Abstract
Drug-resistant Mycobacterium tuberculosis is a major threat to public health. In clinical practice, a limited number of resistance mutations in a short sequence of the beta subunit of RNA polymerase (encoded by rpoB) have been described. Spontaneous resistance to rifampin was induced in vitro in M. tuberculosis H37Rv (ATCC 9360). Only three resistance patterns could be detected by PCR–single-strand conformation polymorphism analysis. Sequence analysis revealed that Ser531→Leu arose most frequently, followed by His526→Arg and then either His526→Tyr or His526→Asp. The relative Darwinian fitness of all but one of the mutant genotypes was less than that of the susceptible parent and, for these mutations, there was a significant correlation between fitness and clinical isolation rate (regression analysis P = 0.026). The fitness deficit in some mutants was small, suggesting that there is little likelihood of a spontaneous reversion to susceptibility.
Mycobacterium tuberculosis remains one of the most important human pathogens. The World Health Organization has estimated that as many as one in every three of the world’s population is infected with tuberculosis (TB), which causes an estimated 3 million deaths per year (22). In recent years, the incidence of TB has been rising throughout the world, as has the prevalence of resistant isolates. The World Health Organization and the International Union Against Tuberculosis and Lung Diseases have initiated directly observed short-course therapy as the preferred regimen for managing cases, but this assumes that the infecting strain of tuberculosis is susceptible to antimicrobial agents. A high rate of resistance in a community would compromise effective standardized chemotherapy (1, 17).
An isolate is defined as a multidrug-resistant M. tuberculosis (MDRTB) isolate if it is resistant to rifampin and isoniazid. Rifampin is central to the effectiveness of short-course chemotherapy because the clinical response to chemotherapy is considerably poorer in patients with initial rifampin resistance (1). MDRTB rates vary from as little as 0.7% in countries with well-managed TB control programs to 22.1% in those with poorly organized health care systems (26).
In part, the rise in tuberculosis infection is due to the human immunodeficiency virus (HIV) epidemic, which is associated with a high tuberculosis attack rate, rapid disease progression, and high mortality (19). Poor compliance among HIV patients in some countries has allowed multiple drug resistance to develop. There have been several reports of outbreaks of infection with MDRTB among hospitalized patients with HIV (5, 8). Multi-drug-resistant tuberculosis strains, such as strain W in New York, have spread widely among HIV-infected individuals but have also been isolated from immunocompetent patients (19). Thus, MDRTB strains might spread more widely in the community but, because of the slow rise of a tuberculosis epidemic curve, an MDRTB epidemic may be delayed for many years (4).
M. tuberculosis lacks plasmids, and although insertion sequences are found, there is no evidence that they are involved in the transfer of DNA among strains. The site of infection (the human macrophage) and the absence in M. tuberculosis of a free-living stage reduce the opportunity for M. tuberculosis to exchange DNA. Thus, M. tuberculosis adapts to antibiotics by spontaneous mutation.
The molecular mechanisms of resistance have been elucidated for many antituberculosis agents, including rifampin, isoniazid, pyrazinamide, streptomycin, quinolones, and ethambutol (20). Isolates have been shown to become resistant through point mutations in chromosomal genes (25), and these mutations arise spontaneously at low frequency (7, 24). In clinical practice, they are selected as a result of inadequate therapeutic regimens or patient failure to comply with adequate treatment (18). The standard three-drug regimen prevents the emergence of resistance; with the low frequency of single point mutations, it is unlikely that a population of resistant clones large enough to make second- and third-drug-resistant mutations likely will emerge (17).
Rifampin resistance arises through mutation in the β subunit of RNA polymerase, which is encoded by the rpoB gene (25). These mutations are centered on a region between codons 507 and 533 of the rpoB gene. More than 35 resistance alleles have been identified in this region (9, 20). Alleles appear in clinical isolates at markedly different frequencies. For example, His526 to Tyr and Ser531 to Leu together account for more than 64% of all reported mutations, whereas Gln509 to His accounts for 0.4%. This study was initiated to investigate the biological basis for this difference. The aim of this study was to test the hypothesis that the variation in frequency of resistant mutations is a function of their Darwinian fitness.
MATERIALS AND METHODS
Bacteria.
M. tuberculosis H37Rv ATCC 9360 (National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom) was used in these experiments. This strain was maintained by subculture on Löwenstein-Jensen media, 7H9 broth (0.1% Tween 80; Difco, Hemel Hempstead, United Kingdom), and BACTEC 12B media (Becton Dickinson, Oxford, United Kingdom).
Viable cell counts were performed by the technique of Miles and Misra (16) on rifampin-containing and rifampin-free plates. M. tuberculosis was cultured in broth culture containing Tween 80 (0.1%) to reduce clumping. The broth culture was passed 8 to 10 times through a 1-ml fine-needle insulin syringe (28-gauge; Sherwood, Crawley, United Kingdom) prior to dilution or subculture.
The dispersed broth was serially diluted 1:10 in Tween-albumin broth (0.01% Tween 80; Merck, Nuneaton, United Kingdom) and 0.2% bovine albumin (Sigma, Poole, United Kingdom) to form a dilution series from 10−1 to 10−7. The Tween-albumin broth was briefly vortexed three times. The total colony count was calculated from the drug-free plate, and the count of resistant cells was calculated from the drug-containing plate. The number of susceptible colonies was calculated by subtracting the number of resistant colonies from the total. Aliquots of these dilutions (50 μl) were inoculated onto Middlebrook 7H10 agar (Difco) containing either 5 mg of rifampin per liter or no drug.
Estimation of mutation rates.
The mutation rate was estimated by the method of Crane et al. (6). This method relies on estimation of the median mutation frequency in a series of broths.
M. tuberculosis H37Rv was cultured in eight 100-ml Middlebrook 7H10 broths for 3 to 4 weeks at 37°C. The broth culture was concentrated by centrifugation at 2,000 × g for 30 min, and the deposit volume was measured. A Miles and Misra viable-cell count was performed on 0.1 ml of the cell deposit to estimate the total number of cells (16). The remaining deposit was spread on the surface of a series of Middlebrook 7H10 agar plates containing 5 or 10 mg of rifampin per liter. The plates were sealed in polyethylene specimen bags to prevent desiccation and incubated at 37°C for 4 weeks. The total number of colonies with dispersed growth on the plates was counted, and the number of resistant cells was estimated as colonies counted/proportion of broth plated. The median number of resistant cells formed by spontaneous mutation in the eight broths (Tm) was estimated and the number of mutation events (Me) occurring in the broths was calculated with the following formula: Me = (Tm − 0.693)/(ln Tm + 0.367) (6). This was used to estimate the mutation rate (M) by using the median broth viable counts as follows: M = Me/median broth CFUs.
Selection of rifampin-resistant mutants for further study.
Samples of approximately 20 colonies were subcultured from each broth that was used to isolate rifampin-resistant mutants. The colonies were selected by drawing a line across a plate and picking all the colonies on that line. Each colony was subcultured onto Löwenstein-Jensen media, and rifampin resistance was confirmed by culture in a Middlebrook 7H9 broth containing 5 mg of rifampin per liter.
Extraction of DNA.
An aliquot of broth was centrifuged (12,000 × g for 5 min) in a 1.5-ml microcentrifuge tube, and the supernatant was discarded. The deposit was heated to 80°C for 20 min in a water bath to kill all M. tuberculosis organisms. To extract the DNA, 100 μl of chloroform was added to the deposit and vortexed for 30 s. The sample was centrifuged at 12,000 × g for 1 min, and the aqueous layer was used as the PCR sample.
PCR-SSCP.
The region of the rpoB gene identified previously by Telenti et al. (25) as a hot spot for rifampin resistance was amplified by PCR. The primers employed were 5′-AGT TCT TCG GCA CCA GC and 5′-CGC TCA CGT GAC AGA CC. The reaction mixture was 1.5 nM MgCl, 150 mM deoxynucleoside triphosphates, 5 U of Taq polymerase enzyme, and 500 μM primers for each PCR in a total volume of 90 μl. An aliquot of 10 μl of M. tuberculosis DNA was added. The optimal PCR cycling conditions were as follows: one cycle of 95°C for 1 min and 30 cycles of 94°C for 1 min, 65°C for 2 min, and 72°C for 3 min. This was followed by strand elongation for 7 min at 72°C. This produced a 120-bp amplimer that was designated PCR120.
The amplimers were analyzed by single-strand conformation polymorphism (SSCP) analysis. The PCR product (6 μl) was denatured at 95°C for 10 min with 3 μl of SSCP loading buffer and 3 μl of stop dye (Promega, Southampton, United Kingdom). Samples were quenched on ice and then loaded directly onto a 0.5% mutation detection enhancement acrylamide analogue gel (Flowgen, Lichfield, United Kingdom). The gel was run for 6 h at 6 W with 0.6× Tris-borate-EDTA (Sigma) at room temperature. DNA bands were visualized by the silver-staining technique, following the manufacturer’s instructions (Promega).
Fitness assay.
Rifampin-resistant mutants with each of the SSCP analysis patterns were selected for fitness assays from separate broth experiments. This was done to reduce the possibility that mutants had a more recent common ancestor than the rifampin-susceptible parent strain.
A 2- to 3-week 4-ml Middlebrook 7H9 broth culture of both the rifampin-resistant and -susceptible H37Rv isolates was used. The Miles and Misra plate technique was used to estimate the viable-cell count (16). A 10-fold (susceptible) or 100-fold (resistant) dilution was prepared; from each, a 50-μl sample was inoculated into 4 ml of fresh Middlebrook 7H9 broth to create a mixed culture of rifampin-susceptible and -resistant cells.
After 2 to 3 weeks of incubation at 37°C, a count of viable cells was performed by the Miles and Misra technique (16) on media containing either 5 mg of rifampin per liter or no drug. The number of colonies growing on drug-free Middlebrook 7H10 agar was used to estimate the total number of cells. The number of rifampin-resistant cells was estimated from the colony count on rifampin-containing media. The number of rifampin-susceptible cells could then be calculated. The number of generations (G) of the mutant that occurred in the mixed broth was calculated with the following formula: G = (log B − log A)/log 2, where A is the number of CFU per milliliter at time 0 and B is the number of CFU per milliliter at the end of the culture period. The relative fitness of each strain was calculated from the ratio of the number of generations of rifampin-resistant to the rifampin-susceptible strains.
PCR for sequence analysis.
A larger PCR amplimer was prepared for direct sequence analysis, and this amplimer was designated PCR411. The previously published primers (25) 5′-TAC GGT CGG CGA GCT CC-3′ and 5′-TAC GGC GTT TCG ATG AAC C-3′ were used; the optimized reaction mixture was 50 mM KCl buffer, 1.5 mM MgCl, 150 mM deoxynucleoside triphosphates, and 5 U of Taq. The cycling conditions described above were also used in this reaction. These primers amplified a 411-bp fragment.
The PCR product was purified with the Wizard PCR cleanup kit, following the manufacturer’s instructions (Promega). Sequencing was performed commercially (Cambridge Bioscience, Cambridge, United Kingdom).
Statistical methods.
Differences in the number of mutant colonies derived and differences between generations formed by resistant mutant and susceptible parents were compared with the paired Student’s t test. Differences in mutation rates were compared with the Mann-Whitney U test. Statistical tests were calculated with Excel, version 4.0. Regression analysis was performed with Unistat, version 1.13.
RESULTS
Results were available from seven of eight broths that had been subcultured on both 5 and 10 mg of rifampin per liter. The mean numbers of colonies detected per milliliter of cell deposit were 209 and 214, respectively. This difference was not significant (P = 0.57) with the matched-pair t test.
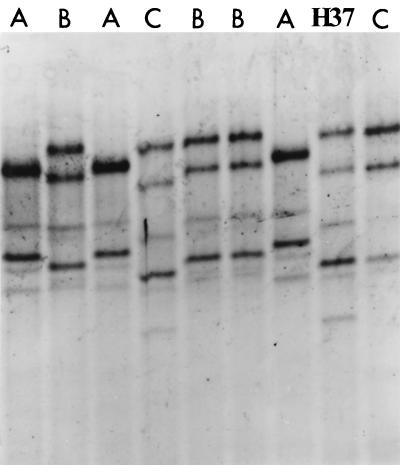
A sample of 20 colonies from each broth experiment was selected, and DNA was extracted and amplified by PCR120. Amplified DNA was examined by SSCP analysis. Only three SSCP patterns could be identified from all of the colonies studied (n = 156). The SSCP patterns are illustrated in Fig. 1, and the frequency of each pattern is shown in Table 1.
FIG. 1.
Mutation detection enhancement gel electrophoresis demonstrating representative resistance genotype SSCP patterns A, B, and C and the sensitive genotype pattern for H37Rv.
TABLE 1.
Relative fitness of induced H37Rv rifampin-resistant mutant alleles
| SSCP pattern | No. of replicates | Mean no. of generations of H37Rv
|
Location of mutation | Relative fitness | |
|---|---|---|---|---|---|
| Sensitive | Resistant | ||||
| A1 | 4 | 8.65 | 6.95 | His526→Tyr | 0.80 |
| A2 | 4 | 5.90 | 2.80 | His526→Tyr | 0.78 |
| A3 | 3 | 6.73 | 4.62 | His526→Asp | 0.42 |
| B1 | 5 | 7.88 | 8.30 | Ser531→Leu | 1.05 |
| B2 | 3 | 7.73 | 7.43 | Ser531→Leu | 0.93 |
| B3 | 3 | 6.83 | 6.29 | Ser531→Leu | 0.89 |
| B4 | 3 | 9.60 | 4.83 | Ser531→Leu | 0.50 |
| C1 | 2 | 5.68 | 10.04 | His526→Arg | 0.56 |
| C2 | 5 | 6.40 | 1.35 | His526→Arg | 0.21 |
The median mutant frequency was used to estimate the mutation rate for each mutant type. The total mutation rate for all rifampin-resistant mutations was 6 × 10−10. For individual SSCP patterns, the mutation rates were 2 × 10−10, 4 × 10−10, and 2 × 10−10 mutations per cell per generation for SSCP patterns A, B, and C, respectively. No significant difference in mutation rate between the three mutant types was found (P > 0.5) (Mann-Whitney U test).
A selection of mutants with each SSCP pattern was sequenced. Examples of each mutant type were taken from separate experiments to ensure that the mutation had arisen independently in each isolate. The locations of mutations in rpoB are given in Table 1. The SSCP pattern B mutants had a Ser531-to-Leu (TCG→TTG) mutation; pattern C mutants had a His526-to-Arg (CAC→CGC) mutation. All three SSCP pattern A mutations were located at codon 526, but two had a His526-to-Tyr (CAC→TAC) mutation and one had a His526-to-Asp (CAC→GAC) mutation.
The results of the fitness experiment are found in Table 1. Mutants A and C do not form as many generations as do their susceptible parents (P = <0.02; Student’s paired t test). There was no significant difference between mutant type B and its susceptible parent (P = 0.09).
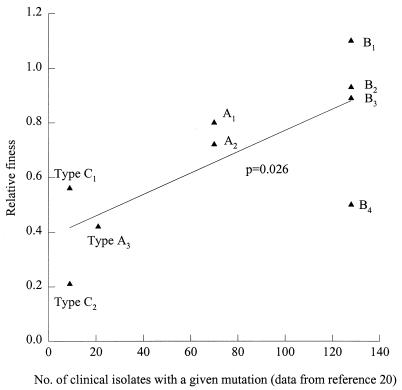
Using data on the clinical frequency of resistance mutation presented in Musser’s review (20), it was possible to correlate the frequency of clinical isolation and the in vitro fitness of different mutations as defined in these experiments (regression analysis P = 0.026).
DISCUSSION
Drug-resistant M. tuberculosis arises in clinical practice when therapy is inadequate. This may arise because of an inadequate prescription or because the patient fails to adhere fully to an appropriate regimen. The molecular basis of resistance is now more fully understood (20). Telenti et al. described an association between mutation in the rpoB gene and a resistance phenotype (25). This mirrors the situation in Escherichia coli, where mutations in rpoB were first reported to be associated with rifampin resistance (10). These mutations result in alterations in termination efficiency (11).
Complex models describing the population dynamics of the evolution of drug resistance have been published (15, 18). Lipsitch and Levin (15) have published a model for the evolution of drug resistance which suggests that drug resistance will emerge only if there is a protected compartment or the host is noncompliant. This model makes assumptions about the relative fitness of resistant mutants, and this study provides data which may be applied to it.
Our study produced only a limited number of resistance mutation types with SSCP analysis. Sequencing a proportion of these mutants confirmed the limited range of mutations expressed in this experimental environment. SSCP patterns B and C each represent a single sequence change (in B, TCG→TTG; in C, CAC→CGC). However, SSCP pattern A was characterized by two point mutations at the same codon. All three of the SSCP patterns occurred with a similar frequency. This data suggests that these mutations are equally likely to arise and that the prevalence of each mutant type depends on its ability to survive, i.e., its fitness.
It is often assumed that drug-resistant strains pay a physiological price for the acquisition of resistance. E. coli strains have a lower growth rate after the acquisition of plasmid-mediated resistance (12, 13). E. coli has also been shown to have a lower chain elongation rate after mutation of the rsl gene to a streptomycin-resistant genotype. This has been calculated to be a fitness cost of up to 18% per generation (23). It is possible that resistance through the acquisition of a plasmid may have different consequences for fitness than do chromosomal point mutations.
Antimicrobial-resistant Salmonella typhimurium organisms were shown to be less virulent in a mouse model than their susceptible parent (3). Isoniazid-resistant strains of M. tuberculosis have been shown to have reduced virulence in guinea pigs (2). More recently, a panel of strains resistant to one or more antimycobacterial drugs have been tested in a mouse model of virulence (21). A range of virulence was demonstrated, with some strains growing more poorly than, some as well as, and some faster than the control strain. As the genetic backgrounds of the organisms in all of these studies are different and uncontrolled, no clear conclusions can be drawn. Moreover, there is no data from previous studies to relate in vitro fitness to virulence.
Our study directly compares strains with identical genetic backgrounds, differing phenotypically with respect to rifampin susceptibility and genotypically with respect to rpoB. The mean fitness of most of the resistant strains tested was lower than that of the susceptible parent, suggesting that this mutation results in a physiological cost. For mutation B, the fitness deficit is smallest (mean relative fitness = 0.85) but includes one mutant with a relative fitness exceeding that of the wild type (relative fitness = 1.05). In intermittent chemotherapy, there are periods when bacteria are exposed to subinhibitory concentrations of antibiotic, and the organisms may regrow (18). During this period, the growth of susceptible organisms would be slowed, but resistant mutants may grow normally. The results of a study in Madras, India (27), suggest that cycles of killing followed by regrowth are likely to occur if once-weekly therapy is used, and this leads rapidly to the emergence of resistance. Strains with the greatest deficit in fitness are least likely to emerge in these circumstances. The mutation Ser531→Leu is the one most frequently seen in clinical practice (26). Mutants with the genotypes that we isolated in vitro make up 74% of the clinically isolated resistant mutants (25). This relationship between fitness as demonstrated in our in vitro model and frequency of isolation in clinical practice is illustrated in Fig. 2. A correlation comparing rates at which mutants are isolated clinically and the fitness of individual mutants is positive (P = 0.026) (Fig. 2). This indicates that mutants isolated more frequently in clinical practice have a higher mean relative fitness on initial isolation.
FIG. 2.
Relationship between relative fitness and the clinical isolation rate for mutants A, B, and C.
Each of the rpoB mutations is associated with different fitness results, although there is some overlap. The more extreme variants, B4 and C2, may represent piggyback mutants: an rpoB mutation that selects for survival may occur at the same time as another mutation that reduces fitness significantly. Experiments in other models suggest that such fitness deficits can be reduced by compensating mutations arising at other sites after further growth (14, 23). The fitness experiments reported here were performed on newly isolated organisms that had no opportunity for compensatory adaptation. Long-term adaptation experiments are now under way. If compensatory mutations were found among rifampin-resistant M. tuberculosis organisms, it would suggest that there was little likelihood of a spontaneous reversion to susceptibility. It is essential, therefore, that every effort be made to ensure that organisms do not become resistant. In the absence of spontaneous reversion, strains with a modest or no fitness deficit would become fixed in the environment, resulting in an epidemic of resistant M. tuberculosis strains. It may be that this process has already begun but is not yet apparent, due to the propensity of the species to cause latent infection and the slow rise in the tuberculosis epidemic curve (4).
Multiple-drug-resistant tuberculosis is a major threat to human health throughout the world. Our data provides an explanation for the pattern of resistance mutations found in clinical practice. They also challenge the assumption that good prescription practices alone will solve the resistance problem. Many of the resistant mutants that were isolated had only a modest decrease in fitness and were likely to survive well in the environment, making spontaneous revertants unlikely. It is possible that, given the long duration of tuberculosis epidemics, the number of resistant mutant strains in human populations will increase.
ACKNOWLEDGMENTS
This work was supported by the Special Trustees of the Royal Free Hospital, London, grant held by S.H.G.
We gratefully acknowledge the secretarial assistance of Therese Donnelly.
REFERENCES
- 1.Aber V R, Nunn A J. Chimiotherapie de courte durée de la tuberculose. Facteurs de rechute dans la chimiotherapie de courte durée. Bull Int Union Tuberc. 1978;53:276–280. [PubMed] [Google Scholar]
- 2.Barnett M, Bushby S R M, Mitchison D A. Tubercle bacilli resistant to isoniazid: virulence and response to treatment with isoniazid in guinea pigs. Br Med J. 1954;1:128–130. doi: 10.1136/bmj.1.4854.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkman J, Hughes D, Andersson D I. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blower S M, McLean A R, Porco T C, Small P M, Hopewell P C, Sanchez M A, Moss A R. The intrinsic transmission dynamics of tuberculosis epidemics. Nat Med. 1995;1:815–821. doi: 10.1038/nm0895-815. [DOI] [PubMed] [Google Scholar]
- 5.Breathnach A S, de Ruiter A, Holdsworth G M, Bateman N T, O’Sullivan D G, Rees P J, Snashall D, Milburn H J, Peters B S, Watson J, Drobniewski F A, French G L. An outbreak of multi-drug-resistant tuberculosis in a London teaching hospital. J Hosp Infect. 1998;39:111–117. doi: 10.1016/s0195-6701(98)90324-3. [DOI] [PubMed] [Google Scholar]
- 6.Crane G J, Thomas S M, Jones M E. A modified Luria-Delbrück fluctuation assay for estimating and comparing mutation rates. Mutat Res. 1996;354:171–182. doi: 10.1016/0027-5107(96)00009-7. [DOI] [PubMed] [Google Scholar]
- 7.David H L. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl Microbiol. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edlin B R, Tokars J I, Grieco M H, Crawford J T, Williams J, Sordillo E M, Ong K R, Kilburn J O, Dooley S W, Castro K G. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. New Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 9.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridisation pattern recognition analysis of generic mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 10.Jin D J, Gross C A. Mapping and sequence of mutations in Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 11.Jin D J, Walter W A, Gross C A. Characterisation of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988;202:245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- 12.Kyslik P, Dobisova M, Maresova H, Sobotkova L. Plasmid burden in chemostat culture of Escherichia coli—its effect on the selection for overproducers of host enzymes. Biotechnol Bioeng. 1993;41:325–329. doi: 10.1002/bit.260410306. [DOI] [PubMed] [Google Scholar]
- 13.Lee S W, Edlin G. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene. 1985;39:173–180. doi: 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- 14.Lenski R E, Simpson S C, Nguyen T T. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsitch M, Levin B R. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Tuber Lung Dis. 1998;2:187–199. [PubMed] [Google Scholar]
- 16.Miles A A, Misra S S. The estimation of the bacteriocidal power of blood. J Hyg Camb. 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchison D A. Basic mechanisms of chemotherapy. Chest. 1979;76:771–781. doi: 10.1378/chest.76.6_supplement.771. [DOI] [PubMed] [Google Scholar]
- 18.Mitchison D A. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int J Tuberc Lung Dis. 1998;2:10–15. [PubMed] [Google Scholar]
- 19.Moss A R, Alland D, Telzak E, Hewlett D, Jr, Sharp V, Chiliade P, LaBombardi V, Kabus D, Hanna B, Palumbo L, Brudney K, Weltman A, Stoeckle K, Chirgwin K, Simberkoff M, Moghazeh S, Eisner W, Lutfey M, Kreiswirth B. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuberc Lung Dis. 1997;1:115–121. [PubMed] [Google Scholar]
- 20.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordway D J, Sonnenberg M G, Donahue S A, Belisle J T, Orme I M. Drug-resistant strains of Mycobacterium tuberculosis exhibit a range of virulence for mice. Infect Immun. 1995;63:741–743. doi: 10.1128/iai.63.2.741-743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raviglione M C, Snider D E, Jr, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 23.Schrag S J, Perrot V, Levin B R. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc R Soc Lond B. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimao T. Drug resistance in tuberculosis control. Tubercle. 1987;68(Suppl. 2):5–18. doi: 10.1016/0041-3879(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 25.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 26.The WHO/IUATLD Global Project on Anti-tuberculosis Drug Resistance Surveillance. Anti-tuberculosis drug resistance in the world. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 27.World Health Organization. A controlled comparison of a twice-weekly and three once-weekly regimens in the initial treatment of pulmonary tuberculosis. Bull W H O. 1970;43:143–206. [PMC free article] [PubMed] [Google Scholar]