on their distributions in natural environment. This pattern needs to be inspected in a broader geographical range. The mechanism lied behind the boosting relationship between trihalomethanes and human viruses remains to be further elaborated through microcosm simulation experiment.
4.4. Glucocorticoids advanced the proliferation of human viruses
Glucocorticoids were potential promoting factors to viability of human virus. Genomes recognized as SARS-Cov-2 RNA taken up a staggering proportion of 36.38% in HuangJia Lake (500 m from LeiShenShan Hospital). Meanwhile, concentration of glucocorticoids peaked at 22.81 ng L-1 in this sample (Table S2). The coincident concurrence hinted the some origin of SARS-Cov-2 RNA and glucocorticoids in environment during COVID-19. To some extent, glucocorticoids could be proxy of hospital related contaminants and embodied the pollution of novel coronavirus. Furthermore, the potential replication vitality of some human viruses i.e., Macacine alphaherpesvirus 1, Sapporo virus, and SARS-Cov-2 rose with glucocorticoids (Fig. 6). This phenomenon signified the promotion effect of glucocorticoids to virus in environment. As a double-edged sword, glucocorticoids was reported to stimulate dormant virus (Seeber et al., 2018) and accelerate virus replication (He et al., 2018). Actually, glucocorticoids directly interfered with these viruses via idiosyncratic responsive element. The expression of herpesvirus replication gene was activated by glucocorticoids via interaction mediated by ES-1 fragment (Yang et al., 2010). SARS-CoV-2 bounded to glucocorticoids through the main protease Mpro based on computational calculation (Fadaka et al., 2022). However, this kind of communication might be advantageous for SARS-CoV-2 to retain (Ma et al., 2020). Thus far, rare researches had referred to glucocorticoids - virus relationship in aquatic environment. On the days when hormone residue in environment has become a widespread problem (Zhong et al., 2021), this research enlightened the environmental interaction between virus and glucocorticoids.
4.5. Threat from human viruses in the environment
To practically evaluate the human health risk from viruses promoted by COVID-19 prevention chemicals, several factors should be taken into consideration in the future research. 1) Source of contaminants. Besides WWTP system that were proved to be hotspot of diverse human viruses (Mehle et al., 2018), surroundings of hospitals where the viability (Fig. 4D) (Fig. 6A–B) of human viruses were high should also be monitored. 2) Persistence and viability of human viruses in the environment. It is necessary to figure out the decay pattern of human viruses in the environment when assessing related health risk. Taking SARS-CoV-2 as an example, the infection of mere RNA was arguable even though it could linger in environment for weeks (Ahmed et al., 2020). 3) Propagation of human viruses in environmental matrices. Exposure pathway of human viruses hinge on their dissemination and circulation in environmental matrix. Viruses were enriched (Adriaenssens et al., 2021) while the infectivity decreased (Fauvel et al., 2017) with water flow. Similarly, a more diverse human virus community was observed in river that received WWTP effluent than in upstream lake (Fig. S3). 4) Integration of epidemiological data. Epidemiological information is the robust evidence of health effect from human viruses in the environment. During COVID-19, co-infection rate of SARS-CoV-2 and other human viruses represented by Influenza A virus could reach up to 50% (Lai et al., 2020). To further corroborate the origin of increasing human health risk during COVID-19, phylogenetic relationship should be identified between clinical and environmental viruses (Ruggeri et al., 2015).
5. Conclusion
This research focused on characteristics of environmental viruses under high-intensity use of pandemic prevention supplies during COVID-19 based on virome. Combined with random forest model, mechanisms between viruses and pandemic prevention chemicals were systematically probed on community, species, and gene levels. The abundance and viability of specific human viruses were potentially promoted by trihalomethanes and glucocorticoids, leading to an overlooked threat to human health. Following up studies should also be launched to assess the subsequent dynamics of virus community and tracking the community succession process. With the vision of environmental virology, this research disclosed the environmental damage under unconventional anthropogenetic activities i.e., excessive usage of pandemic prevention supplies. Scaling up to a boarder extent, this work serve as foundation for formulating ecological management policy in the ongoing pandemic and emphasized the necessity of regular human virus monitoring in the environment.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 22193061), the consulting research project of Chinese Academy of Engineering (2020-ZD-15) and Open Project of State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (No. ES202118).
Author contributions
Z.C.H., L.H.Y. and J.H. conducted the sampling. Z.C.H. L.H.Y. and Y.H.J. carried out experiments. Z.C.H. and Z.S.L. analyzed study data. Y.X.Z. collected environmental data. Z.C.H. visualized research data. Z.C.H. wrote the initial draft. Y.Q.S. coordinated research activity. L.Z.Z. and B.L.H. conceived of the study. Z.S.L. and B.L.H. reviewed and edited the original draft. L.Z.Z. offered fund support. B.L.H. supervised the research.
Handling Editor: Adrian Covaci
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2022.107192.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Adriaenssens E.M., Farkas K., McDonald J.E., et al. Tracing the fate of wastewater viruses reveals catchment-scale virome diversity and connectivity. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117568. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasiri M., Sano D. Specific Interactions between Human Norovirus and Environmental Matrices: Effects on the Virus Ecology. Viruses. 2019;11(3):224. doi: 10.3390/v11030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Morales R., Ripa I., López-Guerrero J.A. Extracellular Vesicles in Viral Spread and Antiviral Response. Viruses. 2020;12(6):623. doi: 10.3390/v12060623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S.A., Bashir O., Bilal M., Ishaq A., Dar M.U., Kumar R., et al. Impact of COVID-related lockdowns on environmental and climate change scenarios. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Sun T., Li G., An T. Traditional and Emerging Water Disinfection Technologies Challenging the Control of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. ACS ES&T Eng. 2021;1(7):1046–1064. [Google Scholar]
- Carducci A., Federigi I., Liu D.S., et al. Making Waves: Coronavirus detection, presence and persistence in the water environment: State of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carratalà A., Bachmann V., Julian T.R., Kohn T. Adaptation of human Enterovirus to warm environments leads to resistance against chlorine disinfection. Environ. Sci. Technol. 2020;54:11292–11300. doi: 10.1021/acs.est.0c03199. [DOI] [PubMed] [Google Scholar]
- Chen X.P., Lei L., Liu S.T., Han J., Li R.W., Men J., et al. Occurrence and risk assessment of pharmaceuticals and personal care products (PPCPs) against COVID-19 in lakes and WWTP-river-estuary system in Wuhan, China. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Fang C., Deng Y., Xu Z. Intensified Disinfection Amid COVID-19 Pandemic Poses Potential Risks to Water Quality and Safety. Environ. Sci. Technol. 2021;55(7):4084–4086. doi: 10.1021/acs.est.0c04394. [DOI] [PubMed] [Google Scholar]
- Debroas D., Siguret C. Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME J. 2019;13(11):2856–2867. doi: 10.1038/s41396-019-0478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Liang T.J. Is SARS-CoV-2 Also an Enteric Pathogen With Potential Fecal-Oral Transmission? A COVID-19 Virological and Clinical Review. Gastroenterology. 2020;159(1):53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaid K., Olabi V., Sayed E.T., Wilberforce T., Abdelkareem M.A. Effects of COVID-19 on the environment: An overview on air, water, wastewater, and solid waste. J. Environ. Manage. 2021;292 doi: 10.1016/j.jenvman.2021.112694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadaka A.O., Sibuyi N.R.S., Madiehe A.M., Meyer M. Computational insight of dexamethasone against potential targets of SARS-CoV-2. J. Biomol. Struct. Dyn. 2022;40(2):875–885. doi: 10.1080/07391102.2020.1819880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Walker D.I., Adriaenssens E.M., et al. Viral indicators for tracking domestic wastewater contamination in the aquatic environment. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvel B., Gantzer C., Cauchie H.-M., Ogorzaly L. In Situ Dynamics of F-Specific RNA Bacteriophages in a Small River: New Way to Assess Viral Propagation in Water Quality Studies. Food Environ. Virol. 2017;9(1):89–102. doi: 10.1007/s12560-016-9266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L.M., Niu B.F., Zhu Z.W., Wu S.T., Li W.Z. CD-HIT: accelerated for clustering the next-generationsequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovina A.Y., Sergiev P.V., Golovin A.V., Serebryakova M.V., Demina I., Govorun V.M., Dontsova O.A. The yfiC gene of E. coli encodes an adenine-N6 methyltransferase that specifically modifies A37of tRNA1Val (cmo5UAC) RNA. 2009;15(6):1134–1141. doi: 10.1261/rna.1494409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. New Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Song X., Huang Y., Huang W., Ye B.o., Luo H., Luo H., Wu L., Wang Z., Chen W., Zhang L.i. Dexamethasone Stimulates Hepatitis B Virus (HBV) Replication Through Autophagy. Med. Sci. Monitor. 2018;24:4617–4624. doi: 10.12659/MSM.906250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.J., Zhao Y.X., Yao X.W., Wang J.Q., Zheng P., Xi C.W., et al. Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146558. [DOI] [PubMed] [Google Scholar]
- Hu Z.C., Liu H., Zhang H., Zhang X., Zhou M., Lou L.P., et al. Temporal discrepancy of airborne total bacteria and pathogenic bacteria between day and night. Environ. Res. 2020;186 doi: 10.1016/j.envres.2020.109540. [DOI] [PubMed] [Google Scholar]
- John S.G., Mendez C.B., Deng L.i., Poulos B., Kauffman A.K.M., Kern S., Brum J., Polz M.F., Boyle E.A., Sullivan M.B. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 2011;3(2):195–202. doi: 10.1111/j.1758-2229.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Thakur A.K., Mazumder P., Kuroda K., Mohapatra S., Rinklebe J., et al. Frontier review on the propensity and repercussion of SARS-CoV-2 migration to aquatic environment. J. Hazardous Mater. Lett. 2020;1 doi: 10.1016/j.hazl.2020.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasam J., Sihag P., Mandal P.K., Sarkar S. Presence offluoroquinolone resistance with persistent occurrence of gyrA gene mutations in a municipal wastewater treatment plant in India. Chemosphere. 2018;211:817–825. doi: 10.1016/j.chemosphere.2018.08.011. [DOI] [PubMed] [Google Scholar]
- Labadie T., Batéjat C., Leclercq I., Manuguerra J.-C. Historical Discoveries on Viruses in the Environment and Their Impact on Public Health. Intervirology. 2020;63(1-6):17–32. doi: 10.1159/000511575. [DOI] [PubMed] [Google Scholar]
- Lai C.-C., Wang C.-Y., Hsueh P.-R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infection. 2020;53(4):505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Wang G., Song D., He W., Zhang D.i., Huang H., Bi J., Gao F., Zhao K. Role of autophagy in cellular response to infection with Orf virus Jilin isolate. Veterinary Microbiol. 2016;193:22–27. doi: 10.1016/j.vetmic.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Langford B.J., So M., Raybardhan S., Leung V., Soucy J.P.R., Westwood D., et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infec. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38(10):1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.-F., Chen E., Wang M., Cheng W., Zee B.-Y., Han X., Yu Z., Sun R., Chong K.C., Wang X. Association between meteorological factors, spatiotemporal effects, and prevalence of influenza A subtype H7 in environmental samples in Zhejiang province, China. Sci. Total Environ. 2019;663:793–803. doi: 10.1016/j.scitotenv.2019.01.403. [DOI] [PubMed] [Google Scholar]
- Li D., Luo R., Liu C.-M., Leung C.-M., Ting H.-F., Sadakane K., Yamashita H., Lam T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hu Z.C., Zhou M., Zhang H.H., Li Z., Zhang H., et al. Airborne microorganisms exacerbate the formation of atmospheric ammonium and sulfate. Environ. Pollution. 2020;263 doi: 10.1016/j.envpol.2020.114293. [DOI] [PubMed] [Google Scholar]
- Liu S., Shen L.D., Lou L.P., Tian G.M., Zheng P., Hu B.L. Spatial Distribution and Factors Shaping the Niche Segregation of Ammonia-Oxidizing Microorganisms in the Qiantang River, China. Appl. Environ. Microbiol. 2013;79(13):4065–4071. doi: 10.1128/AEM.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Guo J.H. Disinfection spreads antimicrobial resistance. Science. 2021;371(6528) doi: 10.1126/science.abg4380. [DOI] [PubMed] [Google Scholar]
- Ma S.Q., Zhang J., Wang Y.S., Xia J., Liu P., Luo H., et al. Glucocorticoids therapy delays the clearance of SARS-CoV-2 RNA in an asymptomatic COVID-19 patient. J. Medical Virol. 2020;92:2396–2397. doi: 10.1002/jmv.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle N., Gutierrez-Aguirre I., Kutnjak D., Ravnikar M. Water-Mediated Transmission of Plant, Animal, and Human Viruses. Adv. Virus Res. 2018;101:85–128. doi: 10.1016/bs.aivir.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Muhammad S., Long X.L., Muhammad S. COVID-19 pandemic and environmental pollution: A blessingin disguise? Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu U., Mainou B.A. Virus interactions with bacteria: Partners in the infectious dance. PLoS Pathogen. 2020;16(2):e1008234. doi: 10.1371/journal.ppat.1008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M., Gan C., Carratalà A., Kohn T., Johnson K.N. Control of Waterborne Human Viruses by Indigenous Bacteria and Protists Is Influenced by Temperature, Virus Type, and Microbial Species. Appl. Environ. Microbiol. 2020;86(3) doi: 10.1128/AEM.01992-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering Earth’s virome. Nature. 2016;536(7617):425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Paul D., Kolar P., Hall S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. npj Clean Water. 2021;4:7. [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revetta R.P., Gomez-Alvarez V., Gerke T.L., Curioso C., Santo Domingo J.W., Ashbolt N.J. Establishment and early succession of bacterial communities in monochloramine-treated drinking water biofilms. FEMS Microbiol. Ecol. 2013;86(3):404–414. doi: 10.1111/1574-6941.12170. [DOI] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri F.M., Bonomo P., Ianiro G., Battistone A., Delogu R., Germinario C., Chironna M., Triassi M., Campagnuolo R., Cicala A., Giammanco G.M., Castiglia P., Serra C., Gaggioli A., Fiore L., Elkins C.A. Rotavirus Genotypes in Sewage Treatment Plants and in Children Hospitalized with Acute Diarrhea in Italy in 2010 and 2011. Appl. Environ. Microbiol. 2015;81(1):241–249. doi: 10.1128/AEM.02695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F., Roux S., Paez-Espino D., Jungbluth S., Walsh D.A., Denef V.J., McMahon K.D., Konstantinidis K.T., Eloe-Fadrosh E.A., Kyrpides N.C., Woyke T. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578(7795):432–436. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber P.A., Quintard B., Sicks F., Dehnhard M., Greenwood A.D., Franz M. Environmental stressors may cause equine herpesvirus reactivation in captive Grévy's zebras (Equus grevyi) PeerJ. 2018;6 doi: 10.7717/peerj.5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Barbieri P., Perrone M.G., Borelli M., et al. SARS-Cov-2 RNA found on particulate matter of Bergamo in Northern Italy: First evidence. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabarova T., Salcher M.M., Porcal P., Znachor P., Nedoma J., Grossart H.-P., Seďa J., Hejzlar J., Šimek K. Recovery of freshwater microbial communities after extreme rain events is mediated by cyclic succession. Nat. Microbiol. 2021;6(4):479–488. doi: 10.1038/s41564-020-00852-1. [DOI] [PubMed] [Google Scholar]
- Shaheen M.N.F. The concept of one health applied to the problem of zoonotic diseases. Rev. Medical Virol. 2022;e2326 doi: 10.1002/rmv.2326. [DOI] [PubMed] [Google Scholar]
- Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., et al. Association Between Administration of Systemic Corticosteroids and Mortality Among Critically Ill Patients With COVID-19 A Meta-analysis. J. Am. Medical Assoc. 2020;324(13):1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.Y., Chen R.H., Jiang W., Chen X., Lin Z.F. QSAR-based investigation on antibiotics facilitating emergence and dissemination of antibiotic resistance genes: A case study of sulfonamides against mutation and conjugative transfer in Escherichia coli. Environ. Res. 2019;173:87–96. doi: 10.1016/j.envres.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Trottier J., Darques R., Mouheb N.A., Partiot E., Bakhache W., Deffieu M.S., et al. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020;10 doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai C., Li Y., Hua M., Wang J., Yang H., Zheng P., Hu B. Denitrifying Anaerobic Methane Oxidation: A Previously Overlooked Methane Sink in Intertidal Zone. Environ. Sci. Technol. 2019;53(1):203–212. doi: 10.1021/acs.est.8b05742. [DOI] [PubMed] [Google Scholar]
- Wang Z., Shin H., Jung S., Yeo D., Park H., Shin S., Seo D.J., Park K.H., Choi C. Effects of Weather and Environmental Factors on the Seasonal Prevalence of Foodborne Viruses in Irrigation Waters in Gyeonggi Province, Korea. Microorganisms. 2020;8(8):1224. doi: 10.3390/microorganisms8081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. Water, Sanitation, Hygiene, and Waste Management for COVID-19. Technical Brief, 227–406.
- Wright R.J., Langille M.G.I., Walker T.R. Food or just a free ride? A meta-analysis reveals the global diversity of the Plastisphere. ISME J. 2020;15:789–806. doi: 10.1038/s41396-020-00814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhan Water Affairs Bureau, 2020. Wuhan put into nearly 2000 tons of disinfectants to promote the strengthening of disinfection in urban drainage and sewage facilities. http://swj.wuhan.gov.cn/swyw/jcss/202004/t20200426_1126979.html.
- Wurtzer S., Waldman P., Ferrier-Rembert A., Frenois-Veyrat G., Mouchel J.M., Boni M., et al. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: Implication for wastewater-based epidemiology and risk assessment. Water Res. 2021;198 doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Huang H., Li N., Li F., Wang D., Luo Q. Occurrence and ecological risk of pharmaceuticals and personal care products (PPCPs) and pesticides in typical surface watersheds, China. Ecotoxicol. Environ. Saf. 2019;175:289–298. doi: 10.1016/j.ecoenv.2019.01.131. [DOI] [PubMed] [Google Scholar]
- Yang E.V., Webster Marketon J.I., Chen M., Lo K.W., Kim S.-J., Glaser R. Glucocorticoids activate Epstein Barr virus lytic replication through the upregulation of immediate early BZLF1 gene expression. Brain Behavior Immunity. 2010;24(7):1089–1096. doi: 10.1016/j.bbi.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.S., Rickinson A.B. EPSTEIN–BARR VIRUS: 40 YEARS ON. Nat. Rev. Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xie J. Systemic Administration of Glucocorticoids in Guizhou Province, China: A Survey Among Hospital Pharmacies. Value Health. 2020;22:S792. [Google Scholar]
- Zhang K., Liu Y., Kong H., Shang Y., Liu X. Comparison and phylogenetic analysis based on the B2L gene of orf virus from goats and sheep in China during 2009–2011. Arch. Virol. 2014;159(6):1475–1479. doi: 10.1007/s00705-013-1946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Forst C.V., Gordon A., Gussin G., Geber A.B., Fernandez P.J., Ding T., Lashua L., Wang M., Balmaseda A., Bonneau R., Zhang B., Ghedin E. Characterization of antibiotic resistance and host-microbiome interactions in the human upper respiratory tract during influenza infection. Microbiome. 2020;8(1) doi: 10.1186/s40168-020-00803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Ghosh S., Kumar M., Santiana M., Bleck C.K.E., Chaimongkol N., Altan-Bonnet N., Shuai D. Emerging Pathogenic Unit of Vesicle-Cloaked Murine Norovirus Clusters is Resistant to Environmental Stresses and UV254 Disinfection. Environ. Sci. Technol. 2021;55(9):6197–6205. doi: 10.1021/acs.est.1c01763. [DOI] [PubMed] [Google Scholar]
- Zhong R.Y., Zou H.Y., Gao J., Wang T., Bu Q.W., Wang Z.L., et al. A critical review on the distribution and ecological risk assessment of steroid hormones in the environment in China. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147452. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.