Abstract
Both feline coronavirus (FCoV) and SARS-CoV-2 are coronaviruses that infect cats and humans, respectively. However, cats have been shown to be susceptible to SARS-CoV-2, and FCoV also had been shown to infect human. To elucidate the relationship between FCoV and SARS-CoV-2, we highlight the main characteristics of the genome, the receptor usage, and the correlation of the receptor-binding domain (RBD) of spike proteins in FCoV and SARS-CoV-2. It is demonstrated that FCoV and SARS-CoV-2 are closely related to the main characteristics of the genome, receptor usage, and RBD of spike proteins with similar furin cleavage sites. In particular, the affinity of the conserved feline angiotensin-converting enzyme 2 (fACE2) receptor to the RBD of SARS-CoV-2 suggests that cats are susceptible to SARS-CoV-2. In addition, cross-species of coronaviruses between cats and humans or other domesticated animals are also discussed. This review sheds light on cats as potential intermediate hosts for SARS-CoV-2 transmission, and cross-species transmission or zoonotic infection of FCoV and SARS-CoV-2 between cats and humans was identified.
Keywords: Feline coronavirus, SARS-CoV-2, Receptor, Receptor-binding domain, Spike protein
Abbreviations: ACE2, angiotensin-converting enzyme 2; Ang, angiotensin; CCoV, canine coronavirus; fAPN, feline aminopeptidase-N; cry-EM, cryo-electron microscopy; FCoV, feline coronavirus; fDC-SIGN, feline dendritic cell-specific intercellular adhesion molecule grabbing non-integrin; FECV, feline enteric coronavirus; FIPV, feline infectious peritonitis virus; HCoV-229E, human coronavirus 229E; NP, nucleocapsid protein; NTD, N-terminal domain; CTD, C-terminal domain; ORFs, open reading frames; PDCoV, porcine delta coronavirus; PRCV, porcine respiratory coronavirus; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome 2; TGEV, transmissible gastroenteritis virus; UCD, University of California, Davis
1. Introduction
The COVID-19 outbreak evolved into a pandemic due to the virulent nature of severe acute respiratory syndrome 2 (SARS-CoV-2), a novel coronavirus. Questions about the origin of this coronavirus and its introduction into human beings have persisted. Due to the detection of a variety of Sarbecoviruses related to SARS-CoV-2 identified among bats and pangolins, it is widely believed that SARS-CoV-2 originated from wild animals and was introduced to humans through an intermediate animal. Thus, coronaviruses from animals, especially animals that come into close contact with humans, have attracted extensive attention.
Feline coronavirus (FCoV) is a coronavirus that is divided into two serotypes according to its antigenicity: FCoV I and FCoV II. The representative strain of FCoV I is feline enteric coronavirus (FECV)-University of California, Davis (UCD), which was isolated by Pedersen (Pedersen et al., 1981). Feline infectious peritonitis virus (FIPV)-UCD1, FIPV-UCD3, FIPV-UCD4 and FIPV-T-TN406 also belong to FCoV I (Pedersen et al., 1981, Motokawa et al., 1996). FCoV I is difficult to isolate in cell culture, as it grows well only in macrophages (Pedersen et al., 1981). The representative strain of FCoV II is FIPV-Norden (NOR) 15, which was isolated by Evermann et al. in the 1980 s (Evermann et al., 1981), as well as FIPV-1683 and FIPV-1146, which were isolated by Pedersen and coworkers (Pedersen et al., 1984). It has been reported that FCoV II originates from FCoV I and canine coronavirus (CCoV) (Herrewegh et al., 1998) and that it can proliferate in many cell lines, generate gene mutations, and produce a high virus yield. FCoV I is the most diffuse among cats in terms of natural infections worldwide, while FCoV II infection is less common. However, to date, most studies on FCoV have focused on FCoV II, while there have been few studies on FCoV I (Amer et al., 2012) due to the difficulty of culturing and isolating the virus in cells.
FCoV is also separated into two biotypes based on pathogenicity: feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV). FECV and FIPV exist in both serotypes I and II. FIPV is more virulent than FECV and can cause death in cats within 1–3 weeks after infection (Wolfe and Griesemer, 1966), while FECV usually causes only mild intestinal symptoms (Desmarets et al., 2016). In addition to differences in pathogenicity in vivo, the two biotypes also display different tropisms (Pedersen, 2009). In general, FECV proliferation is confined to intestinal mucosal epithelial cells or mesenteric lymph nodes during natural infection, causing mild and generally self-limiting infections (Pedersen et al., 1981, Stoddart et al., 1988), whereas FIPV infects feline peritoneal macrophages and can then infect more types of cells, including monocytes, plasma cells, lymphocytes and neurocytes (Pedersen, 2009), resulting in high titers of progeny virus and causing a systemic and fatal disease. FIPV can thus infect other types of organs continuously and more efficiently than FECV (Stoddart and Scott, 1989, Dewerchin et al., 2005).
Comparatively, SARS-CoV-2 can cause patients to develop pathological features with enteric infection and pleural/peritoneal effusion inflammation (Shi et al., 2020, Bennett et al., 2021, Cantley et al., 2021, Mohan et al., 2021). In addition, in April 2020, Newman first reported cases of SARS-CoV-2 infection in companion animals, including two cats in New York (Newman et al., 2020). Later, other researchers reported more cases of cats infected by SARS-CoV-2 or cases of seropositivity in different territories and countries (Barrs et al., 2020, Klaus et al., 2021, Michael et al., 2021, Spada et al., 2021, Mohebali et al., 2022). Then, researchers reported that some feline coronavirus drugs are strong candidates for the treatment of patients with SARS-CoV-2 (Vuong et al., 2020, Paltrinieri et al., 2021). Presently, an increasing number of SARS-CoV-2 drugs have been found or identified by in silico work on natural molecules against SARS-CoV-2, thereby potentially supplying new candidate drugs for treating cats with FCoV in the future (Bhardwaj et al., 2021a, Bhardwaj et al., 2021c, Bhardwaj et al., 2021b, Sharma et al., 2021, Singh et al., 2021a, Singh et al., 2021b, Singh et al., 2021c, Singh et al., 2021d). Based on the above information, the characteristics of FCoV and its infection among cats might shed light on the understanding of human infection with SARS-CoV-2 and vice versa. Thus, it is necessary to provide a comparative review of the potential relationship between FCoV and SARS-CoV-2.
2. The main characteristics of the genomes of FCoV and SARS-CoV-2
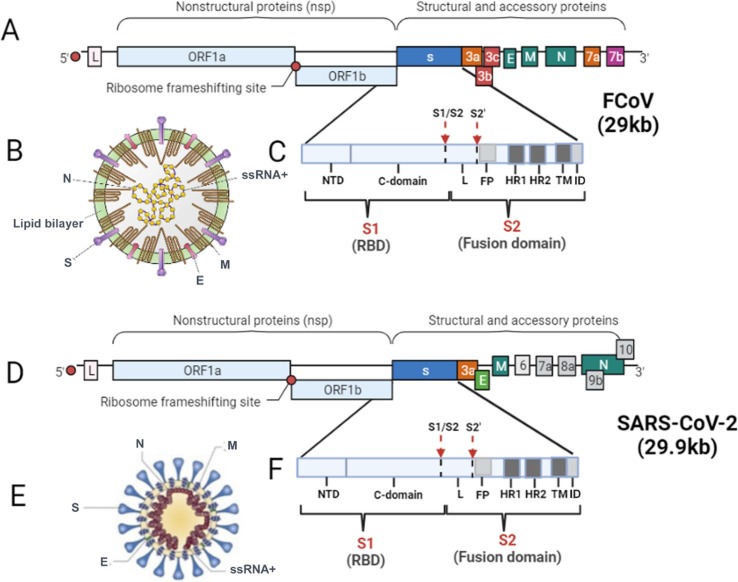
FCoV is a nonsegmented, single-stranded RNA virus that belongs to the order Nidovirales, family Coronaviridae, subfamily Coronavirinae. FCoV is further classified as the genus Alphacoronavirus, species alphacoronavirus 1 (coronavirus group I (Tresnan et al., 1996)) together with canine coronavirus (CCoV) and transmissible gastroenteritis virus (TGEV) of pigs (Kipar and Meli, 2014). The genome of the virus, which is approximately 29 kb in size, includes 11 open reading frames (ORFs) and encodes four major structural proteins: the spike (S) protein, the nucleocapsid (N) protein, the envelope (E) protein and the membrane (M) protein (de Barros et al., 2019). It also encodes seven nonstructural proteins, including two replicase proteins (1a and 1b) and five accessory proteins (3a, 3b, 3c, 7a and 7b) (Tekes and Thiel, 2016) (Fig. 1 A-C). Among the structural proteins, the S protein is a large glycoprotein (180–220 kDa) that forms the peplomers, which project from the surface of virions and are responsible for receptor binding, induction of cell-to-cell fusion and fusion of the viral envelope with target cell membranes, and induction of neutralizing antibody. The S protein is associated with the virulence of FCoV (Jaimes and Whittaker, 2018). The sequence of each S protein can be divided into two regions: S1 and S2. S1 is the N-terminal portion, and S2 is the region between S1 and the transmembrane domain (Olsen, 1993). It has been reported that S1 plays a critical role in receptor binding, while S2 can favor virus–cell fusion (Licitra et al., 2013). The M protein is the most abundant structural protein in the virus. This medium-sized (∼25 to 30 kDa) N-linked glycosylated protein is randomly distributed along the viral envelope and anchored through three transmembrane domains. The small E protein (∼8 to 12 kDa) is a type III membrane protein and is also inserted into the viral envelope but is present at a much lesser extent than M or S. The viral helical nucleocapsid is composed of multiple copies of the RNA binding protein N, a 50-kDa protein that is composed of two domains (N-terminal domain and C-terminal domain) both with the same affinity to bind RNA but through different mechanisms (Jaimes and Whittaker, 2018).
Fig. 1.
Schematic representation of the structure and genome of FCoV and SARS-CoV-2. (A) The genome of FCoV ssRNA+. The FCoV genome is approximately 29 kb in size and contains 11 ORFs encoding four structural proteins (S, M, E and N) and seven nonstructural proteins (replicase proteins 1a and 1b and accessory proteins 3a, 3b, 3c, 7a and 7b). L, leader sequence. (B) The structure and proteins of FCoV. Spike (S), matrix (M), envelope (E) and nucleocapsid (N). (C) The structure of the S protein of FCoV. The S protein consists of two subunits: S1 (the receptor binding domain, RBD) and S2 (the fusion domain, FD). The S1 subunit also includes two functional domains: the N-terminal domain (NTD) and the C-terminal domain (CTD). The S2 subunit consists of a fusion peptide (FP), two heptapeptide repeats (HR1 and HR2), a transmembrane (TM) domain and an internal domain (ID). The arrows indicate the two activation sites, S1/S2 and S2′, and the linker sequence (L) region between S1/S2 and S2′. (D) Genome structure of SARS-CoV-2 ssRNA+. The SARS-CoV-2 genome is approximately 29.9 kb in size and contains 12 ORFs encoding four structural proteins (S, M, E and N) and eight nonstructural proteins (replicase proteins 1a and 1b and accessory proteins 3a, 6, 7a, 8a, 9b and 10). L, leader sequence. (E) The structure and proteins of SARS-CoV-2. Spike (S), matrix (M), envelope (E) and nucleocapsid (N). (F) The structure of the S protein of SARS-CoV-2. The description is the same as that in C.
SARS-CoV-2 is thought to be classified into the order Nidovirales, family Coronaviridae, and subfamily Orthocoronavirinae, the subgenus Sarbecovirus of the genus Betacoronavirus originally (Malik, 2020). SARS-CoV-2 has a different genome structure than FCoV, with a 5′-leader-UTR- replicase-S (Spike)–E (Envelope)-M (Membrane)-N (Nucleocapsid)- 3′UTR-poly (A) tail and accessory genes interspersed within the structural genes at the 3′ end of the genome (Fig. 1D-F). The total length of the genome of SARS-CoV-2 is approximately 29.9 kb, which is slightly larger than that of FCoV. Among them, the S protein (∼150 kDa, smaller than that of FCoV) also plays a key role in mediating attachment of the virus to host cell surface receptors, resulting in fusion and subsequent viral entry (Malik, 2020). The S protein utilizes an N-terminal signal sequence to gain access to the endoplasmic reticulum (ER) and is heavily N-glycosylated. The S protein of SARS-CoV-2 is also composed of S1 and S2. S1 mediates receptor binding, while S2 mediates viral membrane fusion using a highly conserved peptide (Gupta, 2020). Although there is low homology of the S protein between SARS-CoV-2 and FCoV, it seems that the S2 protein of FCoV is more closely related to SARS-CoV-2 in the phylogenetic map than to SARS-CoV or MERS-CoV (Budhraja et al., 2021). The M protein (∼25–30 kDa) of SARS-CoV-2 is a smaller functionally dimeric protein than that of FCoV, with three transmembrane domains that can adopt two different conformations in that this transmembrane protein has an N-terminal ectodomain and a C-terminal endodomain with ion channel activity, and it is responsible for assembly, budding, envelope formation, and pathogenesis of the virus (Malik, 2020, Troyano-Hernáez et al., 2021). The coronavirus nucleocapsid protein (NP) is required for the packaging of viral RNA into the viral particle during viral assembly, similar to the function of the NP protein of FCoV (Gupta, 2020).
3. The receptor of FCoV and SARS-CoV-2
3.1. The receptor of FCoV
Feline aminopeptidase-N (fAPN) has been shown to act as a receptor for feline coronavirus (FCoV), canine coronavirus (CCoV), porcine coronaviruses (transmissible gastroenteritis virus [TGEV] and porcine respiratory coronavirus [PRCV]), and human respiratory coronavirus HCV-229E (HCV-229E) in coronavirus group I (alphacoronavirus 1), which indicates that cats could serve as a “mixing vessel” in these coronavirus infections (Tresnan et al., 1996, Tresnan and Holmes, 1998). fAPN is a ubiquitous and multifunctional glycoprotein of approximately 110 kDa and 967 amino acids (Luan and Xu, 2007, Van Hamme et al., 2011). It is generally accepted that fAPN can act as a receptor for the attachment and entry of FCoV II strains but not for FCoV I (Dye et al., 2007). For serotype II of FECV, the enteric infected coronavirus, APN proteins are supposed to participate in the maturation of small proteins, as well as helping the peptide absorption at the enterocytes. In addition, for all II serotypes of FCoV, including FECV and FIPV, the S protein can bind fAPN, which indicates that the S protein is important for cellular tropism (Jaimes and Whittaker, 2018). Another receptor is feline dendritic cell-specific intercellular adhesion molecule grabbing nonintegrin (fDC-SIGN), which is a type II membrane protein of 44 kDa with an external mannose-binding, C-type lectin domain (Geijtenbeek et al., 2000, Steinman, 2000). fDC-SIGN is associated with FIPV I infection of monocytes. In addition, it has been shown that fDC-SIGN and fAPN are also involved in FIPV II infection, with only fAPN playing a receptor role at the plasma membrane (Van Hamme et al., 2011).
3.2. The receptor of SARS-CoV-2
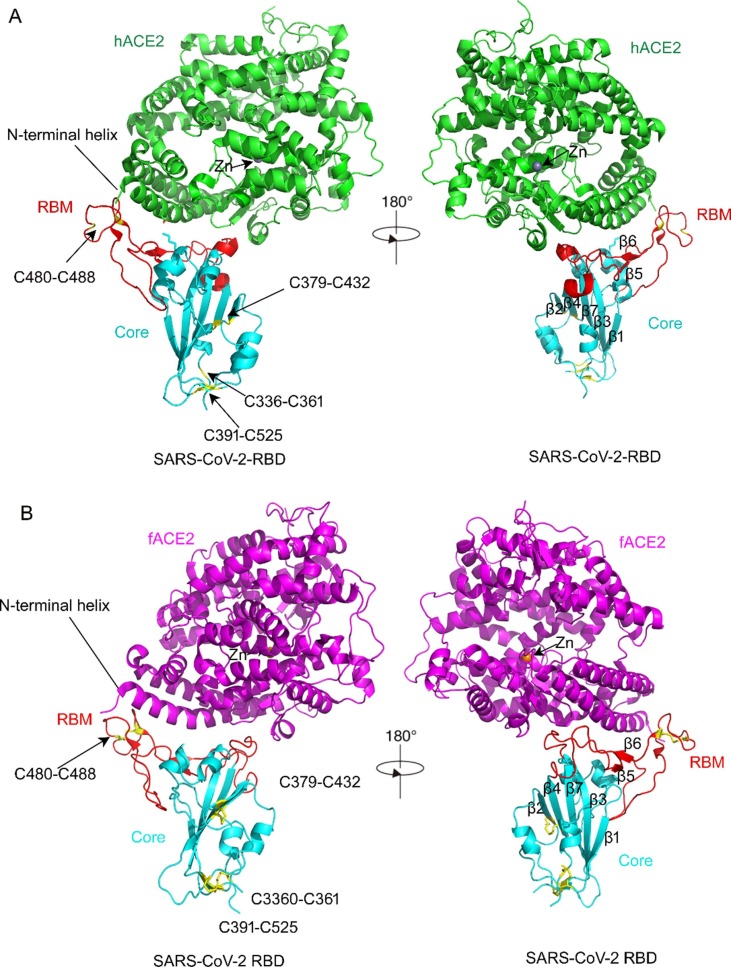
Michael Letko first reported that angiotensin-converting enzyme 2 (ACE2) should be a host cellular receptor for SARS-CoV-2 to enter cells by comparing and analyzing other lineage B Betacoronaviruses, such as SARS-CoV, which emerged in 2002 (Kuiken et al., 2003, Letko et al., 2020). ACE2 is also a kind of aminopeptidase (Bakhshandeh et al., 2021). The 44-kb ACE2 gene is located on chromosome Xp22. This gene has been identified as a kind of zinc metallopeptidase similar to APN (Gheblawi et al., 2020). Zinc plays an important role in the catalytic activity of angiotensin converting enzyme. It has been reported that chelating agents abolish activity by removing the metal ion to yield the metalloprotein inactive, so that it would affect the protein–protein interaction (Bunning and Riordan, 1985). ACE2 possesses only a single catalytic domain, which cleaves C-terminal dipeptide residues from susceptible substrates (a peptidyl dipeptidase). ACE2 also acts as a simple carboxypeptidase able to hydrolyze angiotensin (Ang) I, forming Ang 1–9 and Ang II to Ang 1–7. SARS-CoV-2 has a single region of the spike protein called the receptor-binding domain (RBD), which mediates the interaction with the host-cell ACE 2 receptor. After RBD binds the receptor, a nearby host protease cleaves the spike, which releases the spike fusion peptide, facilitating SARS-CoV-2 entry into host cells (Simmons et al., 2013, Letko et al., 2020). Structural studies of coronaviruses have shown that the spike RBD is capable of folding independent of the rest of the spike protein and contains all of the structural information for host receptor binding in view of a previous study showing that replacing the RBD of the lineage B bat virus Rp3 allowed the virus to enter cells expressing human ACE2 (hACE2) (Becker et al., 2008, Letko et al., 2020). Jun Lan recently determined the crystal structure of the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2 bound to the cell receptor hACE2, which showed that the overall hACE2-binding mode of the SARS-CoV-2 RBD is nearly identical to that of the SARS-CoV RBD (Lan et al., 2020). The structure of the SARS-CoV-2 RBD bound to ACE2 is shown in Fig. 2 A. It has been reported that ACE2 is also expressed in feline tissues and is similar to human ACE2 in sequence and length (Guo et al., 2008). Furthermore, researchers recently resolved the cryo-electron microscopy (cry-EM) structure of the feline ACE2 (fACE2) in complex with the SARS-CoV-2 RBD at a resolution of 3 Å, revealing that the SARS-CoV-2 RBD can bind with fACE2 in a similar binding mode as hACE2 (Wu et al., 2020) (Fig. 2B). This research also revealed that fACE2 retained 16 of 20 key residues of hACE2 responsible for interaction with the SARS-CoV-2 RBD. It was further discovered that the feline ACE2 receptor is 85.2% identical to the human ACE2 receptor (Budhraja et al., 2021), which accounts for why fACE2 can be recognized and bound by SARS-CoV-2 (Fig. 3 ). The only four mutated residues were L24, E30, E38 and T82, whose effect on the interaction between fACE2 and SARS-CoV-2-RBD was negligible because the overall structure of fACE2 molecules bound to SARS-CoV-2 RBD still highly resembles the complex structure of hACE2 binding with SARS-CoV-2 RBD. In addition, the research also indicated that residue D355 of fACE2 acts as an especially crucial amino acid to interact with the SARS-CoV-2 RBD because a point mutation D355A in fACE2 can lead to loss of binding to the SARS-CoV-2 RBD but not in hACE2. It was speculated that these key residues of fACE2 should explain why SARS-CoV-2 can infect cats, although further studies are needed..
Fig. 2.
The structure of SARS-CoV-2 bound to human ACE2 (hACE2) (Lan et al., 2020) and feline ACE2 (fACE2) (Wu et al., 2020). A, The structure of SARS-CoV-2 bound to hACE2 (6m0j). hACE2 is shown in green. The SARS-CoV-2 RBD core is shown in cyan and the receptor-binding motif in red. Disulfide bonds in the SARS-CoV-2 RBD are shown as sticks and indicated by arrows. The N-terminal helix of ACE2 responsible for binding is labeled. There is a Zn ion in the core of hACE2 molecule. B, The structure of SARS-CoV-2 bound to fACE2 (7c8d). fACE2 is shown in magenta. There is a Zn ion shown in the core of the fACE2 molecule, similar to hACE2, as shown in Figure A. The domains of SARS-CoV-2 RBD binding to fACE2 are shown in Figure A. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
The sequence similarity between fACE2 (BAB40370.1) and hACE2 (NP_001034545.1) in amino acids and the key residues of fACE2 and hACE2. The homologous identity was 85.217%. The total amino acids for both fACE2 and hACE2 were 805. The 20 key residue sites of hACE2/fACE2 binding to the SARS-CoV-2 RBD were 19 (S), 24 (Q/L), 27 (T), 28 (F), 30 (D/E), 31 (K), 34 (H), 35 (E), 37 (E), 38 (D/E), 41 (Y), 42 (Q), 45 (L), 82 (M/T), 83 (Y), 330 (N), 353 (K), 354 (G), 355 (D) and 357 (R). The red asterisk denotes the retained residues, and the black asterisk denotes the mutated residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recently, another SARS-CoV-2 candidate receptor named tyrosine-protein kinase receptor UFO (AXL) was reported (Wang et al., 2021). AXL is mostly expressed in human primary lung epithelial cells, while ACE2 is mostly expressed in kidney and gastrointestinal tract cells. It was found that AXL specifically interacts with the N-terminal domain of SARS-CoV-2 spike. The elaborate mechanism of AXL with SARS-CoV-2 has yet to be fully elucidated.
4. The correlation of spike protein RBDs of SARS-CoV-2 and FCoV
Although SARS-CoV and FCoV belong to different subgenera of coronavirus and the cellular receptors are also different, the receptor binding domain (RBD) of spike proteins retains similar features. It is necessary to investigate whether there is a special correlation between SARS-CoV-2′s and FCoV’s RBD of spike proteins that we can elucidate the ability for the viruses to cross species boundaries. The glycoprotein spike carries two distinct domains separated by a linker. A precise proteolytic cut is necessary to separate the receptor binding S1 domain from the fusion competent S2 domain. There are two main proteolytic sites in coronavirus (CoV) spike proteins: one is the S1/S2 site situated at the S1 and S2 domain boundaries, and the other is the S2′ site located close to the S2 domain (Jaimes and Whittaker, 2018) (Fig. 1C, 1F). In the majority of the spike proteins related to SARS-CoV-2, a conserved monobasic sequence (R↓S) preferred by trypsin-like enzymes is normally seen in the S1/S2 site (Budhraja et al., 2021). Furin- and furin-like enzymes can cleave the S1/S2 site and cause the sequence (R↓S) in SARS-CoV-2, MERS and other infectious SARS viruses (Andersen et al., 2020, Hoffmann et al., 2020a). It is speculated that furin- and furin-like cleavage might be one of the major events that resulted in zoonotic transfer, the infectivity of SARS-CoV-2 in pandemic proportions, and lethality based on investigations that furin- and furin-like cleavage could activate the virus to be a widespread pandemic and in zoonotic transfer. Hoffmann first reported that the membrane-bound receptor-activated type II serine protease TMPRSS2 cleaves SARS-CoV-2 and that this cleavage is responsible for viral entry, which indicated that the polybasic insert in experimental model systems could cause virus entry more easily (Hoffmann et al., 2020a). Later, Hoffmann also identified that furin-mediated cleavage of the spike protein at the polybasic site was necessary for the virus to enter human lung cells in culture (Hoffmann et al., 2020b).
In FCoV, it was also found that FECV carried a highly optimized furin cleavage site at S1/S2 (Licitra et al., 2013). However, feline infectious peritonitis virus (FIPV) causes systemic infection and death of the infected cat, and the polybasic insert is either completely lost or mutated. Further studies identified that furin or furin-like polybasic inserts are commonly XRRX amino acid sequences before the site (R↓S). If P2(X) and P5(X) are basic amino acids, such as arginine (R) or lysine (K), the proteolytic activity of furin is high; otherwise, it is low. For example, Budhraja and his coworkers identified that in structural models of furin complexed with peptides, PRRAR↓S binds less well and with distinct differences compared to the all basic RRKRR↓S. They further identified that in FCoV, P1 Arg in FECV when mutated to Gly, Met or Thr results in loss of activity, and His, Leu or Ser at P2 (FIPV) instead of Arg in the canonical sequence (FECV) compromised cleavage efficiency; however, Pro at this position (FIPV) enhanced activity (Budhraja et al., 2021).
In fact, apart from polybasic insert sequences that might be associated with virus entry or even zoonotic transfer, another crucial element is the affinity of the RBD domain to the ACE2 receptor (Andersen et al., 2020). Despite the low sequence homology and distant evolutionary relationship to the novel SARS-CoV-2 spike protein, the RBD domain of the feline coronavirus carries at least six residues that are identical to those at the interface of the SARS-CoV-2 RBD-ACE2 receptor complex (Budhraja et al., 2021). Budhraja and Wu also found that the amino acids of human ACE2 involved in the interaction with the SARS-CoV-2 RBD are conserved in the feline ACE2 receptor (Wu et al., 2020, Budhraja et al., 2021). The above observations support the recent report that cats are susceptible to SARS-CoV-2, especially to airborne transmission (Shi et al., 2020, Gonzales et al., 2021, Fritz et al., 2022), which indicated that the domestic cat, as one of the possible intermediary hosts, may have nurtured SARS-CoV-2 in its zoonotic jump. Therefore, the domestic cat might be a factor in the transmission of SARS-CoV-2.
5. Cross-species of coronaviruses between cats and humans or other domesticated animals
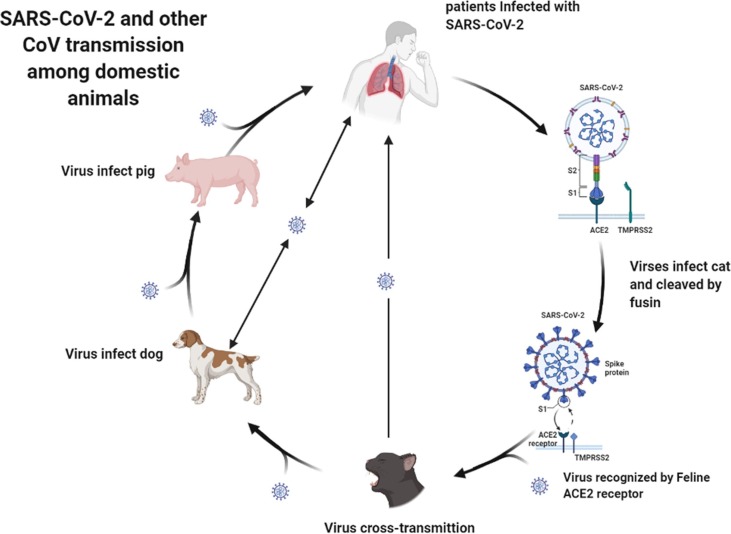
Presently, increasing evidence has shown that cats can be hosts of some human-infecting viruses. Ongradi et al. (Ongradi et al., 2019) first identified that cats can be animal hosts of human adenovirus 1. Zhao et al. (Zhao et al., 2019) reported that cats can be infected by human coronaviruses 229E (HCoV-229E) and (PDCoV) by serological screening, and they revealed the potential role of cats in the cross-species transmission of coronaviruses. Since SARS-CoV-2 emerged at the end of December 2019, researchers have also reported that cats can be infected by common SARS-CoV-2 strains and even (Delta) variant strains from humans (Garigliany et al., 2020, Curukoglu et al., 2021, Hosie et al., 2021, Kang et al., 2021, Mohebali et al., 2022), which revealed that the fACE2 receptor could be recognized by the variant strains. The information also indicates that cats may play a special role in the transmission of SARS-CoV-2, considering the three factors: similar cellular receptor, receptor binding domain (RBD) with similar furin cleavage sites, and especially the third factor, high risk of cross-transmission between humans and cats because cats are usually in close contact with humans, especially companion animals (Stout et al., 2020). In addition, it has been reported that FCoV can infect humans (Silva et al., 2014), which indicates that cat-infecting coronaviruses, including SARS-CoV-2, can also infect humans because of virus variability and evolution, although more in-depth studies are needed. Therefore, it should be pointed out that cats might be potential hosts for producing novel variant strains of SARS-CoV-2. In addition, researchers presently reported that another domestic animal dog can also be infected by a SARS-CoV-2 common strain and B.1.1.7 Variant strain (Barroso-Arévalo et al., 2021, Grandjean et al., 2022, Jairak et al., 2022) indicates that dogs are another potential host of SARS-CoV-2 in domestic animals. In addition, it has been reported that a canine coronavirus can infect humans (Vlasova et al., 2021). Although it has been reported that pigs are not susceptible to SARS-CoV-2 but induce fine immunogenicity, a recent report that emergence of porcine delta-coronavirus pathogenic infections among children strongly implies that coronaviruses in domestic animals could develop to be a potential threat to humans after an independent or cross-transmission (Lednicky et al., 2021). We suggest that the transmission of SARS-CoV-2 and other domestic animals’ coronavirus might occur (Fig. 4 ), and thus, humans should pay more attention to coronavirus infections among cats or other domestic animals.
Fig. 4.
Schematic representation of SARS-CoV-2 and other coronavirus transmission among domestic animals.
6. Conclusions
Although feline coronavirus and SARS-CoV-2 are different in genus classification, they are closely related to the main characteristics of the genome, receptor usage, and RBD of spike proteins. In particular, the RBDs of the S proteins of the two viruses both have polymeric cleavage sites that can cleave the S1/S2 site and cause the sequence (R↓S), which is associated with virus entry or even zoonotic transfer. In addition, the affinity of the human and feline conserved ACE2 receptor for the RBD of SARS-CoV-2 is also crucial in supporting that SARS-CoV-2 can interact with both human ACE2 and feline ACE2 and that cats are susceptible to SARS-CoV-2. However, during the process of SARS-CoV-2 infection in cats, whether FCoV elements serve or which host molecules are associated is still unclear, and further research is ongoing. In conclusion, this review reveals that cats are potential intermediate hosts for SARS-CoV-2 transmission, and cross-species transmission or zoonotic infection of FCoV and SARS-CoV-2 between cats and humans was identified.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This study was financially supported by the National Key Research and Development Plan of China (grant no. 2016YFD0501002).
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Edited by John Doe
References
- Amer A., Siti Suri A., Abdul Rahman O., Mohd H.B., Faruku B., Saeed S., Tengku Azmi T.I. Isolation and molecular characterization of type I and type II feline coronavirus in Malaysia. Virol. J. 2012;9:278. doi: 10.1186/1743-422X-9-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshandeh B., Jahanafrooz Z., Abbasi A., Goli M.B., Sadeghi M., Mottaqi M.S., Zamani M. Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus. Microb. Pathog. 2021;154:104831. doi: 10.1016/j.micpath.2021.104831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Arévalo S., Rivera B., Domínguez L., Sánchez-Vizcaíno J.M. First detection of SARS-CoV-2 B.1.1.7 variant of concern in an asymptomatic dog in spain. Viruses. 2021;13(7):1379. doi: 10.3390/v13071379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs V.R., Peiris M., Tam K.W.S., Law P.Y.T., Brackman C.J., To E.M.W., Yu V.Y.T., Chu D.K.W., Perera R.A.P.M., Sit T.H.C. SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China. Emerg. Infect. Dis. 2020;26(12):3071–3074. doi: 10.3201/eid2612.202786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. USA. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D., Franchi F., De Vita E., Mazzei M.A., Volterrani L., Disanto M.G., Garosi G., Guarnieri A., Cusi M.G., Bargagli E., Scolletta S., Valente S., Gusinu R., Frediani B. SARS-CoV-2 in pleural fluid in a kidney transplant patient. Postgrad. Med. 2021;133(5):540–543. doi: 10.1080/00325481.2020.1838817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Bioactive Molecules of Tea as Potential Inhibitors for RNA-Dependent RNA Polymerase of SARS-CoV-2. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39(10):3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhraja A., Pandey S., Kannan S., Verma C.S., Venkatraman P. The polybasic insert, the RBD of the SARS-CoV-2 spike protein, and the feline coronavirus - evolved or yet to evolve. Biochem. Biophys. Rep. 2021;25:100907. doi: 10.1016/j.bbrep.2021.100907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunning P., Riordan J.F. The functional role of zinc in angiotensin converting enzyme: implications for the enzyme mechanism. J. Inorg. Biochem. 1985;24:183–198. doi: 10.1016/0162-0134(85)85002-9. [DOI] [PubMed] [Google Scholar]
- Cantley R.L., Hrycaj S., Konopka K., Chan M.P., Huang T., Pantanowitz L. Cytologic findings in effusions from patients with SARS-CoV-2 infection. J. Am. Soc. Cytopathol. 2021;10(3):261–269. doi: 10.1016/j.jasc.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curukoglu A., Ergoren M.C., Ozgencil F.E., Sayiner S., Ince M.E., Sanlidag T. First direct human-to-cat transmission of the SARS-CoV-2 B.1.1.7 variant. Aust. Vet. J. 2021;99(11):482–488. doi: 10.1111/avj.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros B.d.C.V., Castro C.M.O.d., Pereira D., Ribeiro L.G., Júnior J.W.B.D., Casseb S.M.M., Holanda G.M., Cruz A.C.R., Júnior E.C.S., Mascarenhas J.D.P., Matthijnssens J. First Complete Genome Sequence of a Feline Alphacoronavirus 1 Strain from Brazil. Microbiol. Resour. Announc. 2019;8(10):e01535-18. doi: 10.1128/MRA.01535-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarets L.M., Vermeulen B.L., Theuns S., Conceicao-Neto N., Zeller M., Roukaerts I.D., Acar D.D., Olyslaegers D.A., Van Ranst M., Matthijnssens J., Nauwynck H.J. Experimental feline enteric coronavirus infection reveals an aberrant infection pattern and shedding of mutants with impaired infectivity in enterocyte cultures. Sci. Rep. 2016;6:20022. doi: 10.1038/srep20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewerchin H.L., Cornelissen E., Nauwynck H.J. Replication of feline coronaviruses in peripheral blood monocytes. Arch. Virol. 2005;150:2483–2500. doi: 10.1007/s00705-005-0598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C., Temperton N., Siddell S.G. Type I feline coronavirus spike glycoprotein fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J. Gen. Virol. 2007;88:1753–1760. doi: 10.1099/vir.0.82666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evermann J.F., Baumgartener L., Ott R.L., Davis E.V., McKeirnan A.J. Characterization of a feline infectious peritonitis virus isolate. Vet. Pathol. 1981;18(2):256–265. doi: 10.1177/030098588101800214. [DOI] [PubMed] [Google Scholar]
- Fritz M., Nesi N., Denolly S., Boson B., Legros V., Rosolen S.G., Briend‐Marchal A., Ar Gouilh M., Leroy E.M. Detection of SARS-CoV-2 in two cats during the second wave of the COVID-19 pandemic in France. Vet. Med. Sci. 2022;8(1):14–20. doi: 10.1002/vms3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigliany M., Van Laere A.-S., Clercx C., Giet D., Escriou N., Huon C., van der Werf S., Eloit M., Desmecht D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. 2020;26(12):3069–3071. doi: 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek T.B.H., Torensma R., van Vliet S.J., van Duijnhoven G.C.F., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100(5):575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020;126(10):1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales J.L., de Jong M.C.M., Gerhards N.M., Van der Poel W.H.M. The SARS-CoV-2 Reproduction Number R0 in Cats. Viruses. 2021;13(12):2480. doi: 10.3390/v13122480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean D., Gallet C., Julien C., Sarkis R., Muzzin Q., Roger V., Roisse D., Dirn N., Levert C., Breton E., Galtat A., Forget A., Charreaudeau S., Gasmi F., Jean-Baptiste C., Petitjean S., Hamon K., Duquesne J.-M., Coudert C., Tourtier J.-P., Billy C., Wurtz J.-M., Chauvin A., Eyer X., Ziani S., Prevel L., Cherubini I., Khelili-Houas E., Hausfater P., Devillier P., Desquilbet L., Chen R.J. Identifying SARS-COV-2 infected patients through canine olfactive detection on axillary sweat samples; study of observed sensitivities and specificities within a group of trained dogs. PLoS ONE. 2022;17(2):e0262631. doi: 10.1371/journal.pone.0262631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Guo A., Wang C., Yan B., Lu H., Chen H. Expression of feline angiotensin converting enzyme 2 and its interaction with SARS-CoV S1 protein. Res. Vet. Sci. 2008;84(3):494–496. doi: 10.1016/j.rvsc.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.P. Progress in Studies on Structural and Remedial Aspects of Newly Born Coronavirus, SARS-CoV-2. Curr. Top. Med. Chem. 2020;20(26):2362–2378. doi: 10.2174/1568026620666200922112300. [DOI] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72(5):4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H. and Pohlmann, S., 2020a. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell 78, 779-784 e5. [DOI] [PMC free article] [PubMed]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.H., Nitsche, A., Muller, M.A., Drosten, C. and Pohlmann, S., 2020b. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271-280 e8. [DOI] [PMC free article] [PubMed]
- Hosie M.J., Epifano I., Herder V., Orton R.J., Stevenson A., Johnson N., MacDonald E., Dunbar D., McDonald M., Howie F., Tennant B., Herrity D., Da Silva Filipe A., Streicker D.G., Willett B.J., Murcia P.R., Jarrett R.F., Robertson D.L., Weir W. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet. Rec. 2021;188(8) doi: 10.1002/vetr.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Whittaker G.R. Feline coronavirus: Insights into viral pathogenesis based on the spike protein structure and function. Virology. 2018;517:108–121. doi: 10.1016/j.virol.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairak W., Charoenkul K., Chamsai E., Udom K., Chaiyawong S., Hangsawek A., Waenkaew S., Mungaomklang A., Tangwangvivat R., Amonsin A. Zoonoses Public Health; Thailand: 2022. Survey of SARS-CoV-2 in dogs and cats in high-risk areas during the second wave of COVID-19 outbreak. (in press) [DOI] [PubMed] [Google Scholar]
- Kang K., Chen Q., Gao Y., Yu K.J. Detection of SARS-CoV-2 B.1.617.2 (Delta) variant in three cats owned by a confirmed COVID-19 patient in Harbin, China. Vet. Med Sci. 2021 doi: 10.1002/vms3.715. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Meli M.L. Feline infectious peritonitis: still an enigma? Vet. Pathol. 2014;51(2):505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- Klaus J., Palizzotto C., Zini E., Meli M.L., Leo C., Egberink H., Zhao S., Hofmann-Lehmann R. SARS-CoV-2 Infection and Antibody Response in a Symptomatic Cat from Italy with Intestinal B-Cell Lymphoma. Viruses. 2021;13(3):527. doi: 10.3390/v13030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.AM., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.KS., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.-C., Stöhr K., Peiris J.S.M., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lednicky J.A., Tagliamonte M.S., White S.K., Elbadry M.A., Alam M.M., Stephenson C.J., Bonny T.S., Loeb J.C., Telisma T., Chavannes S., Ostrov D.A., Mavian C., De Rochars V.M.B., Salemi M., Morris J.G. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv. 2021 Preprint. [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19(7):1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y., Xu W. The structure and main functions of aminopeptidase N. Curr. Med. Chem. 2007;14:639–647. doi: 10.2174/092986707780059571. [DOI] [PubMed] [Google Scholar]
- Malik Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- Michael H.T., Waterhouse T., Estrada M., Seguin M.A. Frequency of respiratory pathogens and SARS-CoV-2 in canine and feline samples submitted for respiratory testing in early 2020. J. Small Anim. Pract. 2021;62:336–342. doi: 10.1111/jsap.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S.V., Hemalatha M., Kopperi H., Ranjith I., Kumar A.K. SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021;405:126893. doi: 10.1016/j.cej.2020.126893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebali M., Hassanpour G., Zainali M., Gouya M.M., Khayatzadeh S., Parsaei M., Sarafraz N., Hassanzadeh M., Azarm A., Salehi-Vaziri M., Sasani F., Heidari Z., Jalali T., Pouriayevali M.H., Shoja Z., Ahmadi Z., Sadjadi M., Tavakoli M., Azad-Manjiri S., Karami C., Zarei Z. SARS-CoV-2 in domestic cats (Felis catus) in the northwest of Iran: evidence for SARS-CoV-2 circulating between human and cats. Virus Res. 2022;310:198673. doi: 10.1016/j.virusres.2022.198673. [DOI] [PubMed] [Google Scholar]
- Motokawa K., Hohdatsu T., Hashimoto H., Koyama H. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer, integral membrane and nucleocapsid proteins of feline, canine and porcine coronaviruses. Microbiol. Immunol. 1996;40(6):425–433. doi: 10.1111/j.1348-0421.1996.tb01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A., Smith D., Ghai R.R., Wallace R.M., Torchetti M.K., Loiacono C., Murrell L.S., Carpenter A., Moroff S., Rooney J.A., Barton Behravesh C. First Reported Cases of SARS-CoV-2 Infection in Companion Animals - New York, March-April 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69(23):710–713. doi: 10.15585/mmwr.mm6923e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993;36(1-2):1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongradi J., Chatlynne L.G., Tarcsai K.R., Stercz B., Lakatos B., Pring-Akerblom P., Gooss D., Sr., Nagy K., Ablashi D.V. Adenovirus Isolated From a Cat Is Related to Human Adenovirus 1. Front. Microbiol. 2019;10:1430. doi: 10.3389/fmicb.2019.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltrinieri S., Giordano A., Stranieri A., Lauzi S. Feline infectious peritonitis (FIP) and coronavirus disease 19 (COVID-19): Are they similar? Transbound Emerg Dis. 2021;68(4):1786–1799. doi: 10.1111/tbed.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg. 2009;11:225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Boyle J.F., Floyd K., Fudge A., Barker J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 1981;42:368–377. [PubMed] [Google Scholar]
- Pedersen N.C., Evermann J.F., McKeirnan A.J., Ott R.L. Pathogenicity studies of feline coronavirus isolates 79–1146 and 79–1683. Am. J. Vet. Res. 1984;45:2580–2585. [PubMed] [Google Scholar]
- Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346:128933. doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C.S., Mullis L.B., Pereira O., Jr., Saif L.J., Vlasova A., Zhang X., Owens R.J., Paulson D., Taylor D., Haynes L.M., Azevedo M.P. Human Respiratory Coronaviruses Detected In Patients with Influenza-Like Illness in Arkansas, USA. Virol. Mycol. 2014;2014(Suppl. 2):004. doi: 10.4172/2161-0517.S2-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135:104555. doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Purohit R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach. Comput. Biol. Med. 2021;139:104965. doi: 10.1016/j.compbiomed.2021.104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Sharma J., Kumar D., Purohit R. Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136:104631. doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J Tradit Complement Med. 2021;12(1):35–43. doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada E., Vitale F., Bruno F., Castelli G., Reale S., Perego R., Baggiani L., Proverbio D. A pre- and during Pandemic Survey of Sars-Cov-2 Infection in Stray Colony and Shelter Cats from a High Endemic Area of Northern Italy. Viruses. 2021;13(4):618. doi: 10.3390/v13040618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M. DC-SIGN: a guide to some mysteries of dendritic cells. Cell. 2000;100(5):491–494. doi: 10.1016/s0092-8674(00)80684-4. [DOI] [PubMed] [Google Scholar]
- Stoddart C.A., Scott F.W. Intrinsic resistance of feline peritoneal macrophages to coronavirus infection correlates with in vivo virulence. J. Virol. 1989;63:436–440. doi: 10.1128/JVI.63.1.436-440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart M.E., Gaskell R.M., Harbour D.A., Pearson G.R. The sites of early viral replication in feline infectious peritonitis. Vet. Microbiol. 1988;18:259–271. doi: 10.1016/0378-1135(88)90092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A.E., André N.M., Jaimes J.A., Millet J.K., Whittaker G.R. Coronaviruses in cats and other companion animals: Where does SARS-CoV-2/COVID-19 fit? Vet. Microbiol. 2020;247:108777. doi: 10.1016/j.vetmic.2020.108777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Thiel H.J. Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis. Adv. Virus Res. 2016;96:193–218. doi: 10.1016/bs.aivir.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan D.B., Holmes K.V. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70(12):8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyano-Hernáez P., Reinosa R., Holguín Á. Evolution of SARS-CoV-2 Envelope, Membrane, Nucleocapsid, and Spike Structural Proteins from the Beginning of the Pandemic to September 2020: A Global and Regional Approach by Epidemiological Week. Viruses. 2021;13(2):243. doi: 10.3390/v13020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hamme E., Desmarets L., Dewerchin H.L., Nauwynck H.J. Intriguing interplay between feline infectious peritonitis virus and its receptors during entry in primary feline monocytes. Virus Res. 2011;160(1-2):32–39. doi: 10.1016/j.virusres.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Diaz A., Damtie D., Xiu L., Toh T.H., Lee J.S., Saif L.J., Gray G.C. Novel Canine Coronavirus Isolated from a Hospitalized Pneumonia Patient, East Malaysia. Clin. Infect. Dis. 2021;74(3):446–454. doi: 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., Sun Z., Liu K., Shan C., Lin A., Jiang S., Xie Y., Zhou Q., Lu L.u., Huang J., Li X.u. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe L.G., Griesemer R.A. Feline Infectious Peritonitis. Pathol. Vet. 1966;3(3):255–270. doi: 10.1177/030098586600300309. [DOI] [PubMed] [Google Scholar]
- Wu L., Chen Q., Liu K., Wang J., Han P., Zhang Y., Hu Y., Meng Y., Pan X., Qiao C., Tian S., Du P., Song H., Shi W., Qi J., Wang H.W., Yan J., Gao G.F., Wang Q. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li W., Schuurman N., van Kuppeveld F., Bosch B.-J., Egberink H. Serological screening for coronavirus infections in cats. Viruses. 2019;11(8):743. doi: 10.3390/v11080743. [DOI] [PMC free article] [PubMed] [Google Scholar]