Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evolution has been characterized by the emergence of sets of mutations impacting the virus characteristics, such as transmissibility and antigenicity, presumably in response to the changing immune profile of the human population. The presence of mutations in the SARS-CoV-2 virus can potentially impact therapeutic and diagnostic test performances. We design and develop here a unique set of DNA probes i.e., antisense oligonucleotides (ASOs) which can interact with genetic sequences of the virus irrespective of its ongoing mutations. The probes, developed herein, target a specific segment of the nucleocapsid phosphoprotein (N) gene of SARS-CoV-2 with high binding efficiency which do not mutate among the known variants. Further probing into the interaction profile of the ASOs reveals that the ASO-RNA hybridization remains unaltered even for a hypothetical single point mutation at the target RNA site and diminished only in case of the hypothetical double or triple point mutations. The mechanism of interaction among the ASOs and SARS-CoV-2 RNA is then explored with a combination of surface-enhanced Raman scattering (SERS) and machine learning techniques. It has been observed that the technique, described herein, could efficiently discriminate between clinically positive and negative samples with ∼100% sensitivity and ∼90% specificity up to 63 copies/mL of SARS-CoV-2 RNA concentration. Thus, this study establishes N gene targeted ASOs as the fundamental machinery to efficiently detect all the current SARS-CoV-2 variants regardless of their mutations.
Keywords: SARS-CoV-2 variants, Mutation resistant probe, Antisense oligonucleotide, Surface-enhanced Raman scattering, Selective and ultrasensitive diagnosis
1. Introduction
The decline in coronavirus disease – 2019 (COVID-19) cases in much of the world in late spring and early summer signaled a new phase in the fight against the disease (Telenti et al., 2021). This progress was made possible by the rapid deployment of vaccines (Mallapaty et al., 2021). However, due to emerging new variants such as Beta, Delta, Lambda, and most recently Omicron, as well as the vaccine reluctance, nations across the globe have witnessed an alarming increase in COVID-19 cases in recent months (Callaway and Ledford, 2021; Karim and Karim, 2021). As a result, it continues to spread rapidly throughout the world, causing unprecedented disruption in modern society. While vaccines continue to be extremely effective at preventing serious illness caused by COVID-19, new data from the United Kingdom (U.K.), Israel, and the U.S. have raised new concerns about their ability to prevent infection from the delta as well as more transmissible and potentially concerning Omicron variant (Janik et al., 2021; Mercatelli and Giorgi, 2020; L. Wang et al., 2021). Furthermore, while the Omicron variant fuels a fresh wave of infections, decline in extensive testing and monitoring together with the unavailability of an effective diagnostic test to selectively detect these emerging variants without any false negativity, makes it impossible to determine how much the virus is circulating in communities and who remains most vulnerable (Daria et al., 2021; Mohiuddin and Kasahara, 2022).
These SARS-CoV-2 genetic variants represent permanent changes in the DNA sequence and may affect three or more nucleotides in a gene (Pachetti et al., 2020). These alterations may result into failure of diagnostics, reduction in vaccine effectiveness, low vaccine-induced protection against severe disease, more severe clinical disease, and significantly diminished susceptibility to diagnostics or approved therapeutics (Barton et al., 2021; Dighe et al., 2021; Liu et al., 2021; P. Wang et al., 2021). As new variants continue to emerge, a deeper understanding of the phenotypes of these variants in terms of infectivity, transmissibility, virulence, and antigenicity must be gained (Copin et al., 2021; Liu et al., 2021). As a result, there is still an unmet requirement for routine large-scale comprehensive testing to prevent COVID–19 spread and provide safe environments for socio-economic activities. And we believe that this can only be done by developing a probe that will interact with all the variants of SARS-CoV-2 independent of their mutations. Unpredicted combinations of mutations will continue to emerge, such perceptions will allow predictions of virus phenotype (Jungreis et al., 2021; Petrova and Russell, 2018) and in turn, help to develop newer probes for mutation independent interaction with SARS-CoV-2 genes.
In this regard, here we have designed and developed several sets of novel DNA Probes, i.e., antisense oligonucleotides, targeted towards nucleocapsid phosphoprotein (N), envelope (E) and RNA-dependent RNA polymerase (RdRp) gene segments of SARS-CoV-2. We hypothesized that a combination of computational and vibrational spectroscopic techniques along with machine learning (ML) algorithm can be applied to study the mutation-independent interaction of these DNA probes with SARS-CoV-2 RNA. Following the successful design and synthesis, these ASOs were connected to gold nanoparticles (AuNPs) and their target binding energies and agglomeration patterns were then investigated in presence of SARS-CoV-2 RNA. We found that N-gene targeted ASOs were the most efficient ones in selectively hybridizing SARS-CoV-2 target RNA with optimum efficiency. The target RNA sites for N-gene ASOs were then examined against the current SARS-CoV-2 variants where it was found that these ASOs should interact with all the known mutated forms of SARS-CoV-2 in a similar manner without any loss in sensitivity and specificity. The mechanism of interaction between these N gene-targeted ASOs and SARS-CoV-2 RNA was then evaluated with surface-enhanced Raman scattering (SERS) and machine learning (ML) techniques (Abid Hasan et al., 2019; Chaichi et al., 2021; Chang et al., 2015; Gartia et al., 2010; Prasad et al., 2020; Xu et al., 2011, 2012). This combinatorial effort discriminated SARS-CoV-2 positive and negative clinical nasal swab samples with ∼100% sensitivity and ∼90% specificity with an analytical detection limit of 63 copies/mL. However, the characterization of different SARS-CoV-2 N-gene mutated forms will continue to evolve and provide extremely useful information on specific mutations or their combinations those may not have been identified yet in circulating viruses (Harvey et al., 2021). Towards this goal, we computationally evaluated next hypothetical single, double, and triple point mutations on the RNA site. Further close monitoring of the ASO-RNA hybridization profile revealed stable interaction between the two, even after a hypothetical single point mutation was introduced at the target RNA site. Thus, this study examined the efficiency of universal DNA probes and their fundamental interaction with the entire known mutated forms of SARS-CoV-2.
There are many attempts to extend or replace nucleic acid-based methods to detect SARS-CoV-2 virus or differentiate its variants (Hu et al., 2021; Kevadiya et al., 2021a). However, numerous challenges still remain with these techniques, such as, extended operation time, requirement of laboratory-based hospital with the accessibility of large stationary equipment and reagents (Carter et al., 2020a; Vandenberg et al., 2021). The major bottleneck that lies with the widely used qPCR technique is the requirement of completely different set of primers and multiple other reagents for the detection of the emerging variants of SARS-CoV-2 (Barreto et al., 2020; Esbin et al., 2020; Vega-Magaña et al., 2021). But the current study relies on the change in Raman spectral signature of the designed N-ASO capped AuNP based probe. This technique does not require any nucleic acid purification or amplification steps to detect its target RNA, rather it involves unique antisense oligonucleotides that can bind to SARS-CoV-2 RNA with similar efficiency irrespective of its current mutations. We showed that the same two N gene targeted ASOs can successfully be used for a point of care (POC), rapid, cost-effective, and selective diagnosis of COVID-19. Further, because of the dependency of the studied technology on Raman spectra, high-throughput screening is quite possible in limited duration. This assay is also found to be quite sensitive (LOD = 63 copies/mL) and better than most of the currently known qPCR-based techniques (Afzal, 2020). Thus, the current study differentiates its signature from the literature known nucleic acid-based studies. We envisage that this study will lead to the real-time multispectral imaging and rapid optical processing of clinical samples towards the selective and sensitive diagnosis of COVID-19 variants.
2. Results and discussion
We designed and developed novel ASOs targeted towards different genetic segments of SARS-CoV-2, e.g., N, E and RdRp gene for the selective and sensitive binding of RNA from the wild type of SARS-CoV-2. More importantly, herein, we investigated the mechanism of interaction of selected ASOs with the various mutated forms of SARS-CoV-2 and studied the ASO-RNA hybridization phenomena through a unique combination of SERS and ML-based techniques.
2.1. Design of SARS-CoV-2 targeted ASOs
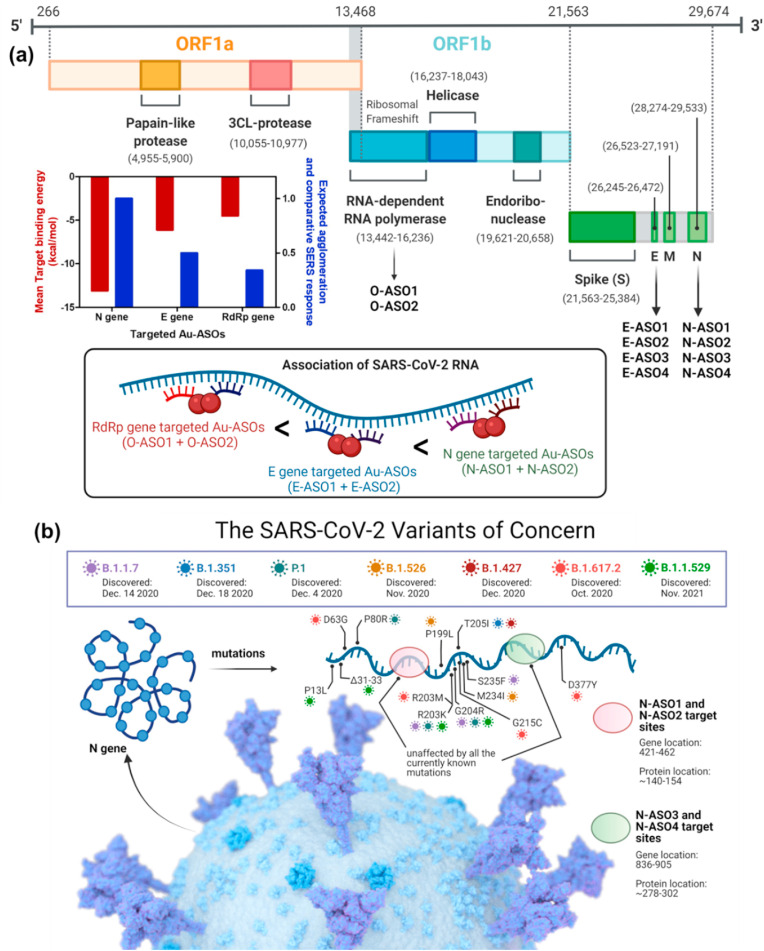
Accordingly, particular gene sequences (RdRp: 13,442–16,236; E: 26,245–26,472 and N: 28,274–29,533) from the whole genome sequence of SARS-CoV-2 (wild type isolate SARS-CoV-2/human/USA/WA-CDC-WA1-A12/2020, MT020880) was chosen and multiple ASO sequences of 20 nucleotides in length were developed (Fig. 1 a). The choice of ASOs were primarily based on the optimum guanine-cytosine (GC) content and theoretically calculated target binding and disruption energies at 37 °C in 1 M NaCl aqueous solution. We propose to select four ASOs, two for the front (ASO1 and ASO2), and two targeting the end region (ASO3 and ASO4) of the gene. The ASOs will also target closely following sequences at each location (Alafeef et al., 2020, 2021a, 2021b, 2020; Dighe et al., 2021; Moitra et al., 2020, 2021a, 2021b). Accordingly, four ASOs were selected both for N and E genes, whereas only two were selected for the RdRp gene as ASOs with high enough binding energies are not available for this target (Table 1 ). Soligo software were used to predict the ASOs (Ding et al., 2004). The detailed methodology is discussed in the method section of supporting information. It was noticed that the mean target binding energy was the highest for N gene-targeted ASOs (N-ASO1-4) and decreased gradually for E gene-targeted ASOs (E-ASO1-4) and was the minimum for RdRp targeted ones (O-ASO1-2). Based on this theoretical observation, it can also be presumed that the effective ASO-RNA hybridization will follow the same trend and will be the highest for N-ASO1 and N-ASO2. We anticipated that once these ASOs were connected to a plasmonic nanoparticle, such as AuNPs, the agglomeration among ASO capped AuNPs would exhibit a strong SERS response (Khan et al., 2018; Misra et al., 2018; Moitra et al., 2020; Pan et al., 2010, 2011) and should also exhibit similar phenomena where a mixture of N-ASO1 and N-ASO2 may be the optimum one (Fig. 1a).
Fig. 1.
(a) Genomic organization of SARS-CoV-2 and its schematic agglomeration pattern with gold nanoparticles differentially functionalized with antisense oligonucleotides targeted towards different genetic segments; (b) Currently known N gene mutation sites for SARS-CoV-2 and their alignment with the targeted sites of the developed ASOs.
Table 1.
Sequences of the differentially functionalized antisense oligonucleotides targeted towards different segments of SARS-CoV-2 isolate 2019-nCoV/USA-WA1-A12/2020 (MT020880). The targeted segments are N gene (28274..29533), E gene (26245..26472) and part of the RDRP gene (13442..16236: a part of the ORF1ab).
| Target position | Target sequence (5p—3p) | ASO sequence (5p—3p) | GC content | Binding site disruption energy (kcal/mol) | Target binding energy (kcal/mol) |
|---|---|---|---|---|---|
| Target gene N | |||||
| 28694–28713 | ACACCAAAAGAUCACAUUGG | HS-C6-CCAATGTGATCTTTTGGTGT (N-ASO1) | 40% | 7.6 | -15.8 |
| 28716–28735 | CCCGCAAUCCUGCUAACAAU | ATTGTTAGCAGGATTGCGGG-C6-SH (N-ASO2) | 50% | 7.6 | -10.4 |
| 29109–29128 | CAGAACAAACCCAAGGAAAU | HS-C6-ATTTCCTTGGGTTTGTTCTG (N-ASO3) | 40% | 6.0 | -14.3 |
| 29159–29178 | ACUGAUUACAAACAUUGGCC | GGCCAATGTTTGTAATCAGT-C6-SH (N-ASO4) | 40% | 8.7 | -10.0 |
| Target gene E | |||||
| 26306–26325 | UUGCUUUCGUGGUAUUCUUG | HS-C6-CAAGAATACCACGAAAGCAA (E-ASO1) | 40% | 12.4 | -5.5 |
| 26331–26350 | UACACUAGCCAUCCUUACUG | CAGTAAGGATGGCTAGTGTA-C6-SH (E-ASO2) | 45% | 15.0 | -6.7 |
| 26366–26385 | CGUACUGCUGCAAUAUUGUU | HS-C6-AACAATATTGCAGCAGTACG (E-ASO3) | 40% | 4.9 | -8.8 |
| 26390–26409 | UGAGUCUUGUAAAACCUUCU | AGAAGGTTTTACAAGACTCA-C6-SH (E-ASO4) | 35% | – | -4.3 |
| Target gene RdRp of ORF1b | |||||
| 15361–15380 | AAACAUACAACGUGUUGUAG | HS-C6-CTACAACACGTTGTATGTTT (O-ASO1) | 35% | – | -6.4 |
| 15389–15408 | ACCGUUUCUAUAGAUUAGCU | AGCTAATCTATAGAAACGGT-C6-SH (O-ASO2) | 35% | – | -2.6 |
2.2. Selective hybridization of ASOs with SARS-CoV-2 RNA
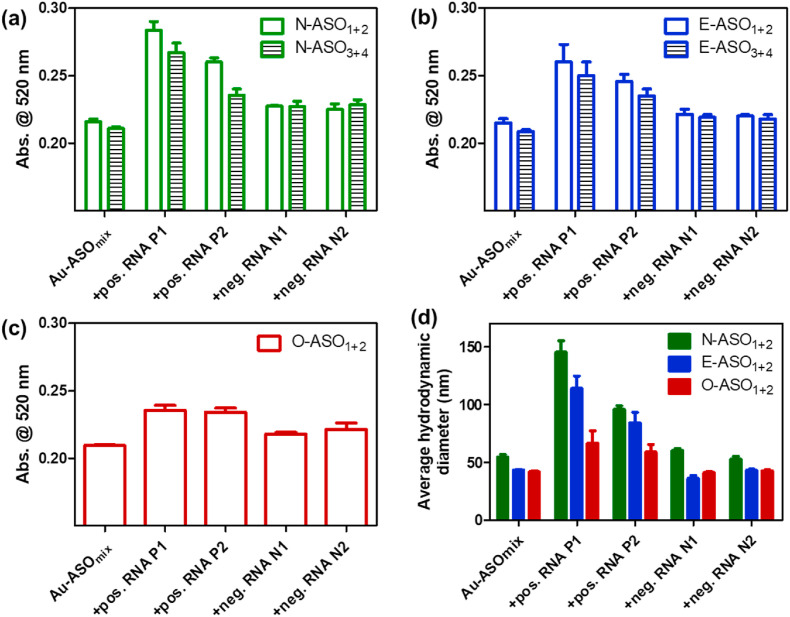
To prove this theoretical assumption, we differentially modified the ASOs, one at the 5′ end and the other at the 3’ end by thiol groups (Table 1). The thiol modified ASOs were then used to cap the citrate stabilized AuNP (Moitra et al., 2020; Zhu et al., 2021). The agglomeration patterns of these ASOs conjugated to AuNPs (Au-ASO NPs) were then investigated in presence of RNA extracted from clinically positive and negative SARS-CoV-2 samples. We employed UV-Visible absorbance spectroscopy and dynamic light scattering (DLS) to initially investigate the aggregation phenomena. SARS-CoV-2 positive nasal swab samples (N = 10) having varying cycle threshold (Ct) numbers from 13 to 28 (Table S1) were considered and RNA was extracted from these samples by standard protocol (Rump et al., 2010; Wozniak et al., 2020). Representative two samples, one having low Ct number (P1), i.e., high viral copy number and the other having high Ct value (P2), i.e., low viral copy number, was considered for this study. It was observed that the absorbance of plasmonic Au-ASO NPs at 520 nm increased with broadening of full-width half maximum (FWHM) when added with SARS-CoV-2 positive RNA. P1 having a high viral copy number increased the absorbance more than P2 which had a low viral copy number. An insignificant increase in absorbance with no change in FWHM was observed when Au-ASO NPs were added with SARS-CoV-2 negative RNAs. This change in surface plasmon band of AuNPs indicated the successful aggregation of Au-ASO NPs only in presence of their target RNAs(Moitra et al., 2020; Ostadhossein et al., 2020; Rump et al., 2010; Srivastava et al., 2020). It was also observed that the mixture of N-ASO1 and N-ASO2 gave a better aggregation response than the mixture having N-ASO3 and N-ASO4 (Fig. 2 a). Similar is the case for E-ASO1 and E-ASO2 compared to the combination of E-ASO3 and E-ASO4 (Fig. 2b). Further, the change in absorbance was found to be the highest for N-ASO1 and N-ASO2, followed by E-ASO1 and E-ASO2 and the minimum for O-ASO1 and O-ASO2 (Fig. S1). This improved agglomeration of N-ASO1 and N-ASO2 mixture in presence of SARS-CoV-2 RNA was further supported by DLS experiments. The average hydrodynamic diameter was found to be the highest for N-ASO1 and N-ASO2 mixture in presence of RNA extracted from P1 compared to the other mixture of Au-ASO NPs (Fig. 2d). Thus, these experiments corroborated with our theoretical observation supporting better binding followed by improved ASO-RNA hybridization in case of N-ASO1 and N-ASO2 mixture.
Fig. 2.
Aggregation induced change in absorbance at 520 nm for the differentially functionalized Au-ASO NPs targeted for (a) N gene; (b) E gene and (c) RdRp gene upon the addition of RNAs (1 μL) extracted from clinically positive and negative SARS-CoV-2 samples. (d) Comparative change in average hydrodynamic diameter among the different Au-ASO NPs upon the addition of RNAs. In each case, the mixture was incubated for 15 min at room temperature prior to recording the change in absorbance. Ct value of positive RNA P1 and P2 are 14.7 and 27 respectively by RT-PCR analyses which correspond to 107186 and 781 copies/μL of SARS-CoV-2 RNA.
2.3. Mutation independent interaction of SARS-CoV-2 RNA with N-gene targeted ASOs
This interesting observation, where we found that N-gene targeted ASOs were more efficient than E gene and RdRp gene-targeted ASOs in hybridizing SARS-CoV-2 RNA, led to the curiosity to investigate these N-ASOs more closely. We asked the question whether these ASOs will still be effective against the current SARS-CoV-2 mutations or not. We realized that the viability of the assay can be improved significantly if a mostly conserved region of N-gene sequence can be targeted by the ASO probes. Accordingly, the design of N-ASOs was revisited and the target RNA sites for these ASOs were examined against the current SARS-CoV-2 variants. Recently, the genetic surveillance of SARS-CoV-2 strains circulating around the world has revealed several variants with one or more mutations that may affect detection by nucleic acid-based testing methods (Carter et al., 2020b; Yu et al., 2021).
It was observed that the currently known mutations associated with SARS-CoV-2 variants mostly occur at 606–617, 702–704 and 1131–1133 of N-gene sequence (Table S2) (Gitman et al., 2021; Kevadiya et al., 2021b; Li et al., 2021). Fig. 1b summarizes known SARS-CoV-2 variants and the impact, if any, on the developed assay based on bioinformatic analysis. It was observed that our DNA probes uniquely targets regions of the N gene (ASO1 starting 421 and ending with 440; ASO2 from 443 to 462; ASO3 from 836 to 855 and ASO4 from 886 to 905) which is unaffected by all the currently known N-gene mutations associated with SARS-CoV-2 variants including Omicron, the most mutated form of SARS-CoV-2. We are, therefore, confident that our ASO-based probes will bind to the RNA of SARS-CoV-2 of any variant without any false-negative signals. In addition to this, the mutations in other regions of the genome, such as in the spike protein, membrane or envelope protein, have no impact on the binding of these probes with the SARS-CoV-2 RNA. Therefore, the reliability and reproducibility of the current probes increase based on its design of simultaneous targeting of two of the N-gene regions of SARS-CoV-2, ensuring the feasibility of using these probes even if one region of the viral gene undergoes mutation during its current spread (Fig. 1).
2.4. Probing the mechanism of ASO-RNA hybridization by surface-enhanced Raman spectroscopy
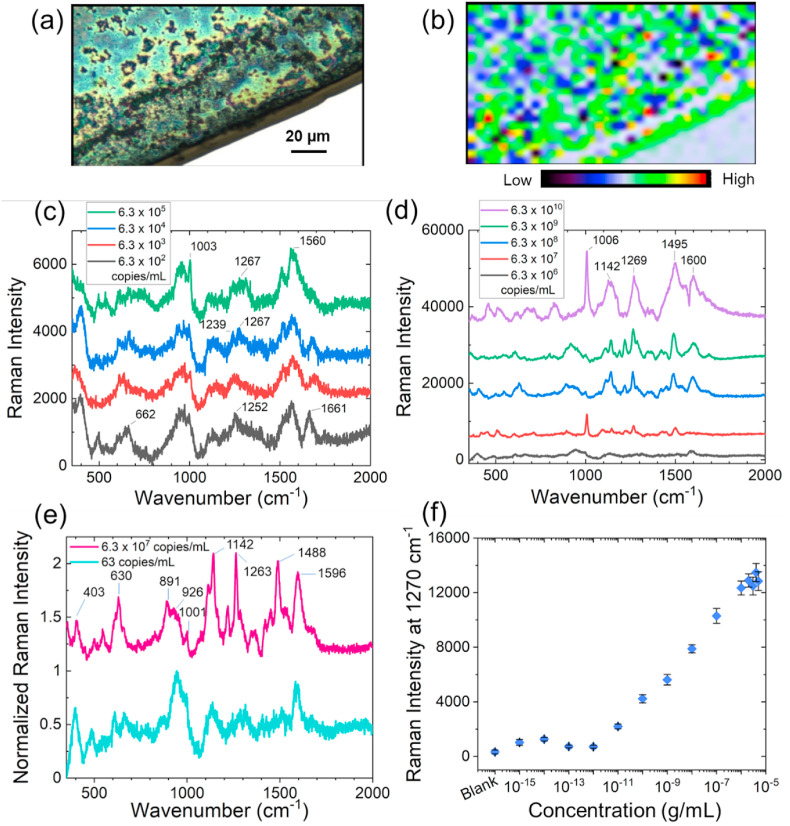
It was understood that the prime reason behind this extraordinary specificity of the probes irrespective of the known SARS-CoV-2 mutations was the befitting interaction of the N-ASOs with their target SARS-CoV-2 RNA samples. We, therefore, became interested to study the fundamental basis of interaction of these N gene-targeted Au-ASO NPs (N-ASO1+2 AuNPs) and SARS-CoV-2 RNA by Raman scattering utilizing the intrinsic surface plasmon resonance of Au-ASO NPs as Raman reporters (Abid Hasan et al., 2019; Chang et al., 2015; Xu et al., 2012). It was envisaged that N-ASO1+2 AuNPs, when targeting particular regions in the gene locations of SARS-CoV-2, will start to agglomerate because of their proximity in the gene sequences (Alafeef et al., 2020; Moitra et al., 2020). This will cause a change in surface plasmon resonance and introduce a change in the SERS response of the AuNPs (Gitman et al., 2021; Kevadiya et al., 2021b; Shrivastav et al., 2021). The N-ASO1+2 capped AuNPs were synthesized and analytically characterized by UV-Visible spectroscopy (Fig. S2) and transmission electron microscopy (Fig. S3a). The nanoparticles were then admixed with SARS-CoV-2 RNA and the corresponding bright field and Raman microscopic images for N-ASO1+2 capped AuNPs are shown in Fig. 3 a and b. The nanogaps between AuNPs are not controllable and hence it is expected to have considerable hot spot variations with intensity fluctuations across the slide. To mitigate this effect, we imaged the slides over a large surface area (120*80 μm2). The size of the aggregated AuNP was found to be ∼170 nm. Hence, the surface area covered by a spherical aggregated nanoparticle would be ∼0.36 μm2. Therefore, the imaging area would have ∼26,666 number of aggregated particles considering their uniform distribution. For each of the sample, we collected at least 1500 individual Raman spectra for the nanoparticles mixed with SARS-CoV-2 RNA. The presented Raman spectra (Fig. 3c) is therefore an average of all the acquired Raman spectra. To clarify, we have now represented the spectrum collected from different spots of the Raman map (Fig. S4).
Fig. 3.
(a) Bright field image of N-ASO1+2 capped AuNPs admixed with SARS-CoV-2 RNA; (b) Raman microscopic image of SARS-CoV-2 RNA associated N-ASO1+2 capped AuNPs in the range of 1495–1602 cm-1 (785 nm laser, 100% power, grating of 1200, 50 XL magnification) with the center of 1200 cm-1. Red means higher concentration and blue indicates lower concentration of sample; SERS spectra of N-ASO1+2 capped AuNPs with (c) lower, and (d) higher SARS-CoV-2 RNA concentrations; (e) Comparison of spectral figures at low and high SARS-CoV-2 RNA concentrations from the normalized Raman spectra of N-ASO1+2 AuNPs. The spectral positions and shapes are found to be similar at low and high concentrations; (f) Limit of detection of N-ASO1+2 capped AuNPs towards the detection of SARS-CoV-2 RNA. Here, 1 fg/mL corresponds to 63 copies/mL of SARS-CoV-2 RNA.
The representative Raman spectra of N-ASO1+2 capped AuNPs at varying concentrations of SARS-CoV-2 RNA ranging from 6.3 × 102 copies/mL to 6.3 × 105 copies/mL (Fig. 3c) and from 6.3 × 106 to 6.3 × 1010 copies/mL (Fig. 3d) are shown where the SERS response of the nanoparticles increased with increase in RNA concentration. The spectral data are consecutively stacked and independently labeled for better understanding. This confirmed the enhanced agglomeration among the N-ASO1+2 capped AuNPs in presence of increasing concentration of SARS-CoV-2 RNA. It was observed that the Raman peaks specific for oligonucleotide backbone, ribose and deoxyribose sugars were visible both at low viral RNA concentration, i.e., 63 copies/mL, and at high viral RNA concentration, i.e., 6.3 × 107 copies/mL (Fig. 3e) indicating efficient hybridization among the ASOs and their target RNA strands (Abid Hasan et al., 2019; Gartia et al., 2010). To better discern the SERS spectral features of N-ASO1+2 AuNPs with low concentrations of SARS-CoV-2 RNA (63 copies/mL), the spectrum was first denoised using Savitzky-Golay method and a higher magnification of the figure was provided (Fig. S5). Raman peaks were accordingly assigned for both concentrations and enlisted in Tables S3 and S4.
With increase in SARS-CoV-2 RNA concentration, there will be increase in agglomeration among N-ASO1+2 AuNPs leading to change in surface plasmon resonance of the gold nanoparticles (Alafeef et al., 2020, 2021a, 2021b, 2020; Dighe et al., 2021; Moitra et al., 2020, 2021b). Further, when the AuNP-RNA conjugate of different concentrations was added onto the surface, the molecule will take certain orientation at the nanostructure surface. It is known that the SERS effect is highly localized, i.e., the electrical field normal to the surface, and therefore, only a signal from part of the molecule, close to the substrate, will be enhanced. Thus, when the molecule is adsorbed on the surface, its symmetry will change, and so do the selection rules which will be followed by the selective enhancement of certain vibrational modes (Pérez-Jiménez et al., 2020). This describes the corresponding symmetry properties of the modified Raman dipole and changes in the relative intensities of the Raman peaks caused by the EM field polarization at the metal surface. Also, because the electromagnetic field enhancement is wavelength dependent, the different spectral regions of the spectrum may be enhanced differently (Kim et al., 2019). Thus. the spatial variation of the SERS intensities can be explained by several processes: (i) differences in local absorption of light by the AuNP-ASO due to the differences in the local concentration of particles; (ii) a difference in local concentration of SARS-CoV-2 RNA molecules, and (iii) local variations of the Raman enhancement effect due to specific microenvironments (orientation of the molecule) (Zeisel et al., 1998). Hence, we propose that there will be change in SERS spectral signature with increase in SARS-CoV-2 RNA concentration. Further, for our analysis, we rely on the area under the peaks over a wavenumber range, and hence small shift in the peak position will not affect the conclusion.
We have further acquired the SERS spectra for SARS-CoV-2 RNA alone without the addition of N-ASO1+2 AuNPs (Fig. S6). This spectrum showed a 1270 cm-1 Raman peak which may be assigned to the in-plane bending of C8–H, N9–H, str N7–C8 of adenine and stretching mode of N3–C4, C4–C5, C6–N1, bending mode of N1–H, C5/6–H of uracil (Madzharova et al., 2016). Hence, we used area under the Raman peak at 1270 cm-1 to calculate the limit of detection. The limit of detection for N-ASO1+2 capped AuNPs towards SARS-CoV-2 RNA was also evaluated and found to be 63 copies/mL which is quite significant and highly comparable to the currently available RT-PCR techniques (Fig. 3f).
2.5. Understanding the degree of AuNP aggregation in presence of SARS-CoV-2 RNA
In presence of SARS-CoV-2 RNA, agglomeration among N-ASO1+2 AuNPs were observed through transmission electron microscopy (Figs. S3a and b). It was observed that the absorbance of AuNPs at 526 nm in absence of the target RNA shifted towards 704 nm in presence of the target SARS-CoV-2 RNA (Fig. S7a). Different sizes of AuNPs with water solvation was then simulated and their corresponding surface plasmon resonance (SPR) band was predicted. It was found that the absorbance peak at 526 nm matched with the AuNP size of 40 nm, while the SPR band at 704 nm corresponded with AuNP size of 170 nm (Fig. S7b). The effect of this difference in AuNP size was then evaluated for change in electric field and extinction properties. According to Mie theory, as the AuNP-size changes, the electromagnetic field of particles will change, and the multipole effect can be observed. It was observed that under 785 nm laser excitation, the electric field intensity varied with increase in sample size for a single spherical AuNP (Cheng et al., 2020). Hextapolar and quadrupolar electromagnetic field was found for the AuNPs with 170 nm and 40 nm respectively when the incident laser beam was propagated along z-direction and focused at the origin (Figs. S8a and b). It can be proposed that increase in size of AuNP, i.e., in presence of SARS-CoV-2 RNA, made it easier to couple with the incident light (laser excitation of 785 nm). Because of this red shift towards higher wavelength, a multipolar distribution has been observed for higher size of AuNPs (Fig. S8b). Next, false color maps, simulated using Mieplot, showing the scattered intensity as a function of scattering angle and the wavelength of light has been shown in Figs. S8c and d for AuNP size of 40 and 170 nm respectively (Laven, 2003). It can be seen that the plots are quite symmetric for smaller size of AuNPs (in absence of SARS-CoV-2 RNA), while they lost their symmetry with increase in size (in presence of SARS-CoV-2 RNA).
2.6. Studying the interaction of direct clinical samples with N-ASO capped AuNPs
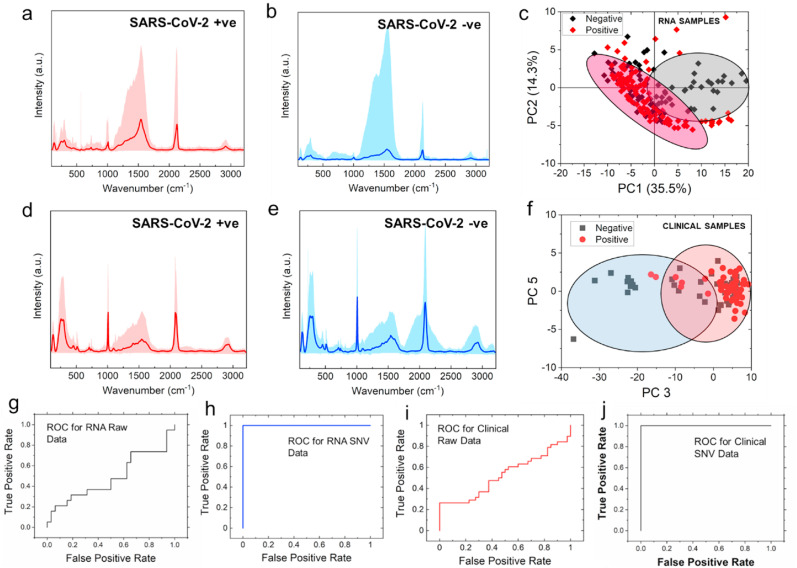
The high sensitivity of N-ASO1+2 capped AuNPs towards the binding of SARS-CoV-2 RNA grew our confidence to study the interaction in clinical nasal swab samples either as COVID-19 positive or negative. We presumed that because of this strong and efficient hybridization among the ASOs and RNA, the binding interaction might be evident even from the direct clinical samples without the extraction and purification of SARS-CoV-2 RNA (Alafeef et al., 2021b; Dighe et al., 2021). Accordingly, samples were treated with lysis buffer containing guanidine isothiocyanate and mixed with N-ASO1+2 capped AuNPs to record their Raman spectra. While for the nanoparticles mixed with SARS-CoV-2 RNA, 1500 individual Raman spectra were acquired, for direct clinical samples, 2500 Raman spectra were acquired for each of the samples. The respective Raman spectra for N-ASO1+2 capped AuNPs added with RNA extracted from SARS-CoV-2 positive, P1 (Fig. 4 a) and negative, N1 (Fig. 4b) clinical samples were shown. The respective Raman spectra of N-ASO1+2 capped AuNPs admixed with direct SARS-CoV-2 positive (Fig. 4d) and negative (Fig. 4e) clinical samples were also shown without the extraction of RNA but with just the addition of lysis buffer. The range of all the acquired spectra are shown as shaded color for each sample while the dark color is representing the mean spectra for each of the sample. The Raman peaks were assigned for the spectra in Fig. 4a and b and enlisted in Table S5. Raman peaks were also assigned for Fig. 4d and e and noted in Table S6.
Fig. 4.
Representative Raman spectra of N-ASO1+2 capped AuNPs added with RNA extracted from SARS-CoV-2 (a) positive, P1 and (b) negative, N1 clinical samples; The respective representative Raman spectra of N-ASO1+2 capped AuNPs admixed with direct SARS-CoV-2 (d) positive and (e) negative clinical samples are also shown without the extraction of RNA but with just the addition of lysis buffer. The range of all the spectra is shown as shaded color for each sample while the dark color is representing the mean spectra. PCA score plots show the separation between negative and positive samples of (c) extracted RNA samples and (f) direct clinical samples; ROC curves for (g) raw data and (h) after performing SNV operation on the SERS data obtained with extracted RNA samples; ROC curves for (i) raw data and (j) after performing SNV operation on the SERS data obtained while analyzing the direct clinical samples without any extraction of RNA from N-ASO1+2 capped AuNPs.
Now, in order to extract useful information from the Raman spectra, multivariate statistical analysis method is generally followed. Principal component analysis (PCA) is the widely applied conventional multivariate statistical analysis technique which by orthogonal transformation converts high dimensional Raman spectra into multiple unrelated independent variables, called principal components (PCs), avoiding any loss of valuable information (Crow et al., 2005; Gautam et al., 2015). Here, most of the important information are distributed over the first few PCs and the contributions of the rest of PCs are negligible. Fig. S9 shows the contribution of eigenvalues of each PCs to the total variance of all SERS spectra for clinical samples. It was observed that the eigenvalues reduced rapidly with increasing PC numbers and only the first few PCs hold the maximum variance of the obtained Raman data. It can be seen that the first five principal components described more than 99% of the variation of the corresponding dataset.
Based on the obtained dataset, we performed the PCA and PCA score plot showing the separation between negative and positive RNA samples (Fig. 4c) and direct clinical samples (Fig. 4f). From these figures, it can be noticed that due to possible interference from sample matrix (background signal), PCA was not able to accurately distinguish negative samples from the positive ones. Though, there were some clusters of points on two sides, but a good amount of overlap was also present. Here, each point is representing the Raman spectra of each sample. However, we calculated several numbers of principal components (PCs) for these analyses where we found that PC1 shifted to negative values for SARS-CoV-2 positive RNA samples. On the other hand, PC3 shifted to positive values for SARS-CoV-2 positive clinical samples (Fig. 4c, f). Figs. S10 and S11 show the loading plots for PCA of clinical samples for PC3 and PC1 respectively. Here, we highlighted the characteristic peaks of our dataset which indicated the differences in the various samples. It was noticeable that in both of these figures the location of the characteristic peaks was very similar. This indicated the variation of all the datasets was not significant enough and thus it led to the overlap in the score plot as observed in Fig. 4c and f. Fig. S12 shows the box plot for the distribution of spectral data for positive and negative groups of clinical samples. The spectral data of the negative samples from the dataset significantly (P < 0.001) shifted towards the negative PC3 range compared to positive samples. Although, PC1 and PC3 could be used to distinguish positive and negative RNA as well as clinical samples, respectively, we further explored the diagnostic capability of these binary classification (positive vs negative) systems. The score plot shown in Fig. 4c and f pointed to a less accurate classification system using the PCA scores alone.
The unsupervised PCA method has thus been used to demonstrate the outlier from the Raman dataset and extract the most useful information to be followed by the supervised support vector machine (SVM) technique. SVM is another multivariate statistical analysis method which can efficiently classify small samples, irrespective of the distributive nature of the samples (Li et al., 2013). With a combination of PCA and SVM methods, we decreased the dimensionality of Raman spectrum in a significant extent which helped us in rapid processing of a large amount of multivariate data (Villa-Manríquez et al., 2017).
2.7. Analyzing the Raman data by machine learning techniques to differentiate among the clinical samples
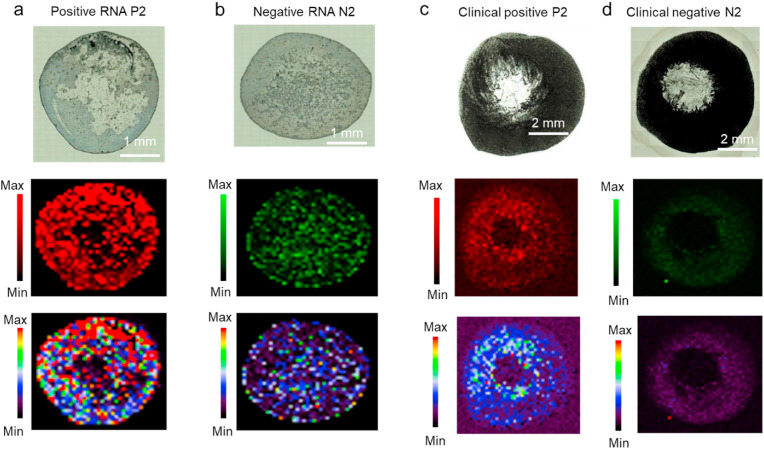
Here, we realized that only the strong binding affinity among the N-ASO1+2 AuNPs with their target RNA sequences obtained through SERS was not enough in selectively differentiating SARS-CoV-2 positive samples from the negative ones with high confidence value (Carlomagno et al., 2021; Huang et al., 2021; Sanchez et al., 2021; Yin et al., 2021). Hence, we used the Support Vector Machine (SVM) algorithm to improve the predictive performance of our assay. At first, we developed a model based on the training dataset and based on that model we predicted the rest of our dataset. Figs. 4g and 4i are showing the receiver operating characteristic (ROC) curve of our developed model based on the raw data for RNA and clinical samples respectively. However, when we applied the standard normal variate (SNV) algorithm to perform preprocessing of our dataset followed by training the model, we have observed significant improvement of our model. The performance of the algorithm can be measured based on the specificity and sensitivity of the results as tabulated in Table S7. Overall, by applying the SNV operation, we have successfully increased the sensitivity and specificity of the probes for the clinical samples, tested directly without the extraction of RNA, from 30% to ∼100% and 50%–90%, respectively. On the other hand, for isolated RNA samples, the specificity was increased from 58% to 95% but, the sensitivity did not improve (went from 67% to 65%). The respective ROC curves for the RNA and direct clinical samples fitted after SNV operation are shown in Figs. 4h and 4j respectively. Thus, we have successfully discriminated SARS-CoV-2 positive and negative samples directly from the clinical nasal swabs with the addition of lysis buffer and N-ASO1+2 AuNPs through a combinatorial effort from SERS and ML-based techniques. Representative Raman microscopic images of N-ASO1+2 capped AuNPs admixed with SARS-CoV-2 positive and negative RNA (Fig. 5 a and b) and direct clinical samples (Fig. 5c and d) has accordingly been acquired. Here, the Raman map for RNA samples had 1500 pixels with 1500 individual Raman spectra, whereas for direct clinical samples, the Raman map had 2500 pixels with 2500 individual Raman spectra.
Fig. 5.
Representative Raman microscopic images of N-ASO1+2 capped AuNPs admixed with SARS-CoV-2 positive and negative (a, b) extracted RNA and (c, d) direct clinical samples at 928 cm-1 using StreamHR acquisition mode and obtained with 785 nm laser, 100% power, grating of 1200, magnification of 50 XL and exposure time of 0.5 s. The pixel size for RNA and clinical samples were 70 μm × 70 μm and 200 μm × 200 μm, respectively. Distribution for whole Raman spectrum is from 200 to 3200 cm-1. For RNA samples, map has around 1500 pixels (that is 1500 Raman spectra) and for clinical samples, map has around 2500 pixels (that is 2500 Raman spectra).
2.8. Hypothesizing the interaction of developed ASOs with imaginary future N-gene mutations
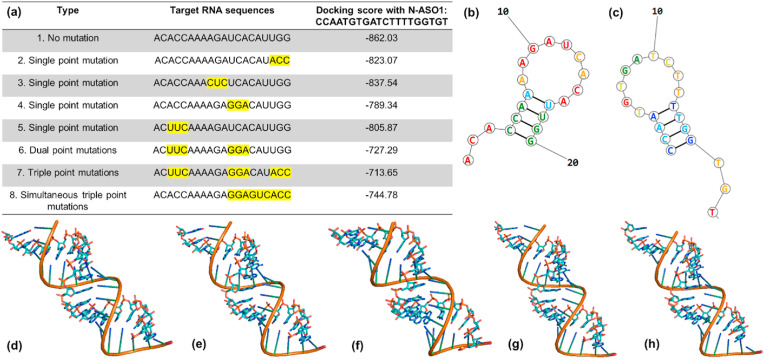
Thus, we have developed herein a combination of SERS and ML-based methods, which can be utilized to study DNA probes with SARS-CoV-2 wild-type genes and current mutated forms. Although the present mutations on the SARS-CoV-2 N gene do not hamper the target RNA sites for the developed ASOs, we were interested to see the effect of the mutation at the target RNA site on the binding efficiency of ASOs. Accordingly, we introduced several hypothetical mutations at the target RNA site for N-ASO1 and observed their effect on binding affinity. Considering the fact that single point mutation in a protein sequence will change three consecutive nucleotides, we modified the original target RNA sequence at four different locations with a change in three nucleotides in each case to represent single mutated forms. In a similar way, dual point and triple point mutations were also introduced at the target RNA site. N-ASO1 was then docked with these RNA sequences in HNADOCK server (He et al., 2019), and their respective docking scores were mentioned in Fig. 6 a. The docked geometries of N-ASO1 with the original non-mutated target RNA sequence and their mutated forms were shown in Fig. 6. Close monitoring of the ASO-RNA hybridization profiles revealed stable interaction between the two, even after a hypothetical single point mutation was introduced at the target RNA site. We observed a drop in docking score and hence in binding affinity when a dual point or triple point mutations were introduced at the target RNA sites.
Fig. 6.
(a) List of hypothetical single, double, and triple point mutations on the RNA site targeted by N-ASO1. The hypothetical mutation sites are highlighted with yellow background; (b) Theoretical model of RNA with no mutation; (c) Theoretical model of N-ASO1; Docked model of N-ASO1 with RNA having (d) no mutation sites; (e) single point mutation as shown in no. 4; (f) double point mutations as shown in no. 6; (g) triple point mutations as shown in no. 7 and (h) simultaneous triple point mutations as shown in no. 8.
Thus, this study examined the efficiency of a DNA-based universal probe and its fundamental interaction with the entire known mutated forms of SARS-CoV-2 without any false negative signals through a unique combination of SERS and ML-based approaches. This study also demonstrated that the developed assay will still be effective in case of a future single point mutation at the target site and would only have reduced activity in case of dual or triple point mutations.
3. Conclusions
Briefly, we have developed herein a unique combination of Surface Enhanced Raman Spectroscopy (SERS) and machine learning (ML) based approaches for studying selective and sensitive interaction with SARS-CoV-2 having any of its current mutations including the recently known Omicron. Several antisense oligonucleotides (ASOs) were developed against N, E and RdRp gene segments of SARS-CoV-2 where N gene-targeted ones were found to be most effective. The thiolated N-ASO1+2 capped AuNPs agglomerated in presence of its target RNA sequence and demonstrated a change in its surface plasmon resonance (SPR). The agglomeration of Au-ASO NPs also tuned the size of the generated nanoparticles which was followed by the change in their SERS response. The SERS signal was then evaluated with ML techniques to selectively distinguish among COVID-19 positive and negative samples with high accuracy, sensitivity and specificity. The limit of detection of SARS-CoV-2 RNA was found to be 63 copies/mL. The developed quantitative assay was also validated with a range of clinical samples having varying Ct numbers. Thus, this study establishes N gene-targeted ASOs as the critical part of the assay to efficiently detect all the current SARS-CoV-2 variants regardless of their mutations. Further probing into the hybridization profile of the ASOs revealed that the ASO-RNA interaction remained unaltered for any hypothetical single point mutations at the original target RNA site and diminished only in case of the hypothetical double or triple point mutation cases. This approach could therefore have an immediate impact on understanding different mutations associated with SARS-CoV-2, especially during the spread of its current variant, Omicron. To the best of our knowledge, there is no literature report for an effective DNA probe that can selectively hybridize with all the different genomic variants of SARS-CoV-2 with similar sensitivity and specificity without any false negative signals which is one of the most concerning limitations of the current COVID-19 diagnosis. However, this is a proof-of-concept study before the developed technology can be taken over for the pre-clinical study with larger cohort of samples.
4. Materials and methods
All the chemicals were procured from reputable commercial vendors and used without any further purification steps. The custom designed and thiol-modified ASOs were procured from Sigma Aldrich and stored at −20 °C until further use. All the experiments were carried out at constant room temperature of 25 °C unless otherwise specified.
4.1. Design of antisense oligonucleotides (ASOs)
Gene sequences (RdRp: 13,442–16,236; E: 26,245–26,472 and N: 28,274–29,533) from the whole genome sequence of SARS-CoV-2 (wild type isolate SARS-CoV-2/human/USA/WA-CDC-WA1-A12/2020, MT020880) was chosen and multiple ASO sequences of 20 nucleotides in length were selected from Soligo software output (Ding et al., 2004). The choice of ASOs were primarily based on the optimum GC content and theoretically calculated target binding and disruption energies at 37 °C in 1 M NaCl aqueous solution. We propose to select four ASOs, two for the front (ASO1 and ASO2), and two targeting the end region (ASO3 and ASO4) of the gene. The ASOs will also target closely following sequences at each location. Accordingly, four ASOs were selected both for N and E gene, whereas only two were selected for RdRp gene (Table 1). We were not able to select more than two ASOs for RdRp gene target as ASOs with high enough binding energies are not available for this target.
4.2. Synthesis of gold nanoparticles (AuNPs)
The gold nanoparticles were prepared from a solution of chloroauric acid and sodium citrate following a previously published literature protocol (Kumar et al., 2020; Moitra et al., 2020; Schwartz-Duval et al., 2020). Briefly, 150 mL solution of 2.2 mM sodium citrate was refluxed for 15 min under continuous stirring. 1 mL solution of 25 mM chloroauric acid was then added the boiling solution of sodium citrate. This mixture was kept under reflux for an additional 15 min over which the color of the solution changed from yellow to wine red. The resultant mixture was centrifuged at 25,000 rpm for 10 min, precipitate collected and washed with milli-Q water for at least two times to remove any reaction contaminations. Finally, the precipitate containing citrate stabilized gold nanoparticles were collected and resuspended in water for future use.
4.3. Conjugation of ASOs to AuNPs
The thiolated ASOs, at a concentration of 1 μM, were added to a solution of citrate stabilized AuNPs having ∼3 × 1010 particles/mL concentration. The mixture was stirred at room temperature for ∼1 h, centrifuged to remove any excess of uncapped ASO from the supernatant, and the pellet was then resuspended in a similar volume of milli-Q water. The nanoparticles were kept at 4 °C for future use.
4.4. UV-visible absorbance spectroscopy
The absorbance spectra were acquired on a Biotek Synergy Neo2Microplate Reader.
4.5. Dynamic light scattering
The hydrodynamic diameters of the Au-ASO NPs in presence and absence of their target RNA were monitored on a Malvern Nano S system.
4.6. Transmission electron microscopy (TEM)
The N-ASO1+2 AuNPs were investigated under the transmission electron microscope (FEI tecnai T12). The tungsten filament was used as the electron optics, and the voltage was kept constant at 80 kV. A 20 μL sample droplet was spotted onto a carbon-coated copper grid (400 mesh) and allowed to stay there for about 10 min before being removed.
4.7. Sample preparation for Raman analyses
RNA from the clinical samples were extracted and purified following standard commercial technique. The RNAs were stored in multiple aliquots at -80 °C for future use. This was done to prevent denaturation of the stock RNA samples from excessive freeze thaw.
2 μL of Au-ASO NPs were then mixed with 2 μL of SARS-CoV-2 RNA by gentle pipetting a few times on an ice rack. 2 μL of this mixture were dropped on a clean stainless-steel slide and left to dry at room temperature. Raman spectra of the dried spots were recorded immediately after using a Renishaw inVia Reflex Raman Spectroscope.
For the direct analyses of clinical samples without the extraction of RNA, the samples were added with lysis buffer having guanidine isothiocyanate with a sample: lysis buffer at 2:1 M ratio. 2 μL of these samples were then added with 2 μL of Au-ASO NPs and mixed properly as discussed earlier for RNA sample before recording Raman spectra.
4.8. Raman spectroscopy
The stock concentration of N-ASO1+2 AuNPs was 1012 nanoparticles/mL as measured by Zetaview. This stock solution was diluted five times before being added with different concentrations of RNA samples. Raman experiments were performed on the samples with 785 (grating of 1200) nm laser (laser spot ∼ 1.1 μm; penetration depth ∼ a few microns depending on the samples) with 10% of power and 10 s exposure time on mirror-like surface stainless steel slides. Higher magnification with 50× long working distance dry objective was used for all the experiments with free working distance of 8.2 and numerical aperture of 0.5. At least ten spectra were acquired from each sample for statistical analysis in the range of 100–3200 cm-1. Raman images were acquired in StreamHR image acquisition mode by utilizing 785 nm laser (100% power, grating of 1200, 50 XL magnification) with the center of 1200 cm-1 (609-1728 cm-1) by step size (resolution) of 200 × 200 μm for clinical samples and 70 × 70 μm for RNA samples. During the Raman imaging, exposure time was set to be 0.5 s. Renishaw WiRE 4.4 was used for date processing and analysis of Raman signals and images. For baseline correction, we used intelligent fitting of WiRE 4.4 with polynomial order of 11 and noise tolerance of 1.5. Subsequent data processing was also performed by OriginLab 2018.
4.9. Calculation of limit of detection
Further to calculate the limit of detection from the Raman calibration curve, N-ASO1+2 AuNPs (concentration of 2*1011 particles/mL) were added with SARS-CoV-2 RNA samples having the concentration in the range from 1 fg/mL (63 copies/mL) to 1 μg/mL (63 × 109 copies/mL). Five spectra were collected for each of the samples and the average spectra were reported. Limit of detection (LoD) and the limit of blank (LoB) was then calculated from these spectra. LoB was estimated by measuring replicates of a blank sample and calculating the mean and standard deviation (SD). For each concentration, as mentioned before, five data points were collected. The LoB was calculated as follows (Armbruster and Pry, 2008):
| LoB = mean blank + 1.645*(SD blank) |
Assuming a Gaussian distribution of the raw analytical signals from blank samples, the LoB represents 95% of the observed values. Thus, with mean intensity of 325 from blank sample (no molecule present) and SD of 142, the LoB was calculated to be 558.6.
On the other hand, LoD is the lowest analyte concentration likely to be reliably distinguished from the LoB and at which detection is feasible. It is therefore greater than LoB. It is generally calculated using the following equation (Armbruster and Pry, 2008):
| LoD = LoB + 1.645*(SD low concentration sample) |
The lowest concentration used was 1 fg/mL and the SD of the 1 fg/mL sample was found to be 212. Hence, the sample at LoD must have intensity > or = 907.3. From our experiment, we found that the lowest concentration whose corresponding Raman intensity >907.3, was 1 fg/mL (or 63 copies/mL). Hence, the LoD of the current sensor was noted as 63 copies/mL.
4.10. FDTD simulation and Mie plot calculation
The extinction spectra were simulated using nanoHUB (Juluri et al., 2016). The electromagnetic field were calculated using nanoDDSCAT program in nanoHUB (Jain et al., 2019). The dispersion curves (wavelength vs Scattering angle) were generated using MiePlot program 3.4 (http://www.philiplaven.com/mieplot.htm).
4.11. Statistical analyses (Principal Component Analysis)
After gathering the Raman spectra from the samples, it was corrected by Renishaw's WiRE 4.4 (Windows-based Raman Environment) software. Baseline subtraction was performed for all the data by using intelligent fitting of Wire 4.4 and a polynomial value of 11 was set as default for the fitting. For the further analysis of spectral data we performed Principal Component Analysis (PCA) as we have found it very useful for Raman spectroscopy analysis previously. This multivariate analysis was completed with the “Principal Component Analysis for Spectroscopy” toolbox of Origin Lab 2020 (Origin Lab, Northampton, MA)). The spectral differences were distinguished with Principal components (PC). The PCs were calculated based on the covariance matrix and 8 components were extracted. These parameters were set before performing the analysis and based on the variation of the dataset we received several PCs. Though, mean centering is a common step for calculating principal components, but we didn't have to perform it separately as while computing the covariance matrix the software performed mean centering implicitly. To visualize the results, we plotted score plot where each point is representing a Raman spectrum separately while selecting two or more PCs. The clustering of each Raman spectra from the score plot shows the separation of the data. Here, we used 95% confidence level to show the clustering of the Raman spectra. In order to find the vibrational fingerprint, we plotted the loading plot calculated from PCA. From the loading plot, we also found the peak locations which dominated the PCA. Besides performing these analyses on raw data, we also used min-max normalization through Origin where the normalization was performed in [0, 1] range. Later, a smoothing operation was also performed to reduce the noise of the dataset with the help of Savitzky-Golay method. During this analysis, window size was selected as 5 and polynomial order was set to 2. Based on these posts processed dataset, PCA was performed in a similar fashion.
4.12. Machine learning analysis of Raman data (SVM analysis)
Besides performing PCA, we also developed an algorithm based on Support vector machine (SVM) analysis. This supervised analysis helps to classify groups. Here based on the Raman spectra we tried to separate our dataset between positive and negative group of samples. Support vector machine classification model by MATLAB was used in our method. Here at first, we trained a model and then predict rest of our dataset. Based on the classification results we calculated the specificity and sensitivity of our approach. Later, we used similar approach on a selected range of dataset. Here, the selected range of the wavelength was 400–1800 nm. The reasoning for selecting this range was, we noticed most of the peaks of the dataset located within this range (Raman fingerprint region) and thus contribution for separation was much higher. Besides this selected range, we performed the Standard Normal Variate (SNV) analysis to improve the results we gathered from previous stage. To get the SNV results we calculated mean and standard deviation of each of the dataset. The mean value was subtracted from the raw spectra and then it was divided by the standard deviation of that particular dataset. These operations were performed through MATLAB and each dataset was considered as a matrix while performing the calculation.
4.13. Docking analyses
The single stranded models for N-ASO1 and RNA strands (non-mutated and mutated ones) were made in Web 3DNA 2.0 software (Li et al., 2019) and then docked on a HNADOCK server (He et al., 2019) to obtain the docking score and conformationally stable models.
CRediT authorship contribution statement
Parikshit Moitra: Conceptualization, Methodology, Investigation, Writing – original draft, revision and editing. Ardalan Chaichi: Investigation. Syed Mohammad Abid Hasan: Investigation. Ketan Dighe: Investigation, Writing – original draft. Maha Alafeef: Investigation. Alisha Prasad: Investigation. Manas Ranjan Gartia: Supervision, Methodology, Conceptualization, Writing – review & editing. Dipanjan Pan: Supervision, Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
D.P. is the founder or co-founder for four University based start ups. None of these entities, however, supported this work.
Acknowledgements
The authors gratefully acknowledge the receipt of funding from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) R03EB028026, R03 EB028026-02S2, and R03 EB028026-02S1, the University of Maryland, Baltimore (UMB), and University of Maryland, Baltimore County (UMBC). MRG is supported by National Science Foundation (NSF CAREER award number: 2045640).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114200.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abid Hasan S.M., He Y., Chang T.-W., Wang J., Gartia M.R. Detecting DNA methylation using surface-enhanced Raman spectroscopy. J. Phys. Chem. C. 2019;123:698–709. doi: 10.1021/acs.jpcc.8b10178. [DOI] [Google Scholar]
- Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Moitra P., Dighe K., Pan D. Hyperspectral mapping for the detection of SARS-CoV-2 using nanomolecular probes with Yoctomole sensitivity. ACS Nano. 2021;15:13742–13758. doi: 10.1021/acsnano.1c05226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alafeef M., Moitra P., Dighe K., Pan D. RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19. Nat. Protoc. 2021;16:3141–3162. doi: 10.1038/s41596-021-00546-w. [DOI] [PubMed] [Google Scholar]
- Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008;29(Suppl. 1):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- Barreto H.G., de Pádua Milagres F.A., de Araújo G.C., Daúde M.M., Benedito V.A. Diagnosing the novel SARS-CoV-2 by quantitative RT-PCR: variations and opportunities. J. Mol. Med. 2020;98:1727–1736. doi: 10.1007/s00109-020-01992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.I., MacGowan S.A., Kutuzov M.A., Dushek O., Barton G.J., van der Merwe P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10 doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600:197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- Carlomagno C., Bertazioli D., Gualerzi A., Picciolini S., Banfi P.I., Lax A., Messina E., Navarro J., Bianchi L., Caronni A., Marenco F., Monteleone S., Arienti C., Bedoni M. COVID-19 salivary Raman fingerprint: innovative approach for the detection of current and past SARS-CoV-2 infections. Sci. Rep. 2021;11:4943. doi: 10.1038/s41598-021-84565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.v., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.v., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichi A., Hasan S.M.A., Mehta N., Donnarumma F., Ebenezer P., Murray K.K., Francis J., Gartia M.R. Label-free lipidome study of paraventricular thalamic nucleus (PVT) of rat brain with post-traumatic stress injury by Raman imaging. Analyst. 2021;146:170–183. doi: 10.1039/D0AN01615B. [DOI] [PubMed] [Google Scholar]
- Chang T.-W., Wang X., Hsiao A., Xu Z., Lin G., Gartia M.R., Liu X., Liu G.L. Bifunctional nano Lycurgus Cup array plasmonic sensor for colorimetric sensing and surface-enhanced Raman spectroscopy. Adv. Opt. Mater. 2015;3:1397–1404. doi: 10.1002/adom.201500092. [DOI] [Google Scholar]
- Cheng L., Zhu G., Liu G., Zhu L. FDTD simulation of the optical properties for gold nanoparticles. Mater. Res. Express. 2020;7 doi: 10.1088/2053-1591/abd139. [DOI] [Google Scholar]
- Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., Coppola A., Chiu J., Ni M., Wei Y., Atwal G.S., Hernandez A.R., Saotome K., Zhou Y., Franklin M.C., Hooper A.T., McCarthy S., Hamon S., Hamilton J.D., Staples H.M., Alfson K., Carrion R., Ali S., Norton T., Somersan-Karakaya S., Sivapalasingam S., Herman G.A., Weinreich D.M., Lipsich L., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184:3949–3961. doi: 10.1016/j.cell.2021.06.002. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow P., Barrass B., Kendall C., Hart-Prieto M., Wright M., Persad R., Stone N. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br. J. Cancer. 2005;92:2166–2170. doi: 10.1038/sj.bjc.6602638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daria S., Bhuiyan M.A., Islam MdR. Detection of highly muted coronavirus variant Omicron (B.1.1.529) is triggering the alarm for South Asian countries: associated risk factors and preventive actions. J. Med. Virol. 2021 doi: 10.1002/jmv.27503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe K., Moitra P., Alafeef M., Gunaseelan N., Pan D. A rapid RNA extraction-free lateral flow assay for molecular point-of-care detection of SARS-CoV-2 augmented by chemical probes. Biosens. Bioelectron. 2021 doi: 10.1016/j.bios.2021.113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:135–141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartia M.R., Xu Z., Behymer E., Nguyen H., Britten J.A., Larson C., Miles R., Bora M., Chang A.S.-P., Bond T.C., Logan Liu G. Rigorous surface enhanced Raman spectral characterization of large-area high-uniformity silver-coated tapered silica nanopillar arrays. Nanotechnology. 2010;21 doi: 10.1088/0957-4484/21/39/395701. [DOI] [PubMed] [Google Scholar]
- Gautam R., Vanga S., Ariese F., Umapathy S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Techn. Instrument. 2015;2:8. doi: 10.1140/epjti/s40485-015-0018-6. [DOI] [Google Scholar]
- Gitman M.R., Shaban M.v., Paniz-Mondolfi A.E., Sordillo E.M. Laboratory diagnosis of SARS-CoV-2 Pneumonia. Diagnostics. 2021;11:1270. doi: 10.3390/diagnostics11071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Wang J., Tao H., Xiao Y., Huang S.-Y. HNADOCK: a nucleic acid docking server for modeling RNA/DNA–RNA/DNA 3D complex structures. Nucleic Acids Res. 2019;47:W35–W42. doi: 10.1093/nar/gkz412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wen J., Zhou M., Ni S., Le W., Chen G., Wei L., Zeng Y., Qi D., Pan M., Xu J., Wu Y., Li Z., Feng Y., Zhao Z., He Z., Li B., Zhao S., Zhang B., Xue P., He S., Fang K., Zhao Y., Du K. On-site detection of SARS-CoV-2 antigen by deep learning-based surface-enhanced Raman spectroscopy and its biochemical Foundations. Anal. Chem. 2021;93:9174–9182. doi: 10.1021/acs.analchem.1c01061. [DOI] [PubMed] [Google Scholar]
- Jain P.K., Nahil S., Smith J., Sobh A.N., White S., Faucheaux J., Feser J. 2019. nanoDDSCAT.https://nanohub.org/resources/dda [DOI] [Google Scholar]
- Janik E., Niemcewicz M., Podogrocki M., Majsterek I., Bijak M. The emerging concern and interest SARS-CoV-2 variants. Pathogens. 2021;10:633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juluri B.K., Huang J., Jensen L. 2016. Extinction, Scattering And Absorption Efficiencies Of Single And Multilayer Nanoparticles.https://nanohub.org/resources/nmie [DOI] [Google Scholar]
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:2642. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.S., Misra S.K., Dighe K., Wang Z., Schwartz-Duval A.S., Sar D., Pan D. Electrically-receptive and thermally-responsive paper-based sensor chip for rapid detection of bacterial cells. Biosens. Bioelectron. 2018;110:132–140. doi: 10.1016/j.bios.2018.03.044. [DOI] [PubMed] [Google Scholar]
- Kim J., Jang Y., Kim N.-J., Kim H., Yi G.-C., Shin Y., Kim M.H., Yoon S. Study of chemical enhancement mechanism in non-plasmonic surface enhanced Raman spectroscopy (SERS) Front. Chem. 2019;7 doi: 10.3389/fchem.2019.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Moitra P., Bashir M., Kondaiah P., Bhattacharya S. Natural tripeptide capped pH-sensitive gold nanoparticles for efficacious doxorubicin delivery both: in vitro and in vivo. Nanoscale. 2020;12:1067–1074. doi: 10.1039/c9nr08475d. [DOI] [PubMed] [Google Scholar]
- Laven P. Simulation of rainbows, coronas, and glories by use of Mie theory. Appl. Opt. 2003;42:436–444. doi: 10.1364/ao.42.000436. [DOI] [PubMed] [Google Scholar]
- Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- Li S., Olson W.K., Lu X.-J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019;47:W26–W34. doi: 10.1093/nar/gkz394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-X., Zeng Q.-Y., Li L.-F., Zhang Y.-J., Wan M.-M., Liu Z.-M., Xiong H.-L., Guo Z.-Y., Liu S.-H. Study of support vector machine and serum surface-enhanced Raman spectroscopy for noninvasive esophageal cancer detection. J. Biomed. Opt. 2013;18 doi: 10.1117/1.JBO.18.2.027008. [DOI] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Costa Clemens S.A., Gomes Naveca F., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., da Silva K., Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzharova F., Heiner Z., Gühlke M., Kneipp J. Surface-enhanced hyper-Raman spectra of adenine, guanine, Cytosine, Thymine, and uracil. J. Phys. Chem. C. 2016;120:15415–15423. doi: 10.1021/acs.jpcc.6b02753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S., Callaway E., Kozlov M., Ledford H., Pickrell J., van Noorden R. How COVID vaccines shaped 2021 in eight powerful charts. Nature. 2021 doi: 10.1038/d41586-021-03686-x. [DOI] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S.K., Dighe K., Schwartz-Duval A.S., Shang Z., Labriola L.T., Pan D. In situ plasmonic generation in functional ionic-gold-nanogel scaffold for rapid quantitative bio-sensing. Biosens. Bioelectron. 2018;120:77–84. doi: 10.1016/j.bios.2018.08.019. [DOI] [PubMed] [Google Scholar]
- Mohiuddin M., Kasahara K. Investigating the aggressiveness of the COVID-19 Omicron variant and suggestions for possible treatment options. Respir. Med. 2022;191 doi: 10.1016/j.rmed.2021.106716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Ray P., Chang J., Thole A., Punshon-Smith B., Tolosa M., Ramamurthy S.S., Ge X., Frey D.D., Pan D., Rao G. Rapid and low-cost sampling for detection of airborne SARS-CoV-2 in dehumidifier condensate. Biotechnol. Bioeng. 2021:3029–3036. doi: 10.1002/bit.27812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Sheffield Z., Dahal D., Pan D. Synthesis and characterisation of N-gene targeted NIR-II fluorescent probe for selective localisation of SARS-CoV-2. Chem. Commun. 2021;57:6229–6232. doi: 10.1039/D1CC01410B. [DOI] [PubMed] [Google Scholar]
- Ostadhossein F., Sar D., Tripathi I., Soares J., Remsen E.E., Pan D. Oligodots: structurally defined fluorescent nanoprobes for multiscale dual-color imaging in vitro and in vivo. ACS Appl. Mater. Interfaces. 2020;12:10183–10192. doi: 10.1021/acsami.0c00705. [DOI] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Pramanik M., Senpan A., Wickline S.A., Wang L.v., Lanza G.M. A facile synthesis of novel self-assembled gold nanorods designed for near-infrared imaging. J. Nanosci. Nanotechnol. 2010;10:8118–8123. doi: 10.1166/jnn.2010.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Pramanik M., Wickline S.A., Wang L.v., Lanza G.M. Recent advances in colloidal gold nanobeacons for molecular photoacoustic imaging. Contrast Media Mol. Imaging. 2011;6:378–388. doi: 10.1002/cmmi.449. [DOI] [PubMed] [Google Scholar]
- Pérez-Jiménez A.I., Lyu D., Lu Z., Liu G., Ren B. Surface-enhanced Raman spectroscopy: benefits, trade-offs and future developments. Chem. Sci. 2020;11:4563–4577. doi: 10.1039/D0SC00809E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- Prasad A., Chaichi A., Mahigir A., Sahu S.P., Ganta D., Veronis G., Gartia M.R. Ripple mediated surface enhanced Raman spectroscopy on graphene. Carbon. 2020;157:525–536. doi: 10.1016/j.carbon.2019.09.078. [DOI] [Google Scholar]
- Rump L. v, Asamoah B., Gonzalez-Escalona N. Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res. Notes. 2010;3:211. doi: 10.1186/1756-0500-3-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J.E., Jaramillo S.A., Settles E., Velazquez Salazar J.J., Lehr A., Gonzalez J., Rodríguez Aranda C., Navarro-Contreras H.R., Raniere M.O., Harvey M., Wagner D.M., Koppisch A., Kellar R., Keim P., Jose Yacaman M. Detection of SARS-CoV-2 and its S and N proteins using surface enhanced Raman spectroscopy. RSC Adv. 2021;11:25788–25794. doi: 10.1039/D1RA03481B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz-Duval A.S., Konopka C.J., Moitra P., Daza E.A., Srivastava I., Johnson E.v., Kampert T.L., Fayn S., Haran A., Dobrucki L.W., Pan D. Intratumoral generation of photothermal gold nanoparticles through a vectorized biomineralization of ionic gold. Nat. Commun. 2020;11:4530. doi: 10.1038/s41467-020-17595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav A.M., Cvelbar U., Abdulhalim I. A comprehensive review on plasmonic-based biosensors used in viral diagnostics. Commun. Biol. 2021;4:70. doi: 10.1038/s42003-020-01615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava I., Misra S.K., Bangru S., Boateng K.A., Soares J.A.N.T., Schwartz-Duval A.S., Kalsotra A., Pan D. Complementary oligonucleotide conjugated multicolor carbon dots for intracellular recognition of biological events. ACS Appl. Mater. Interfaces. 2020;12:16137–16149. doi: 10.1021/acsami.0c02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Arvin A., Corey L., Corti D., Diamond M.S., García-Sastre A., Garry R.F., Holmes E.C., Pang P.S., Virgin H.W. After the pandemic: perspectives on the future trajectory of COVID-19. Nature. 2021;596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Magaña N., Sánchez-Sánchez R., Hernández-Bello J., Venancio-Landeros A.A., Peña-Rodríguez M., Vega-Zepeda R.A., Galindo-Ornelas B., Díaz-Sánchez M., García-Chagollán M., Macedo-Ojeda G., García-González O.P., Muñoz-Valle J.F. RT-qPCR Assays for Rapid Detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 Mutations: a Screening Strategy to Identify Variants With Clinical Impact. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Manríquez J.F., Castro-Ramos J., Gutiérrez-Delgado F., Lopéz-Pacheco M.A., Villanueva-Luna A.E. Raman spectroscopy and PCA-SVM as a non-invasive diagnostic tool to identify and classify qualitatively glycated hemoglobin levels in vivo. J. Biophot. 2017;10:1074–1079. doi: 10.1002/jbio.201600169. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloniniyi O.K., Henry A.R., Darko S., Narpala S.R., Hatcher C., Martinez D.R., Tsybovsky Y., Phung E., Abiona O.M., Antia A., Cale E.M., Chang L.A., Choe M., Corbett K.S., Davis R.L., DiPiazza A.T., Gordon I.J., Hait S.H., Hermanus T., Kgagudi P., Laboune F., Leung K., Liu T., Mason R.D., Nazzari A.F., Novik L., O'Connell S., O'Dell S., Olia A.S., Schmidt S.D., Stephens T., Stringham C.D., Talana C.A., Teng I.-T., Wagner D.A., Widge A.T., Zhang B., Roederer M., Ledgerwood J.E., Ruckwardt T.J., Gaudinski M.R., Moore P.L., Doria-Rose N.A., Baric R.S., Graham B.S., McDermott A.B., Douek D.C., Kwong P.D., Mascola J.R., Sullivan N.J., Misasi J. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373:759. doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wozniak A., Cerda A., Ibarra-Henríquez C., Sebastian V., Armijo G., Lamig L., Miranda C., Lagos M., Solari S., Guzmán A.M., Quiroga T., Hitschfeld S., Riveras E., Ferrés M., Gutiérrez R.A., García P. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-73616-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gartia M.R., Choi C.J., Jiang J., Chen Y., Cunningham B.T., Liu G.L. Quick detection of contaminants leaching from polypropylene centrifuge tubes with surface-enhanced Raman spectroscopy and ultraviolet absorption spectroscopy. J. Raman Spectrosc. 2011;42:1939–1944. doi: 10.1002/jrs.2950. [DOI] [Google Scholar]
- Xu Z., Jiang J., Gartia M.R., Liu G.L. Monolithic integrations of slanted silicon nanostructures on 3D microstructures and their application to surface-enhanced Raman spectroscopy. J. Phys. Chem. C. 2012;116:24161–24170. doi: 10.1021/jp308162c. [DOI] [Google Scholar]
- Yin G., Li L., Lu S., Yin Y., Su Y., Zeng Y., Luo M., Ma M., Zhou H., Orlandini L., Yao D., Liu G., Lang J. An efficient primary screening of COVID‐19 by serum Raman spectroscopy. J. Raman Spectrosc. 2021;52:949–958. doi: 10.1002/jrs.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.Y., Chan K.G., Yean C.Y., Ang G.Y. Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics. 2021;11:53. doi: 10.3390/diagnostics11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel D., Deckert V., Zenobi R., Vo-Dinh T. Near-field surface-enhanced Raman spectroscopy of dye molecules adsorbed on silver island films. Chem. Phys. Lett. 1998;283:381–385. doi: 10.1016/S0009-2614(97)01391-2. [DOI] [Google Scholar]
- Zhu X., Wang X., Li S., Luo W., Zhang X., Wang C., Chen Q., Yu S., Tai J., Wang Y. Rapid, ultrasensitive, and highly specific diagnosis of COVID-19 by CRISPR-based detection. ACS Sens. 2021;6:881–888. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.