Summary
Human pluripotent stem cells (hPSCs) show promise for studying diseases affecting cell populations that are not easily available, including sensory neurons (SNs). Here, we present a differentiation protocol in chemically defined conditions to generate peripheral SNs from hPSCs. We describe four main steps: expansion of hPSCs, neural crest cell (NCC) differentiation, NCC dissociation and replating, and sensory neuron (SN) differentiation. This protocol enables generation of a mechanoreceptor-enriched culture or a population containing all three SN subtypes (nociceptors, mechanoreceptors, and proprioceptors).
For complete details on the use and execution of this protocol, please refer to Saito-Diaz et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell Differentiation, Neuroscience, Stem Cells
Graphical abstract
Highlights
-
•
Protocol for differentiation of hPSCs into different types of SNs from one culture
-
•
Step-by-step protocol for in vitro differentiation of NCCs
-
•
In vitro differentiation into mechanoreceptor-enriched culture
-
•
Differentiation of three main SN subtypes that mimic in vivo composition
Human pluripotent stem cells (hPSCs) show promise for studying diseases affecting cell populations that are not easily available, including sensory neurons (SNs). Here, we present a differentiation protocol in chemically defined conditions to generate peripheral SNs from hPSCs. We describe four main steps: expansion of hPSCs, neural crest cell (NCC) differentiation, NCC dissociation and replating, and sensory neuron (SN) differentiation. This protocol enables generation of a mechanoreceptor-enriched culture or a population containing all three SN subtypes (nociceptors, mechanoreceptors, and proprioceptors).
Before you begin
The protocol below describes a method to differentiate sensory neurons (SNs) from human embryonic stem cells (hESCs, H9, WA09) (Figure 1). The differentiated SNs undergo all developmental stages and can be used to obtain a mechanoreceptor-enriched culture, or DRG-like composition (nociceptors, mechanoreceptors, and proprioceptors). SNs obtained in this protocol can be replated to maintain the neurons for long periods of time (>70 days) if necessary. This replated step can be also used as an axotomy model (Saito-Diaz et al., 2021). Moreover, this protocol has also been successfully tested in patient-derived induced pluripotent stem cells (iPSCs). All the steps described here are carried out in a laminar flow Class II biological hood using proper aseptic technique to avoid contamination.
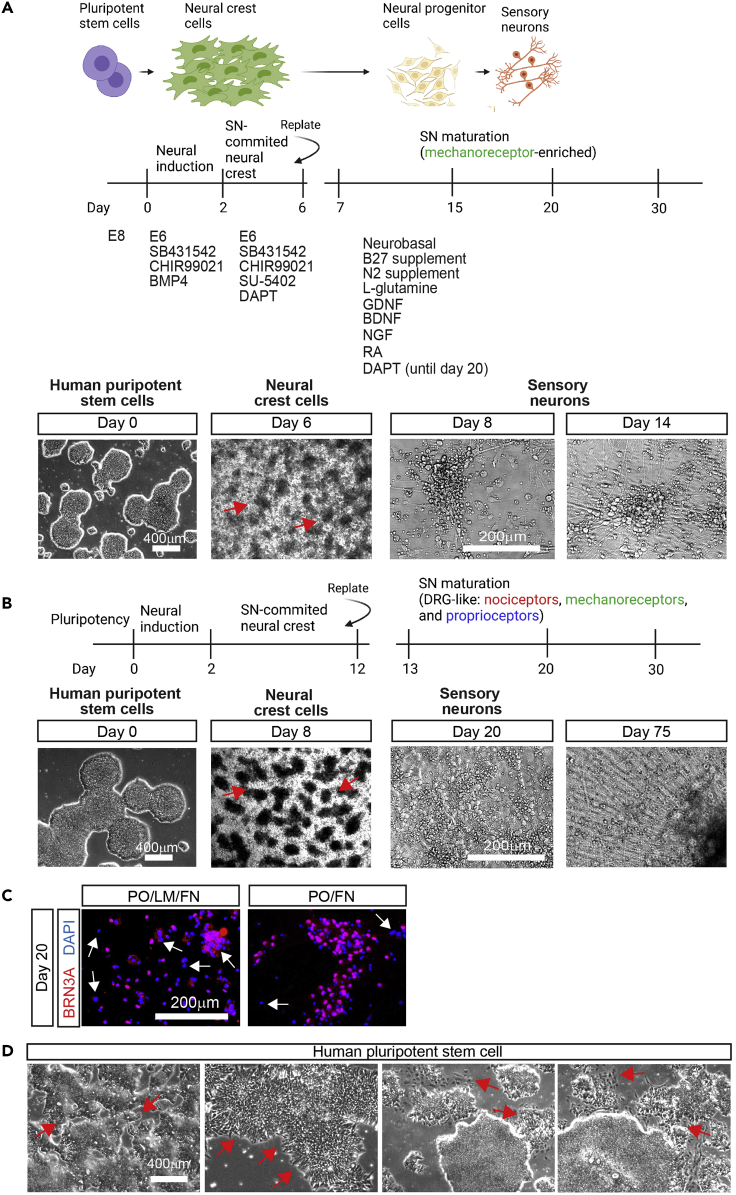
Figure 1.
Differentiation strategy
(A) Differentiation timeline of SNs replated on day 6 (to obtain mainly a population of mechanoreceptors). Developmental steps (top), growth factors used at each step of differentiation (middle) and representative brightfield images of each step (bottom) are shown. Red arrows indicated typical NCC aggregations (“ridges”).
(B) Differentiation timeline of SNs replated on day 12 (to obtain a DRG-like SN composition). Developmental steps (top) and representative brightfield images (bottom) are shown. Red arrows indicated typical “ridges” of NCCs.
(C) Effects of different coats on day 20 SNs. Day 12 NCCs were replated on day 12 on plates coated with PO/LM/FN or PO/FN and differentiated until day 20. SNs were then fixed and stained using the indicated antibodies. Note that the number of BRN3A- nuclei (arrows) decreases in PO/FN coated plates, indicating low number of contaminant cells.
(D) Phenotypes of hPSC colonies not suitable for differentiations. We observed that the following colonies will not yield a robust differentiation (arrows): small colonies very close to each other (left image), colonies with no defined edges (middle-left image), and colonies showing differentiated cells on the edges (middle-right and right images).
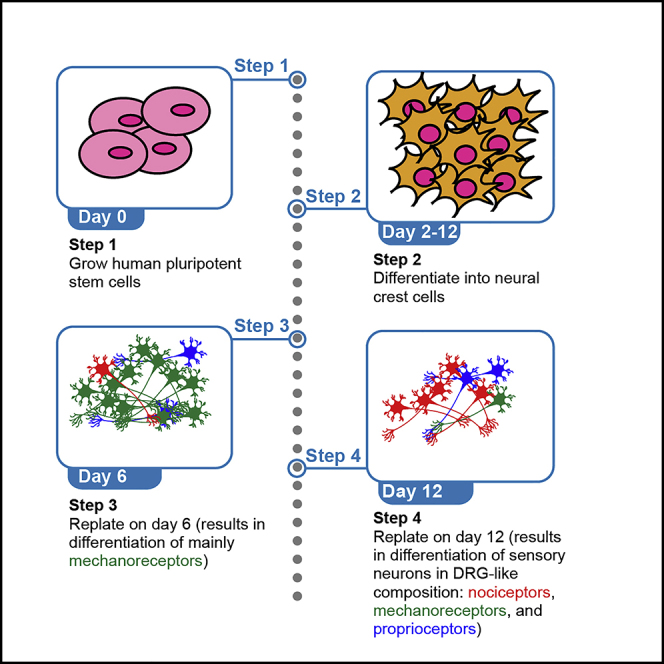
The main steps consist of: 1) Expansion of hPSCs, 2) Neural crest cell (NCC) differentiation stage, 3) NCC dissociation and replating stage, and 4) sensory neuron (SN) differentiation. NCCs can be dissociated and replated on day 6 to generate mainly mechanoreceptor SNs, or on day 12 to generate a DRG-like (with higher percentage of nociceptors) SN composition (Saito-Diaz et al., 2021) (Figures 1A and 1B).
We start the culture from 10 cm dishes with H9 cells. Each dish yields ∼12 × 106 cells. Thus, it is possible to differentiate one 6-well plate (10.8 × 106 cells) and one 4-well plate (1.52 × 106 cells) of NCCs from a single 10 cm dish. Similarly, one 6-well plate of NCCs yields ∼30 × 106 cells, which can be used to differentiate two 24-well plates of SNs (12 × 106 cells each). We recommend differentiating H9 cells in 6-well plates (for the NCC stage) for further replating and SN differentiation, and doing a differentiation in parallel in a 4-well plate to measure expression of NCC markers (such as SOX10) as a quality control (QC) check.
This protocol is detailed for one 6-well plate of NCCs. Three wells will be replated on day 6 to grow one 24-well plate of mechanoreceptors and the remaining wells will be replated on day 12 to grow a pool of nociceptors, mechanoreceptors, and proprioceptors. You will need to scale up the volumes for multiple plates or a different format.
Preparation of vitronectin (VTN) 10 cm dishes for hPSC expansion
Timing: 1 h
To prepare one 10 cm dish:
-
1.
Thaw a vial of VTN (0.5 mg/mL) in a water bath
-
2.
Dilute 7 μL VTN into 7 mL DPBS.
-
3.
Mix vigorously.
-
4.
Add mixture to a 10 cm dish.
-
5.
Incubate for 1 h at room temperature (RT; 20°C–22°C).
-
6.
Aspirate the mix.
-
7.
Use the plate immediately or add 8 mL DPBS and store at 37°C for up to 7 days. Make sure the dish does not dry out during this time by adding additional DPBS if necessary.
Preparation of VTN plates for NCC differentiation
Timing: 1 h
-
8.
Prepare one 6-well plate for differentiation.
-
9.
Thaw an aliquot of VTN (0.5 mg/mL) in a 37°C water bath.
Alternatives: Geltrex (ThermoFisher, Cat# A1413301) can be used instead of VTN. However, differentiation efficiency decreases (Saito-Diaz et al., 2021).
-
10.
Add 12 μL VTN to 12 mL DPBS.
-
11.
Mix and add 2 mL of the mixture to each well.
-
12.
Incubate for 1 h at RT.
-
13.
Use the plate immediately or add 2 mL DPBS to each well and store at 37°C for up to 7 days. Make sure the dish does not dry out during this time by adding additional DPBS if necessary.
Preparation of poly-L-ornithine/laminin/fibronectin (PO/LM/FN) plates for SN differentiation
Timing: 2 days
-
14.
Prepare one 24-well plate for differentiation.
-
15.
Add 6 μL of poly-L-ornithine hydrobromide (PO, 7.5 μg/mL) to 12 mL DPBS.
-
16.
Add 0.5 mL of the mixture to each well.
-
17.
Incubate at 37°C for 16–20 h.
-
18.
The next day wash each well twice with 0.5 mL DPBS.
-
19.
Add 12 μL Laminin (LM, 1 μg/mL) and 12 μL Fibronectin (FN, 1 μg /mL) to 12 mL DPBS.
-
20.
Mix and add 0.5 mL of the mixture to each well.
-
21.
Incubate at 37°C for ≥ 16 h.
Note: Coated plates can be stored for ∼7 days at 37°C. For longer storage (>1 week), seal them with Parafilm and store at 4°C. Make sure that the wells do not dry out by adding DPBS. These plates can be used on step 34 and step 52.
CRITICAL: SNs can grow in only PO/FN-coated plates which will reduce the presence of contaminant cell types (Figure 1C). Please note that SNs do not attach strongly on the PO/FN plates, thus should be handled with care to avoid detachment.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SOX10 (dilution: 1:100) | Santa Cruz | Cat# sc-365692, RRID:AB_10844002 |
| BRN3A (dilution: 1:100) | Merck Millipore | Cat# MAB1585, RRID:AB_94166 |
| TUJ1 (dilution: 1:1500) | BioLegend | Cat# 802001, RRID:AB_2564645 |
| TUJ1 (dilution: 1:1500) | BioLegend | Cat# 801201, RRID:AB_2313773 |
| TFAP2A (dilution: 1:500) | Abcam | Cat# ab108311, RRID:AB_10861200 |
| TRKB (dilution: 1:100) | R&D Systems | Cat# MAB3971, RRID:AB_2155144 |
| RET (dilution: 1:100) | Sigma-Aldrich | Cat# SAB1409600 |
| Chemicals, peptides, and recombinant proteins | ||
| Essential 8 Medium | Gibco | Cat# A1517001 |
| Essential 8 Medium supplement | Gibco | Cat# A1517001 |
| Essential 6 Medium | Gibco | Cat# A1516401 |
| Neurobasal Medium | Gibco | Cat# 21103-049 |
| Recombinant human BMP4 | R&D Systems | Cat# 314-BP |
| CHIR99021 | R&D Systems | Cat# 4423 |
| SB431542 | R&D Systems | Cat# 1614 |
| DAPT | R&D Systems | Cat# 2634 |
| SU5402 | Biogems | Cat## 2159233 |
| Y-27632 | Biogems | Cat# #1293823 |
| N2 Supplement | Gibco | Cat# 17502-048 |
| B-27 Supplement | Gibco | Cat# 12587-010 |
| L-glutamine | Thermo Fisher Scientific | Cat# 25030-081 |
| BDNF | R&D Systems | Cat# 248-BD |
| GDNF | PeproTech | Cat# 450-10 |
| NGF | PeproTech | Cat# 450-01 |
| Retinoic acid | Sigma-Aldrich | Cat# R2625 |
| Antibiotic-Antimycotic | Gibco | Cat# 15240096 |
| Poly-L-ornithine hydrobromide | Sigma-Aldrich | Cat# P3655 |
| EDTA (0.5 M, pH 8.0, RNase-free) | Thermo Fisher Scientific | Cat# AM9262 |
| NaCl (BioReagent, suitable for cell culture, ≥99%) | Sigma-Aldrich | Cat# S5886-500G |
| Dulbecco’s Phosphate-Buffered Saline (DPBS) | Corning | Cat# 21-031-CM |
| Recombinant human Vitronectin | Thermo Fisher Scientific | Cat# A31804 |
| Laminin-1 | Cultrex | Cat# 3401-010-02 |
| Fibronectin | Corning | Cat# 47743-654 |
| Trypan Blue Solution (0.4% w/v in PBS, pH 7.5) | Corning | Cat# 45000-717 |
| TRIZOL | Invitrogen | Cat# 15-596-018 |
| Accutase | Innovative Cell Technologies | Cat# NC9464543 |
| DNase I | Roche | Cat# 10104159001 |
| Nuclease-free H2O | Thermo Fisher Scientific | Cat# 10977015 |
| Critical commercial assays | ||
| Sso Advanced Universal SYBR Green Supermix | Bio-Rad | Cat# 1725272 |
| Experimental models: Cell lines | ||
| Human embryonic stem cells (H9) | WiCell | WA-09 |
| Oligonucleotides | ||
| RT-qPCR Primer: SOX10 F | Sigma-Aldrich; Saito-Diaz et al., 2021 | 5′-CCAGGCCCACTACAAGAGC-3′ |
| RT-qPCR Primer: SOX10 R | Sigma-Aldrich; Saito-Diaz et al., 2021 | 5′-CTCTGGCCTGAGGGGTGC-3′ |
| RT-qPCR Primer: BRN3A F | Sigma-Aldrich; Saito-Diaz et al., 2021 | 5′-AGTACCCGTCGCTGCACTCCA-3′ |
| RT-qPCR Primer: BRN3A R | Sigma-Aldrich; Saito-Diaz et al., 2021 | 5′-TTGCCCTGGGACACGGCGATG-3′ |
| Software and algorithms | ||
| Prism 9.2.0 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| Tissue culture dishes (10 cm) | Corning | Cat# 353003 |
| Tissue culture plates (6-well) | Fisher Scientific | Cat# FB012927 |
| Tissue culture plates (24-well) | Corning | Cat# 353047 |
| Vacuum filtration system (500 mL, 0.22 μm filter) | Corning | Cat# 28199-778 |
| CFX96 Touch Real-Time PCR Detection System | Bio-Rad | Cat# 1855195 |
| Hard-Shell Thin-Wall 96-Well Skirted PCR plates | Bio-Rad | Cat# HSP9601 |
Materials and equipment
EDTA Dissociation solution (500 mL stock):
| Reagent | Storage | Stock concentration | Final concentration | Volume |
|---|---|---|---|---|
| EDTA | RT | 0.5 M | 0.5 mM | 500 μL |
| NaCl | RT | 3.0801 M | 30 mM | 5 mL |
| DPBS | RT | n/a | n/a | Up to 500 mL |
| Total | 500 mL |
Filter the solution using a 0.22 μm filter, aliquot, and store at RT. It is stable for > 3 months.
NCC Differentiation media 1:
| Reagent | Storage | Stock concentration | Final concentration | Volume |
|---|---|---|---|---|
| SB431542 | −20°C | 10 mM | 10 μM | 50 μL |
| BMP4 | −80°C | 10 μg/mL | 1 ng/mL | 5 μL |
| CHIR99021 | −20°C | 6 mM | 300 nM | 2.5 μL |
| Y-27632 | −20°C | 10 mM | 10 μM | 50 μL |
| E6 Essential Medium | 4°C | n/a | n/a | Up to 50 mL |
| Total | 50 mL |
Store at 4°C. It is recommended to prepare fresh but can be stored for up to 7 days.
Alternatives: Y-27632 can also be purchased from R&D Systems (cat# 1254).
Alternatives: E6 Essential Medium can be made in house using stock components (Chen et al., 2011). All components need to be as fresh as possible when the media is made to ensure high quality. We recommend making single-use aliquots and store them at their recommended storage temperature.
Note: BMP4 concentrations might need to be adjusted depending on the cell line used (See troubleshooting problem 1).
NCC Differentiation media 2:
| Reagent | Storage | Stock concentration | Final concentration | Volume |
|---|---|---|---|---|
| SB431542 | −20°C | 10 mM | 10 μM | 100 μL |
| CHIR99021 | −20°C | 6 mM | 0.75 μM | 12.5 μL |
| DAPT | −80°C | 40 mM | 2.5 μM | 6.25 μL |
| SU5402 | −80°C | 40 mM | 2.5 μM | 6.25 μL |
| E6 Essential Medium | 4°C | n/a | n/a | Up to 100 mL |
| Total | 100 mL |
Store at 4°C for up to 14 days. It is recommended to prepare the exact volume required for the duration of the experiment (10 days).
Alternatives: SU5402 can also be purchased from Biovision (cat# 1645-1)
SN Differentiation media:
| Reagent | Storage | Stock concentration | Final concentration | Volume |
|---|---|---|---|---|
| N2 Supplement | −20°C | 100X | 1X | 1 mL |
| B-27 Supplement | −20°C | 50X | 1X | 2 mL |
| L-Glutamine | −20°C | 200 mM | 2 mM | 1 mL |
| GDNF | −80°C | 10 μg/mL | 20 ng/mL | 200 μL |
| BDNF | −80°C | 10 μg/mL | 20 ng/mL | 200 μL |
| NGF | −80°C | 25 μg/mL | 25 ng/mL | 100 μL |
| Laminin-1 | −80°C | 1 mg/mL | 0.6 μg/mL | 60 μL |
| Fibronectin | −80°C | 1 mg/mL | 0.6 μg/mL | 60 μL |
| Retinoic acid | −80°C | 1 mM | 0.125μM | 12.5 μL |
| Antibiotic-Antimycotic | −20°C | 100X | 1X | 1 mL |
| DAPT (see note) | −80°C | 40 mM | 1 μM | 2.5 μL |
| Neurobasal medium | 4°C | n/a | n/a | Up to 100 mL |
| Total | 100 mL |
Store at 4°C. It is recommended to prepare fresh with each differentiation. It can be stored for up to 14 days.
CRITICAL: Retinoic acid is light sensitive and unstable. Aliquot in small volumes (25 μL) and protect it from light. Add fresh in every feeding. Do not use aliquots that are older than 1 year and avoid using the same aliquot more than 5 times.
Note: DAPT promotes differentiation of NCCs into SNs. By day 20, we do not see SOX10+ proliferating cells (which represent NCCs) in culture so DAPT is not necessary after that.
Note: The media do not need to be warmed up prior to using.
Antibodies to be used:
| Antibody | Storage | Species | Working dilution |
|---|---|---|---|
| SOX10 | 4°C | Mouse | 1:100 |
| BRN3A | −20°C | Mouse | 1:100 |
| TUJ1 | 4°C | Mouse | 1:1500 |
| TUJ1 | 4°C | Rabbit | 1:1500 |
| TFAP2A | −20°C | Rabbit | 1:500 |
| TRKB | −20°C | Mouse | 1:100 |
| RET | −20°C | Mouse | 1:100 |
RT-qPCR: Prepare the RT-qPCR master mix by adding the following components (except cDNA) for a 10 μL reaction:
| Reagent | Stock concentration | Final concentration | Volume |
|---|---|---|---|
| SYBR mix | 2X | 1X | 5 μL |
| Primer F | 10 μM | 500 nM | 0.5 μL |
| Primer R | 10 μM | 500 nM | 0.5 μL |
| cDNA | 1 μg | 10 ng | 1 μL |
| Nuclease-free H2O | 3 μL | ||
| Total | 10 μL |
The master mix should be prepared at 4°C.
Mix the reaction thoroughly, add into the wells of a qPCR plate and add the cDNA to each of the wells containing the RT-qPCR master mix.
Program the following protocol on a real-time PCR thermocycler:
| Step | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95°C | 30 s |
| 2 | 40 | 95°C | 30 s |
| 3 | 40 | 60°C | 10 s |
| 4 | Melting curve | variable | |
| 5 | 1 | 4°C | keep |
Step-by-step method details
hPSCs expansion
Timing: ∼5 days
-
1.
Grow undifferentiated H9 hESCs (WiCell Research Institute) in 10 mL E8 Essential Medium (according to Thermo Fisher instructions: https://www.thermofisher.com/order/catalog/product/A1517001) in a VTN-coated 10 cm dish. To differentiate one 6-well plate, you will need 10.8 × 106 cells. Each 10 cm dish yields ∼12 × 106 undifferentiated hPSCs.
-
2.Expand the hPSCs to as many dishes as needed. To split cells:
-
a.Aspirate media.
-
b.Add 6 mL DPBS to the cells.
-
c.Aspirate DBPS.
-
d.Add 4 mL EDTA Dissociation solution. Incubate for 2 min at 37°C.
-
e.Aspirate EDTA Dissociation solution.
-
f.Resuspend cells by adding 10 mL E8 Essential Medium directly to the cells.
-
g.Pipet mixture of media and cells relatively harsh (no more than five times) while rotating the dish to remove all the cells.
-
h.Add cells to a 15 mL conical tube.
-
i.Split the cells at a 1:20 ratio using E8 Essential Medium and seed them into a new VTN-coated 10 cm dish.
-
a.
CRITICAL: Cell colonies must have clearly defined bright edges with minimal spontaneous differentiation, visualized under a brightfield microscope (Figures 1A, 1B, and 1D, day 0). Differentiating efficiency might decrease if cells are cultured for >5 consecutive passages (See troubleshooting problem 1).
CRITICAL: Check the cultures at least every two weeks to rule out presence of mycoplasma and other contaminants. We recommend using MycoAlert Mycoplasma Detection Kit (Lonza, Cat# LT07-318).
Note: Research involving hPSCs must be done in accordance with legal and ethical guidelines.
Alternatives: E8 Essential Medium can be made in house using stock components (Chen et al., 2011). All components need to be as fresh as possible when the media is made to ensure high quality. We recommend making single-use aliquots and store them at their recommended storage temperature.
Differentiation into neural crest cells (NCCs)
Timing: up to 12 days
In this section, we describe in detail the steps to differentiate hPSCs into NCCs with a SN fate. NCCs start to aggregate on day 4, and by day 8 dark aggregates (“ridges”) are visible under a brightfield microscope (Figures 1A, 1B, and 2A). Additionally, NCCs start expressing SN markers as early as day 4 (Figure 2A).
Note: It is important to carry out a parallel differentiation at the NCC stage for quality control (QC) purposes in a small format (4-well plate). This allows isolation of RNA and/or perform immunofluorescence (on day 6 or 8) from a plate that will be treated in the same way as the main plate.
-
3.
Aspirate E8 media from the 10 cm dish of undifferentiated, ready to split hPSCs.
-
4.
Wash once with ∼10 mL DPBS.
-
5.
Add 4 mL EDTA Dissociation solution to a 10-cm dish, incubate for 15 min at 37°C.
Note: Do not aspirate the EDTA Dissociation solution. The hPSCs colonies will be dissociated and resuspended into single cells and the EDTA Dissociation solution will be cloudy.
-
6.
Use 10 mL DPBS to wash off the cells. Most or all cells should be resuspended or detach from the plate rather easily at this point.
-
7.
Transfer the cells into a 50 mL conical tube and add an additional 10 mL DPBS.
-
8.
Spin down at 200×g for 4 min.
-
9.
Resuspend in 5 mL NCC Differentiation media 1.
-
10.Count cells using a Countess II or hematocytometer:
-
a.Mix 10 μL Trypan blue (Corning) with 10 μL of cell suspension.
-
b.Mix by pipetting and put 10 μL in a counting slide (ThermoFisher, Cat# C10312).
-
c.Count the cells.
-
a.
Note: Resuspend 0.5 − 1 × 106 cells in 500 μL TRIZOL for RNA for day 0.
-
11.
Aspirate VTN from the 6-well plate (NCC differentiation plate).
-
12.
Transfer 10.8 × 106 cells (to seed one 6-well plate at a density of 200,000 cells/cm2) to a 15 mL conical tube and fill with NCC Differentiation media 1 up to 12 mL.
Note: A total of 10.8 × 106 cells are needed for a one 6-well plate, thus we recommend growing one 10 cm dish of H9 cells per 6-well plate.
-
13.
Plate 2 mL of the cell suspension directly onto each of the VTN-coated wells (do not let it dry before seeding the cells).
-
14.
Incubate at 37°C for 16–20 h.
-
15.
On the next day (day 1), replace media with 3 mL per well of NCC Differentiation media 1.
-
16.
Take a picture for your records of cell density. High density is desired (>80%) (Figure 2B).
-
17.
Incubate at 37°C for 16–20 h.
-
18.
The following day (day 2), replace media (3 mL/well) of NCC Differentiation media 2.
-
19.Change media every 2 days until day 6 for inducing mechanoreceptors or day 12 for obtaining all three neuronal types:
-
a.To generate mainly mechanoreceptors, on day 6 follow the steps in the section “Differentiation into SNs (mechanoreceptor enrichment)”. Step 22.
-
b.To generate nociceptors, mechanoreceptors, and proprioceptors, on day 12 follow the steps in the section “Differentiation into SNs (nociceptors, mechanoreceptors, and proprioceptors)”. Step 40.
-
a.
-
20.On day 8, harvest RNA from 4-well quality control plate:
-
a.Aspirate the media from one well.
-
b.Lyse the cells in 500 μL TRIZOL.
-
a.
CRITICAL: do not wash cells with PBS as this could cause mRNA degradation.
Alternatives: The quality control plate can be fixed for immunofluorescence staining.
-
21.
Take pictures on every feeding day to follow proper progression (Figures 1A and 1B).
Note: It is normal to see some cell aggregates (“ridges”) starting on day 4 (Figure 2A), which overlaps with an increase of SOX10 expression at the mRNA and protein levels (Figures 2A and 2C) (See troubleshooting problem 2).
CRITICAL: If there is no clear formation of NCC ridges (Figure 2D), it is best to stop the differentiation because the SN yield will be too low or the presence of contaminant cells will be too high to perform any subsequent analysis (Figure 3D). See troubleshooting problem 1.
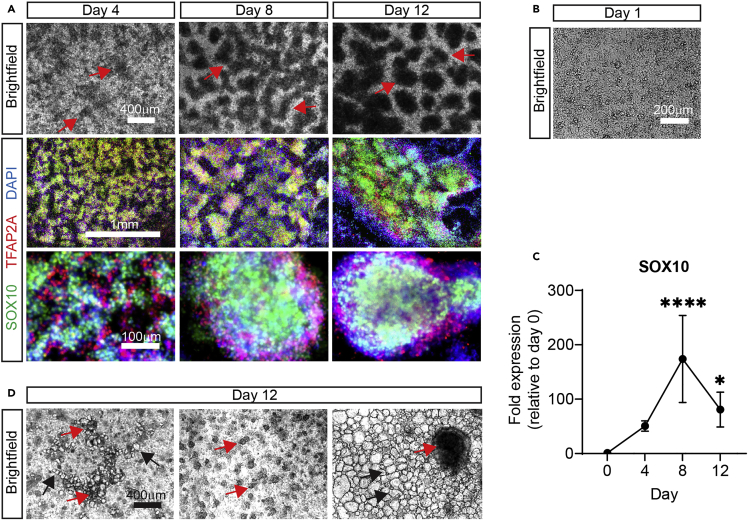
Figure 2.
Characterization of NCC stage
(A) Expected phenotypes of properly differentiated NCCs. Brightfield images of NCCs were taken on the indicated days (top panel). Arrows show growth and progression of NCC “ridges”, which start to form on day 4. Expression of NCC markers. NCCs were fixed on the indicated days and stained for SOX10 (green), TFAP2A (red), and DAPI (blue). Images were taken at two different magnifications (middle and bottom panels).
(B) Appropriate seeding density. H9 cells were seeded at a density of 200,000 cell/cm2 (day 0). The next day (day 1) brightfield images were taken to show the expected density (>80% confluency).
(C) Expression of SOX10 mRNA by RT-qPCR. n=4 (biological replicates), one-way ANOVA followed by Dunnett’s multiple comparisons. Graph show mean ± S.D. ∗p < 0.05, ∗∗∗∗p < 0.0001.
(D) Expected phenotypes of improper NCC differentiation. Brightfield images were taken on day 12. Note small size of the NCC “ridges” (red arrows left and middle images) or the formation of a single large NCC cluster (red arrow, right image), which will cause a very low differentiation efficiency. Also, note the presence of large blister-like cells/aggregates (black arrows, left and right images), the identity of which we do not know at this point.
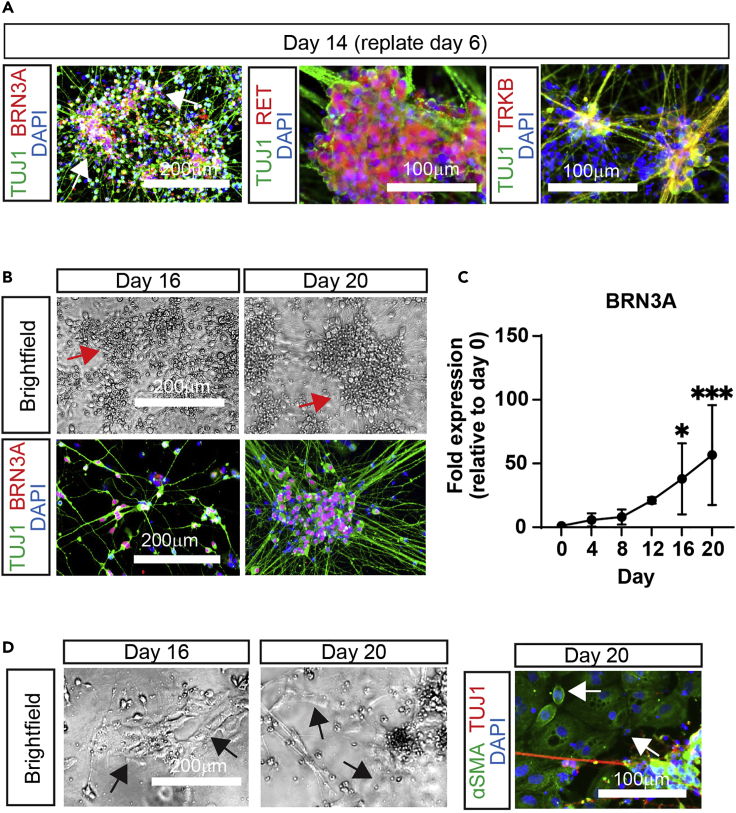
Figure 3.
Characterization of SNs
(A) Expected phenotypes of SNs obtained from day-6 NCCs. Cells were fixed 8 days post-replated and stained for TUJ1, the panSN-marker BRN3A, and the mechanoreceptor-specific markers RET and TRKB.
(B) Expected phenotypes of SNs obtained from day-12 NCCs. Brightfield images (top) and immunofluorescent (bottom) images of SNs on day 16 and 20. For immunofluorescence, SNs were fixed on the indicated days and stained for TUJ1 (green), BRN3A (red), and DAPI (blue).
(C) Expression of the pan-SN marker BRN3A mRNA by RT-qPCR. n=4 (biological replicates), one-way ANOVA followed by Dunnett’s multiple comparisons. Graph show mean ± S.D. ∗p < 0.05, ∗∗∗p < 0.001.
(D) Representative images of failed differentiations. On day 12, NCCs showing suboptimal phenotypes (low number or complete lack of ridges) were replated. On the indicated days, brightfield images were taken (left) or fixed and stained (right). Fixed SNs were stained for αSMA (green), TUJ1 (red), and DAPI (blue). Note the low number of dendrites and the presence of a high number of non-neural ectoderm cells (arrows).
Differentiation into SNs (mechanoreceptor enrichment)
Timing: ≥ 8 days
Here we describe the steps to differentiate NCCs into sensory neurons with an enrichment of mechanoreceptors. By replating on day 6, you will obtain a majority of mechanoreceptors which aggregate in ganglia-like structures. SNs express pan-SN marker BRN3A as well as the mechanoreceptor markers RET and TRKB (Figure 3A). In these conditions, SNs can be maintained for at least 30 days.
Note: This protocol details the differentiation of one 24-well plate of SNs from three wells of the 6-well plate of differentiated NCCs. The volumes will need to be scaled up if more plates are needed.
-
22.
On day 6 of the NC differentiation, wash each of the three designated wells of the 6-well plate with 1 mL DPBS.
Note: The remaining three wells will be processed on step 40.
-
23.
Aspirate DPBS.
-
24.
Add 1 mL/well Accutase and incubate at 37°C for 20 min (or until the cells start to detach).
-
25.
Add 1 mL DPBS to each well to resuspend the cells and transfer them to a 50 mL conical tube using a p1000.
-
26.
Fill up with DPBS.
-
27.
Centrifuge, 200×g, 5 min, RT.
-
28.
Resuspend the cells in 30 mL DPBS by pipetting up to six times using a 5 mL pipette, trying to gently break up the clumps.
-
29.
Add 150 μL DNaseI (from a 10 mg/mL stock) and mix by inversion.
-
30.
Centrifuge, 200×g, 5 min, RT.
-
31.
Resuspend in 5 mL of SN Differentiation media + 1 μM DAPT.
-
32.
Gently break all the clumps by pipetting with a p1000 (<6 times).
-
33.
Count cells.
-
34.
Aspirate LM/FN mixture from the PO/LM/FN-coated 24-well plate.
-
35.
Transfer 12 × 106 cells (to seed one 24-well plate at a density of 250,000 cells/cm2) and fill up to 12 mL with SN Differentiation media + 1 μM DAPT.
-
36.
Add 0.5 mL of the cell suspension to each well of the 24-well plate.
CRITICAL: replating is necessary to increase the differentiation efficiency.
Pause Point: Resuspended NCCs can be cryopreserved at a concentration of 5 × 106 cell in 1 mL NCC Differentiation media 2 containing 10% DMSO. The viability using this method is ∼15%. Commercially available freezing reagent (such as STEM-CELLBANKER, amsbio, cat# 11890) provide a better viability (∼70%).
-
37.
Incubate cells at 37°C for 16–20 h.
-
38.
On the next day, aspirate the media and add 1 mL SN Differentiation media + 1 μM DAPT per well of the 24-well plate.
CRITICAL: Media should be added to the walls of the wells slowly. Avoid adding the media directly to the SNs as this can cause detachment of the newly plated cells (See troubleshooting problem 2).
-
39.
Replace the media every 2–3 days until day ∼30.
Note: After day 14 (eight days post-replating), the number of SOX10+ progenitor cells will decrease so DAPT can be removed from the SN Differentiation media (See troubleshooting problem 3). If a high number of non-neural ectoderm cells arise, please see troubleshooting problem 4 and troubleshooting problem 5.
Differentiation into SNs (nociceptors, mechanoreceptors, and proprioceptors)
Timing: ≥ 8 days
Here, we describe the steps to differentiate NCCs into SNs at a DRG-like ratio. By replating on day 12, you will obtain a mixed peripheral SN population of nociceptors, mechanoreceptors, and proprioceptors. SNs aggregate in ganglia-like structures (Figure 3B). In these conditions, SNs can be maintained for over 90 days. SNs can also be replated a second time after day 16 for additional experiments or to maintain them for a longer time period (Saito-Diaz et al., 2021).
Note: This protocol details the differentiation of one 24-well plate of SNs from the three remaining wells of one 6-well plate of differentiated NCCs.
Note: these steps need to be carried out on day 12 and continue from step 21
-
40.
On day 12 of the NC differentiation, wash each well with 1 mL DPBS.
-
41.
Add 1 mL/well Accutase and incubate at 37°C for 20 min (or until the cells start to detach).
-
42.
Add 1 mL DPBS to each well to resuspend the cells and transfer them to a 50 mL conical tube using a p1000.
-
43.
Fill up with DPBS.
-
44.
Centrifuge, 200×g, 5 min, RT.
-
45.
Aspirate the supernatant and add 30 mL DPBS.
-
46.
Resuspend the cells by pipetting up to six times using a 5 mL pipette, trying to gently break up the clumps.
-
47.
Add 150 μL DNaseI (from a 10 mg/mL stock) and mix by inversion.
-
48.
Centrifuge, 200×g for 5 min at RT.
-
49.
Resuspend in 5 mL of SN Differentiation media + 1 μM DAPT using a 5 mL pipette.
-
50.
Gently break all the clumps by pipetting with a p1000 (<6 times).
-
51.
Count cells.
-
52.
Aspirate LM/FN mix from the PO/LM/FN-coated 24-well plate.
-
53.
Transfer 12 × 106 cells (for one 24-well plate at a density of 250,000 cells/cm2) to a 15 mL conical tube and fill up to 12 mL with SN Differentiation media + 1 μM DAPT.
-
54.
Add 0.5 mL to the cell suspension (from step 49) to each well of the 24-well plate.
CRITICAL: replating is necessary to increase the differentiation efficiency.
Pause Point: Resuspended NCCs can be cryopreserved using NCC Differentiation media 2 (see above for details on how to prepare the media) containing 10% DMSO or commercially available freezing reagents (see step 36). We recommend freezing at a concentration of 5 × 106 cells/mL.
-
55.
Incubate the cells at 37°C for 16–20 h.
-
56.
Next day, aspirate the media and add 1 mL SN Differentiation media + 1 μM DAPT to each well.
CRITICAL: Media should be added to the walls of the wells slowly. Avoid adding the media directly to the SNs as this can cause detachment of the newly plated cells (See troubleshooting problem 2).
-
57.
Replace the media every 2–3 days until day 30. After 30 days, media can be changed every five days.
Note: After day 20 (eight days post-replating), DAPT is not necessary in the SN Differentiation media (see above for details). See troubleshooting problem 3. If a high number of non-neural ectoderm cells arise, please see troubleshooting problem 4 and troubleshooting problem 5.
Note: SNs can be replated a second time after day 16. This can be used as an axotomy model (Saito-Diaz et al., 2021). To do so: 1) Wash SNs with DPBS, followed by incubation at 37°C with Accutase for at least 1 h. 2) Transfer the cells to a 50 mL conical tube with 30 mL DPBS and centrifuge it at 200x g for 4 min at RT. 3) Resuspend the pellet in 1 mL of SN Differentiation media using a p1000 micropipette to break all the clumps. 4) Add 4 mL of SN Differentiation media. 5) Count the cells and replate at a density of 250,000 cells/cm2 on a PO/LM/FN plate (see above). 6) Change the media the next day.
Expected outcomes
In the NC stage of the differentiation, multiple circular or irregular-shaped ridges of different sizes appear starting on day 4 (Figure 2A). These ridges are SOX10 positive as shown by immunofluorescence (Figure 2A). Furthermore, SOX10 expression measured by RT-qPCR should be approximately 100-fold higher compared to hPSCs (Figure 2C). Additionally, the number of NCCs peaks on day 8 (∼90% of viable cells) as measured by the number of cells expressing the surface marker CD49d (which correlates with SOX10 expression (Zeltner et al., 2016)) by flow cytometry. Finally, by day 12 we observe expression of the pan-neuronal marker TUJ1 (Saito-Diaz et al., 2021). It is important to note that a lack of NCC ridges is an indicative of a failed differentiation (Figure 2D) (Troubleshooting 1).
On day 6, NCCs can be replated to generate SNs that by day 14 express the pan-sensory neuronal marker BRN3A as well as the mechanoreceptor-specific markers RET and TRKB (Figure 3A). Furthermore, approximately ∼80% of all cells are mechanoreceptors (TRKB+) (Saito-Diaz et al., 2021).
Alternatively, NCCs can be replated on day 12 to generate SNs that also BRN3A measured by immunofluorescence and RT-qPCR (Figures 3B and 3C). By day 20, the earliest SNs appear, at approximately 70% of the total cell population (Saito-Diaz et al., 2021). Furthermore, all SN subtypes are differentiated at a ratio of roughly 70% nociceptors (TRKA+), 30% mechanoreceptors (TRKB+), and 30% proprioceptors (TRKC+) (Saito-Diaz et al., 2021). It is important to note that in this protocol some SNs (i.e., mechanoreceptors) are double labeled for TRKB+/C+, which explains why the total proportion is higher than 100%. Additional characterization could include analysis of expression of additional SN markers such as ISL1, PRPH, and RET. A failed differentiation will be characterized by the low number of SNs and presence of non-neural cells, mostly expressing smooth muscle actin (αSMA) (Figure 3D). We have observed the same outcomes using healthy iPSC-derived SNs (∼70% from total population). In contrast, SNs differentiated from iPSCs derived from patients with the peripheral neuropathy familial Dysautonomia comprise ∼20% of the total population (Saito-Diaz et al., 2021). This correlates with the phenotypes observed in patients which demonstrates that this protocol is a powerful tool to model diseases of the PNS.
Limitations
We have observed that approximately one out of every five differentiations does not yield high NCC ridge formation and SOX10 expression, resulting in a low number of SNs (Troubleshooting 1). This number (below the projected 70%), may result in an increased number of non-neural ectoderm cells which express αSMA (Figure 3D). Also, due to the very similar developmental pathway, it is possible that the cultures contain trigeminal neurons, which we haven’t been able to completely rule out due to the lack of specifically differentiating markers between the two lineages. Finally, a limited number of Schwann cells, (expressing the markers myelin basic protein (MBP), myelin protein zero (MPZ), and SOX10) sometimes can be obtained in this protocol and can be found along the axons of the SNs.
Troubleshooting
Problem 1
No formation of NCCs ridges or appearance of undetermined cell types and blister-like aggregates (step 21, Figure 2D).
Potential solution
Some hPSCs (including iPSCs) have high levels of intrinsic BMP4 expression, which can cause a failure to generate NCCs. We recommend titrating BMP4, starting at 0 ng/mL up to 1 ng/mL, to identify the concentration that results in the highest SOX10 expression and formation of NCC ridges and might remove blister-like aggregates. Also, different sources of BMP4 have different activity, which introduce variability to the efficiency.
Use a lower passage number of hPSCs. We have found that maintaining the cells in culture for over 4–5 consecutive passages reduces the rate of successful differentiations. Additionally, using the CryoPause method drastically improved differentiation efficiency (Wong et al., 2017).
Problem 2
NCCs and SNs in culture are lifting off the plate (steps 19, 38, and 56).
Potential solution
At the NCC stage we would recommend performing the differentiation in Geltrex instead of VTN. For SNs cultured for longer periods of time, we recommend replating them on a later date following the steps described on step 57. We have successfully replated SNs from day 16 up to day 50 of differentiation.
Problem 3
Presence of SOX10+ cells in SN culture even after adding DAPT to SN Differentiation media after eight days post-replating (steps 39 and 57).
Potential solution
SOX10+ might indicate presence of proliferating undifferentiated NCCs. This can be confirmed by co-staining of SOX10 and a proliferation marker (e.g., Ki67). If you identify multiple co-stained cells, titrate DAPT concentration (up to 10 μM) in the SN Differentiation media. It would also be advisable to continue the treatment with DAPT beyond eight days post-replating. Additionally, you can use antimitotic agents, such as mitomycin C or Cytosine β-D arabinofuranoside after replating (on day 6 or day 12) to remove proliferating cells. If there is a population of SOX10+ Ki67- cells, this might suggest the presence of Schwann cells. RT-qPCR of additional Schwann cell markers such MBP and MPZ can confirm the presence of Schwann cells.
Problem 4
Presence of a high number of non-neural ectoderm cells and few SNs (steps 39 and 57, Figure 3D).
Potential solution
Replating NCCs at a low density (<100,000 cells/cm2) causes differentiation into αSMA+ non-neural ectoderm cells. Titrate the number of cells per cm2 that best suits your applications. We recommend testing a range between 100,000 – 400,000 cells/cm2.
Problem 5
There is a large variability between differentiations (steps 39 and 57).
Potential solution
We have observed that hPSCs at different passage numbers tend to cause variable differentiation efficiencies. If this is the case, we recommend using the CryoPause method (Wong et al., 2017), which will allow the differentiation from a well characterized hPSC population. This will decrease variability between differentiations.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Nadja Zeltner (nadja.zeltner@uga.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We want to thank Dr. Lorenz Studer for his support at the initial stages of this work and Jessica McAlpine for critical reading of the manuscript. This work was funded by faculty start-up funds from the University of Georgia to N.Z. and NIH/NINDS 1R01NS114567-01A1 to N.Z. Schematics were done using Biorender.com.
Author contributions
N.Z. and K.S-D. conceived and designed the experiments; K.S-D. conducted the experiments; K.S-D. and N.Z. analyzed and interpreted the data; K.S-D. and N.Z. wrote the manuscript; N.Z. provided mentoring, financial, and administrative support and approved the final version of manuscript.
Declaration of interests
The authors declare no competing interests. This work is linked to the patent ‘Compositions and methods for making sensory neurons’, Serial number is 63/127,9, held by the University of Georgia.
Contributor Information
Kenyi Saito-Diaz, Email: saitod@uga.edu.
Nadja Zeltner, Email: nadja.zeltner@uga.edu.
Data and code availability
This study did not generate any datasets or code.
References
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Diaz K., Street J.R., Ulrichs H., Zeltner N. Derivation of peripheral nociceptive, mechanoreceptive, and proprioceptive sensory neurons from the same culture of human pluripotent stem cells. Stem Cell Rep. 2021;16:446–457. doi: 10.1016/j.stemcr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.G., Ryan S.D., Ramnarine K., Rosen S.A., Mann S.E., Kulick A., De Stanchina E., Muller F.J., Kacmarczyk T.J., Zhang C., et al. CryoPause: a new method to immediately initiate experiments after cryopreservation of pluripotent stem cells. Stem Cell Rep. 2017;9:355–365. doi: 10.1016/j.stemcr.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeltner N., Fattahi F., Dubois N.C., Saurat N., Lafaille F., Shang L., Zimmer B., Tchieu J., Soliman M.A., Lee G., et al. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 2016;22:1421–1427. doi: 10.1038/nm.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any datasets or code.