Abstract
Steroid-induced osteonecrosis of the femoral head (ONFH) is a common and devastating bone disorder, which often results in progressive collapse of the femoral head and subsequent osteoarthritis. The proliferation ability and osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) play critical roles in maintaining the structural and functional integrity of the femoral head to prevent ONFH. Until now, little has been known about the underlying mechanism of BMSCs differentiation disorder during ONFH progression. Circular RNAs (circRNAs) are considered to be vital non-coding RNAs functionally involved in various human diseases. However, whether and how circRNA regulates the proliferation and osteogenic differentiation of BMSCs in ONFH remain unclear. In this study, we analyzed the circRNA expression profile of five samples of BMSCs in ONFH and five samples of control by using circRNA microarray assays. We identified 182 differentially expressed circRNAs, among which 108 circRNAs were upregulated. We further investigated the effects of a significantly upregulated circRNA, circHGF, on the proliferation and osteogenic differentiation of BMSCs in vitro. Results showed that circHGF suppressed the proliferation and osteogenic differentiation of BMSCs in ONFH by targeting miR-25-3p/SMAD7 axis. Our findings provided a potential diagnostic and therapeutic strategy for ONFH.
Keywords: MT: Non-coding RNAs, bone marrow mesenchymal stem cells, osteogenic differentiation, osteonecrosis of the femoral head, circular RNA, SMAD7
Graphical abstract
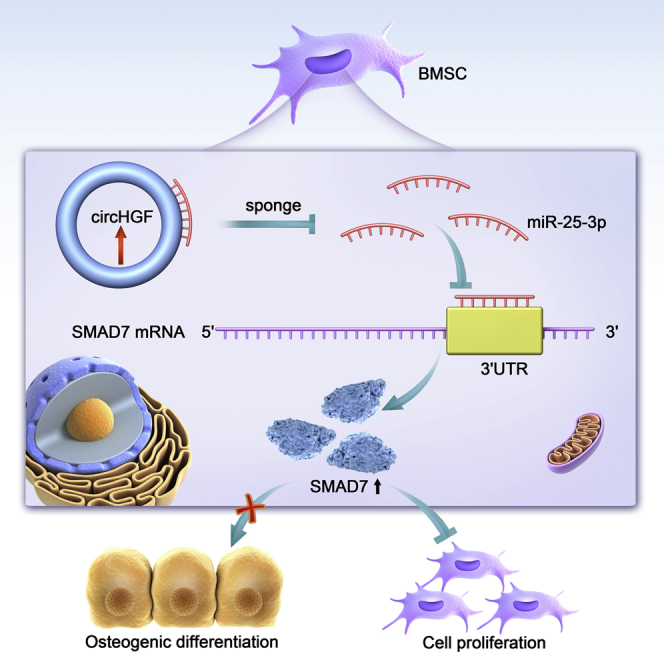
Steroid-induced osteonecrosis of the femoral head (ONFH) is a common devastating bone disease. CircRNAs are a novel class of non-coding RNAs functionally involved in various human diseases. This study reveals the key roles of circRNAs in ONFH and provides a potential diagnostic and therapeutic strategy for ONFH.
Introduction
Osteonecrosis of the femoral head (ONFH) is a common and devastating bone disorder, which usually results in femoral head collapse and subsequent damage to the hip joint.1 Generally, ONFH can be divided into traumatic ONFH and non-traumatic ONFH. There are numerous risk factors related to non-traumatic ONFH, including corticosteroid therapy, alcohol abuse, infection, hyperbaric events, etc. Corticosteroid drugs are widely used in the treatment of autoimmune and rheumatic as well as many other kinds of human diseases, and steroid-induced ONFH accounts for the majority of clinical non-traumatic ONFH.2 However, the underlying mechanism of steroid-induced ONFH remains not entirely clear so far.
Bone marrow mesenchymal stem cells (BMSCs) are a kind of adult and fibroblast-like multi-potent cells, which have strong potential in multi-lineage differentiation and self-renewal capacity.3 They can be induced to differentiate into chondrocytes, adipocytes, or osteoblasts when placed in different conditions in vitro. BMSCs differentiation regulation have important effects on many pathological processes of human diseases. Accumulating evidence has reported that imbalance between adipogenic and osteogenic differentiation of BMSCs, including the weakened osteogenic differentiation and enhanced adipogenic differentiation, contributed significantly to ONFH pathogenesis.4 Especially, it has been reported that the deficiency in osteogenic differentiation of BMSCs would result in ONFH.5 BMSCs are widely used in regenerative medicine, for the reason that they are easily isolated and cultured with weak immunogenicity.6 BMSCs transplantation has been demonstrated as an ideal candidate for early ONFH treatment, and BMSCs survival in bone necrotic areas is critical during the treating process.7 The mesenchymal stem cells differentiate into osteoblasts to mediate bone formation mainly involve bone morphogenetic protein (BMP)/drosophila mothers against decapentaplegic (SMAD) and Wnt signaling pathways.8 SMAD7 is an essential negative regulator in SMAD family and can antagonize the transforming growth factor β (TGF-β)/BMP signaling pathway through various molecular mechanisms.9,10 SMAD7 has been reported to play crucial roles in the multi-differentiation process of mesenchymal stem cells, including the osteogenic differentiation of BMSCs during ONFH development.11,12
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and circular RNAs (circRNAs) as well as long ncRNAs (lncRNAs), have been reported to be functionally involved in various biological processes, such as cell proliferation, metabolism, stem cell differentiation, etc.13, 14, 15, 16 Accumulating studies have indicated that ncRNAs play crucial roles in bone development and regeneration.14,15,17 MiRNAs are a class of short, single-stranded, and highly conserved endogenous ncRNAs and they mainly negatively regulate gene expression post-transcriptionally through interacting with its 3′-untranslated region (3′-UTR).18 It has been demonstrated that miRNAs participated functionally in the initiation and progression of ONFH.15 For instance, a previous study suggested that a dysregulated miRNA, termed miR-15b, decreased the mRNA and protein expression levels of SMAD7 in BMSCs, and over-expressing miR-15b could obviously relieve ONFH by targeting SMAD7 and inhibiting osteogenic differentiation of BMSCs.12 CircRNAs are a newly identified class of endogenous ncRNAs, which can mediate biological functions by regulating their target miRNAs.19 They can affect miRNAs and corresponding downstream gene expression through the competing endogenous RNA (ceRNA) mechanism, in which circRNAs contain miRNA response elements (MREs) and act as miRNA sponges.19 CircRNAs have been reported to play crucial regulating roles in the pathogenesis of various orthopedic conditions by functioning as miRNA sponges, including osteoporosis, osteoarthritis, intervertebral disc degeneration, etc.20, 21, 22 To the best of our knowledge, however, the biological roles and therapeutic potential of circRNAs in ONFH remain obscure.
In this study, we aimed to detect the circRNAs expression profile in BMSCs of ONFH by using circRNA microarray assays. We also performed pathway enrichment and molecular function prediction for the differentially expressed circRNAs by applying bioinformatics analysis. We further investigated the effects of a most significantly upregulated circRNA, circHGF, on the proliferation and osteogenic differentiation of BMSCs. We unveiled that circHGF suppressed the proliferation and osteogenic differentiation of BMSCs in ONFH by targeting the miR-25-3p/SMAD7 axis. Our research might provide a novel diagnostic and therapeutic strategy for ONFH.
Results
Identification of differentially expressed circRNAs in ONFH-BMSCs
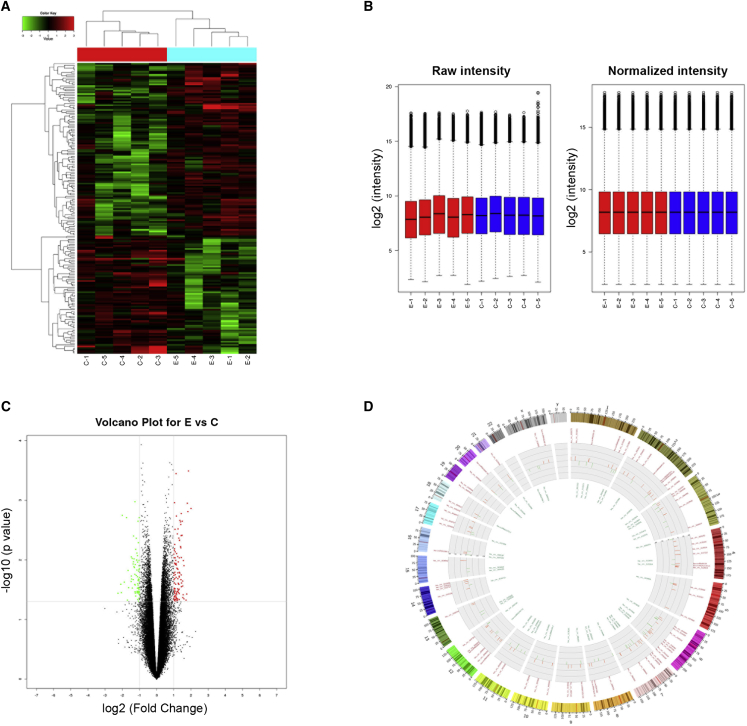
In order to investigate the potential involvement of circRNAs in the cell proliferation and osteogenic differentiation of BMSCs in ONFH, we first screened out the differentially expressed circRNAs in ONFH-BMSCs. Total RNA was extracted from each of the five ONFH-BMSCs samples versus five control BMSCs samples to perform circRNA microarray analysis. Results of the microarray assay demonstrated there were 182 differentially expressed circRNAs between the two BMSCs groups, among which 108 circRNAs were upregulated while 74 circRNAs were downregulated in the ONFH-BMSCs group compared with the control group. As shown in the hierarchical cluster heat map, the expression pattern of circRNAs between the two groups was distinguishable and a majority of them were upregulated in ONFH-BMSCs (Figure 1A). The box diagram suggested that the expression signal intensity after normalization in the two groups is relatively consistent (Figure 1B). The fold change and p value for each circRNA expression were shown on the volcano plots (Figure 1C). The distribution of the circRNAs on the human chromosomes was shown in the Circos plot, and we found the differentially expressed circRNAs were derived from all chromosomes (Figure 1D).
Figure 1.
Differentially expression profile of circRNAs in ONFH-BMSCs
(A) Hierarchical cluster heat map for the expression pattern of circRNAs between the groups from five experimental ONFH-BMSCs samples versus five control BMSCs samples. The green rows indicated downregulated circRNAs, and the red rows indicated upregulated circRNAs. Columns represented cells samples. Experimental group samples: E1, E2, E3, E4, and E5, and control group samples: C1, C2, C3, C4, and C5. (B) The box diagram suggested the consistency of fluorescent signals as well as the signals difference between samples from the two groups. (C) The volcano plot for the differentially expressed pattern of circRNAs between the two groups. Vertical lines represented 2.0 fold changes (log2 scaled); horizontal line suggested a p 0.05 determined by t test. (D) Circos plot for the differentially expressed circRNAs, indicating the gene distribution on the human chromosomes. The circRNAs in green indicated an expressed pattern of downregulation, and circRNAs in red indicated an upregulation pattern.
Function analysis for the genes of differentially expressed circRNAs
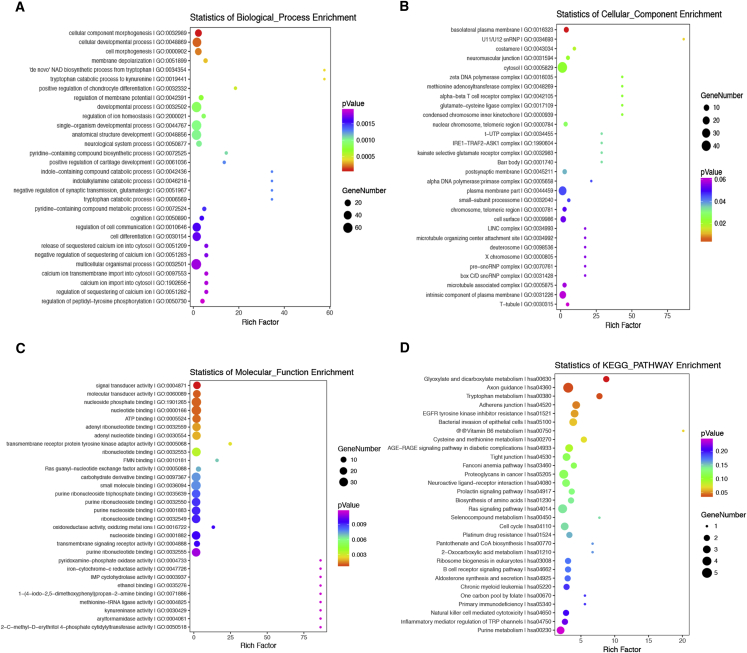
To predict to the potential functions of these differentially expressed circRNAs, we performed GO (gene ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis for the circRNAs host genes. The GO enrichment analysis consisted of three parts, namely biological process (BP), cellular component (CC), and molecular function (MF) (Figures 2A–2C). As shown in Figure 2A, the most significantly and meaningful enriched GO terms were cellular developmental process, CC morphogenesis, and cell differentiation regarding BP. As for the CC part of GO analysis, the differentially expressed circRNA genes were enriched in cell surface, cytosol, plasma membrane part, and so on (Figure 2B). As for the MF, the differentially expressed circRNA genes were enriched in signal transducer activity, molecular transducer activity, nucleotide binding, etc. (Figure 2C). As shown in Figure 2D, the most significantly and meaningful enriched KEGG pathways to which the differentially expressed circRNAs were related were to cell cycle, Ras signaling pathway, and so on.
Figure 2.
Function analysis for the differentially expressed circRNAs
(A–C) The top 30 predicted functions of the differentially expressed circRNAs host genes were obtained by gene ontology (GO) analysis, which were categorized based on biological process (BP), cellular component (CC), and molecular function (MF). (D) The top 30 enrichment pathways of the differentially expressed circRNAs host genes were obtained by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
CircHGF was identified as an ONFH related circular RNA
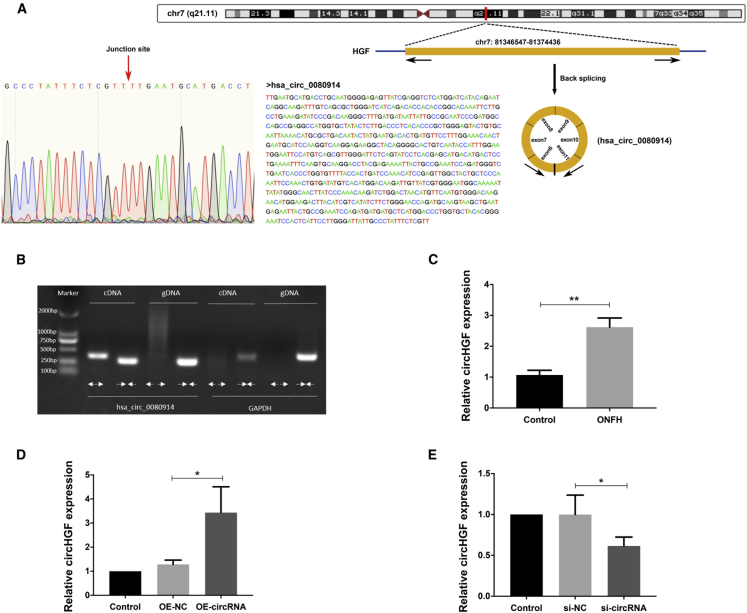
In the circRNAs microarray analysis, a majority of the differentially expressed circRNAs were upregulated. We further investigated a most significantly upregulated circRNA hsa_circ_0080914 among the top 20 upregulated circRNAs, considering its potential function of rich miRNA binding sites. As annotated in the circBase database using human reference genome GRCh37/hg19, hsa_circ_0080914 was generated by the exons 6 to 10 of HGF (hepatocyte growth factor) gene, which was located on chr7: 81346547-81374436 (Figure 3A). We termed this circRNA, circHGF, and the spliced length of circHGF was 780 nucleotides. The PCR assay with agarose gel electrophoresis analysis showed that the divergent primers amplified the cirRNA circHGF in cDNA but not in genomic DNA (gDNA), and convergent primers amplified both circHGF and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Figure 3B). As revealed in Figure 3A (left), Sanger sequencing was conducted to verify the predicted back-splicing junction in the amplified products by divergent primers, with a red arrow indicating the junction site. These findings demonstrated that circHGF had a circular structure. The upregulated expression pattern of hsa_circ_0080914 in ONFH-BMSCs was validated by RT-qPCR analysis (Figure 3C). Next, we transfected the BMSCs with the circHGF over-expression (OE) vector or with small interfering RNA (siRNA) targeting the junction site of circHGF to regulate its expression. Results validated that the BMSCs transfected with circHGF OE vector had a significantly upregulated circHGF expression level compared with the negative control (NC) (Figure 3D), while the circHGF siRNA transfection group had a significantly downregulated circHGF expression level (Figure 3E).
Figure 3.
CircHGF was characterized as an ONFH-related circular RNA
(A) Schematic of the genomic loci of HGF gene as well as circHGF. The genomic coordinates were reported using the the human genome assembly GRCh37 (hg19). The sequence of circHGF was shown and the expression of circHGF was validated by Sanger sequencing, with the red arrow indicating the junction site of this circRNA. (B) The products amplified by divergent or convergent primers were vilidated using agarose gel electrophoresis. Divergent primers detected circular circHGF in cDNA rather than in genomic DNA (gDNA). Convergent primers detected both the circular circHGF and negative control GAPDH. (C) The relative expression levels of circHGF in ONFH-BMSCs and control BMSCs were assessed by RT-qPCR analysis. (D) The BMSCs were treated with circHGF over-expression vector (OE-circRNA) or its control (OE-NC), and the relative expression levels of circHGF were examined by RT-qPCR analysis. (E) The BMSCs were treated with circHGF siRNA (si-circRNA) or its control (si-NC), and the relative expression levels of circHGF were detected by RT-qPCR analysis. Data were shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, n = 3. Data between two groups were analyzed by Student's t test. Data among multiple groups were analyzed by one-way ANOVA test.
CircHGF suppressed the proliferation and osteogenic differentiation of BMSCs in ONFH
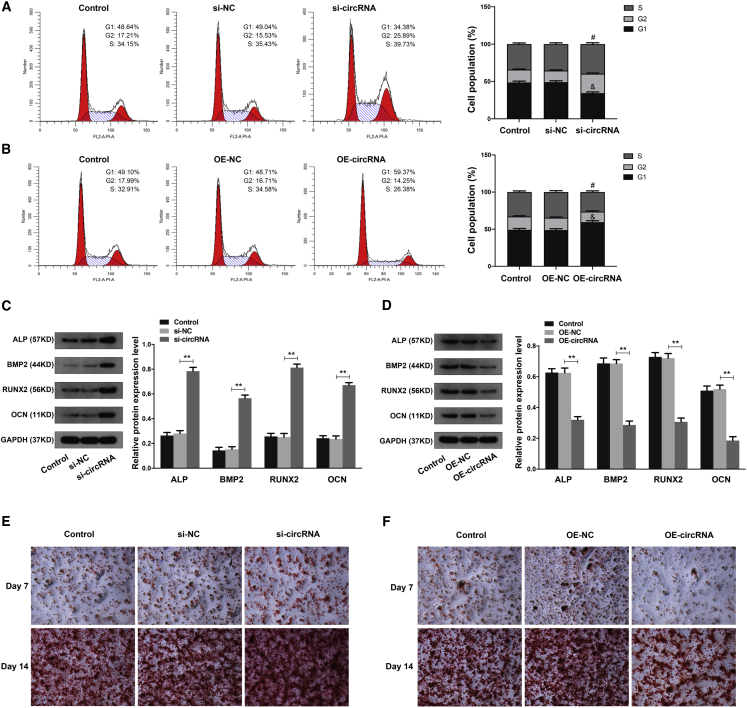
Next, we investigated the effects of circHGF on the proliferation and osteogenic differentiation of BMSCs. The cell-cycle analysis demonstrated that circHGF downregulation obviously increased the percentage of S phase cells and decreased the percentage of G1 phase cells (Figure 4A), while circHGF OE decreased the percentage of S phase cells and increased the percentage of G1 phase cells (Figure 4B). The above results indicated that circHGF could suppressed the proliferation ability of BMSCs in ONFH. By using western blot analysis, we found that circHGF downregulation significantly enhanced the expression of osteogenic marker genes, including ALP, BMP2, RUNX2, and OCN in BMSCs, after osteogenesis induction (Figure 4C). However, circHGF OE could obviously inhibit the expression levels of these osteogenic marker genes (Figure 4D). Meanwhile, by using alizarin red staining at the 7th and 14th day after circHGF siRNA transfection, we found that the osteogenic differentiation of BMSCs was markedly increased compared with the control group (Figure 4E), while the results of alizarin red staining at day 7 and 14 after OE-circHGF transfection demonstrated that the osteogenic differentiation of BMSCs was markedly attenuated (Figure 4F).
Figure 4.
CircHGF inhibited the proliferation and osteogenic differentiation of BMSCs in ONFH
(A) The BMSCs were treated with si-circRNA or its control si-NC, and the cell proliferation ability was revealed by the cell-cycle analysis, with the cell population of G1, G2, and S cell-cycle phase indicated. #p < 0.05 versus si-NC group for comparing cell percentage in S phase, &p < 0.05 versus si-NC group for comparing cell percentage in G1 phase, n = 3. (B) The BMSCs were treated with circHGF over-expression vector OE-circRNA or its control OE-NC, and the cell proliferation ability was revealed by the cell-cycle analysis. #p < 0.05 versus OE-NC group for comparing cell percentage in S phase, &p < 0.05 versus OE-NC group for comparing cell percentage in G1 phase, n = 3. (C and D) The protein expression levels of osteogenic marker genes in BMSCs were detected by western blot analysis. GAPDH served as an internal control correspondingly. The quantitative analysis showed the protein expression levels of ALP, BMP2, RUNX2, and OCN were all increased in the si-circRNA group compared with the si-NC group and were decreased in the OE-circRNA group compared with the OE-NC group. (E) The osteogenic differentiation ability of BMSCs at the 7th and 14th day after si-circRNA or control treatment was determined by alizarin red staining. (F) The osteogenic differentiation ability of BMSCs at the 7th and 14th day after OE-circRNA or OE-NC treatment was evaluated by alizarin red staining. Data were shown as mean ± SD. ∗∗p < 0.01, n = 3. Data among multiple groups were analyzed by one-way ANOVA test.
MiR-25-3p facilitated the proliferation and osteogenic differentiation ability of BMSCs
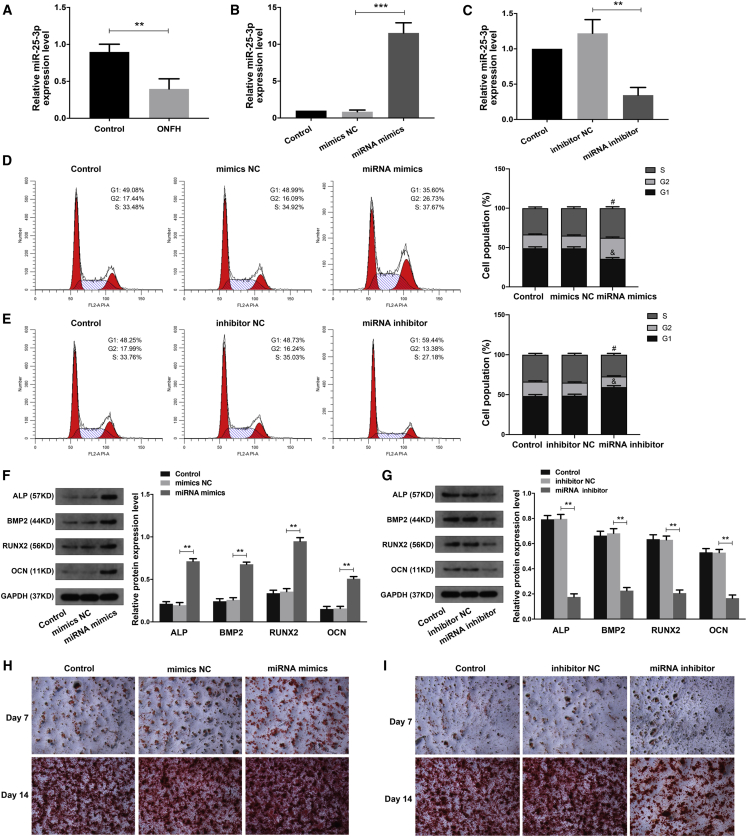
Considering miR-25-3p was predicted as a critical targeting miRNA for circHGF using the starBase online tool (http://starbase.sysu.edu.cn), we then explored the expression pattern and the function of miR-25-3p in BMSCs. First, the RT-qPCR analysis indicated that the expression of miR-25-3p in ONFH-BMSCs was significantly downregulated compared with the control group (Figure 5A). Next, we transfected BMSCs with miRNA mimics to over-express miR-25-3p or with miRNA inhibitor to knock down miR-25-3p. The RT-qPCR analysis validated that the miR-25-3p expression level was obviously increased after miRNA mimics treatment (Figure 5B) and decreased after miRNA inhibitor treatment (Figure 5C). Then, by using the cell-cycle analysis, we found that miR-25-3p mimics significantly increased the percentage of S phase cells and decreased the percentage of G1 phase cells (Figure 5D). In contrast, miR-25-3p inhibitor treatment decreased the percentage of S phase cells and increased the percentage of G1 phase cells (Figure 5E). These results suggested that miR-25-3p could facilitate the proliferation ability of BMSCs in ONFH. The western blot analysis showed that miR-25-3p mimics obviously increased the protein expression levels of osteogenic marker genes (ALP, BMP2, RUNX2, and OCN) in BMSCs after osteogenesis induction (Figure 5F). Conversely, the miR-25-3p inhibitor exerted the opposite effects on BMSCs (Figure 5G). Meanwhile, using alizarin red staining at day 7 and 14, we found that the osteogenic differentiation ability of BMSCs was markedly increased in the miR-25-3p mimics group (Figure 5H) and was evidently decreased in the miR-25-3p inhibitor group (Figure 5I).
Figure 5.
MiR-25-3p enhanced the proliferation and osteogenic differentiation ability of BMSCs
(A) The relative expression levels of miR-25-3p in ONFH-BMSCs and control BMSCs were examined by RT-qPCR analysis. (B) The BMSCs were transfected with miR-25-3p mimics or its control mimics NC, and the relative expression levels of miR-25-3p were determined by RT-qPCR analysis. (C) The BMSCs were treated with miR-25-3p inhibitor or its control inhibitor NC, and the relative expression levels of miR-25-3p were determined by RT-qPCR analysis. (D and E) The cell proliferation ability of BMSCs with corresponding treatment was revealed by the cell-cycle analysis, with the cell population of G1, G2, and S cell-cycle phase indicated. #p < 0.05 versus the corresponding NC group for comparing cell percentage in S phase; &p < 0.05 versus the corresponding NC group for comparing cell percentage in G1 phase; n = 3. (F and G) The protein expression levels of osteogenic marker genes in BMSCs of different groups were detected by western blot analysis. The quantitative analysis showed the protein expression levels of ALP, BMP2, RUNX2, and OCN were increased in the miRNA mimics group compared with the mimics NC group and were decreased in the miRNA inhibitor group compared with the inhibitor NC group. (H and I) The osteogenic differentiation ability of BMSCs at the 7th and 14th day after corresponding treatment was evaluated by alizarin red staining. Data were shown as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001, n = 3. Data between two groups were analyzed by Student's t test. Data among multiple groups were analyzed by one-way ANOVA test.
MiR-25-3p enhanced the proliferation and osteogenic differentiation of BMSCs by targeting SMAD7
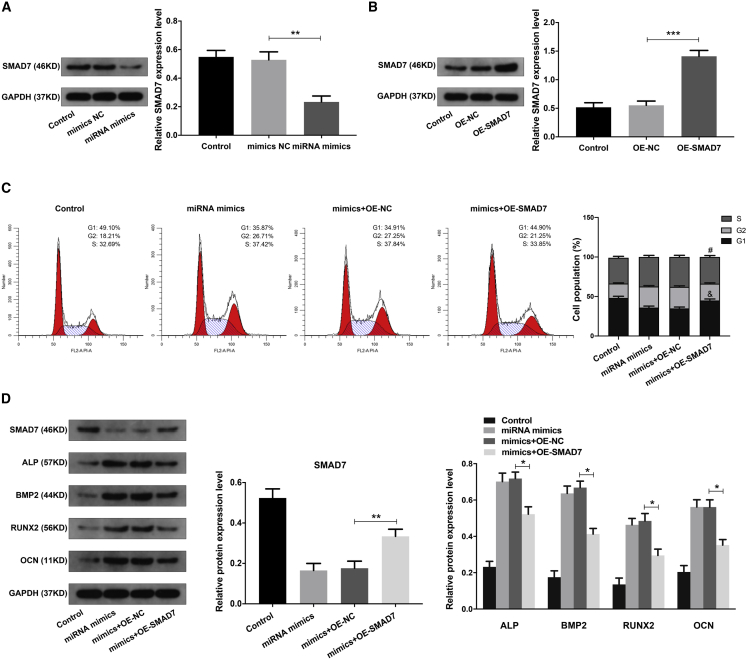
It has been reported that SMAD7 played a critical role in the osteogenic differentiation of BMSCs and ONFH.12 Using RT-qPCR analysis, we found that the expression of SMAD7 mRNA was obviously upregulated in ONFH-BMSCs (Figure S1). In addition, the upregulated expression pattern of circHGF and SMAD7 and the downregulated expression pattern of miR-25-3p were also validated in the the blood samples of ONFH patients (Figure S1). Using the TargetScan 7 online tool and the miRDB online database, we found that the SMAD7 was the direct target gene for miR-25-3p. Next, we explored the effects and interaction between miR-25-3p and SMAD7 in BMSCs. The western blot analysis showed that miR-25-3p could obviously inhibit SMAD7 expression in BMSCs (Figure 6A). Next, the OE vector (OE-SMAD7) was further used to investigate the critical role of SMAD7, and the SMAD7 expression was validated by western blot analysis (Figure 6B). The combination treatment of miR-25-3p mimics and OE-SMAD7 in BMSCs was validated by RT-qPCR analysis (Figure S1). The cell-cycle analysis demonstrated that miR-25-3p increased the cell proliferation of BMSCs, and this effect could be partially counteracted by SMAD7 (Figure 6C). Using western blot analysis, we found that the function of miR-25-3p in the osteogenic differentiation of BMSCs could also be partially counteracted by SMAD7 (Figure 6D). It was suggested that miR-25-3p increased the proliferation and osteogenic differentiation of BMSCs by interacting with SMAD7.
Figure 6.
MiR-25-3p enhanced the proliferation and osteogenic differentiation of BMSCs via SMAD7
(A) The BMSCs were transfected with miR-25-3p mimics or mimics NC, and the protein expression level of SMAD7 was examined by western blot analysis. The quantitative analysis showed the SMAD7 protein expression level was significantly decreased in the miRNA mimics group compared with the mimics NC group. (B) The BMSCs were transfected with SMAD7 over-expression vector OE-SMAD7 or the control OE-NC, and the SMAD7 protein expression was validated by western blot analysis. The quantitative analysis showed the SMAD7 protein expression level was significantly increased in the OE-SMAD7 group compared with the OE-NC group. (C) The BMSCs were treated with miR-25-3p mimics, mimics + OE-NC, or mimics + OE-SMAD7, and cell proliferation ability was determined by cell-cycle analysis. #p < 0.05 versus mimics + OE-NC group for comparing cell percentage in S phase, &p < 0.05 versus mimics + OE-NC group for comparing cell percentage in G1 phase, n = 3. (D) The protein expression levels of SMAD7, and the osteogenic marker genes in BMSCs of different groups were detected by western blot analysis. GAPDH served as an internal control correspondingly. The quantitative analysis showed the SMAD7 expression level was increased, and the expression levels of ALP, BMP2, RUNX2, and OCN were decreased, in mimics + OE-SMAD7 group compared with mimics + OE-NC group. Data were shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n = 3. Data among multiple groups were analyzed by one-way ANOVA test.
CircHGF functioned in BMSCs by targeting miR-25-3p/SMAD7 axis
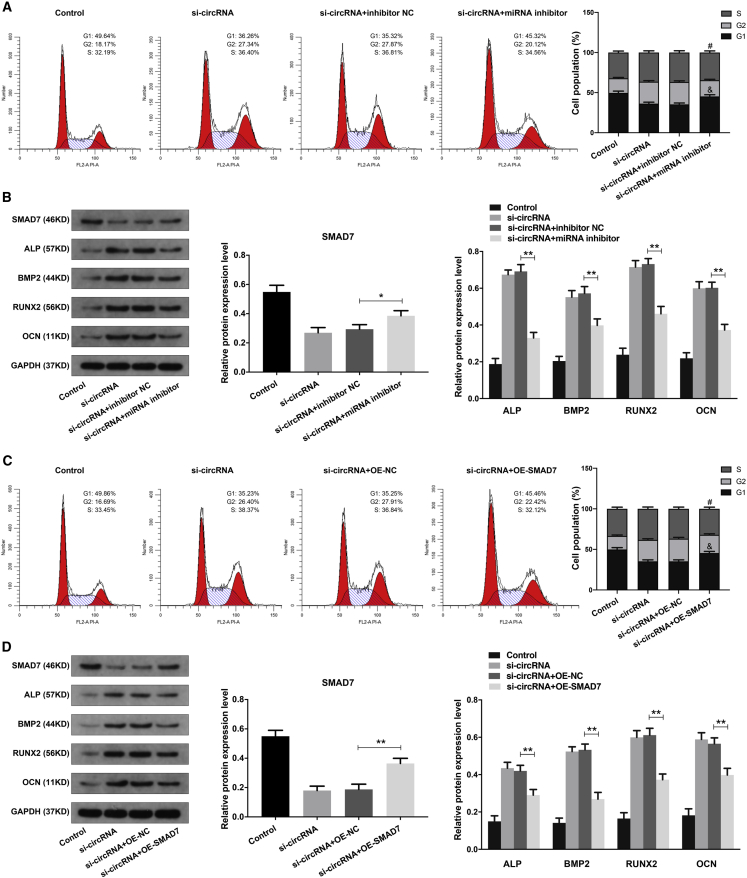
Based on the above results, we next explored whether circHGF function in BMSCs by interacting with miR-25-3p/SMAD7 axis. The combination treatment of circHGF siRNA and miR-25-3p inhibitor in BMSCs was first validated by RT-qPCR analysis (Figure S1). Using the cell-cycle analysis, we found that the elevated cell proliferation of BMSCs caused by circHGF knocking down could be counteracted by miR-25-3p inhibitor (Figure 7A). The western blot analysis showed that miR-25-3p inhibitor could counteract the effects of circHGF siRNA on the osteogenic differentiation ability of BMSCs (Figure 7B). The combination treatment of circHGF siRNA and OE-SMAD7 in BMSCs was validated by RT-qPCR analysis (Figure S1). We found that the SMAD7 OE treatment markedly counteracted the effects of circHGF siRNA on the proliferation and osteogenic differentiation ability of BMSCs (Figures 7C and 7D). Moreover, further experiments demonstrated that either miR-25-3p mimics or SMAD7 siRNA could obviously counteract the effects of circHGF OE on the cell proliferation and osteogenic differentiation ability of BMSCs (Figures S2 and S3). These results demonstrated that circHGF exerted its functions by targeting miR-25-3p/SMAD7 axis.
Figure 7.
CircHGF functioned in BMSCs by targeting miR-25-3p and SMAD7
(A) The BMSCs were treated with si-circRNA, si-circRNA + inhibitor NC, or si-circRNA + miRNA inhibitor, and cell proliferation ability was determined by cell-cycle analysis, with the cell population of G1, G2, and S cell-cycle phase indicated. #p < 0.05 versus si-circRNA + inhibitor NC group for comparing cell percentage in S phase, &p < 0.05 versus si-circRNA + inhibitor NC group for comparing cell percentage in G1 phase, n = 3. (B) The protein expression levels of SMAD7, ALP, BMP2, RUNX2, and OCN in BMSCs of different groups were evaluated by western blot analysis. The quantitative analysis showed the SMAD7 expression level was increased, and the expression levels of ALP, BMP2, RUNX2, and OCN were decreased, in the si-circRNA + miRNA inhibitor group compared with the si-circRNA + inhibitor NC group. (C) The BMSCs were treated with si-circRNA, si-circRNA + OE-NC, or si-circRNA + OE-SMAD7 vector, and cell proliferation ability was examined by cell-cycle analysis. #p < 0.05 versus si-circRNA + OE-NC group for comparing cell percentage in S phase, &p < 0.05 versus si-circRNA + OE-NC group for comparing cell percentage in G1 phase, n = 3. (D) The protein expression levels of SMAD7, and the osteogenic marker genes in BMSCs were detected by western blot analysis. GAPDH served as an internal control correspondingly. The quantitative analysis showed the SMAD7 expression level was increased, and the expression levels of ALP, BMP2, RUNX2, and OCN were decreased, in the si-circRNA + OE-SMAD7 group compared with the si-circRNA + OE-NC group. Data were shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, n = 3. Data among multiple groups were analyzed by one-way ANOVA test.
CircHGF could directly interact with miR-25-3p and miR-25-3p could directly target SMAD7 3′UTR
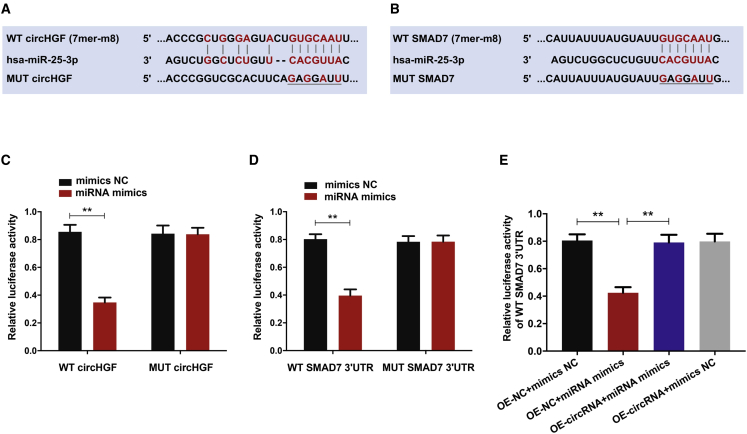
We carried out the luciferase reporter gene assays to validate the binding site of circHGF and SMAD7 with miR-25-3p, as predicted by bioinformatics analysis (Figures 8A and 8B). Luciferase reporter vector with mutant (MUT) or wild-type (WT) circHGF, which possessed the miR-25-3p binding sites, were transfected to HEK293T cells treated with miR-25-3p mimics. As expected, miR-25-3p markedly suppressed the luciferase activity of WT-circHGF reporter, while there was no significant difference in MUT-circHGF reporter activity between miR-25-3p mimics and the control group (Figure 8C). Similarly, miR-25-3p markedly suppressed the luciferase activity of WT-SMAD7 3′UTR reporter, while there was no significant difference in MUT-SMAD7 3′UTR reporter activity between miR-25-3p mimics and the control group (Figure 8D). Moreover, the luciferase activity assay revealed that miR-25-3p markedly repressed WT-SMAD7 3′UTR luciferase activity while circHGF could partially reverse this inhibitory effect (Figure 8E). These results revealed that circHGF could directly interact with miR-25-3p, and miR-25-3p could directly target SMAD7 3′UTR.
Figure 8.
CircHGF served as a sponge for miR-25-3p and miR-25-3p could directly target SMAD7 3′UTR
(A) The binding sequence between circHGF and miR-25-3p as predicted by bioinformatics analysis based on the starBase online database. (B) The binding site between SMAD7 and miR-25-3p as predicted by bioinformatics analysis based on the TargetScan 7 online tool and miRDB online database. (C) Relative luciferase reporter activity of WT or MUT circHGF in 293T cells co-transfected with miR-25-3p mimics or the control mimics NC. (D) Relative luciferase reporter activity of WT or MUT SMAD7 3′UTR in 293T cells co-transfected with miR-25-3p mimics or mimics NC. (E) The WT SMAD7 3′UTR luciferase reporter vector was co-transfected into 293T cells with miR-25-3p mimics or mimics NC, and OE-circRNA or OE-NC vector, and the luciferase activity was detected in corresponding groups. Data were shown as means with error bars representing SD. ∗∗p < 0.01, n = 3. Data between two groups were analyzed by Student's t test. Data among multiple groups were analyzed by one-way ANOVA test.
Discussion
Glucocorticoid-induced ONFH is a common metabolic disorder of bone, which is often caused by the serious side effects of long-term use or overdose of glucocorticoid drugs. There have been proposed several potential hypotheses to account for the glucocorticoid-induced ONFH initiation and progression, including the differentiation imbalance of BMSCs, apoptosis of osteocytes and osteoblasts, abnormal intraosseous pressure, increased oxidative stress, etc.,23, 24, 25, 26 although until now, the specific underlying mechanism involved in ONFH pathogenesis and development remains unclear, limiting the diagnosis and treatment strategies for ONFH. BMSCs are a subset of adult stem cells derived from mesodermal cell lineages, and they have strong potential in multi-lineage differentiation and self-renewal capacity.3 Accumulating evidence has corroborated that BMSCs participate functionally in the ONFH pathogenesis, which is closely related to the balance of BMSC adipogenic and osteogenic differentiation.4 It has been reported that glucocorticoids exerted significant influence on the osteogenic differentiation of BMSCs and thus led to ONFH progression.27 Previous research indicated that overdose of glucocorticoid could suppress the bone formation of function of BMSCs by reciprocally regulating miRNA-34a-5p.28 Therefore, facilitating the proliferation and osteogenic differentiation of BMSCs could be a promising strategy for the prevention of glucocorticoid-induced ONFH. However, the molecular mechanisms of steroid-induced ONFH still need further investigation.
NcRNAs have been reported to play critical roles in various human diseases, and attempts have been made to uncover the expression profiles as well as the function of ncRNAs in ONFH initiation and progression. Previous studies have demonstrated that lncRNAs were found differentially expressed in BMSCs of ONFH as well as bone microvascular endothelial cells of ONFH patients.14 Some specific lncRNAs have been found to play critical regulating roles in BMSCs of ONFH. For example, Xiang et al.29 reported that lncRNA RP11-154D6 significantly decreased in BMSCs of patients with ONFH, accelerated the development of steroid-induced ONFH via mediating the differentiation behavior of BMSCs. MiRNAs are a class of short, single-stranded, and highly conserved endogenous ncRNAs, which regulate targeting genes post-transcriptionally in a way of binding to the 3′-UTR of the targeting mRNAs.30 MiRNAs have been known to function as important regulators in many bone-associated diseases, including steroid-induced ONFH. For instance, miR-217 was found markedly decreased in glucocorticoid-associated ONFH, and it could promote cell proliferation and osteogenic differentiation of BMSCs via suppressing DKK1 expression.31 Another study demonstrated that miR-181d expression was significantly upregulated in the bone marrow of patients with steroid-induced ONFH, and it suppressed the differentiation of BMSCs into osteoblasts via targeting and negatively regulating SMAD3.32 Our previous study also revealed that miR-17-5p promoted the osteogenesis of BMSCs through directly interacting with SMAD7 to repress its expression in non-traumatic osteonecrosis.33 In recent years, miRNAs have emerged as a powerful regulator for gene expression and function in the steroid-induced ONFH pathogenesis.34
CircRNAs are another novel class of endogenous ncRNAs, which have a covalently closed and single-stranded structure specially.35 CircRNAs are widely discovered in the eukaryotic transcriptome and function as miRNA sponges and thus to play key roles in BPs and pathogenesis.19,36,37 Although more and more circRNAs have been identified in different species, only a minority of them have been uncovered and investigated about the biological implication.38 In the past few years, there has been increasing interest in the roles and mechanisms of this special class of ncRNA in steroid-induced ONFH.34 A previous study reported that circRNA circUSP45 expression was elevated in glucocorticoid-induced ONFH patients, and it could inhibit the osteogenesis and proliferation of BMSCs via miR-127-5p/PTEN/AKT pathway.39 Xin et al.40 revealed that circRNA hsa_circ_0066523 increased in the osteogenic induction process of BMSCs, and knocking down hsa_circ_0066523 repressed the proliferation and osteogenic differentiation of BMSCs by epigenetically modulating PTEN. Recently, Chen and colleagues 17 detected 820 differentially expressed circRNAs in the BMSCs from steroid-induced ONFH and they investigated the function of a famous initially identified circRNA called CDR1as, which harbors a great number of miR-7 binding sites.41 It was suggested that CDR1as has exerted significant effects on the adipogenic and osteogenic differentiation disorder of BMSCs by interacting wtih miR-7-5p/WNT5B axis.
In the present study, we screened the circRNAs expression profile in steroid-induced ONFH using five samples of BMSCs-ONFH and five samples of control by means of circRNA microarray analysis. Up to 182 differentially expressed circRNAs were identified, among which 108 circRNAs were significantly upregulated. Then, we further investigated the function of a most markedly upregulated circRNA called circHGF on the proliferation and osteogenic differentiation of BMSCs in steroid-induced ONFH. The circRNA identified in our study was a novel circRNA, and it played crucial roles in the pathogenesis of steroid-induced ONFH. Experimental results demonstrated that circHGF inhibited the proliferation and osteogenic differentiation of BMSCs, while knocking down circHGF exerted the opposite effects on BMSCs in ONFH. Using bioinformatics analysis and dual luciferase reporter assay, we further uncovered that miR-25-3p was a critical targeting miRNA for circHGF, and they could directly interact. Moreover, we found that the miR-25-3p expression level was markedly elevated in ONFH-BMSCs compared with the control group, as validated by the RT-qPCR analysis. Subsequent experiments revealed that over-expressing miR-25-3p enhanced the proliferation and osteogenic differentiation ability of BMSCs, and downregulating miR-25-3p could inhibited these effects conversely. Then, by using bioinformatics analysis of miRNA targets prediction, we found SMAD7 was the direct target gene for miR-25-3p. SMAD7 was classified as a negative regulator among the SMAD family, which significantly suppressed BMP as well as TGF-β signals and inhibited osteogenesis of BMSCs and bone formation.33,42 We then validated that miR-25-3p could directly target 3′UTR of SMAD7 to negatively modulate its expression in BMSCs, which played an important part in the proliferation and osteogenic differentiation of BMSCs. Subsequently, we found that inhibiting miR-25-3p could neutralize the enhanced cell proliferation and osteogenic differentiation ability of BMSCs induced by knocking down circHGF. Over-expressing SMAD7 obviously counteracted the effects of circHGF siRNA exerted on BMSCs. Notably, our results revealed that the key upregulated circRNA circHGF suppressed the proliferation and osteogenic differentiation of BMSCs via directly targeting miR-25-3p/SMAD7 axis. Nevertheless, the possibilities of other crucial targeting genes for circHGF to function in BMSCs and other specific dysregulated circRNAs involving in steroid-induced ONFH pathogenesis were still not excluded.
Collectively, we successfully mapped the circRNAs expression profile of BMSCs in steroid-induced ONFH and identified a novel and critical circRNA, termed circHGF. By performing systematical experiments, we unveiled that circHGF suppressed the proliferation and osteogenic differentiation of BMSCs in ONFH by interacting with miR-25-3p to modulate SMAD7. In conclusion, the present research suggested that circHGF targeting miR-25-3p/SMAD7 axis could provide a novel diagnostic and therapeutic strategy against steroid-induced ONFH.
Materials and methods
Patient samples and ethics statement
The bone marrow specimens were harvested from 10 patients with steroid-induced ONFH (7 men and 3 women, aged 25–68 years, mean 50.0 years) and 10 patients with femoral neck fracture as a control group (6 men and 4 women, aged 33–68 years, mean 54.6 years). Each donor's medical record was collected and the diagnostic criteria for steroid-induced ONFH were confirmed by the preoperative X-ray and/or magnetic resonance image (MRI) based on the Steinberg or University of Pennsylvania system. Steroid-induced ONFH was defined as a history of a mean daily dose of 16.6 mg or highest daily dose of 80 mg of prednisolone equivalent within 1 year.43 Patients with severe congenital diseases, cancer, or alcohol consumption were excluded. The study protocol was approved by the clinical research ethics committee of Tongji Medical College, Huazhong University of Science and Technology, and informed consent was obtained from all the enrolled patients.
BMSCs isolation and culture
The BMSCs were isolated and purified using gradient density centrifugation, as reported previously.44,45 Briefly, the bone marrow samples were resuspended in PBS. Next, the cell suspension was transferred to a centrifuge tube containing an equal volume of lymphocyte separation solution (Corning, Manassas, VA, USA). After density gradient centrifugation at 2500 rpm for 30 min, the mononuclear cells in the white layer were harvested. Subsequently, the cells were cultured in DMEM (Gibco, New York) with 15% fetal bovine serum (FBS) (Gibco) and streptomycin (100 mg/mL; Gibco) as well as penicillin (100 U/mL; Gibco) in 5% CO2 at 37°C. The culture medium was replaced three times a week. The flow cytometry analysis was used for identification of the surface marker of BMSCs, which has been reported in our previous work.46 The cells were passaged at a 1:2 when the culture reached 90% confluency and cells from passage two or three were used in the subsequent experiments.
RNA extraction and qPCR assay
After corresponding treatments for the cells, the total RNA was extracted by using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA), as previously described.47 The RNA purity and concentration were examined by a microspectrophotometer. For the qPCR analysis, the RNA was reverse-transcribed to cDNA using a Transcriptor 1st Strand cDNA Synthesis Kit (Takara Bio, Japan). Then the PCR amplification was performed with SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) using an QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). The cycling conditions were set as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 60 s; followed by a melt curve stage of 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s; and finally hold at 4°C. GAPDH and U6 were used as corresponding internal controls. The RT-qPCR primers used in this study are listed as follows (5′-3′): SMAD7 Forward, TACTGGGAGGAGAAGACGAGAGTGG; SMAD7 Reverse, GTCGAAAGCCTTGATGGAGAAACCG; hsa_circ_0080914 Forward, CACTCATTCCTTGGGATTAT; hsa_circ_0080914 Reverse, GGTGTGAGGGTCAAGAGTAT; GAPDH Forward, TCAAGAAGGTGGTGAAGCAGG; GAPDH Reverse, TCAAAGGTGGAGGAGTGGGT; miR-25-3p loop primer, GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGACCG; miR-25-3p Forward, TGCGCCATTGCACTTGTCTCGG; U6 Forward, CGCTTCGGCAGCACATATAC; U6 Reverse, AAATATGGAACGCTTCACGA; Divergent-hsa_circ_0080914 Forward, TGCCGAAATCCAGATGGGTC; Divergent-hsa_circ_0080914 Reverse, GCAGGTCATGCATTCAAAACGAGA; Convergent-hsa_circ_0080914 Forward, ACCTACGAGAAAATTACTGCCG; Convergent-hsa_circ_0080914 Reverse, CGGCAGTAATTCTCATTCAGCT; Divergent-GAPDH Forward, CACTTTGTCAAGCTCATTTCC; Divergent-GAPDH Reverse, TTGGCAGTGGGGACACGGAAG; Convergent-GAPDH Forward, TCCATGCCATCACTGCCAC; and Convergent-GAPDH Reverse, GTGGTCGTTGAGGGCAATG.
Western blot
The cell specimens from various groups were lysed by RIPA lysis buffer (Beyotime, Shanghai, China), and the corresponding protein contents were determined by the enhanced BCA protein analysis kit (Beyotime), according to the manufacturer's guide. Subsequently, equal quantities of protein of each sample was separated by using 10%–12% SDS-PAGE and were transferred onto the polyvinylidene fluoride membrane. The membrane was then blocked at 25°C for 1 h with 5% non-fat dried milk in Tris-buffered saline with Tween (TBST) and incubated with the specific primary antibody (1:500–1:1000) overnight at 4°C. Then the samples were incubated with the horseradish peroxidase (HRP)-conjugated secondary antibody (1:2000; Abcam, Cambridge, MA, USA) at 25°C for 2 h. The protein expression was observed with enhanced chemiluminescence reagents (Amersham, Piscataway, NJ, USA). The primary antibodies used in the present study are listed as follows: SMAD7 (25840-1-AP, Proteintech, Wuhan, China), ALP (DF6225, Affinity, Cincinnati, OH, USA), BMP2 (AF5163, Affinity, Cincinnati, OH, USA), RUNX2 (AF5186, Affinity, Cincinnati, OH, USA), and Osteocalcin (DF12303, Affinity, Cincinnati, OH, USA).
CircRNA microarray and bioinformatics analysis
The BMSCs samples for circRNA microarray were chosen randomly from the group of 10 ONFH patients and the group of 10 femoral neck fracture patients, respectively, by using simple random sampling. The samples were from 5 patients with steroid-induced ONFH (3 men and 2 women, aged 25–55 years, mean age 39.2 years) and 5 patients with femoral neck fracture as a control group (3 men and 2 women, aged 33–68 years, mean age 54.6 years). Total RNA from the ONFH-BMSCs samples and control BMSCs samples was isolated for microarray assay, which was carried out by CapitalBio (Beijing, China) in standard procedures. The microarray data was analyzed using the GeneSpring GX software. The threshold in this study was made as follows: fold change >2 and p < 0.05, as examined by Student's t test. The hierarchical clustering analysis were performed by using Cluster 3.0 software to present the profiling of differentially expressed circRNAs between the ONFH-BMSCs group and the control BMSCs group. The upregulated and downregulated circRNAs were also presented on the volcano plot filtering. Then the predicted functions of the differentially expressed circRNAs were obtained by the GO and KEGG pathway enrichment analysis of their host genes. The targeting miRNA for circRNA was predicted via the starBase online database (http://starbase.sysu.edu.cn). The targeting mRNA for miR-25-3p was predicted by the TargetScan 7 online tool (www.targetscan.org) and miRDB online database (http://www.mirdb.org).
Transfection vector construction and cell transfection
The circHGF siRNA targeting the back-splicing region of hsa_circ_0080914 and its control vector si-NC were designed and synthesized by GenePharma (Suzhou, China). The target sequence for circHGF was as follows: 5′-CTATTTCTCGTTTTGAATG-3′ and NC: 5′-TTCTCCGAACGTGTCACGT-3′. The circHGF OE vector pcDNA3.1(+)-S-circRNA(hsa_circ_0080914) and the control vector were designed and constructed by Obio Technology (Shanghai, China). The hsa-miR-25-3p mimics, hsa-miR-25-3p inhibitor, and corresponding NCs were purchased from GenePharma. The corresponding sequence was as follows: mimics sequence 5′-CAUUGCACUUGUCUCGGUCUGAAGACCGAGACAAGUGCAAUGUU-3′; mimic NC sequence 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′; inhibitor sequence 5′-UCAGACCGAGACAAGUGCAAUG-3′; and inhibitor NC sequence 5′-CAGUACUUUUGUGUAGUACAA-3′. The SMAD7 OE vector pcDNA3.1-Hygro(+)-homo-SMAD7 (OE-SMAD7) as well as its control vector was designed and synthesized by Biofavor Biotech (Wuhan, China). The BMSCs were seeded and cultured in 6-well plates until 70%–80% confluence, and then the cell transfection was performed by using Lipofectamine 2000 transfection reagent, according to the manufacturer's instructions. The transfection was performed up to 48 h, unless otherwise specified, followed by subsequent cell treatments. Experiments were performed three times independently.
Flow cytometric analysis of cell cycle
The BMSCs were seeded at a density of 2 × 105 cells per well in 6-well plates. At 48 h after the corresponding transfection treatment, the cells were assayed with flow cytometric analysis after propidium iodide (PI) staining. BMSCs were detached with 0.25% trypsin for 3 min at room temperature and fixed with ice-cold 70% ethanol at 4°C for 4 h. After centrifugation at 1500 rpm for 5 min, the cells were washed twice with PBS and then stained with 50 μg/mL of PI (100 μg/mL RNase A) at room temperature for 0.5 h away from light. The resulting cell suspensions were examined by flow cytometry on a CytoFLEX flow cytometer (Beckman Coulter, CA, USA), and the data were analyzed by using ModFit LT software.
Osteogenic differentiation of BMSCs
BMSCs were seeded in 6-well plates and cultured in DMEM with 10% FBS and 1% penicillin/streptomycin to 80% confluence for the corresponding transfection treatment. To evaluate the osteogenic differentiation ability of BMSCs, the infected cells were induced by osteogenic induction medium containing 10% FBS, 5 mM β-glycerophosphate, 0.1 μM dexamethasone, and 0.1 mM ascorbate-2-phosphate. The osteogenic induction medium was replaced every three days.
Alizarin red staining
Alizarin red staining was used to evaluate the osteogenic differentiation ability of BMSCs at the 7th and 14th day after corresponding transfection treatment. The cells were cultured in osteogenic differentiation medium in 6-well plates. For alizarin red staining, cells were fixed with 4% paraformaldehyde, rinsed with deionized water, and then stained with alizarin red stain at pH = 4.3 for 10 min. The staining results were examined by microscopy (Olympus, Tokyo, Japan).
Dual luciferase reporter assay
The human embryonic kidney (HEK) 293T cells were used for dual luciferase activity analysis in this experiment. When cultured to 60%–70% confluence; the cells were co-transfected by the corresponding luciferase reporter plasmid and miR-25-3p mimics or its control. The corresponding luciferase reporter plasmid of WT pYr-MirTarget-hsa_circ_0080914 and MUT pYr-MirTarget-hsa_circ_0080914, WT pYr-MirTarget-homo smad7-3′UTR as well as MUT pYr-MirTarget-homo smad7-3′UTR were obtained from Biofavor Biotech. At 48 h post-transfection using Lipofectamine 2000, the cells were assayed with the Dual Luciferase Reporter Gene Assay Kit (RG027, Beyotime, Shanghai, China) following the manufacturer's instructions.
Statistical analysis
The data were presented as the mean ± SD. Each experiment was repeated independently at least three times. Difference between groups was analyzed by using unpaired Student's t test or one-way ANOVA test. The statistical significance levels were expressed as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Acknowledgments
This study was supported by the grants from the National Natural Science Foundation of China (81902260, 81702193 and 82001788), Natural Science Foundation of Hubei Province (2020CFB410).
Author contributions
X.F., Q.X., J.J., X.C., and X.L. conceived the study protocol. X.F., Q.X., J.J., and T.G. performed the experiments and collected and analyzed data. Q.X. and Z.L. wrote the manuscript. X.L. and X.C. proofread the manuscript and made revisions. All authors approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.02.017.
Contributor Information
Xianyi Cai, Email: xianyi_cai@hust.edu.cn.
Xianzhe Liu, Email: liuxianzhe@hust.edu.cn.
Supplemental information
References
- 1.Zhou W., Qu M., Lv Y., Zhu J. New advances in stem cell therapy for osteonecrosis of the femoral head. Curr. Stem Cell Res. Ther. 2019;14:226–229. doi: 10.2174/1574888X13666181025120252. [DOI] [PubMed] [Google Scholar]
- 2.Assouline-Dayan Y., Chang C., Greenspan A., Shoenfeld Y., Gershwin M.E. Pathogenesis and natural history of osteonecrosis. Semin. Arthritis Rheum. 2002;32:94–124. [PubMed] [Google Scholar]
- 3.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F., Peng W., Zhang J., Dong W., Wu J., Wang T., Xie Z. P53 and Parkin co-regulate mitophagy in bone marrow mesenchymal stem cells to promote the repair of early steroid-induced osteonecrosis of the femoral head. Cell Death Dis. 2020;11:42. doi: 10.1038/s41419-020-2238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houdek M.T., Wyles C.C., Packard B.D., Terzic A., Behfar A., Sierra R.J. Decreased osteogenic activity of mesenchymal stem cells in patients with corticosteroid-induced osteonecrosis of the femoral head. J. Arthroplasty. 2016;31:893–898. doi: 10.1016/j.arth.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 6.MacFarlane R.J., Graham S.M., Davies P.S., Korres N., Tsouchnica H., Heliotis M., Mantalaris A., Tsiridis E. Anti-inflammatory role and immunomodulation of mesenchymal stem cells in systemic joint diseases: potential for treatment. Expert Opin. Ther. Targets. 2013;17:243–254. doi: 10.1517/14728222.2013.746954. [DOI] [PubMed] [Google Scholar]
- 7.Fan L., Zhang C., Yu Z., Shi Z., Dang X., Wang K. Transplantation of hypoxia preconditioned bone marrow mesenchymal stem cells enhances angiogenesis and osteogenesis in rabbit femoral head osteonecrosis. Bone. 2015;81:544–553. doi: 10.1016/j.bone.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa K., Miyazono K. Regulation of TGF-β family signaling by inhibitory smads. Cold Spring Harb. Perspect. Biol. 2017;9:a022095. doi: 10.1101/cshperspect.a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao P., Zhu Z., Du C., Zeng Y., Liao J., Cheng Q., Chen H., Zhao C., Huang W. Silencing Smad7 potentiates BMP2-induced chondrogenic differentiation and inhibits endochondral ossification in human synovial-derived mesenchymal stromal cells. Stem Cell Res. Ther. 2021;12:132. doi: 10.1186/s13287-021-02202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estrada K.D., Wang W., Retting K.N., Chien C.T., Elkhoury F.F., Heuchel R., Lyons K.M. Smad7 regulates terminal maturation of chondrocytes in the growth plate. Dev. Biol. 2013;382:375–384. doi: 10.1016/j.ydbio.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang S.H., Chen L., Chen H.H., Li Y.F., Luo H.B., Hu D.Q., Chen P. MiR-15b ameliorates SONFH by targeting Smad7 and inhibiting osteogenic differentiation of BMSCs. Eur. Rev. Med. Pharmacol. Sci. 2019;23:9761–9771. doi: 10.26355/eurrev_201911_19539. [DOI] [PubMed] [Google Scholar]
- 13.Razin S.V., Gavrilov A.A. Non-coding RNAs in chromatin folding and nuclear organization. Cell Mol. Life Sci. 2021;78:5489–5504. doi: 10.1007/s00018-021-03876-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Huang C., Yang B., Hu W., Chan M.T., Wu W.K.K. Emerging roles of long non-coding RNAs in osteonecrosis of the femoral head. Am. J. Transl. Res. 2020;12:5984–5991. [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Yang B., Weng X., Tse G., Chan M.T.V., Wu W.K.K. Emerging roles of MicroRNAs in osteonecrosis of the femoral head. Cell Prolif. 2018;51:e12405. doi: 10.1111/cpr.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verduci L., Tarcitano E., Strano S., Yarden Y., Blandino G. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12:468. doi: 10.1038/s41419-021-03743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G., Wang Q., Li Z., Yang Q., Liu Y., Du Z., Zhang G., Song Y. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone. 2020;133:115258. doi: 10.1016/j.bone.2020.115258. [DOI] [PubMed] [Google Scholar]
- 18.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Zhi F., Ding Y., Wang R., Yang Y., Luo K., Hua F. Exosomal hsa_circ_0006859 is a potential biomarker for postmenopausal osteoporosis and enhances adipogenic versus osteogenic differentiation in human bone marrow mesenchymal stem cells by sponging miR-431-5p. Stem Cell Res. Ther. 2021;12:157. doi: 10.1186/s13287-021-02214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao X., Cao Y., Guo Z., Wang L., Xiang C. Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol. Ther. Nucleic Acids. 2021;24:856–867. doi: 10.1016/j.omtn.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Q., Kang L., Wang J., Liao Z., Song Y., Zhao K., Wang K., Yang C., Zhang Y. CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine. 2020;53:102679. doi: 10.1016/j.ebiom.2020.102679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X., Alqwbani M., Luo Y., Chen C., Ge A., Wei Y., Li D., Wang Q., Tian M., Kang P. Glucocorticoids decreased Cx43 expression in osteonecrosis of femoral head: the effect on proliferation and osteogenic differentiation of rat BMSCs. J. Cell Mol. Med. 2021;25:484–498. doi: 10.1111/jcmm.16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hungerford D.S., Lennox D.W. The importance of increased intraosseous pressure in the development of osteonecrosis of the femoral head: implications for treatment. Orthop. Clin. North Am. 1985;16:635–654. [PubMed] [Google Scholar]
- 25.Li J., Ge Z., Fan L., Wang K. Protective effects of molecular hydrogen on steroid-induced osteonecrosis in rabbits via reducing oxidative stress and apoptosis. BMC Musculoskelet. Disord. 2017;18:58. doi: 10.1186/s12891-017-1431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng S., Dai G., Chen S., Nie Z., Zhou J., Fang H., Peng H. Dexamethasone induces osteoblast apoptosis through ROS-PI3K/AKT/GSK3β signaling pathway. Biomed. Pharmacother. 2019;110:602–608. doi: 10.1016/j.biopha.2018.11.103. [DOI] [PubMed] [Google Scholar]
- 27.Zhou D.A., Zheng H.X., Wang C.W., Shi D., Li J.J. Influence of glucocorticoids on the osteogenic differentiation of rat bone marrow-derived mesenchymal stem cells. BMC Musculoskelet. Disord. 2014;15:239. doi: 10.1186/1471-2474-15-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H., Chen H., Huang P., Qi J., Qian N., Deng L., Guo L. Glucocorticoids impair bone formation of bone marrow stromal stem cells by reciprocally regulating microRNA-34a-5p. Osteoporos. Int. 2016;27:1493–1505. doi: 10.1007/s00198-015-3381-x. [DOI] [PubMed] [Google Scholar]
- 29.Xiang S., Li Z., Weng X. The role of lncRNA RP11-154D6 in steroid-induced osteonecrosis of the femoral head through BMSC regulation. J. Cell Biochem. 2019;120:18435–18445. doi: 10.1002/jcb.29161. [DOI] [PubMed] [Google Scholar]
- 30.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Dai Z., Jin Y., Zheng J., Liu K., Zhao J., Zhang S., Wu F., Sun Z. MiR-217 promotes cell proliferation and osteogenic differentiation of BMSCs by targeting DKK1 in steroid-associated osteonecrosis. Biomed. Pharmacother. 2019;109:1112–1119. doi: 10.1016/j.biopha.2018.10.166. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y., Hu J.Z., Shi Z.Y. MiR-181d promotes steroid-induced osteonecrosis of the femoral head by targeting SMAD3 to inhibit osteogenic differentiation of hBMSCs. Eur. Rev. Med. Pharmacol. Sci. 2018;22:4053–4062. doi: 10.26355/eurrev_201807_15393. [DOI] [PubMed] [Google Scholar]
- 33.Jia J., Feng X., Xu W., Yang S., Zhang Q., Liu X., Feng Y., Dai Z. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp. Mol. Med. 2014;46:e107. doi: 10.1038/emm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X., Sun W., Tan M. Noncoding RNAs in steroid-induced osteonecrosis of the femoral head. Biomed. Res. Int. 2019;2019:8140595. doi: 10.1155/2019/8140595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 36.Ni T., Zhang Q., Li Y., Huang C., Zhou T., Yan J., Chen Z.J. CircSTK40 contributes to recurrent implantation failure via modulating the HSP90/AKT/FOXO1 axis. Mol. Ther. Nucleic Acids. 2021;26:208–221. doi: 10.1016/j.omtn.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian T., Zhao Y., Zheng J., Jin S., Liu Z., Wang T. Circular RNA: a potential diagnostic, prognostic, and therapeutic biomarker for human triple-negative breast cancer. Mol. Ther. Nucleic Acids. 2021;26:63–80. doi: 10.1016/j.omtn.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Kuang M.J., Xing F., Wang D., Sun L., Ma J.X., Ma X.L. CircUSP45 inhibited osteogenesis in glucocorticoid-induced osteonecrosis of femoral head by sponging miR-127-5p through PTEN/AKT signal pathway: experimental studies. Biochem. Biophys. Res. Commun. 2019;509:255–261. doi: 10.1016/j.bbrc.2018.12.116. [DOI] [PubMed] [Google Scholar]
- 40.Xin W., Yuan S., Wang B., Qian Q., Chen Y. Hsa_circ_0066523 promotes the proliferation and osteogenic differentiation of bone mesenchymal stem cells by repressing PTEN. Bone Joint Res. 2021;10:526–535. doi: 10.1302/2046-3758.108.BJR-2020-0127.R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 42.Yano M., Inoue Y., Tobimatsu T., Hendy G., Canaff L., Sugimoto T., Seino S., Kaji H. Smad7 inhibits differentiation and mineralization of mouse osteoblastic cells. Endocr. J. 2012;59:653–662. doi: 10.1507/endocrj.ej12-0022. [DOI] [PubMed] [Google Scholar]
- 43.Koo K.H., Kim R., Kim Y.S., Ahn I.O., Cho S.H., Song H.R., Park Y.S., Kim H., Wang G.J. Risk period for developing osteonecrosis of the femoral head in patients on steroid treatment. Clin. Rheumatol. 2002;21:299–303. doi: 10.1007/s100670200078. [DOI] [PubMed] [Google Scholar]
- 44.Otsuru S., Hofmann T.J., Olson T.S., Dominici M., Horwitz E.M. Improved isolation and expansion of bone marrow mesenchymal stromal cells using a novel marrow filter device. Cytotherapy. 2013;15:146–153. doi: 10.1016/j.jcyt.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Fu D., Yang S., Lu J., Lian H., Qin K. LncRNA NORAD promotes bone marrow stem cell differentiation and proliferation by targeting miR-26a-5p in steroid-induced osteonecrosis of the femoral head. Stem Cell Res. Ther. 2021;12:18. doi: 10.1186/s13287-020-02075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Z., Luo R., Li G., Song Y., Zhan S., Zhao K., Hua W., Zhang Y., Wu X., Yang C. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics. 2019;9:4084–4100. doi: 10.7150/thno.33638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang L., Xiang Q., Zhan S., Song Y., Wang K., Zhao K., Li S., Shao Z., Yang C., Zhang Y. Restoration of autophagic flux Rescues oxidative damage and mitochondrial dysfunction to protect against intervertebral disc degeneration. Oxid Med. Cell Longev. 2019;2019:7810320. doi: 10.1155/2019/7810320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.