Abstract
The sudden increase of the COVID-19 outbreak and its continued growth with mutations in various forms has created a global health crisis as well as devastating social and economic effects over the past two years. In this study, a screen-printed carbon electrode reinforced with boron nitride quantum dots/flower-like gold nanostructures (BNQDs/FGNs/SPCE) and functionalized by highly specific antisense DNA oligonucleotide presents an alternative and promising solution for targeting SARS-CoV-2 RNA without nucleic acid amplification. The platform was tested on 120 SARS-CoV-2 RNA isolated from real clinical samples (60 positive and 60 negative confirmed by conventional RT-PCR method). Based on obtained quantitative results and statistical analysis (box-diagram, cutoff value, receiver operating characteristic curve, and t-test), the biosensor revealed a significant difference between the two positive and negative groups with 100% sensitivity and 100% specificity. To evaluate the quantitation capacity and detection limit of the biosensor for clinical trials, the detection performance of the biosensor for continuously diluted RNA isolated from SARS-CoV-2-confirmed patients was compared to those obtained by RT-PCR, demonstrating that the detection limit of the biosensor is lower than or comparable to that of RT-PCR. The ssDNA/BNQDs/FGNs/SPCE showed negligible cross-reactivity with RNA fragments isolated from Influenza A (IAV) clinical samples and also remained stable for up to 14 days. In conclusion, the fabricated biosensor may serve as a promising tool for point-of-care applications.
Keywords: COVID-19 pandemic, Virus diagnosis, SARS-CoV-2 RNA, Electrochemical biosensor
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2 coronavirus, is a respiratory infectious disease which first discovered in 2019 (Kilic et al., 2020; Orooji et al., 2021). In the past two years, this severe acute respiratory syndrome leading to millions of deaths around the world and has become a global public health threat because of its rapid spread and significant mortality. It has also caused serious economic and social consequences and losses (Pan et al., 2021; Svobodova et al., 2021; Verma et al., 2020). The efficient and smart diagnostic tests can have a significant impact on the management of the Covid-19 epidemic, enable rapid detection and isolation even before symptoms manifest (Khan et al., 2020; Qiu et al., 2020; Udugama et al., 2020). Currently, real-time reverse transcription−polymerase chain reaction (RT-PCR) due to its high sensitivity is the preferred testing method for the detection of SARS-CoV-2 (Corman et al., 2020; Lai et al., 2021). Nevertheless, RT-PCR test for SARS-CoV-2 detection is prone to false-negative results as well as the need for well-trained professionals, expensive instruments and equipped laboratories, limits of flexibility, slow diagnostic (3–4h), labor-intensive, and costly make the RT-PCR method vulnerable (Surkova et al., 2020; Xie et al., 2020). Apart from that, enzyme-linked immunosorbent analysis (ELISA) has also been established for rapid recognition of SARS-CoV-2 (Laidoudi et al., 2021; Larsen et al., 2021). Serological analysis require minimal equipment and are readily available. However, low accuracy and sensitivity limit their applicability because antibodies may not be detected in the early steps of infection (Scohy et al., 2020; Zhang et al., 2020). For timely diagnosis of COVID-19, isothermal amplification techniques (Choi et al., 2021; John et al., 2021) and lateral flow immunoassay (LFA) (Zhou et al., 2021) have also been widely reported. Despite great advancements, they do not offer the ideal solution for reliable and fast screening of COVID-19 because there are still cost and performance shortcomings that limit laboratory research and widespread applicability of these methods (Hwang et al., 2021). Given the aforementioned challenges, still, a specific, accurate, sensitive, rapid, and cost-effective method to point-of-care (POC) diagnosis of COVID-19 has to be developed. Novel biomedical assays enabling simple, fast, reliable, and sensitive detection and can overcome the major drawbacks of commonly used viral infection diagnosis methods, such as laborious amplification strategies, expensive and complicated instruments, and long analysis times, provide an impressive solution to this problem and facilitate early diagnosis and transmission prevention. Electrochemical analytical devices are now in the spotlight to achieve such goals. In this context, the development of electrochemical biosensors could be an ideal scheme due to their superior selectivity and sensitivity, possibility of portability, short analysis time, and simplicity at a low cost (Feng et al., 2021; Gattani et al., 2019; Wang et al., 2021). To date, electrochemical biosensors have been successfully fabricated to detect a variety of viruses such as dengue virus (DENV) (Navakul et al., 2017), Ebola virus (EBV) (Ilkhani and Farhad, 2018), human immune deficiency virus (HIV) (Hu et al., 2018), Zika virus (ZIKV) (Faria and Mazon, 2019), hepatitis A virus (HAV) (Manzano et al., 2018), and most recently SARS-CoV-2 (Farzin et al., 2021; Yakoh et al., 2021; Zhao et al., 2021). Currently, the biosensors for SARS-CoV-2 can be classified into two categories, immunological biosensors and nucleic acid-based biosensors. Compared to immunological diagnostic biosensors, nucleic acid-based biosensors indicate excellent performance in DNA/RNA analysis because they provide speedy molecular diagnostics based on specific DNA/RNA sequences detection when the immune responses are still too low (Kudr et al., 2021). Therefore, nucleic acid-based biosensors can become the most efficient applicant to the instantaneous monitoring of COVID-19. Especially, with the continuous development of nanomaterial and signal amplification strategies, powerful biosensors with high sensitivity have emerged for clinical diagnosis and POC testing.
In the present study, an ultrasensitive and nucleic acid amplification-free electrochemical biosensor for sensitive and specific diagnosis of SARS-CoV-2 was designed using a SPCE reinforced by boron nitride quantum dots/flower-like gold nanostructures (BNQDs/FGNs/SPCE). The BNQDs, an emerging zero-dimensional nanomaterial with a structure similar to graphene, due to its outstanding capabilities such as chemical stability, nontoxicity, and high quantum yield (Lee et al., 2021; Yang et al., 2021), and FGNs because of their favorable biocompatibility, easy functionalization, and high electrical/thermal conductivities (Chen et al., 2019a; Theerthagiri et al., 2021) were used to amplify the electrical signal. The ssDNA probe specific for SARS-CoV-2 RNA (designed based on analysis of SARS-CoV-2 genomes) was directly immobilized onto the modified electrode surface by covalently attaching the thiolated DNA probe to gold nanoparticles through thiol-gold chemistry. The fabricated biosensor was incubated with an artificial target sequence/RNA isolated from clinical samples at room temperature for 30 min. The turn-off and turn-on detection strategies were employed based on electrical signal of redox marker [Fe(CN)6]3-/4- and electrical signal of electroactive dye methylene blue (MB), respectively. Electrochemical impedance spectroscopy (EIS), cyclic voltammetry (CV), and atomic force microscope (AFM) were used to study the electrochemical characterization and surface characteristics of the biosensor. Under optimal conditions, the sensitivity, specificity, reproducibility, stability, and accuracy of the biosensor were evaluated. Strong agreement between biosensor results and conventional RT-PCR clearly demonstrated the ability of the biosensor to sensitively and specifically diagnose SARS-CoV-2 in real clinical samples. The fabricated biosensor may be a potential candidate for accurate, sensitive, and timely diagnosis of SARS-CoV-2 in clinical applications.
2. Experimental section
2.1. Reagents and instrumentation
Details of the reagents and instrumentation are included in the supplementary data file.
2.2. Synthesis of BNQDs
Details of the synthesis of BNQDs are included in the supplementary data file.
2.3. Antisense oligonucleotide design
In the present study, the specific antisense oligonucleotide (ssDNA capture probe) was selected based on the sequence of the Orf1ab gene within the SARS-CoV-2 genome. According to research, this gene has shown significant sensitivity for molecular diagnosis of SARS-CoV-2 (Liu et al., 2021b; Moitra et al., 2020). First, we collected a set of the Orf1ab gene sequences from different SARS-CoV-2 genotypes (Table S1) at NCBI SARS-CoV-2 database. Then, alimenting of selected viral sequences was done by ClustalW algorithm of Bio Edit sequence alignment editor v 7.2.5 (Hall, 1999). Several candidate probe sequences from the highly conserved regions of Orf1ab gene were chosen. Next, the hotspots and topologic-thermodynamics parameters of sequences were evaluated by the GISAID database (global initiative on sharing avian flu data, www.gisaid.org) and Gene Runner software version 6.0.11, respectively (Maxmen, 2021; Wong et al., 2004). The specificity performance of DNA probe was analyzed by “BLASTn” search algorithm for the potential hybridization to the unintended targets (Eklund et al., 2010). The results of BLASTn, guaranteed the high specificity of the designed DNA probe (Table S2). The sequences of the oligonucleotides used in this study are given in Table S3.
2.4. Sensing platform fabrication
First, 10 μL of BNQDs suspension was dropped onto the clean screen-printed carbon electrode (SPCE) surface and dried completely at room temperature to construct BNQDs/SPCE. Then, the BNQDs/SPCE was immersed in 2.0 mM HAuCl4 prepared in 0.1 M KCl and flower-like gold nanostructures (FGNs) were electrodeposited on the BNQDs/SPCE by applying a constant potential of +0.3 V for 1800 s. This process gave FGNs/BNQDs/SPCE.
For ssDNA probe immobilization, the thiolated-probe DNA was initially activated using TCEP solution (the concentration ratio of TCEP to thiolated DNA was 100:1) for 1 h at room temperature (Ge et al., 2019). Thereafter, 10 μL of the 5.0 μM activated thiolated DNA prepared in phosphate buffer (0.05 M, pH 7.4) containing 0.10 M NaCl was added onto the FGNs/BNQDs/SPCE surface and incubated in a humidified atmosphere chamber at 25 °C for 90 min. In this way, the ssDNA probe was attached to the electrode surface via dative binding between FGNs and free-SH groups of the DNA. The as prepared ssDNA/FGNs/BNQDs/SPCE was then thoroughly rinsed with phosphate buffer (PB, 0.05M, pH 7.4) to get rid of unlinked ssDNA probe. Next, the nonspecific binding sites were closed with 10 μL of 3% bovine serum albumin (BSA) for 15 min at room temperature.
2.5. Electrochemical detection procedures
In this study, the turn-off and turn-on strategies were employed for diagnosis of SARS-CoV-2 RNA by differential pulse voltammetry (DPV) technique. The turn-off strategy was designed based on measuring the electrical signal of redox marker [Fe(CN)6]3-/4- to survey the hybridization event. In this assay, the decrease in the peak current intensity of ferrocyanide after hybridization is the result of electrostatic repulsion phenomenon of DNA/DNA or DNA/RNA duplexes and redox couple. For hybridization, a 10 μL droplet of phosphate buffer saline (PBS, pH 7.4) containing a certain concentration of detection samples (synthetic complementary ssDNA target/SARS-CoV-2 RNA isolated from clinical samples) was dropped onto the electrode surface followed by incubated in a humidified atmosphere chamber at 25 °C for 30 min. Subsequently, the electrode was washed with PB to remove non-hybridized DNA/RNA molecules. DPV measurements were done between - 0.5 V and +0.6 V with a scan rate of 50 mV s−1 in 5.0 mM [Fe(CN)6]3-/4- containing 0.1 M KCl.
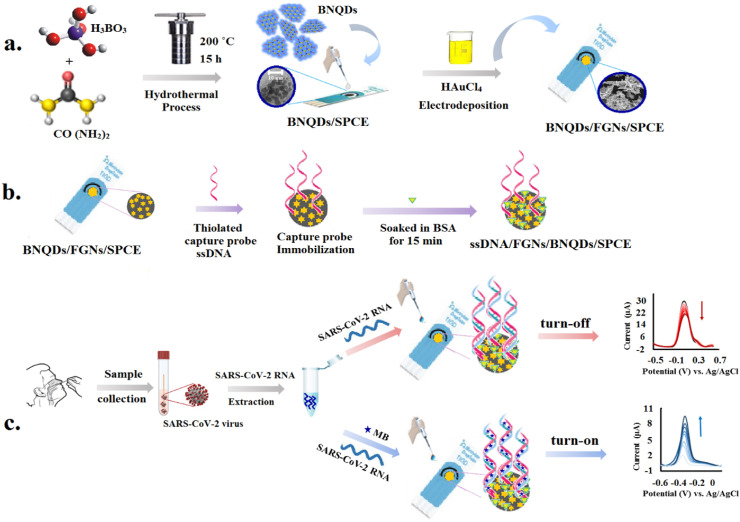
Likewise, the turn-on strategy was designed based on measuring the electrical signal of electroactive dye methylene blue (MB) to monitor the hybridization event. MB, as a redox indicator, can be embedded into the dsDNA and DNA/RNA skeleton (Cinti and Arduini, 2017; Li et al., 2021). When complementary ssDNA target or SARS-CoV-2 RNA is present in the detection system, hybridization causes more MB molecules to be adsorbed onto the electrode surface. Consequently, a strong electrochemical signal is generated. In this system, hybridization was performed by depositing 10 μL of the PBS containing 300 nM MB as redox marker and a certain concentration of detection samples (synthetic complementary/SARS-CoV-2 RNA isolated from clinical samples) and followed by incubation in a humidified atmosphere chamber at 25 °C for 30 min. Afterward, the electrode was washed with PB to eliminate unfixed sequences. DPV measurements were conducted between - 0.6 V and +0.1 V with a scan rate of 50 mV s−1 in PB. For electrochemical characterization, CV and EIS measurements were done in 1 mM [Fe(CN)6]3-/4- containing 0.1 M KCl. The CV voltammograms were recorded between - 0.6 V and +0.6 V with scan rates of 10–200 mV s−1. EIS measurements were recorded at a frequency range of 10–100 kHz with an amplitude of 10 mV. The construction process and sensing mechanism of the biosensors were presented in Scheme 1 .
Scheme. 1.
Schematic representation of SARS-CoV-2 detection using the electrochemical biosensor. (a) Synthesis of nanostructures; (b) Capture probe immobilization and electrode preparation; (c) Process of electrochemical detection using turn-off and turn-on strategies.
2.6. Preparation of clinical samples
To verify the preliminary practicability and reliability of the biosensing strategy, 120 RNA samples isolated from anonymized patients were evaluated simultaneously by the proposed electrochemical biosensor and RT-PCR method. Clinical RNA samples were obtained from Imam Reza Hospital (Mashhad, Iran) in the form of purified RNA extracted by QIAamp Viral RNA Mini Kit (QIAGEN, Germany) and used without requiring pre-treatment. The RT-PCR experiments were performed using a commercial Pishtaz Teb Diagnostic COVID-19 RT-PCR Kit (Pishtaz Teb, Iran). Amplification conditions comprised reverse transcription at 50 °C for 20 min, cDNA initial denaturation at 95 °C for 3 min, denaturation at 94 °C for 10 s, annealing, extension, and fluorescence measurement at 55 °C for 40 s, followed by cooling at 25 °C for 60 s.
3. Results and discussion
3.1. SARS-CoV-2 biosensor characterization
3.1.1. Morphological characterization
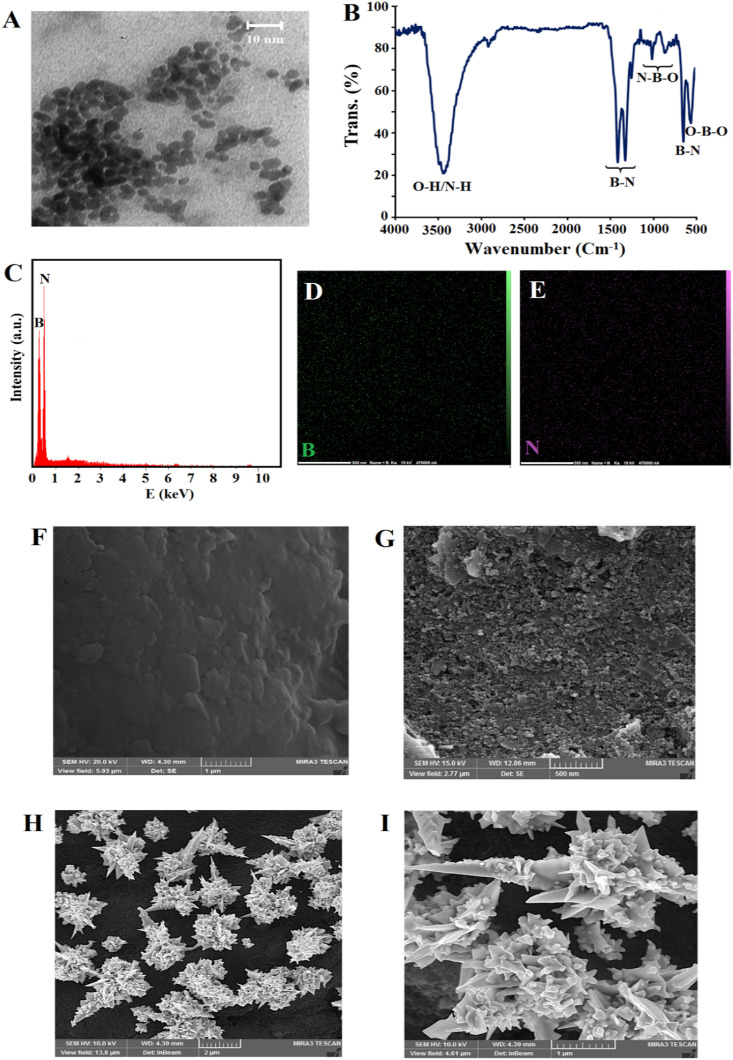
At first, the morphology, functional groups, and chemical compositions of synthesized BNQDs was studied by TEM, FT-IR, and EDX analysis. Fig. 1 A exhibits a TEM image of BNQDs with uniform size distribution, good dispersibility, and average diameter of ∼5 nm.
Fig. 1.
TEM image (A), FT-IR spectra (B), EDX analysis (C), and corresponding elemental mapping data of BNQDs (D), (E), FE-SEM images of bare SPCE (F), BNQDs/SPCE (G), FGNs/BNQDs/SPCE in different magnitudes; 2 μm (H) and 1 μm (I).
In the FT-IR spectra of BNQDs (Fig. 1B), broad bands at 3450 and 3140 cm−1 were attributed to stretching modes of O–H and N–H groups (Ding et al., 2021). The bands at 1340, 1378, and 770 cm−1 were ascribed to the stretching and bending vibration of B–N (Hatamluyi et al., 2020a). The characteristic bands at 1170, 890, and 610 cm−1 related to the N–B–O and O–B–O bonds, respectively, proving the oxygen-containing functional groups were attached on the BNQDs surface (Li et al., 2015).
Further evidence for the formation of BNQDs was obtained by EDX analysis, as shown in Fig. 1C, D, and E. The EDX spectrum of BNQDs indicated the strong peaks of boron and nitrogen and corresponding elemental mapping confirmed the two elements B and N were evenly distributed over all BNQDs.
Secondly, the morphology of modified electrodes were investigated via FE-SEM. Comparison of the FE-SEM images of bare SPCE (Fig. 1F) and BNQDs/SPCE (Fig. 1G), confirms that a uniform layer of BNQDs was formed on the SPCE surface. FE-SEM tests were also conducted to explore the morphology and the size of the gold nanostructures. As shown in Fig. 1H, gold nanostructures were successfully formed on the BNQDs/SPCE surface by electrosynthesis strategy. A higher magnification image clearly shows that these nanostructures have flower-like morphology (Fig. 1I).
3.1.2. Electrochemical characterization
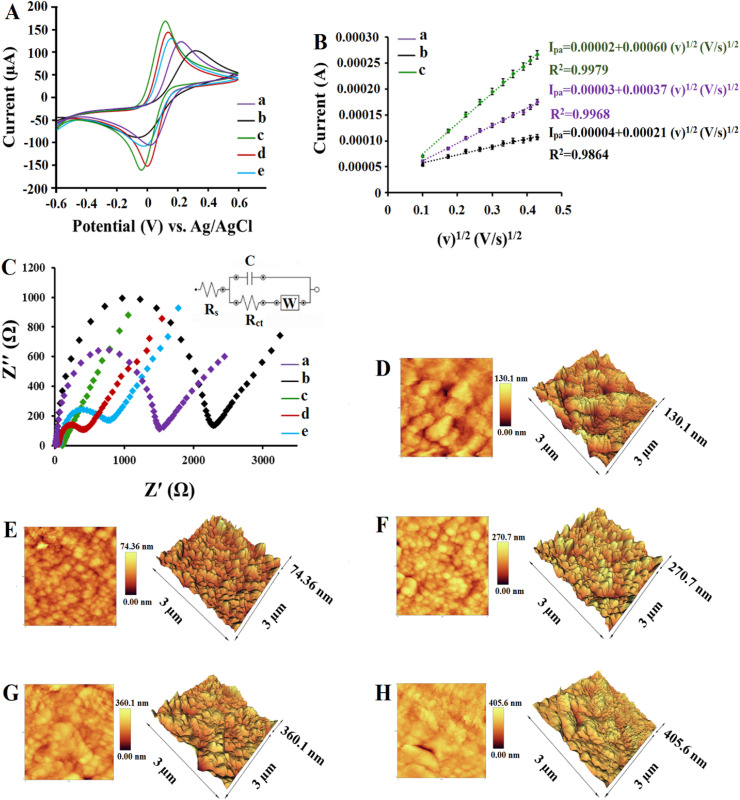
CV and EIS techniques were employed to verify the entire process of SARS-CoV-2 biosensing platform fabrication utilizing [Fe(CN)6]3−/4− ions as a reversible redox probe (Chen et al., 2019d). The impedance spectra for all steps were studied utilizing a Randles equivalent circuit. The Nyquist plots contain a semicircular portion and a linear portion. The diameter of the semicircular portion in the high-frequency region represents the electron transfer resistance (Ret) that controls the charge transfer at the electrode surface (Chen et al. 2019b, 2019c). Compared with the bare SPCE, BNQDs/SPCE presented decreasing redox currents (Fig. 2 A, curve a and b) and increasing Ret value from 1508 to 2259 Ω (Fig. 2C, curve a and b). These observations can be due to BN's insulator character, which prevents the electron transport and clearly shows the electroactivity of the electrode was influenced by BNQDs (Angizi et al., 2018; Öndeş et al., 2021). When FGNs were electrodeposited on the BNQDs/SPCE surface, the significantly increased redox currents (Fig. 2A, curve c) and decreased Ret value to 126 Ω (Fig. 2C, curve c) proved that the electron transfer process was facilitated, due to expansion of the active surface area of the electrode, and improvement of the conductivity. With the introduction of the ssDNA probe, the negatively charged phosphate skeletons of the DNA probe was assembled on the surface of the FGNs/BNQDs/SPCE, and electron transfer was restricted due to the electrostatic repulsion so that the redox currents declined (Fig. 2A, curve d) and the value of Ret increased to 428 Ω (Fig. 2C, curve d). By performing hybridization between ssDNA probe and target DNA sequence, the redox currents further decreased (Fig. 2A, curve e) and Ret value increased to 918 Ω (Fig. 2C, curve e) due to blocking effects of negative charges and reduced access of the charged species to the sensing patform surface (Rizi et al., 2021; Zare et al., 2022). These CV and EIS experiments verified the successful construction of SARS-CoV-2 biosensing platform.
Fig. 2.
(A) CVs of different modified electrodes in 1.0 mM [Fe(CN)6]3-/4- containing 0.1 M KCl, (a) bare SPCE, (b) BNQDs/SPCE, (c) HFGNs/BNQDs/SPCE, (d) ssDNA/HFGNs/BNQDs/SPCE, (e) dsDNA/HFGNs/BNQDs/SPCE. (B) Linear curves of (a) bare SPCE, (b) BNQDs/SPCE, and (c) HFGNs/BNQDs/SPCE in 1.0 mM [Fe(CN)6]3-/4- containing 0.1 M KCl at different scan rates (10–200 mV s−1). (C) EIS of different modified electrodes in1.0 mM [Fe(CN)6]3-/4- containing 0.1 M KCl, (a) bare SPCE, (b) BNQDs/SPCE, (c) HFGNs/BNQDs/SPCE, (d) ssDNA/HFGNs/BNQDs/SPCE, (e) dsDNA/HFGNs/BNQDs/SPCE. AFM images of (D) bare SPCE, (E) BNQDs/SPCE, (F) HFGNs/BNQDs/SPCE, (G) ssDNA/HFGNs/BNQDs/SPCE, and (H) dsDNA/HFGNs/BNQDs/SPCE.
In order to further study the effective surface area values of different electrodes, a set of CV measurements with different scan rates (10–200 mV s−1) were conducted according to the Randles-Sevcik equation (Hatamluyi and Es'haghi 2017). All three bare SPCE, BNQDs/SPCE, and FGNs/BNQDs/SPCE electrodes showed a linear dependency between resulting anodic peak currents and the square root of scan rate (Fig. 2B), which proved that the electrochemical reaction is under a diffusion control process (Hatamluyi et al., 2018). Based on the Randles-Sevich equation, the effective surface area of bare SPCE, BNQDs/SPCE, and FGNs/BNQDs/SPCE were calculated to be 0.49 Cm-2, 0.27 Cm-2, and 0.81 Cm-2, respectivly. These results are in good agreement with the presented results in Fig. 2A and C and confirm that FGNs have significantly improved the effective surface area of the electrode to assemble DNA probes for subsequent hybridization detection.
3.1.3. Atomic force microscopy analysis
To study the SPCE surface modifications upon functionalization by the BNQDs, GFNs, ssDNA probe and after hybridization, atomic force microscopy (AFM) was applied. Tapping mode three-dimensional AFM topographic and two-dimensional AFM phase profiles of bare and modified electrodes are displayed in Fig. 2. AFM images of the bare SPCE (Fig. 2D) and BNQDs/SPCE (Fig. 2E) revealed an obvious decrease in the average roughness (Ra) and root mean square roughness (Rq), from 87.86 nm to 30.03 nm and 102.01 nm–37.76 nm upon BNQDs casting, respectively, explained by the small and uniform size of the BNQDs and regular surface morphology compared with bare SPCE. After electrodeposition of FGNs, the electrode surface was coated with a large amount of flower-like gold nanostructures which had assembled in random organizing giving Ra and Rq increased to 82.60 nm and 101.05 nm, respectively (Fig. 2F).
After the immobilization of the ssDNA probe, the surface morphology as well as Ra and Rq values changed (Fig. 2G), revealing the successful formation of covalent bonds between FGNs and free –SH groups of DNA (Manzano et al., 2018). The Ra and Rq values of ssDNA/FGNs/BNQDs/SPCE were estimated at 138.41 nm and 177.35 nm, respectively. The change in Ra and Rq values to 197.63 nm and 235.73 nm upon incubation of the electrode with complementary strand of DNA confirmed hybridization on the surface of the biosensor, to the good extent.
3.2. Optimization of the experimental conditions
Optimizing experimental conditions is essential to achieve the best detecting efficiency. In this study, the optimization experiments were performed in two steps. In the first step, the factors affecting the construction of the biosensor platform were optimized and in the second step, the factors affecting the performance of the biosensor were studied and optimized.
3.2.1. Construction process optimization
During the construction of the biosensing platform, the influence of four important factors including the amount of BNQDs, the concentration of HAuCl4, deposition time, and deposition potential on sensor performance was investigated and optimized. These factors were analyzed at four levels via Taguchi standard orthogonal array methodology using L16 matrix (Hatamluyi et al., 2020b). The factors and their levels are shown in Table S4. Using [Fe(CN)6]3-/4- as a redox probe, the electrochemical performances of various electrodes were examined by checking their CV responses. In all cases, higher ΔI (ΔI = the anodic peak current of FGNs/BNQDs/SPCE minus the anodic peak current of ssDNA/FGNs/BNQDs/SPCE) values evidenced a higher sensitivity of the fabricated biosensor. The design matrix for 16 experiments and the average responses of each experiment are summarized in Table S5 (n = 3). Table S6 shows the average signal-to-noise (S/N) ratio for each level of considered factors. In this table, the highest S/N ratio reveals the optimal level of that factor, and the highest delta value determines the most effective factor among all the considered factors (Hatamluyi and Es'haghi 2018). Accordingly, it can be understood that deposition potential is the most significant factor, and the concentration of HAuCl4, deposition time, and amount of BNQDs are at the next levels of importance. This is because the deposition potential affects the size and morphology of the formed gold nanostructures on the electrode surface thus affecting the kinetics and thermodynamics of the assembly, bonding, and electron transfer (Bin et al., 2010; Soleymani et al., 2009; Su et al., 2016). The main effects diagram of the S/N ratio as a function of the examined factors is shown in Fig. S1. In this diagram, the highest S/N ratio indicates the optimal level of that factors (Hatamluyi et al., 2021). As it is obvious, the results of Table S6 are confirmed by the results of Fig. S1. As a result, the biosensor was modified under the following optimal conditions: 10 μL of BNQDs, 2.0 mM of HAuCl4, 1800 s for deposition time, and +0.3 V for deposition potential was applied. The analysis of variance (ANOVA) was also employed to further validate and confirm the experimental design. In Table S7, a lower P-value implies that the factor is significant. Therefore, the ANOVA results are also in good agreement with the results of Table S6.
3.2.2. Sensing process optimization
Upon successful modification of electrode surface with BNQDs/FGNs nanocomposite, several parameters including ssDNA probe concentration, probe immobilization time, and hybridization time were optimized to favor the hybridization between ssDNA probe and target sequence.
In the turn-off method, hybridization between the probe and complementary target DNA causes a significant reduction in the ferrocyanide redox signal, due to DNA double helix hold more negative charges and the electrostatic repulsion would reduce the ability of electrons transfer (Zhou et al., 2019). Therefore, changes in DPV peak current intensity (ΔI) of ferrocyanide before and after hybridization was used to survey the DNA hybridization event and quantify the results accurately.
In the turn-on method, MB molecules, as an electrochemical indicator, intercalate into the DNA double helix significantly amplifying the electrochemical signal (Li et al., 2021). Thus, the electrochemical signal of MB was used in the detection of DNA hybridization.
The ssDNA probe concentration is an important factor affecting the performance of the biosensor.
The effect of probe concentration on signal response was studied in the range of 0.5–10 μM. As depicted in Figs. S2A and D, in both methods signal response, reached the top when the probe concentration was 5.0 μM. Weak responses at lower 5.0 μM may be related to insufficient amount of immobilized ssDNA probe on the electrode surface and the slight decrease in response signals at concentrations greater than 5.0 μM is due to the blocking effect of extra mass of immobilized DNA single strands. Therefore, 5.0 μM was selected as the optimum concentration of ssDNA probe.
The incubation time for ssDNA probe immobilization on the electrode surface was examined between 15 and 150 min. In both methods, signal response increased with increasing immobilization time up to 90 min; however, no considerable increase in the signal response was observed over this time (Figs. S2B and E). Therefore, we chose an optimal immobilization time of 90 min to ensure favorable immobilization of the ssDNA probe on the electrode surface.
In the case of biosensors for point-of-care systems, it is well known the importance of hybridization time. Consequently, to achieve a short time with a sensitive response, the response signals at different incubation times including 10, 15, 20, 30, 45, 60, and 90 min were measured. Results in Figs. S2C and F demonstrate a minimum hybridization time of 30 min necessary to obtain a reproducible and stable hybridization, obviously less than most of the previous reports.
3.3. Performance of the biosensing strategy
The analytical performance of the developed assay was initially evaluated using the artificial ssDNA target in standard solutions by recording DPVs before and after 30 min incubation of the ssDNA/FGNs/BNQDs/SPCE with different concentrations of the target sequence. Following that, the practical application value of the biosensor in clinical samples was investigated.
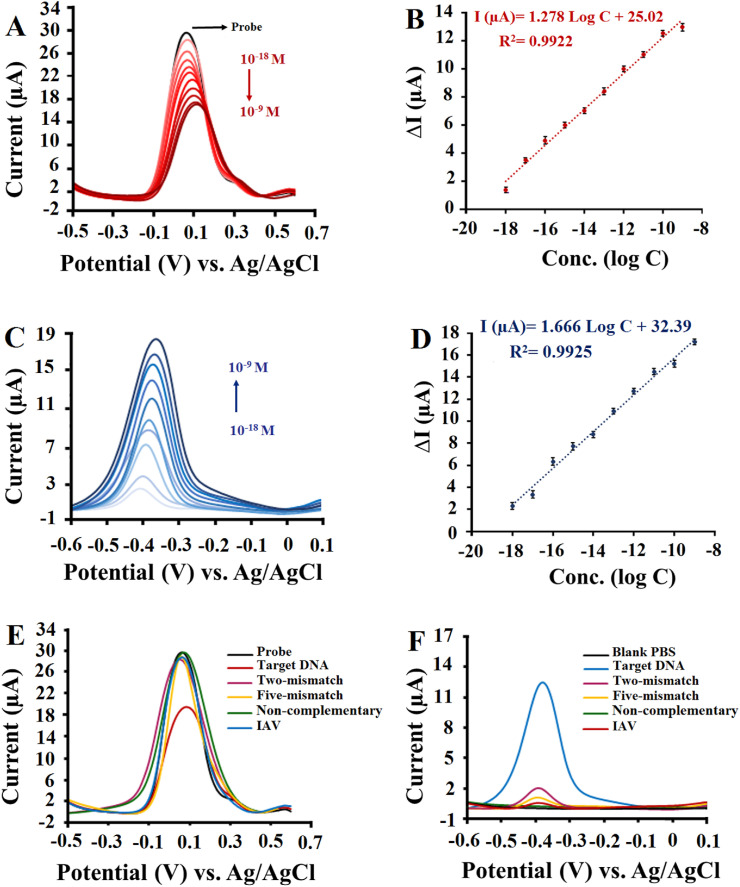
For this purpose, the ability of quantitative analysis of proposed biosensor based on both turn-off and turn-on methods were examined individually. Fig. 3 A demonstrates DPVs of the biosensor based on the turn-off analysis method. As can be seen the change in current (ΔI), ΔI is equal to the difference between the peak height of ssDNA probe modified electrode and the peak height after hybridization reaction, presented an increasing trend with increasing target sequence concentration and revealed a good linear dependence with the logarithm concentration of ssDNA target in the range from 10−18 to 10−9 M (Fig. 3B). For this response, the linear equation was: ΔI (μA) = 1.278 log C + 25.02 and limit of detection (LOD) was 0.48 aM, calculated as 3Sb/m.
Fig. 3.
(A) DPV signals based on turn-off method in 1.0 mM [Fe(CN)6]3-/4- containing 0.1 M KCl for different concentrations of ssDNA Target. (B) The resulting calibration plots for log [C] vs. DPV responses in the range of 10−18 -10−9 M. (C) DPV signals based on turn-on method in 0.01 M PBS (pH 7.4) for different concentrations of ssDNA Target. (D) The resulting calibration plots for log [C] vs. DPV responses in the range of 10−18 -10−9 M. The error bars represent the standard deviations measured by three independent measurements. (E) DPV signals based on turn-off method for the complementary target, two-mismatch, five-mismatch, Non-complementary of 10−12 M, and IAV isolated from clinical sample (F) DPV signals based on turn-on method for the complementary target, two-mismatch, five-mismatch, Non-complementary of 10−12 M, and IAV isolated from clinical sample.
The resulting DPVs with the corresponding standard curve based on the turn-on method are shown in Fig. 3C and D. It can be seen that the MB peak current intensity increases with the increasing target sequence concentration from 10−18 to 10−9 M. Obviously, a good linear dependence was obtained between the MB oxidation peak current and logarithm concentration of ssDNA target with a corresponding linear equation of I (μA) = 1.666 log C + 32.39 and a limit of detection of 0.27 aM. From Fig. 3, both turn-off and turn-on methods demonstrated similar performance. Upon the introduction of the complementary sequences, the successful formation of dsDNA was confirmed by a significant increase in ΔI values in the turn-off method and MB peak current intensity in the turn-on method.
The performance comparison of different techniques for SARS-CoV-2 detection based on different gene fragments (i.e., E, N, S, and Orf1ab) are listed in Table S8. The detailed comparison results in Table S8 obviously reveal that the introduced biosensor has wide linear range and a comparable or even lower detection limit versus other biosensors for SARS-CoV-2 quantitative analysis.
3.4. Selectivity, reproducibility, and stability of the biosensing strategy
To evaluate the specificity property of the proposed biosensor, DPV responses of the biosensor incubated with 10−12 M complementary target, two-base-mismatched, five-base-mismatched, and non-complementary DNA sequences were investigated. The results in Fig. 3E and F shows that based on both turn-off and turn-on methods, only the complementary DNA sequence can induce a significant response. As expected, the electrochemical response for the non-complementary DNA sequence was equivalent to or less than the blank signal, whereas with a decrease in the number of mismatches the electrode response gradually increased. To further verify the selectivity of the biosensor in the diagnostic of SARA-CoV-2 in actual samples, the biosensor was challenged with the RNA fragments isolated from Influenza A (IAV) clinical sample. As revealed in Fig. 3E and F, no notable response was found neither for turn-off method nor for turn-on method.
Figs. S3 and S4, shows the results of reproducibility tests obtained at 10−12 M target sequence concentration for inter-day and intra-day measurements with six different biosensors. The inter-day relative standard deviation (RSD) values of six measurements were calculated to be 3.39% and 3.48% based on turn-off and turn-on methods, respectively, and intra-day RSD of six measurements were obtained 4.59% and 5.44% based on turn-off and turn-on methods, respectively, demonstrating appropriate reproducibility of the sensing strategies (Figs. S3 and S4).
The long-term stability of the proposed biosensor was also investigated. For studying this feature, a series of fabricated biosensors were stored at 4 °C for up to 20 days and used to test 10−12 M target sequence concentration. The fabricated biosensors exhibited stable readings over a period of 14 days. Based on turn-off method, the ΔI remained 94.7% and 89.6% of the initial value after 14 days and 20 days, respectively (Fig. S5 A), and based on turn-on method, the MB oxidation peak current remained 95.1% and 90.4% of the initial value after 14 days and 20 days, respectively (Fig. S5 B), suggesting a good stability.
3.5. Analytical application of the biosensing strategies
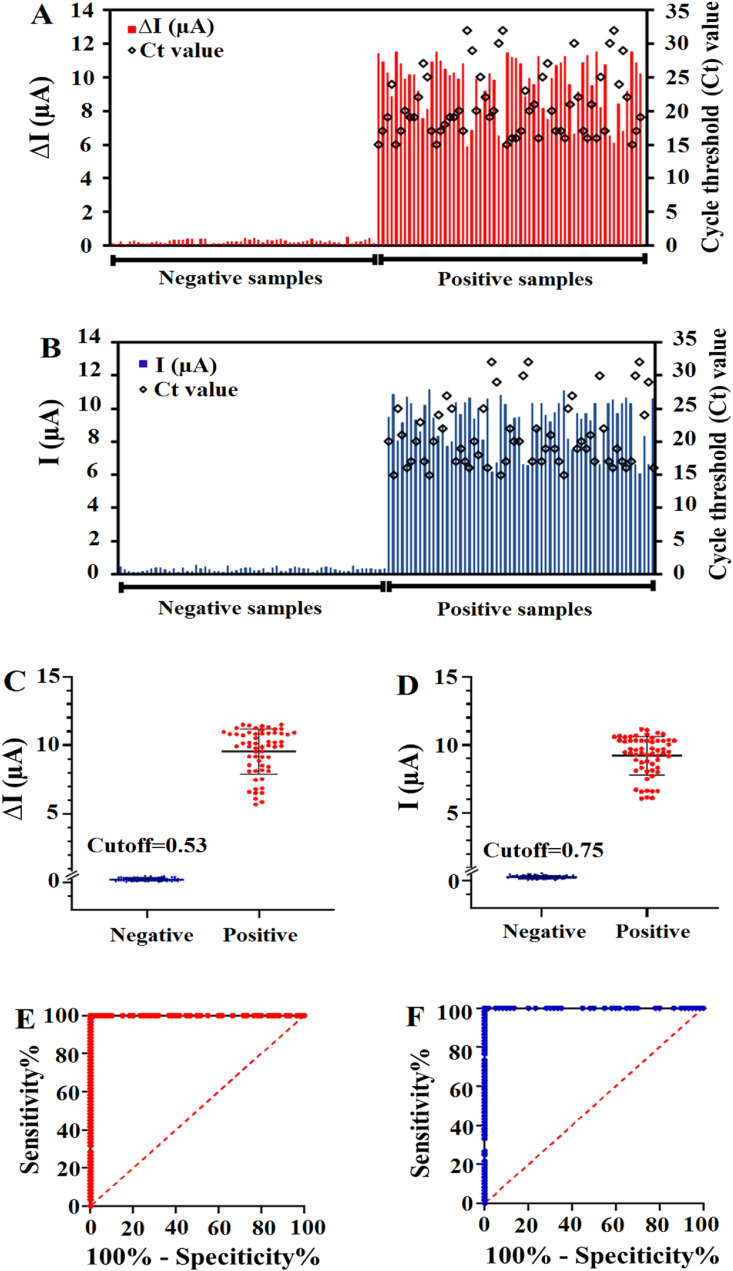
The applicability of the biosensor for clinical trials was examined by testing the 120 RNA sequences extracted from clinical nasopharyngeal swab specimens (60 SARS-CoV-2 positive and 60 SARS-CoV-2 negative samples confirmed by the RT-PCR). For this purpose, 60 SARS-CoV-2 negative samples were firstly analyzed based on both turn-off and turn-on strategies. These samples did not significantly change the electrochemical signals. The ΔI values and the MB peak current intensities of all 60 negative samples were rather low (<0.46 in the turn-off method and <0.53 in the turn-on method). In contrast, all the 60 SARS-CoV-2 positive samples exhibited a strong change in electrochemical signals with minimum intensity values that were greater than 5.86 and 6.11 in the turn-off and turn-on methods, respectively. This means that electrochemical biosensor revealed negative results for SARS-CoV-2 negative samples and positive results for SARS-CoV-2 positive samples (Fig. 4 A and B). For further investigation, the results of the analysis of 120 RNA samples were recorded in the box-diagram (Fig. 4C and D) and the cutoff values were calculated to be 0.53 and 0.75 for the turn-off and turn-on strategies, respectively, based on the IUPAC method (Liu et al., 2021a). Receiver operating characteristic (ROC) curves were plotted to evaluate the discrimination ability of proposed strategies. Using both turn-off and turn-on strategies, the biosensor yielded area under curve (AUC) values of 1, proving higher specificity and accuracy of the proposed biosensor for distinguishing SARS-CoV-2 negative samples from positive samples (Fig. 4E and F). Moreover, a T-test was performed to detect the significance of difference between the negative and positive groups. According to the obtained results, significant differences between the two groups were found (p < 0.0001, Fig. S6) for both applied strategies. Therefore, in the turn-off method the ΔI values higher than 0.53 were defined as SARS-CoV-2 positive, whereas those with ΔI values lower than 0.53 were considered as SARS-CoV-2 negative. As well in the turn-on method the MB peak current higher than 0.75 was defined as SARS-CoV-2 positive, whereas those with intensities lower than 0.75 were considered as SARS-CoV-2 negative.
Fig. 4.
The detection of SARS-CoV-2 RNA in 120 clinical specimens using DPV and RT-PCR methods. The electrochemical signals are depicted in bar charts based on turn-off (A) and turn-on (B) strategies. Box-diagram for the quantitative detection of SARS-CoV-2 RNA in 120 clinical samples by ssDNA/FGNs/BNQDs/SPCE based on turn-off (C) and turn-on (D) strategies. ROC analysis to assess the detection capability of the biosensor based on turn-off (E) and turn-on (F) strategies.
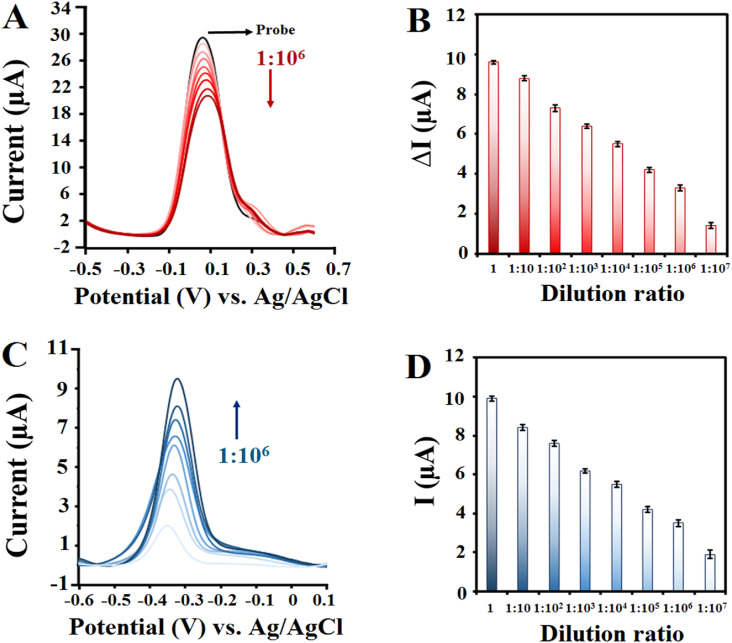
Further experiments of this study focused onto determine the absolute quantitation capacity of the biosensor. For this purpose, the detection performance of the biosensor and conventional RT-PCR for continuously diluted RNA sequences isolated from clinical specimens was evaluated. These included SARS-CoV-2 RNAs extracted from 3 positive patients with cycle threshold = 20. Serial dilutions of isolated RNA samples (dilution ratio from 1:10 to 1:1010) were analyzed by the ssDNA/FGNs/BNQDs/SPCE and RT-PCR, simultaneously. As shown in Fig. 5 , based on both turn-off and turn-on assays, RNA samples almost showed concentration gradient-dependent signal responses. Owing to the high sensitivity, ΔI values (based on turn-off method) and MB peak current intensities (based on turn-on method) all exceeded the cut-off values, and SARS-CoV-2 RNA is still detectable even after more than 107-fold dilution, while the RT-PCR method showed up to 105-fold dilution as positive samples and reported other dilutions as negative samples. Compared with the RT-PCR results presented in Table S9, the LOD of our proposed biosensor is 100-fold lower than RT-PCR. Considering that the LOD of the Pishtaz Teb Diagnostic COVID-19 RT-PCR Kit, which was used in this study, was 200 copies/mL as claimed by the manufacturer, it can be found that our biosensor has a LOD of 2 copies/mL.
Fig. 5.
DPV signals and corresponding current intensities for SARS-CoV-2 RNA extracted from positive patient (CT = 20) with different dilution ratios based on turn-off (A), (B) and turn-on (C), (D) strategies.
In similar previous studies in the field of electrochemical biosensors for the detection of SARS-CoV-2, the concentration of transcribed fragments of the gene was first determined and the copy number concentration was calculated based on the length and weight of the fragment. Then serial dilutions were prepared from that and the LOD of the biosensor was calculated by using transcribed fragments as templates. However, the novelty of this study is that we have achieved the LOD of the biosensor in the matrices of RNAs isolated from real patient samples, which, to the best of our knowledge, have not been reported before. The obtained results of our biosensor proved that the presence of each class of interfering species in the extracted RNA matrices did not significantly interfere with the detection of SARS-CoV-2. This suggested that low-concentrated SARS-CoV-2 RNA from real clinical samples could be accurately recognized with as short as 30 min of incubation time by our proposed biosensor.
Overall, the proposed biosensor offers several inherent advantages over other electrochemical biosensors reported for SARS-CoV-2 detection. First, the biosensor showed quantitative detection ability of SARS-CoV-2 with low-expression level, with detection limit down to attogram level in synthetic target and 2 copies/mL in real RNA samples. Second, the biosensor revealed a significant difference between the two positive and negative groups with 100% sensitivity and 100% specificity. Third, the biosensor allowed for ultrahigh sensitive quantification of SARS-CoV-2 in the presence of interfering species in the extracted RNA matrices.
4. Conclusions
In the present study, we developed a novel ssDNA/FGNs/BNQDs/SPCE platform and then demonstrated its ability to accurately and quickly analyze the SARS-CoV-2 RNAs isolated from 120 real clinical samples (60 positive and 60 negative samples confirmed by RT-PCR COVID-19 diagnostic kit) without the need for nucleic acid amplification. Two commonly used methods in electrochemical assays, turn-off and turn-on strategies, were applied to determine the SARS-CoV-2 using the DPV technique after 30 min incubation with extracted RNAs. The good agreement of the obtained data with the conventional RT-PCR method and successful diagnosis of SARS-CoV-2 in real clinical samples with different virus loads revealed a promising prospect for the suggested biosensor to POC applications.
CRediT authorship contribution statement
Behnaz Hatamluyi: Investigation, Conceptualization, Methodology, Data curation, Re-sources, Software, Writing – original draft. Majid Rezayi: Conceptualization, Methodology, Validation. Saeid Amel Jamehdar: Investigation, Conceptualization, Methodology, Validation, Writing – review & editing. Kobra Salimian Rizi: Investigation, Software. Majid Mojarrad: Conceptualization, Methodology, Validation. Zahra Meshkat: Conceptualization, Methodology, Validation. Hamzeh Choobin: Conceptualization, Methodology, Validation. Saman Soleimanpour: Conceptualization, Methodology, Validation. Mohammad Taher Boroushaki: Project administration, Supervision, Conceptualization, Methodology, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was financially supported by; 1) Iran National Science Foundation (INSF No. 99004067), 2) The Deputy of Research, Mashhad University of Medical Sciences (Grant No. 990524), Mashhad, Iran.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114209.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Angizi S., Hatamie A., Ghanbari H., Simchi A. ACS Appl. Mater. Interfaces. 2018;10(34):28819–28827. doi: 10.1021/acsami.8b07332. [DOI] [PubMed] [Google Scholar]
- Bin X., Sargent E.H., Kelley S.O. Anal. Chem. 2010;82(14):5928–5931. doi: 10.1021/ac101164n. [DOI] [PubMed] [Google Scholar]
- Chen J., Hu W., Wei J., Yu F., Wu L., Wang C., Wang W., Zuo S., Shang B., Chen Q. Mikrochim. Acta. 2019;186(6):388. doi: 10.1007/s00604-019-3497-3. [DOI] [PubMed] [Google Scholar]
- Chen Y., Mei L.-P., Feng J.-J., Yuan P.-X., Luo X., Wang A.-J. Biosens. Bioelectron. 2019;145:111638. doi: 10.1016/j.bios.2019.111638. [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang A.-J., Yuan P.-X., Luo X., Xue Y., Feng J.-J. Biosens. Bioelectron. 2019;132:294–301. doi: 10.1016/j.bios.2019.02.057. [DOI] [PubMed] [Google Scholar]
- Chen Y., Yuan P.-X., Wang A.-J., Luo X., Xue Y., Zhang L., Feng J.-J. Biosens. Bioelectron. 2019;126:187–192. doi: 10.1016/j.bios.2018.10.057. [DOI] [PubMed] [Google Scholar]
- Choi M.H., Lee J., Seo Y.J. Anal. Chim. Acta. 2021;1158:338390. doi: 10.1016/j.aca.2021.338390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S., Arduini F. Biosens. Bioelectron. 2017;89:107–122. doi: 10.1016/j.bios.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Euro Surveill. 2020;25(3):2000045. [Google Scholar]
- Ding Y., He P., Li S., Chang B., Zhang S., Wang Z., Chen J., Yu J., Wu S., Zeng H., Tao L. ACS Nano. 2021;15(9):14610–14617. doi: 10.1021/acsnano.1c04321. [DOI] [PubMed] [Google Scholar]
- Eklund A.C., Friis P., Wernersson R., Szallasi Z. Nucleic Acids Res. 2010;38(4):e27. doi: 10.1093/nar/gkp1116. e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria A.M., Mazon T. Talanta. 2019;203:153–160. doi: 10.1016/j.talanta.2019.04.080. [DOI] [PubMed] [Google Scholar]
- Farzin L., Sadjadi S., Sheini A., Mohagheghpour E. Mikrochim. Acta. 2021;188(4):121. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.-G., Zhu J.-H., Wang A.-J., Mei L.-P., Luo X., Feng J.-J. Sens. Actuators, B. 2021;346:130501. [Google Scholar]
- Gattani A., Singh S.V., Agrawal A., Khan M.H., Singh P. Anal. Biochem. 2019;579:25–34. doi: 10.1016/j.ab.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Ge L., Li B., Xu H., Pu W., Kwok H.F. Biosens. Bioelectron. 2019;132:210–216. doi: 10.1016/j.bios.2019.02.051. [DOI] [PubMed] [Google Scholar]
- Hall T. 1999. Nucleic Acids Symp. Ser; pp. 95–98. [Google Scholar]
- Hatamluyi B., Es'haghi Z. J. Electroanal. Chem. 2017;801:439–449. [Google Scholar]
- Hatamluyi B., Es'haghi Z. Electrochim. Acta. 2018;283:1170–1177. [Google Scholar]
- Hatamluyi B., Hashemzadeh A., Darroudi M. Sens. Actuators, B. 2020;307:127614. [Google Scholar]
- Hatamluyi B., Lorestani F., Es’haghi Z. Biosens. Bioelectron. 2018;120:22–29. doi: 10.1016/j.bios.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Hatamluyi B., Rezayi M., Beheshti H.R., Boroushaki M.T. Sens. Actuators, B. 2020;318:128219. [Google Scholar]
- Hatamluyi B., Sadeghian R., Malek F., Boroushaki M.T. Food Chem. 2021;357:129782. doi: 10.1016/j.foodchem.2021.129782. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li H., Li J. J. Electroanal. Chem. 2018;822:66–72. [Google Scholar]
- Hwang C., Park N., Kim E.S., Kim M., Kim S.D., Park S., Kim N.Y., Kim J.H. Biosens. Bioelectron. 2021;185:113177. doi: 10.1016/j.bios.2021.113177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilkhani H., Farhad S. Anal. Biochem. 2018;557:151–155. doi: 10.1016/j.ab.2018.06.010. [DOI] [PubMed] [Google Scholar]
- John A.J.U.K., He P.J.W., Katis I.N., Galanis P.P., Iles A.H., Eason R.W., Sones C.L. Anal. Chim. Acta. 2021;1185:339002. doi: 10.1016/j.aca.2021.339002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.Z.H., Hasan M.R., Hossain S.I., Ahommed M.S., Daizy M. Biosens. Bioelectron. 2020;166:112431. doi: 10.1016/j.bios.2020.112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. iScience. 2020;23(8):101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. TrAC Trends Anal. Chem. (Reference Ed.) 2021;136:116192. doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Wang C.-Y., Ko W.-C., Hsueh P.-R. J. Microbiol. Immunol. Infect. 2021;54(2):164–174. doi: 10.1016/j.jmii.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidoudi Y., Sereme Y., Medkour H., Watier-Grillot S., Scandola P., Ginesta J., Andréo V., Labarde C., Comtet L., Pourquier P., Raoult D., Marié J.-L., Davoust B. One Health. 2021;13:100293. doi: 10.1016/j.onehlt.2021.100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S.E., Berube B.J., Pecor T., Cross E., Brown B.P., Williams B.D., Johnson E., Qu P., Carter L., Wrenn S., Kepl E., Sydeman C., King N.P., Baldwin S.L., Coler R.N. J. Immunol. Methods. 2021;499:113160. doi: 10.1016/j.jim.2021.113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Kshetri T., Park K.R., Kim N.H., Park O.-K., Lee J.H. Compos. B Eng. 2021;222:109089. [Google Scholar]
- Li H., Tay R.Y., Tsang S.H., Zhen X., Teo E.H. Small. 2015;11(48):6491–6499. doi: 10.1002/smll.201501632. [DOI] [PubMed] [Google Scholar]
- Li Y., Cai Y., Yang Y., Miao J., Xu Y. Sens. Actuators, B. l. 2021;335:129702. [Google Scholar]
- Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Sens. Actuators, B. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Li H., Jia Y., Mak P.-I., Martins R.P. Micromachines. 2021;12(2):197. doi: 10.3390/mi12020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- Maxmen A. Nature. 2021;593(7857):21. doi: 10.1038/d41586-021-01069-w. 21. [DOI] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navakul K., Warakulwit C., Yenchitsomanus P.-t., Panya A., Lieberzeit P.A., Sangma C. Nanomed. Nanotechnol. Biol. Med. 2017;13(2):549–557. doi: 10.1016/j.nano.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Öndeş B., Evli S., Uygun M., Aktaş Uygun D. Biosens. Bioelectron. 2021;191:113454. doi: 10.1016/j.bios.2021.113454. [DOI] [PubMed] [Google Scholar]
- Orooji Y., Sohrabi H., Hemmat N., Oroojalian F., Baradaran B., Mokhtarzadeh A., Mohaghegh M., Karimi-Maleh H. Nano-Micro Lett. 2021;13(1):18. doi: 10.1007/s40820-020-00533-y. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., He Y., Liu Z., Chen J. Anal. Chem. 2021;94(2):714–722. doi: 10.1021/acs.analchem.1c03051. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Rizi K.S., Hatamluyi B., Rezayi M., Meshkat Z., Sankian M., Ghazvini K., Farsiani H., Aryan E. Talanta. 2021;226:122099. doi: 10.1016/j.talanta.2021.122099. [DOI] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. J. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani L., Fang Z., Sargent E.H., Kelley S.O. Nat. Nanotechnol. 2009;4(12):844–848. doi: 10.1038/nnano.2009.276. [DOI] [PubMed] [Google Scholar]
- Su S., Wu Y., Zhu D., Chao J., Liu X., Wan Y., Su Y., Zuo X., Fan C., Wang L. Small. 2016;12(28):3794–3801. doi: 10.1002/smll.201601066. [DOI] [PubMed] [Google Scholar]
- Surkova E., Nikolayevskyy V., Drobniewski F. Lancet Respir. Med. 2020;8(12):1167–1168. doi: 10.1016/S2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova M., Skouridou V., Jauset-Rubio M., Viéitez I., Fernández-Villar A., Cabrera Alvargonzalez J.J., Poveda E., Bofill C.B., Sans T., Bashammakh A., Alyoubi A.O., O'Sullivan C.K. ACS Omega. 2021;6(51):35657–35666. doi: 10.1021/acsomega.1c05521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri J., Lee S.J., Karuppasamy K., Park J., Yu Y., Kumari M.L.A., Chandrasekaran S., Kim H.-S., Choi M.Y. J. Hazard Mater. 2021;420:126648. doi: 10.1016/j.jhazmat.2021.126648. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Verma N., Henderson J.A., Shen J. J. Am. Chem. Soc. 2020;142(52):21883–21890. doi: 10.1021/jacs.0c10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-Y., Feng Y.-G., Wang A.-J., Mei L.-P., Yuan P.-X., Luo X., Feng J.-J. Sens. Actuators, B. 2021;331:129460. [Google Scholar]
- Wong C.W., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D. Genome Res. 2004;14(3):398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.Y., Wang Y.Z., Wang L.L., Zhu J.W., Zhao J., Zong H.L., Chen C.X. Biosens. Bioelectron. 2021;191:113393. doi: 10.1016/j.bios.2021.113393. [DOI] [PubMed] [Google Scholar]
- Zare H., Meshkat Z., Hatamluyi B., Rezayi M., Ghazvini K., Derakhshan M., Sankian M., Neshani A., Aryan E. Talanta. 2022;238:123049. doi: 10.1016/j.talanta.2021.123049. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Emerg. microbes & infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Sens. Actuators, B. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Yang C., Xu H., Luo J., Zhang W., Tang X., Yang S., Fu W., Chang K., Chen M. Biosens. Bioelectron. 2019;126:657–663. doi: 10.1016/j.bios.2018.11.028. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wu Y., Ding L., Huang X., Xiong Y. TrAC Trends Anal. Chem. (Reference Ed.) 2021;145:116452. doi: 10.1016/j.trac.2021.116452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.