Abstract
Endothelial cells (ECs) lining blood vessels are exposed to mechanical forces, such as shear stress. These forces control many aspects of EC biology, including vascular tone, cell migration and proliferation. Despite a good understanding of the genes responding to shear stress, our insight into the transcriptional regulation of these genes is much more limited. Here, we set out to study alterations in the chromatin landscape of human umbilical vein endothelial cells (HUVEC) exposed to laminar shear stress. To do so, we performed ChIP-Seq for H3K27 acetylation, indicative of active enhancer elements and ATAC-Seq to mark regions of open chromatin in addition to RNA-Seq on HUVEC exposed to 6 h of laminar shear stress. Our results show a correlation of gained and lost enhancers with up and downregulated genes, respectively. DNA motif analysis revealed an over-representation of KLF transcription factor (TF) binding sites in gained enhancers, while lost enhancers contained more ETV/ETS motifs. We validated a subset of flow responsive enhancers using luciferase-based reporter constructs and CRISPR-Cas9 mediated genome editing. Lastly, we characterized the shear stress response in ECs of zebrafish embryos using RNA-Seq. Our results lay the groundwork for the exploration of shear stress responsive elements in controlling EC biology.
Subject terms: Computational biology and bioinformatics, Gene regulatory networks
Introduction
The formation of properly patterned blood vessels needs to be tightly controlled during embryonic development as they provide growing tissues with signaling molecules and the means of gas and metabolite exchange. Early vascular development is often stereotyped and depends on hard-wired genetic programs, while at later stages blood flow patterns provide additional mechanical cues1. These include tangential fluid shear stress and circumferential cyclic strain2. During angiogenesis stages, the formation of new sprouts is initiated in areas with lower shear stress3, suggesting that high levels of shear stress tone down angiogenesis and induce endothelial quiescence4,5. Mechanical forces are also instrumental in vascular plexus remodeling that occurs through the pruning of excess blood vessels, ultimately resulting in the formation of mature and efficient vascular networks6. In this setting, shear stress induces the migration of ECs from vessels with low blood flow into vessels with high flow, as observed in the mouse retina7 and yolk sac vasculature8, zebrafish brain9, eye10 and subintestinal vessels11, as well as in quail embryos12.
Besides its role in vascular development and remodeling, shear stress is critical for blood vessel maintenance13. Unidirectional laminar shear stress affects proliferation and survival of ECs14 and has anti-thrombotic15 and anti-inflammatory effects16. It also controls endothelial barrier function17 and the cytoskeleton18. In contrast, oscillatory shear stress observed in areas of vessel curvature and vessel branching induces expression of pro-inflammatory endothelial markers19 and thus contributes to atherosclerosis development20. ECs can respond to changes in shear stress by regulating vessel size21. We have shown that in early embryonic development in zebrafish shear stress can induce changes in EC shape and orientation leading to vessel constriction22. In adult vessels, increases in blood flow can either result in vessel dilation through the release of nitric oxide (NO) from ECs23 or in vasoconstriction, mediated by the mechanosensing ion channel Piezo124. The interplay of these two processes is likely important for blood pressure regulation.
Previous studies identified TFs of the Krüppel-like factor family as master regulators of the shear stress response25,26, in particular KLF227 and KLF428. Fledderus et al. suggested that KLF2 together with nuclear factor erythroid-2 (Nrf2) regulates the expression of 70% of flow responsive genes29. However, other studies showed a lower magnitude of KLF2-dependent regulation30 and the association of other members of the KLF family or Sp1 to KLF binding sites31. Single cell ATAC-Seq identified binding sites for KLF4 and Nrf1 in ECs exposed to stable flow in carotid arteries, while ECs in arteries with disturbed flow showed enrichment of TEAD1 and ETV3 TF binding sites, among others32. We previously identified shear stress responsive elements in ECs exposed to atheroprone oscillatory shear stress and showed that activation of YAP/TAZ signaling was involved in mediating the EC response to this type of shear stress33. Work investigating histone modifications in response to pulsatile shear stress identified KLF4 as a downstream TF34. Furthermore, early growth response (EGR1) and SMAD1/5 have been implicated in the regulation of shear stress-induced gene expression changes35,36, while the Snail TF responds to low shear stress in mediating endothelial to mesenchymal transition37. Therefore, our understanding of the transcriptional regulation of the shear stress response is far from complete and might involve the use of distinct sets of TFs depending on the flow regime and/or EC type examined5.
In this study, we used chromatin immunoprecipitation sequencing (ChIP-Seq) for acetylated H3K27, which marks active enhancers38, in combination with assay for transposase-accessible chromatin using sequencing (ATAC-Seq) to identify and characterize laminar shear stress responsive enhancer elements in cultured ECs. Our results show that proximity to gained or lost enhancers can faithfully predict shear stress induced changes in gene expression. We further identified binding sites for members of the KLF family of TFs in gained enhancers, while ETS binding sites were overrepresented in lost enhancers. We also identified several novel DNA motifs. Finally, deleting shear stress responsive enhancer elements using CRISPR-Cas9 mediated genome editing resulted in a reduction in shear stress regulation of SEMA7A and CXCR4 expression. Together, our results identified a suite of shear stress responsive enhancer elements and will thus serve as a resource for future studies investigating shear stress-mediated regulation of gene expression.
Results
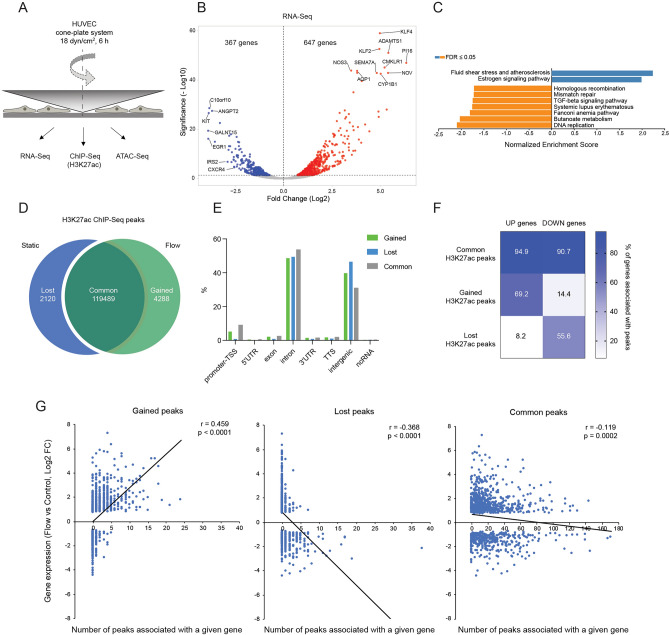
Shear stress induces gene expression changes in ECs
To understand the regulation of gene expression by shear stress at the level of cis-regulatory elements we first performed RNA-Sequencing (RNA-Seq) in HUVEC exposed to shear stress in a BioTechFlow system (Fig. 1A) and compared gene expression to cells cultured under static conditions. To determine the optimal timepoint for our analysis, we initially exposed HUVEC to 18 dyn/cm2 for 30 min and for 6 h. After 30 min of shear stress, 59 genes were upregulated and 17 were downregulated (Suppl. Fig. 1, Suppl. Data 1). Several chaperones and co-chaperones (HSPA1A, HSPA1B, HSP6, DNAJB1) were the most highly upregulated genes. In addition, the known shear stress-regulated TFs KLF2 and KLF4 were upregulated, pointing to the onset of a shear stress-specific transcriptional program already after a short exposure. After 6 h of shear stress, 1014 genes were differentially expressed (DE) with 647 genes up-regulated and 367 down-regulated when compared to control (Fig. 1B, Suppl. Data 1). We detected differential expression of known flow-responsive genes, such as KLF2, KLF4, NOS3, CYP1B1, WNT9B, which were upregulated by shear stress, and EGR1 and CXCR4, which were downregulated by shear stress (Fig. 1B, Suppl. Fig. 2A,B). Based on the results of these initial experiments and a separate longitudinal study39, we decided to continue our analysis with 6 h of shear stress exposure. We first validated our RNA-Seq results using several methods. Quantitative polymerase chain reaction (qPCR) on the most up- and downregulated DE genes and of non-regulated genes with a Log2 fold change close to 0 revealed a high degree of correlation between RNA-Seq and qPCR results (Suppl. Fig. 2C). Pathway analysis of DE genes using gene set enrichment analysis (GSEA, Fig. 1C) and overrepresentation analysis (ORA, Suppl. Fig. 2D) revealed that “fluid shear stress and atherosclerosis” was the most enriched pathway in our dataset. We also compared our RNA-Seq dataset with published studies on shear stress-induced gene expression changes in ECs (Suppl. Fig. 2E). This analysis showed a 34.5% overlap with all DE genes from an RNA-Seq dataset of HUVEC exposed to 20 dyn/cm2 for 3 days by Maleszewska et al.40 and a 32.5% overlap with all DE genes from a meta-analysis of multiple microarray-based reports study by Maimari et al.41. Additionally, we detected a substantial overlap (15.7%) between our data and the dataset generated by Dekker et al.4, mimicking the shear stress response through KLF2 overexpression (Suppl. Fig. 2E). Together, our RNA-Seq data are highly reproducible and partially match previously published datasets, while at the same time revealing considerable differences in the transcriptional response of ECs to different flow regimes. They furthermore suggest that TFs other than KLF2 play important roles in regulating EC flow responses.
Figure 1.
Shear stress regulates the expression of a defined set of genes that correlates with enhancer activation. (A) HUVEC exposed to unidirectional shear stress (18 dyn/cm2) for 6 h were used for RNA-Seq, ATAC-Seq and H3K27ac ChIP-Seq. (B) Volcano plot showing gene expression changes in HUVEC under shear stress. Differentially expressed (DE) genes (FDR < 0.05) are labelled in red (upregulated) and blue (downregulated under shear stress). (C) Gene Set Enrichment Analysis (GSEA) of KEGG pathways regulated in the RNA-Seq dataset. (D) The number of H3K27ac ChIP-Seq peaks in HUVEC under static conditions (“lost” after shear stress) or exposed to shear stress (“gained” after shear stress) and the peaks “common” for both conditions. (E) Annotation of gained, lost and common H3K27ac peaks to defined regions of the genome. (F) Heatmap representing the spatial association of the genes up- or down-regulated by shear stress with identified common, gained or lost H3K27ac ChIP-Seq peaks. Numbers represent the percentage of up-or down-regulated genes, associated with at least one peak. (G) Comparison of gene expression (log2 fold change (FC) from RNA-Seq) to the number of gained, lost or common H3K27ac peaks spatially associated with a particular gene. Pearson correlation coefficient (r) and p-value of the correlation are shown.
ChIP-Seq identification of flow responsive regulatory elements marked by H3K27ac
To interrogate which enhancer elements might be responsible for the observed changes in gene expression, we performed ChIP-Seq analysis of shear stress (18 dyn/cm2 for 6 h) exposed and unexposed static control HUVEC using an antibody specific for histone 3 acetylated at the lysine 27 residue (H3K27ac), which marks active enhancers38. To validate our data, we compared our data with previously generated datasets from the ENCODE consortium42. Our analysis revealed a high degree of correlation between our replicates, both for static and shear-stress-exposed conditions (Suppl. Fig. 3A). Our static replicates correlated well with HUVEC samples from ENCODE (Pearson coefficient 0.72–0.82), similar to the correlation between individual ENCODE replicates (0.78–0.89). The analysis of our ChIP-Seq data identified 119,489 peaks (DNA regions with significantly enriched H3K27ac signal compared to input control) common for both flow and static conditions (Fig. 1D). Furthermore, we identified 4288 peaks specific for shear stress-exposed cells (“gained” peaks) and 2120 peaks present only in control cells (peaks “lost” in shear stress condition; Fig. 1D). Annotation of gained and lost H3K27ac peaks to defined regions of the genome revealed that most of the peaks were in introns of coding genes and in intergenic regions, while a relatively low number of peaks was associated with gene promoters (Fig. 1E). To further validate our results, we performed qPCR with primers for selected regions showing a high enrichment for H3K27ac. These data on independent biological replicates confirmed the results from our ChIP-Seq dataset (Suppl. Fig. 3B). Thus, we obtained a validated dataset of flow-responsive gene regulatory elements.
Flow responsive cis-regulatory elements associate with flow regulated genes
As proximity favors enhancer-promoter choice to some extent43,44, we associated H3K27ac peaks to neighboring genes using the GREAT algorithm45. This algorithm identifies the nearest genes of a given peak within a range of 1 Mb. We then compared these genes to the set of DE genes from our RNA-Seq data. This analysis revealed that upregulated genes were more often associated with gained peaks, while downregulated genes were more often associated with lost peaks. Hence, 69% of upregulated genes associated with at least one gained peak, while this number was only 14% for downregulated genes. Conversely, 56% of downregulated genes associated with at least one lost peak, while only 8% of upregulated genes had associated lost peaks (Fig. 1F). Upregulated genes had more gained peaks per gene associated with them, while downregulated genes associated with more lost peaks per gene. Additionally, the number of associated peaks correlated with gene expression levels (Fig. 1G). This correlation was not observed for common H3K27ac peaks (Fig. 1F,G). These data are in line with our hypothesis that shear stress activates enhancers associated with upregulated genes and inhibits enhancers associated with downregulated genes. They furthermore show that our ChIP-Seq analysis has predictive power to anticipate shear stress-mediated regulation of gene expression.
Analysis of chromatin accessibility associated with shear stress-responsive regulatory elements
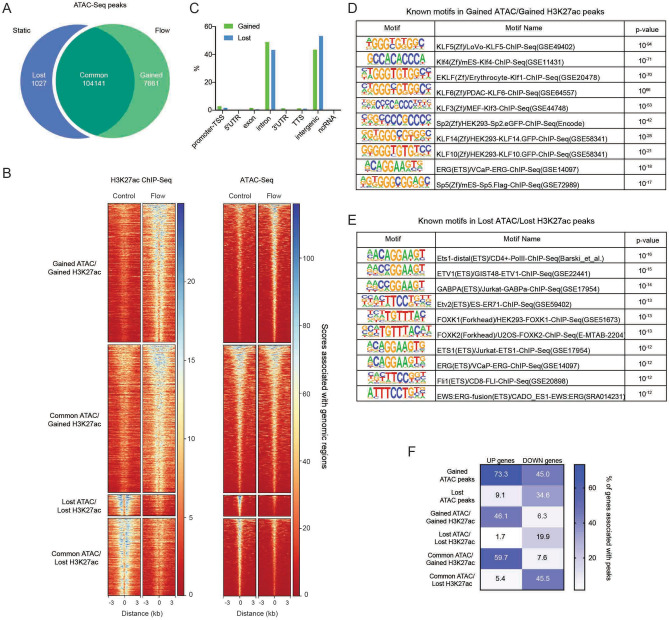
To analyze chromatin accessibility, which is often associated with TF binding, we performed ATAC-Seq in HUVEC exposed to shear stress and compared the data to our sets of DE genes and H3K27ac peaks. This analysis identified over 104,000 common ATAC peaks, while 7661 peaks were gained, and 1027 peaks were lost upon shear stress exposure of the cells (Fig. 2A). Like H3K27ac peaks, gained and lost ATAC-Seq peaks were preferentially located in gene introns and intergenic regions (Suppl. Fig. 4A). To understand how chromatin accessibility correlated with histone acetylation, we analyzed the overlap between ATAC-Seq and H3K27ac peaks (located within 100 bp of each other, Fig. 2B). 21% of gained ATAC-Seq peaks were associated with gained H3K27ac peaks, while 24% of lost ATAC-Seq peaks overlapped with lost H3K27ac peaks (Suppl. Fig. 4B), indicating a similar mode of activation/deactivation for this subset of peaks. The numbers of ATAC-Seq peaks associated with H3K27ac peaks showing an opposite mode of regulation was lower. Only 0.1% of gained ATAC-Seq peaks overlapped with lost H3K27ac peaks and only 0.2% of lost ATAC-Seq peaks overlapped with gained H3K27ac peaks. A substantial amount of both gained and lost ATAC-Seq peaks corresponded with common H3K27ac peaks or showed no overlap. A reverse analysis of the overlap between H3K27ac peaks and ATAC-Seq peaks showed a similar tendency. The overlap was higher for gained H3K27ac and ATAC-Seq peaks and for lost H3K27ac and ATAC-Seq peaks (45% and 15%, respectively) than vice versa (0.1% for both, Suppl. Fig. 4C). This analysis also revealed that many H3K27ac peaks were associated with common ATAC-Seq peaks (48% of the gained and 51% of the lost H3K27ac peaks, Suppl. Fig. 4C). Therefore, our comparative analysis of ATAC-Seq and H3K27Ac peaks revealed the presence of distinct subsets of H3K27ac peaks. In the first set, changes in histone acetylation coincide with changes in chromatin accessibility, suggesting enhancer activation/deactivation in response to shear stress. A second, large subset of peaks contains regions of open chromatin, which do not change with changes in histone acetylation (Suppl. Fig. 4B). This could suggest the presence of poised enhancers that get activated by shear stress and active enhancers that get deactivated but remain poised. Lastly, there is a subset of H3K27ac peaks which do not overlap with ATAC-Seq peaks. It remains to be determined, whether these are functional enhancers or whether they are the result of limitations in our study.
Figure 2.
ATAC-Seq identifies DNA motifs that associate with shear stress responsive enhancers. (A) Number of ATAC-Seq peaks specific for HUVEC under static conditions (“lost” after shear stress) or exposed to shear stress (“gained” after shear stress), as well as “common” for both conditions. (B) Heatmap representing scores associated with genomic regions centered on the indicated H3K27ac- and ATAC-Seq peaks in control and flow (18 dyn/cm2 shear stress for 6 h) conditions. (C) Annotation of gained and lost ATAC-Seq peaks that overlap with gained and lost H3K27ac ChIP-Seq peaks, respectively, to defined regions of the genome. (D,E) Analysis of known DNA motifs enriched in gained (D) or lost (E) ATAC-Seq peaks that overlap with gained or lost H3K27ac ChIP-Seq peaks, respectively. The 10 most enriched motifs are shown. (F) Heatmap representing spatial association of genes up- or down-regulated by shear stress with different subsets of ATAC-Seq peaks (all gained and lost ATAC-Seq peaks, gained and lost ATAC-Seq peaks that overlap with gained or lost H3K27ac ChIP-Seq peaks, respectively, or common ATAC-Seq peaks that overlap with gained or lost H3K27ac ChIP-Seq peaks). The numbers represent the percentage of up-or down-regulated genes, associated with at least one peak.
To identify TF binding site motifs in flow responsive enhancers, we analyzed the subset of H3K27ac peaks, which showed similar changes in H3K27ac and chromatin accessibility. These peaks were mostly located in introns and intergenic regions (Fig. 2C). In line with the role of KLF TFs in EC responses to shear stress, a number of KLF motifs were enriched in gained peaks (e.g. KLF5, KLF4, KLF1, KLF3, KLF6, KLF10, KLF14, Fig. 2D, Suppl. Fig. 4D; of note, a motif for KLF2 is not included in the database). Lost peaks were enriched in motifs for endothelial lineage TFs ETS1, ETV1, ETV2 and FLI1 (Fig. 2E, Suppl. Fig. 4E). Interestingly, common ATAC peaks that overlapped with gained H3K27ac peaks also contained several enriched KLF motifs (Suppl. Fig. 4F). However, other motifs such as JUNB, FOSL2 and ETV2 were also highly enriched in this subset. Motifs found in common ATAC peaks that overlapped with lost H3K27ac peaks were similar to those found in lost ATAC peaks overlapping with lost H3K27ac peaks (compare motifs in Suppl. Fig. 4G with those in Fig. 2E). Together, our analysis uncovered several known and novel DNA motifs enriched in shear stress activated enhancers.
Changes in gene expression correlate with chromatin accessibility
To understand how chromatin accessibility predicts changes in gene expression, we associated ATAC-Seq peaks with nearby genes using the GREAT algorithm. Comparing genes associated with ATAC-Seq peaks with DE genes from our RNA-Seq dataset revealed that upregulated genes associated more with gained than lost ATAC-Seq peaks and vice versa (Fig. 2F). This association was more obvious for upregulated genes, and it was higher when considering all ATAC-Seq peaks compared to the ones that overlap with H3K27ac peaks. Interestingly, analysis of the genes associating with H3K27ac peaks that overlap with common ATAC peaks revealed that they also had a high degree of association with DE genes. Hence, 60% of upregulated genes had gained H3K27ac peaks that overlapped with common ATAC peaks in their vicinity and 46% of downregulated genes associated with lost H3K27ac peaks that overlapped with common ATAC-Seq peaks (Fig. 2F). Together, our data might indicate the existence of different modes of regulation of shear stress-induced gene expression.
Functional analysis of identified putative enhancers
Previous studies have shown that only a fraction (26%) of bioinformatically identified enhancer elements are functionally relevant in reporter assays46, while even fewer showed activity in large scale functional screens using the KRAB repressor44,47. Therefore, the flow responsiveness of identified putative enhancers and their regulation of flow-responsive genes need to be validated experimentally. We tested flow responsiveness of several putative enhancers using luciferase assays. To do so, we cloned putative enhancers upstream of a minimal promoter regulating the expression of firefly luciferase (Suppl. Fig. 5A). Out of 14 constructs tested, 3 showed a significant increase in luciferase activity in response to shear stress (18 dyn/cm2 for 24 h; Suppl. Fig. 5B). This correlated with higher H3K27ac in these regions and with their association with upregulated DE genes. Most DE genes associated with multiple peaks. Since their cooperation might be necessary for shear stress-induced gene expression, putative enhancers may not function in isolation in a luciferase assay.
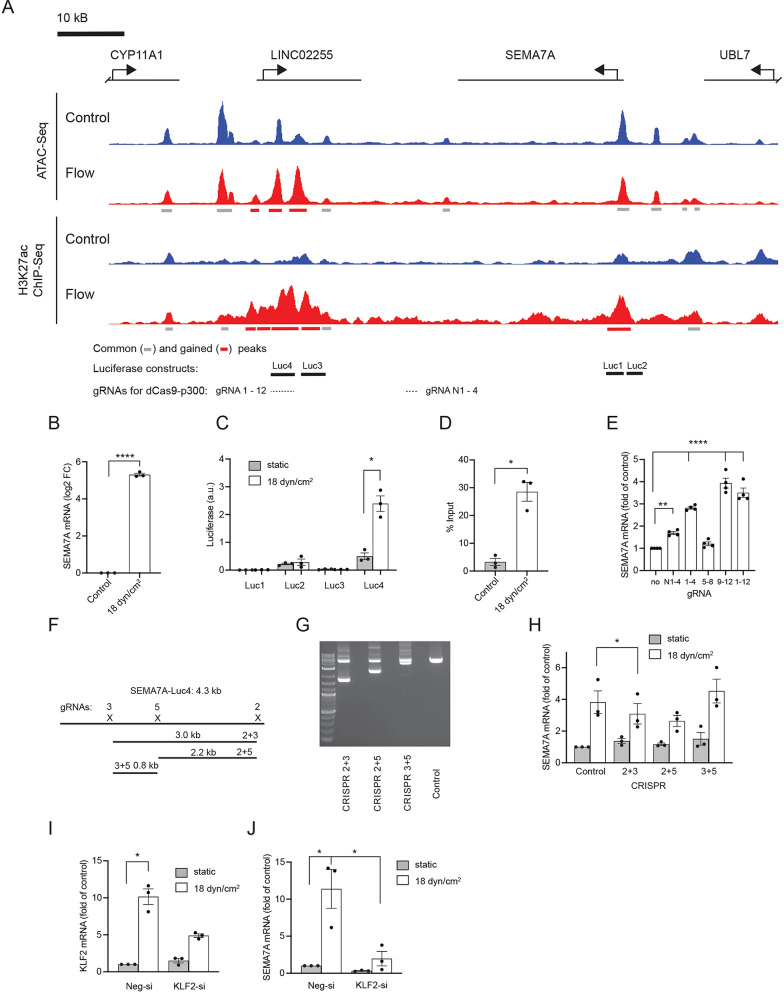
We then concentrated on the SEMA7A gene, which is associated with several gained H3K27ac peaks (in the promoter region and 45–55 kb downstream of the transcription start site). These overlapped with both gained and common ATAC peaks (Fig. 3A). This correlated with SEMA7A expression, which was induced 35-fold in shear stress exposed HUVEC (Fig. 1B, Suppl. Fig. 2A), while qPCR showed a 41-fold induction (Fig. 3B). SEMA7A was also upregulated in several published datasets of flow-regulated genes40,41. To confirm enhancer activity of putative SEMA7A enhancers, we cloned 4 luciferase constructs (Luc1 − 4; Fig. 3A) and tested them for flow response. Construct Luc2, containing the sequence in the promoter region, showed stronger activity compared to an empty vector (data not shown), but no flow response (Fig. 3C). In contrast, Luc4 showed a strong activation in response to shear stress with relatively high basal activity observed also under static conditions confirming its flow-responsive enhancer activity (Fig. 3C). We confirmed H3K27ac enrichment in this flow-responsive enhancer by ChIP-qPCR (Fig. 3D). Luc1 and 3 showed neither basal, nor flow-induced activity.
Figure 3.
Identification and characterization of a shear stress responsive enhancer in the SEMA7A gene. (A) Normalized ATAC-Seq and H3K27ac ChIP-Seq tracks in the vicinity of the SEMA7A gene. Bars below ATAC and ChIP-Seq tracks show identified common (grey) and gained (red) ATAC-Seq or H3K27ac ChIP-Seq peaks. In addition, the regions used for luciferase constructs in (C) and locations targeted by gRNAs in (E) are shown underneath the tracks. (B) SEMA7A mRNA expression in HUVEC exposed to 18 dyn/cm2 for 6 h (Log2 fold of control, mean ± sem, n = 3, ****p < 0.001, two-tailed paired t-test). (C) Luciferase assay showing luciferase activity of the constructs containing DNA regions indicated in (A) upstream of firefly luciferase in HUVEC exposed to shear stress (18 dyn/cm2 for 24 h, data are presented in arbitrary units (a.u.) after normalization to the signal of a renilla luciferase construct; mean ± sem, n = 3, *p < 0.05, two-tailed paired t-test). (D) ChIP-qPCR showing higher H3K27ac ChIP-Seq enrichment in the region corresponding to Luc4 (see (A) in HUVEC exposed to shear stress (18 dyn/cm2 for 6 h, data is presented as % input, mean ± sem, n = 3, *p < 0.05, two-tailed paired t-test). (E) SEMA7A mRNA expression in HEK293T17 cells transfected with plasmids encoding dCas9-p300 and different gRNAs (combination of 4 or 12 gRNAs) targeting genomic regions downstream of the SEMA7A gene (marked in (A); data is shown as fold of control, mean ± sem, n = 4, ****p < 0.0001, **p < 0.01, one-way ANOVA with Dunnet’s multiple comparison test). (F) Schematic representation of the genomic region corresponding to the Luc4 construct with pairs of CRISPR/Cas9 constructs designed to remove parts of the region. (G) Ethidium bromide gel showing PCR products of the genomic region corresponding to the Luc4 construct. Shorter PCR products result from the removal of targeted genomic regions. Uncropped agarose gel is shown in Suppl. Fig. 10. (H) SEMA7A mRNA expression in HUVEC infected with lentiviral constructs encoding Cas9 and gRNAs targeting genomic regions downstream of the SEMA7A gene and exposed to shear stress (18 dyn/cm2 for 6 h, data is shown as fold of control, mean ± sem, n = 3, * p < 0.05, repeated measures one-way ANOVA with Dunnet’s multiple comparison test). (I,J) KLF2 (I) and SEMA7A (J) mRNA expression in HUVEC transfected with KLF2 or control siRNA (Neg-si) and exposed to shear stress (18 dyn/cm2 for 6 h, data are presented as fold of control, mean ± sem, n = 3, *p < 0.05, one-way ANOVA with Tukey’s multiple comparison test).
Of note, the Luc4 enhancer is located within the long intergenic non-protein coding RNA LINC02255. Future studies will be needed to understand the relationship between Luc4 activity and LINC02255, as we did not detect LINC02255 expression in response to shear stress or Luc4 enhancer manipulations (data not shown). Since the activity of EC-specific enhancers has been reported to be higher in conjunction with their own promoters compared to minimal promoters48, we combined Luc2 and Luc4 in one construct (Luc2 + 4). Indeed, the flow response of Luc2 + 4 was higher than that of Luc4 alone (Suppl. Fig. 5E), suggesting cooperative interactions between this enhancer and the SEMA7A promoter. To further investigate this enhancer, we used dCas9-p300 to target and activate the Luc4 enhancer using sets of gRNAs (Suppl. Fig. 5C, see Fig. 3A for gRNA targeting location) specific for different regions of the enhancer in HEK293T cells. These cells are easier to transfect compared to ECs and they have lower SEMA7A expression. HEK293T cells were transfected with corresponding gRNA-coding plasmids together with dCas9-p300 and the Luc4 plasmid. We detected a 50-fold increase in luciferase activity when using 12 gRNAs targeting the whole length of the Luc4 sequence (4 kb) compared to cells transfected with Luc4 and control plasmids indicating successful targeting of the SEMA7A enhancer (Suppl. Fig. 5F). Targeting the endogenous enhancer with the same gRNAs resulted in 3.5-fold SEMA7A mRNA upregulation in HEK293T cells without shear stress (Fig. 3E), with different sets of gRNAs showing similar activities as in luciferase assays. Importantly, targeting another region downstream of SEMA7A with no H3K27ac ChIP-Seq and ATAC-Seq peaks led to a lower change in SEMA7A expression (gRNAs N1-4; Fig. 3A). In a complementary approach, we deactivated the SEMA7A enhancer using Cas9 and different pairs of gRNAs to remove parts of the enhancer (Suppl. Fig. 5D, Fig. 3F,G). Exposing targeted cells to shear stress resulted in a small, but statistically significant induction in SEMA7A expression (Fig. 3H). The relatively weak effect might be attributed to variations in the efficiency of enhancer deletion, as we used pooled primary cells without clonal selection in our experiments. Together, several lines of evidence suggest that we identified a gained flow-responsive enhancer that can regulate SEMA7A expression.
To test the function of KLF2 in regulating SEMA7A, we analyzed SEMA7A expression after KLF2 knockdown. We achieved a more than 50% knockdown of flow mediated KLF2 induction after siRNA transfection (Fig. 3I). This knockdown led to a strong decrease of SEMA7A expression in shear stress exposed ECs (Fig. 3J). Together, these results suggest that KLF2 mediates the observed shear stress regulation of SEMA7A expression.
Most reports focus on the positive effects enhancers have on gene expression. However, it is equally important that genes are being turned off. To validate a flow-responsive lost enhancer, we examined the known shear stress responsive gene CXCR4, which was strongly down-regulated upon exposure to shear stress (Fig. 1B, Suppl. Fig. 2B), which we confirmed by qPCR (Suppl. Fig. 5H). This gene was associated with a lost H3K27ac peak located 125 kb upstream of its TSS that overlapped with common ATAC peaks (Suppl. Fig. 5G). To confirm that this enhancer regulates CXCR4 expression, we used CRISPR/Cas9 to remove parts of the enhancer (Suppl. Fig. 5I,J). Removal of parts of the enhancer with CRISPR pairs 4 + 6 and 6 + 7 resulted in reduction of CXCR4 mRNA expression (Suppl. Fig. 5K), mimicking the shear stress response. We therefore identified and validated a shear stress responsive lost enhancer that regulates CXCR4 expression.
Identification of flow-induced transcriptional program in zebrafish embryos
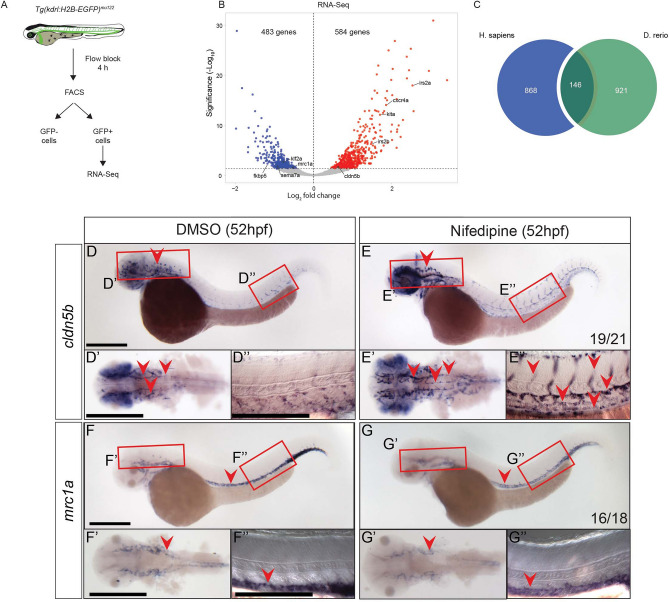
To study whether the transcriptional program we identified in adult human ECs is conserved during vascular development in other species, we compared gene expression in ECs in zebrafish embryos without blood flow with ECs from embryos experiencing normal blood flow. For this, we blocked blood flow in 48 hpf Tg(kdrl:H2B-EGFP)mu122 embryos for 4 h and sorted ECs (Fig. 4A). RNA-Seq analysis revealed 1067 differentially expressed genes in ECs experiencing flow block compared to control zebrafish embryos. 584 genes were up- and 483 genes were downregulated (Fig. 4B, Suppl. Fig. 6A,B, Suppl. Data 1). We validated our RNA-Seq data by qPCR for select most up- and downregulated DE genes (Suppl. Fig. 6C), which showed a strong correlation between the two methods. GSEA pathway analysis revealed MAPK and Notch signaling as some of the most enriched pathways after flow block (Suppl. Fig. 6D). Importantly, the orthologues of the genes that were among the most DE genes in the shear stress exposed HUVEC dataset were also regulated by blood flow in zebrafish embryos. Hence, cxcr4a, kita, irs2a and irs2b were upregulated, while klf2a and sema7a were downregulated by flow block (Fig. 4B). Expression changes were opposite to those we observed in HUVEC exposed to shear stress (Fig. 1B). Comparing the datasets with such opposite regulation in zebrafish and HUVEC revealed an 8% overlap between the genes upregulated in zebrafish ECs after flow block and downregulated in HUVEC exposed to shear stress (Suppl. Fig. 6E). Conversely, there was a 6.2% overlap of the genes downregulated in zebrafish ECs and upregulated in HUVEC (Suppl. Fig. 6E). This is a substantial overlap considering that only distinct EC populations in zebrafish embryos might show a flow response, which in addition might only occur at select developmental time points. Also, the experimental approach to interrogate for shear stress responses within ECs varied in these two systems (flow increase in the case of HUVEC, while we blocked blood flow in zebrafish embryos). Furthermore, the overlap was higher (13.7%) when we took into consideration all DE genes (Fig. 4C, Suppl. Fig. 6E). This highlights the possibility that some genes might show differential flow responses, depending on the timing and the magnitude of shear stress. For example, ELMSAN1 is upregulated in HUVEC after 6 h of shear stress exposure and elmsan1b is also upregulated in zebrafish ECs after stopping blood flow. However, shorter shear stress exposure (30 min) resulted in downregulation of ELMSAN1 in HUVEC (Suppl. Fig. 1B) in line with our observations in zebrafish. DNA acetylation/de-acetylation will also require different sets of enzymes (histone acetylases and de-acetylases, respectively), which might not be available to the same extent in all cell populations.
Figure 4.
Blocking blood flow in zebrafish reveals genes regulated by shear stress in developing embryos. (A) Schematic representation of EC FACS sorting from Tg(kdrl:H2B-EGFP)mu122 embryos treated with nifedipine for 4 h at 48 hpf to induce flow block. Sorted GFP + cells were used for RNA-Seq. As control, embryos were treated with a corresponding amount of DMSO. (B) Volcano plot showing gene expression changes in ECs after flow block in zebrafish embryos. Differentially expressed (DE) genes (FDR < 0.05) are labelled in red (upregulated) and blue (downregulated after flow block). (C) Overlap of flow-regulated genes in human and zebrafish datasets. All DE genes were used for this analysis. (D,E) Whole mount in situ hybridization for cldn5b in 52 hpf embryos after 4 h of treatment with DMSO (D) or nifedipine (E). Regions in the head and the trunk highlighted with red boxes in (D,E) are magnified in (D′,E′,D′′,E′′), respectively. Vascular expression is highlighted with red arrowheads. (F,G) Whole mount in situ hybridization for mrc1a after flow block from 48 to 52 hpf. Regions in the head and the trunk highlighted with red boxes in (F,G) are magnified in (F′,G′,F′′,G′′), respectively. Vascular expression is highlighted with red arrowheads. Numbers represent the number of embryos with the depicted expression pattern out of the total number of embryos analyzed. Scale bar is 300 μm.
We further validated vascular expression and flow responsiveness of cldn5b (Fig. 4D,E) and mrc1a (Fig. 4F,G) by whole mount in situ hybridization in zebrafish embryos. cldn5b was expressed in brain vasculature of 52 hpf embryos (Fig. 4D, arrowhead), while expression was weak in trunk vasculature (Fig. 4D, enlarged in Dʹ and Dʹʹ). Flow block increased the expression of cldn5b in brain and trunk vasculature (Fig. 4E, arrowheads). This suggests that blood flow inhibits cldn5b expression, an observation that is in line with increased cldn5b expression after flow block in our RNA-Seq dataset (Fig. 4B). In contrast, expression of mrc1a in both brain and trunk vessels was reduced after flow block (Fig. 4F,G, enlarged in Fʹ, Fʹʹ and Gʹ, Gʹʹ). This correlated with mrc1a down-regulation by flow block in RNA-Seq (Fig. 4B) and reveals blood flow as a positive regulator of mrc1a expression. We furthermore confirmed upregulation of adma and lyve1a (Suppl. Fig. 7) and downregulation of gpr182 and id1 (Suppl. Fig. 8) upon flow block. These genes were not regulated in HUVEC but were differentially expressed on RNA-Seq of zebrafish ECs after flow block. In addition, we validated flow responsiveness of sox18 and klf2a (Suppl. Fig. 9), which were regulated in zebrafish embryos and in HUVEC. In line with RNA-Seq results, flow block induced sox18 expression in zebrafish blood vessels (Suppl. Fig. 9B,C). Interestingly, sox18 was expressed in the dorsal aorta and posterior cardinal vein in 24 hpf embryos, but its expression was reduced in 36 and 48 hpf embryos, which experience increased blood flow (Suppl. Fig. 9A). Conversely, the expression of klf2a was higher in intersegmental blood vessels of 48 hpf embryos, compared to an earlier time point (36 hpf), which could be attributed to higher blood flow (Suppl. Fig. 9D). Blocking blood flow reduced intersegmental blood vessel expression of klf2a in accordance with our RNA-Seq results (Suppl. Fig. 9E,F).
Discussion
It is widely appreciated that changes in hemodynamics have profound effects on EC biology. However, the underlying gene regulatory programs mediating these effects are poorly understood. In this report, we characterized the laminar shear stress response of HUVEC on the transcriptional and chromatin accessibility level. In addition, we provide evidence that reciprocal changes in gene expression occur in ECs of zebrafish embryos subjected to flow-block. Our analysis shows correlation between gene expression patterns and identified enhancers, with upregulated genes more commonly clustering with gained peaks, while downregulated genes associate with lost peaks. Thus, comparing ChIP-Seq and ATAC-Seq data from HUVEC revealed the presence of distinct sets of gene regulatory elements. We find a considerable overlap between gained H3K27ac ChIP-Seq and gained ATAC-Seq peaks. However, there is also an overlap between gained H3K27ac ChIP-Seq peaks and common ATAC-Seq peaks. This might suggest the existence of poised enhancers, which are positioned in regions of open chromatin, but have not obtained H3K27ac marks38,49.
We find that shear stress upregulates the expression of the transcription factors KLF2 and KLF4, which previous studies identified as master regulators of the endothelial shear stress response5,50. We find KLF2 and KLF4 upregulation already after 30 min of shear stress exposure, identifying these genes being among the earliest transcriptional responses to shear stress39. Loss of KLF2 has been reported to reduce vascular smooth muscle cell and pericyte coverage51,52, lead to high cardiac output failure53 and cardiac valve defects in zebrafish54 and mouse55. High klf2a expression in developing veins in zebrafish is needed to restrict smooth muscle cells to the dorsal aorta56. Further studies showed that klf2a controls VEGF signaling during aortic arch development57. In addition to these requirements of KLF2 during embryonic development, KLF2 also regulates vascular tone58, represses inflammatory responses59 and induces endothelial quiescence and atheroprotection60. Studies analyzing the human disease condition cerebral cavernous malformations (CCM) also showed that increases in KLF2 mRNA levels can contribute to disease progression61, illustrating the need of a tight control of KLF2 expression. Given the multitude of processes KLF2 functions in, additional work is needed to separate primary from secondary effects within the vasculature. Our work provides an inroad to answering these questions through the identification of KLF binding sites in regions of the chromatin that show marks of active enhancers upon flow exposure. Functionally interrogating these regions will help in delineating direct KLF2 target genes in addition to other transcription factors which might bind to the novel motifs we identified.
We initiated these studies by analyzing putative enhancer elements driving the expression of SEMA7A. We chose SEMA7A due to its regulation by shear stress and reported role in blood vessel homeostasis62. Our bioinformatic analysis showed the presence of putative enhancer elements about 50 KB downstream of the SEMA7A TSS. We confirmed the flow response of this enhancer element using luciferase reporter constructs and through deleting it from the HUVEC genome. Therefore, our approach can lead to the identification of shear stress responsive enhancer elements. However, about 80% of the enhancers we tested did not change the expression of luciferase reporter constructs. This is within the predicted range46. Several reasons can account for the lack of reporter activity. Enhancers often function together with the promoters of the genes they regulate48,63. Thus, using a gene’s endogenous promoter instead of a generic minimal promoter might increase the number of enhancers functioning in reporter assays. The genomic context of a given enhancer might also influence its activity64. In addition, a growing body of evidence suggests that genes are being regulated through the combinatorial action of several “weak” enhancers47,65. In the future, interrogating these interactions within our dataset will likely contribute significant insights into shear stress mediated gene regulation. It will also be of importance to understand the effects different flow regimes exert on endothelial cells. We previously characterized enhancer elements and transcription factors in ECs exposed to oscillatory shear stress33, while He et al. generated a similar dataset for ECs exposed to pulsatile shear stress34. Furthermore, Andueza et al. generated datasets for disturbed flow32. Comparing these data to our data investigating laminar shear stress responses will help in elucidating how ECs might adapt to changes in flow during embryonic development or disease settings.
Our data significantly expand previous studies that focused more on the activating effects of shear stress on gene expression and the transcription factors mediating these effects32. In this study, we also analyzed lost enhancers upon flow exposure, which might be equally important for regulating vascular morphogenesis. For instance, during angiogenesis in zebrafish, expression of flt4 and cxcr4a needs to be downregulated in order to prevent ectopic blood vessel sprouting66–69, while prolonged ETV2 expression during embryogenesis can lead to vascular defects70. Our data now provide a resource to investigate the influence of lost enhancers on endothelial gene expression. An unexpected finding was the enrichment of ETV2/ETS binding sites in this enhancer subset. During embryogenesis ETV2 is required for EC differentiation, but is strongly downregulated at later stages of blood vessel development71–75. In undifferentiated progenitor cells in culture, ETV2 expression is needed to commit these cells to the endothelial lineage76, while overexpression in HUVECs allows these cells to obtain a more embryonic EC state77. This overexpression leads to an upregulation of KLF2 and KLF4 expression with ETV2 being bound to KLF2 and KLF4 TSS. By contrast, continuous ETV2 expression in mouse embryos reduces KLF2 and KLF4 levels and leads to vascular anomalies70. Of note, analysis of several vein specific enhancers revealed the presence of multiple ETS binding sites. Reporter constructs using these enhancers displayed robust expression during early embryonic stages but were downregulated after E13 in mice and did not show expression in the mature micro-vasculature78. Together, these results suggest that suppression of ETV2 and ETV2 target gene expression might be required for proper shear stress responses and that expression of KLF2 and KLF4 might initially rely on ETV2, but that continuous expression requires ETV2 downregulation. Our results showing a loss of binding sites for ETS and FLI transcription factors in response to shear stress support this notion and will enable the discovery of further ETV2 target genes. How KLF and ETS TFs function in concert and the mechanisms controlling their regulatory interdependence will provide important future insights into shear stress mediated endothelial differentiation and vascular maturation.
Zebrafish have advanced as a widely used model system to study cardiovascular development and disease79. Our findings that many genes are similarly regulated by shear stress when compared to HUVEC underscore the conserved nature of shear stress responses. Importantly, zebrafish can be used to investigate tissue and temporal differences in these responses. Future studies will be needed to reveal how gene regulatory circuits change during blood vessel development and decipher the contribution shear stress has on these processes and thereby vascular morphogenesis.
Materials and methods
Cell culture
To collect samples for ChIP-Seq, ATAC-Seq and RNA-Seq, HUVEC were isolated and cultured as described before80. The isolation was approved by the ethical board of the Westfälische Wilhelms-Universität Münster and was in accordance with the Declaration of Helsinki. Within this framework, informed written consent was obtained from human subjects prior to HUVEC isolation. HUVEC were exposed to fluid shear stress of 18 dyn/cm2 in a BioTechFlow system (BTF system; MOS Technologies) at 37 °C and under constant CO2 perfusion, as previously described81. HUVEC were seeded on 6-cm BTF glass plates coated with cross-linked gelatin and cultured to confluence (approximately 105 cells/cm2) in EC growth medium (Promocell, Heidelberg, Germany). The maximum flow of 18 dynes/cm2 was achieved by computer-controlled cone rotation in culture medium containing 3% polyvinylpyrrolidone (PVP) within ten minutes (ramp function) and then kept constant for the required time, as described before33. Static controls were incubated in a cell culture incubator. For all other experiments, HUVEC were purchased (Gibco, # C0035C) and cultured in EBM-2 medium (Lonza, # CC-3156) with EGM-2 supplements (Lonza, # CC-4176). HUVEC in passage 4 were seeded on ibiTreat 0.4 mm μ-slide I Lure (ibidi, # 80176) pre-coated with fibronectin (5 μg/ml; Sigma, # F2006), and exposed to shear stress (18 dyn/cm2) using an ibidi Pump System (ibidi, # 10902).
Zebrafish experiments
All zebrafish experiments were conducted on animals prior to the third day of embryonic development and are therefore not considered animal experiments. The maintenance of adult zebrafish for breeding purposes was in accordance with local and international guidelines and approved by the Landesamt fuer Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV)82. Zebrafish were maintained as described previously83. Tg(kdrl:H2B-EGFP)mu122 embryos10 were dechorionated at 48 h post fertilization (hpf) using pronase and treated for 4 h with nifedipine (12.5 nM, Biomol, # Cay11106) and tricaine (1.3 mM, Sigma-Aldrich, # A5040) to stop the heart beat or with a corresponding amount of DMSO as a control. Embryos were used for RNA-Seq or in situ hybridization. For RNA-Seq 300 embryos per condition were collected in ice cold 10% DMSO and kept at − 80 °C for 2 min. Afterwards, embryos were washed twice with 1× Hanks’ Balanced Salt Solution (HBSS, Gibco, # 14185-052) and deyolked with calcium-free Ringer’s solution. Embryos were then dissociated with trypsin–EDTA (0.5%) and collagenase type IV (50 mg/ml). The reaction was stopped by addition of FCS (5%) and cells were collected by centrifugation at 350×g for 5 min at 4 °C and washed twice with 1× HBSS followed by centrifugation. After a final wash, cells were resuspended in 1× HBSS, passed through a 40 μm nylon filter to obtain a single cell suspension, which was subjected to fluorescence-activated cell sorting (FACS). EGFP positive ECs were collected in RLT buffer (RNeasy Micro kit, Qiagen).
RNA-Seq
HUVEC or FACS-sorted zebrafish ECs were lysed with RLT buffer containing β-mercaptoethanol and RNA was isolated with the RNeasy Plus Mini or Micro Kit (Qiagen), respectively. RNA was quantified with NanoDrop (Thermo Fisher Scientific) and quality assessment was performed with a Bioanalyzer (Agilent). Libraries were prepared using TruSeq Stranded Total RNA Sample Prep with RiboZero depletion (Illumina), quantified with Qubit (Thermo Fisher Scientific), analyzed for fragment distribution with a Bioanalyzer and sequenced with paired-end settings (2 × 75 cycles) on a MiSeq System (Illumina).
ChIP-Seq
ChIP was performed as previously described33. Briefly, HUVEC were cross-linked with formaldehyde (final concentration 1%) for 5 min. After neutralization with glycine (0.125 M), cells were resuspended in nuclei lysis buffer and sonicated using an AFA sonicator (Covaris). Immunoprecipitation was performed using a H3K27ac antibody (Abcam, # ab4729) pre-bound to M-280 sheep anti-rabbit Dynabeads (Thermo Fisher Scientific, # 11204D). DNA was purified with phenol/chloroform/isoamyl alcohol and precipitated with ethanol. DNA was quantified using Qubit and fragment distribution was controlled with a Bioanalyzer. Libraries were prepared using TruSeq ChIP Sample Preparation Kit (Illumina), quantified by Qubit and analyzed for fragment distribution with a Bioanalyzer and used for paired-end sequencing (2 × 75 cycles) on a MiSeq System (Illumina).
ATAC-Seq
HUVEC were collected via trypsinization and slowly frozen in FCS containing 10% DMSO in an isopropanol chamber at − 80 °C according to a published protocol84. After thawing, cells were resuspended in EBM-2 medium and 50,000 cells were used for nuclei isolation and ATAC reaction and library preparation using the OMNI-ATAC protocol84,85. Libraries were quantified with a NanoDrop and fragment distribution was analyzed on a Bioanalyzer. Libraries were used for paired-end sequencing (2 × 35 cycles) on a NextSeq500 system (Illumina).
Data analysis
Sequencing reads were aligned to hg19 (human) or danRer10 (zebrafish) genomes. Two replicates per condition were analyzed.
RNA-Seq
Data was mapped using TopHat2 v2.1.1.86 and differential gene expression analysis between flow-treated samples and static controls was performed using DESeq287. Read counts were obtained with HTSeq88. Genes were considered as differentially expressed when the FDR-adjusted p-value was < 0.05. Gene Set Enrichment Analysis (GSEA) was done at http://www.webgestalt.org and overrepresentation analysis (ORA) was performed with g:Profiler (https://biit.cs.ut.ee/gprofiler/gost).
ChIP-Seq
Sequencing reads from ChIP and input samples were mapped with Bowtie 2 v2.3.5.89 and Samtools v1.9.90 was used to edit the alignments. Reads were downsampled to equivalent read depth. Peak calling was performed with Genrich (https://github.com/jsh58/Genrich) applying the following parameters: genrich -v -e Y,MT -E blacklist.bed -t Sample1ChIP.bam, Sample2ChIP.bam -c Sample1Input.bam, Sample2Input.bam -o Sample.narrowPeak -k Sample.bedGraphish. Differential peak analysis was performed with DiffBind91 and peaks with FDR < 0.05 were used for further analysis. To analyse correlation between our and ENCODE replicates, Deeptools v2.29.292 multiBamSummary was used to compute read coverages for genomic regions that were directly used to calculate and visualize pairwise Pearson’s correlation values.
ATAC-Seq
Sequencing reads were mapped with Bowtie2, downsampled to equivalent read depth and peak calling was performed with Genrich in ATAC-Seq mode using the following parameters: genrich -j -v -y -r -e Y,MT -E blacklist.bed -t Sample1.bam, Sample2.bam -o Sample.narrowPeak -k Sample.bedGraphish. Differential peak analysis was performed with DiffBind91 and peaks with FDR < 0.05 were used for further analysis.
ChIP-Seq and ATAC-Seq tracks were produced with deepTools bamCoverage, normalized to sequencing depth and visualized with UCSC Genome Browser. The overlap between ChIP-Seq and ATAC-Seq peaks was analysed with bedtools closest function and peaks within 100 bp of each other were considered to overlap. Heatmaps of overlapping regions were produced with deepTools. Peak association with neighbouring genes was performed with GREAT45. Annotations of the peaks to defined genomic regions was done with HOMER93 annotatePeaks.pl and DNA motif analysis was performed with findMotifsGenome.pl function using default region size and corresponding common peaks as background regions.
Luciferase assay
Putative enhancers were amplified from genomic DNA with Phusion High-Fidelity DNA Polymerase (New England Biolabs, # M0530). Betaine (1 M final concentration, Sigma Aldrich, # B0300) was added to the PCR reactions. Primers contained overhangs for cloning (primer sequences and overhangs are listed in Suppl. Table 1). PCR products were cloned into pCR4Blunt-TOPO vector (Invitrogen, # 450031) and the sequence of the cloned DNA was confirmed by Sanger sequencing. Inserts were cut out with appropriate restriction enzymes (Suppl. Table 1) and ligated with pre-digested pGL4.23[luc2/minP] (Promega, # E8411) plasmid upstream of a minimal promoter and a firefly luciferase gene. HUVEC were co-transfected using Lipofectamin 2000 (Invitrogen, # 11668027) and PLUS reagent (Invitrogen, # 11514015) with a firefly plasmid and the pRL-CMV plasmid (Promega, # E2261) containing the Renilla luciferase gene for normalization. HUVEC were exposed to shear stress (18 dyn/cm2 for 24 h) 2 h after transfection. Afterwards, cells were lysed and activity of firefly and renilla luciferase was measured with a Dual-Luciferase Reporter Assay System (Promega, # E1910).
Enhancer activation with dCas9-p300
gRNA sequences were designed with chopchop (https://chopchop.cbu.uib.no) to target putative enhancers. Corresponding DNA oligonucleotides with BbsI overhangs (sequences are listed in Suppl. Table 1) were annealed and ligated with pre-digested pSPgRNA plasmid (Addgene, # 47108). HEK293T17 cells (ATCC, # CRL11268) were co-transfected using Lipofectamin 2000 (Invitrogen, # 11668027) and PLUS reagent (Invitrogen, # 11514015) with combinations of pSPgRNA plasmids and pcDNA-dCas9-p300Core plasmid (Addgene, # 61357) to activate putative SEMA7A enhancers94. SEMA7A expression was measured by qPCR 4 days after transfection. Alternatively, cells were co-transfected with SEMA7A-Luc4 and pRL-CMV (Promega, # E2261) constructs and enhancer activity was measured with a luciferase assay.
CRISPR-Cas9-mediated enhancer removal
Putative enhancers were removed using CRISPR pairs. gRNA sequences were designed with chopchop (https://chopchop.cbu.uib.no) and corresponding DNA oligonucleotides containing BsmBI overhangs were annealed and ligated with lentiCRISPR v2 plasmid (Addgene, # 52961), which contains both Cas9 and sgRNA sequences95. The sequences targeting putative enhancers are listed in Suppl. Table 1. HEK293T17 cells (ATCC, # CRL11268) were transfected with specific lentiCRISPR v2 plasmids, as well as pMD2.G (Addgene, # 12259) and psPAX2 (Addgene, # 12260) plasmids using Fugene 6 Transfection Reagent (Promega, #E2311). 72 h after transfection, the supernatant was collected and filtered through a 0.45 μm filter (Sarstedt, # 83.1826) and virus was collected with LentiX Concentrator (Takara, # 631232). HUVEC were infected with combinations of two viruses and used 96 h after infection. Enhancer removal was confirmed by PCR, using Phusion High-Fidelity DNA Polymerase (New England Biolabs, # M0530), 1 M betaine (Sigma Aldrich, # B0300) and the primers listed in Suppl. Table 1. PCR products were visualized on ethidium bromide agarose gels.
qPCR
RNA was isolated with an RNeasy Micro Kit (Qiagen) and reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad, # 1708891). Power SYBR Green PCR Master Mix (Applied Biosystems, # 4367659) was used to run qPCR in a 7900HT Fast Real-Time PCR System (Applied Biosystems). ddCt method was used to calculate relative expression using RPL13A or TBP as endogenous controls. Statistical analysis was performed on dCt data. Primer sequences are listed in Suppl. Table 1.
ChIP-qPCR
DNA isolated after H3K27ac ChIP was used for qPCR with Power SYBR Green PCR Master Mix and primers listed in Suppl. Table 1. In addition, qPCR was performed on DNA from input control and H3K27ac enrichment was calculated as % of input.
siRNA
HUVEC were transfected with 50 nM of siRNA using Oligofectamine (Invitrogen, # 12252011) and used for flow experiments in an ibidi system 48 h after transfection. Silencer Select Negative Control No. 1 siRNA (Invitrogen, # 4390843) and Silencer Select KLF2 siRNA (Invitrogen, # 4392420, ID # s20270) were used.
Statistical analysis
Statistical analysis was performed with GraphPad Prism.
Supplementary Information
Acknowledgements
We would like to thank Gerd Blobel for critical reading of the manuscript.
Author contributions
R.T. and A.F.S. conceived the experiments. A.F.S., H.S., J.M.V. and Z.A. supervised the work. R.T. performed cell culture experiments and performed data analysis. E.V.L. performed zebrafish experiments. N.D. and N.Y. performed data analysis. M.O.-S., O.B. and R.T. performed shear stress experiments. R.T. and A.F.S. wrote the manuscript. All authors provided manuscript input and discussed and commented on the work.
Funding
This work was supported from funds provided by the Max-Planck society (A.F.S.), Deutsche Forschungsgemeinschaft (DFG) grant SI 1374/4-1, SI 1374/5-1, SI 1374/ 3-2 (A.F.S.) and the Perelman School of Medicine and the Cardiovascular Research Institute of the University of Pennsylvania (A.F.S.). We would also like to acknowledge the following funding bodies: BMBF 03ZZ0906E (H.S.), DFG SCHN 430/6-2 (H.S.), SCHN 430/9-1 (H.S.) and INST 2105/241 (H.S.). The funding was also provided by National Institutes of Health (R01HL152086).
Data availability
All data from this study are available under GEO accession number GSE198221.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08645-8.
References
- 1.Yin J, Heutschi D, Belting HG, Affolter M. Building the complex architectures of vascular networks: Where to branch, where to connect and where to remodel? Curr. Top. Dev. Biol. 2021;143:281–297. doi: 10.1016/bs.ctdb.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Hahn C, Schwartz MA. Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 2009;10:53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaffari S, Leask RL, Jones EA. Flow dynamics control the location of sprouting and direct elongation during developmental angiogenesis. Development. 2015;142:4151–4157. doi: 10.1242/dev.128058. [DOI] [PubMed] [Google Scholar]
- 4.Dekker RJ, Boon RA, Rondaij MG, Kragt A, Volger OL, Elderkamp YW, Meijers JC, Voorberg J, Pannekoek H, Horrevoets AJ. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima H, Mochizuki N. Flow pattern-dependent endothelial cell responses through transcriptional regulation. Cell Cycle. 2017;16:1893–1901. doi: 10.1080/15384101.2017.1364324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korn C, Augustin HG. Mechanisms of vessel pruning and regression. Dev. Cell. 2015;34:5–17. doi: 10.1016/j.devcel.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Franco CA, Jones ML, Bernabeu MO, Geudens I, Mathivet T, Rosa A, Lopes FM, Lima AP, Ragab A, Collins RT, Phng LK, Coveney PV, Gerhardt H. Dynamic endothelial cell rearrangements drive developmental vessel regression. PLoS Biol. 2015;13:e1002125. doi: 10.1371/journal.pbio.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udan RS, Vadakkan TJ, Dickinson ME. Dynamic responses of endothelial cells to changes in blood flow during vascular remodeling of the mouse yolk sac. Development. 2013;140:4041–4050. doi: 10.1242/dev.096255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Jiang L, Li C, Hu D, Bu JW, Cai D, Du JL. Haemodynamics-driven developmental pruning of brain vasculature in zebrafish. PLoS Biol. 2012;10:e1001374. doi: 10.1371/journal.pbio.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kochhan E, Lenard A, Ellertsdottir E, Herwig L, Affolter M, Belting HG, Siekmann AF. Blood flow changes coincide with cellular rearrangements during blood vessel pruning in zebrafish embryos. PLoS ONE. 2013;8:e75060. doi: 10.1371/journal.pone.0075060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenard A, Daetwyler S, Betz C, Ellertsdottir E, Belting HG, Huisken J, Affolter M. Endothelial cell self-fusion during vascular pruning. PLoS Biol. 2015;13:e1002126. doi: 10.1371/journal.pbio.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y, Poynter G, Huss D, Filla MB, Czirok A, Rongish BJ, Little CD, Fraser SE, Lansford R. Dynamic analysis of vascular morphogenesis using transgenic quail embryos. PLoS ONE. 2010;5:e12674. doi: 10.1371/journal.pone.0012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA. Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Investig. 2016;126:821–828. doi: 10.1172/JCI83083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadohama T, Nishimura K, Hoshino Y, Sasajima T, Sumpio BE. Effects of different types of fluid shear stress on endothelial cell proliferation and survival. J. Cell Physiol. 2007;212:244–251. doi: 10.1002/jcp.21024. [DOI] [PubMed] [Google Scholar]
- 15.Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Jr, Balasubramanian V, Garcia-Cardena G, Jain MK. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ. Res. 2005;96:e48–e57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 16.Berk BC, Min W, Yan C, Surapisitchat J, Liu Y, Hoefen R. Atheroprotective mechanisms activated by fluid shear stress in endothelial cells. Drug News Perspect. 2002;15:133–139. doi: 10.1358/dnp.2002.15.3.704684. [DOI] [PubMed] [Google Scholar]
- 17.Seebach J, Donnert G, Kronstein R, Werth S, Wojciak-Stothard B, Falzarano D, Mrowietz C, Hell SW, Schnittler HJ. Regulation of endothelial barrier function during flow-induced conversion to an arterial phenotype. Cardiovasc. Res. 2007;75:596–607. doi: 10.1016/j.cardiores.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Taha M, Aldirawi M, Marz S, Seebach J, Odenthal-Schnittler M, Bondareva O, Bojovic V, Schmandra T, Wirth B, Mietkowska M, Rottner K, Schnittler H. EPLIN-alpha and -beta Isoforms modulate endothelial cell dynamics through a spatiotemporally differentiated interaction with actin. Cell Rep. 2019;29:1010–1026. doi: 10.1016/j.celrep.2019.09.043. [DOI] [PubMed] [Google Scholar]
- 19.Chappell DC, Varner SE, Nerem RM, Medford RM, Alexander RW. Oscillatory shear stress stimulates adhesion molecule expression in cultured human endothelium. Circ. Res. 1998;82:532–539. doi: 10.1161/01.res.82.5.532. [DOI] [PubMed] [Google Scholar]
- 20.Souilhol C, Serbanovic-Canic J, Fragiadaki M, Chico TJ, Ridger V, Roddie H, Evans PC. Endothelial responses to shear stress in atherosclerosis: A novel role for developmental genes. Nat. Rev. Cardiol. 2020;17:52–63. doi: 10.1038/s41569-019-0239-5. [DOI] [PubMed] [Google Scholar]
- 21.Gifre-Renom L, Jones EAV. Vessel enlargement in development and pathophysiology. Front. Physiol. 2021;12:639645. doi: 10.3389/fphys.2021.639645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugden WW, Meissner R, Aegerter-Wilmsen T, Tsaryk R, Leonard EV, Bussmann J, Hamm MJ, Herzog W, Jin Y, Jakobsson L, Denz C, Siekmann AF. Endoglin controls blood vessel diameter through endothelial cell shape changes in response to haemodynamic cues. Nat. Cell Biol. 2017;19:653–665. doi: 10.1038/ncb3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjan V, Xiao Z, Diamond SL. Constitutive NOS expression in cultured endothelial cells is elevated by fluid shear stress. Am. J. Physiol. 1995;269:H550–555. doi: 10.1152/ajpheart.1995.269.2.H550. [DOI] [PubMed] [Google Scholar]
- 24.Rode B, Shi J, Endesh N, Drinkhill MJ, Webster PJ, Lotteau SJ, Bailey MA, Yuldasheva NY, Ludlow MJ, Cubbon RM, Li J, Futers TS, Morley L, Gaunt HJ, Marszalek K, Viswambharan H, Cuthbertson K, Baxter PD, Foster R, Sukumar P, Weightman A, Calaghan SC, Wheatcroft SB, Kearney MT, Beech DJ. Piezo1 channels sense whole body physical activity to reset cardiovascular homeostasis and enhance performance. Nat. Commun. 2017;8:350. doi: 10.1038/s41467-017-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang E, Nayak L, Jain MK. Kruppel-like factors in endothelial cell biology. Curr. Opin. Hematol. 2017;24:224–229. doi: 10.1097/MOH.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 26.Sangwung P, Zhou G, Nayak L, Chan ER, Kumar S, Kang DW, Zhang R, Liao X, Lu Y, Sugi K, Fujioka H, Shi H, Lapping SD, Ghosh CC, Higgins SJ, Parikh SM, Jo H, Jain MK. KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight. 2017;2:e91700. doi: 10.1172/jci.insight.91700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 28.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J. Biol. Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 29.Fledderus JO, Boon RA, Volger OL, Hurttila H, Yla-Herttuala S, Pannekoek H, Levonen AL, Horrevoets AJ. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2008;28:1339–1346. doi: 10.1161/ATVBAHA.108.165811. [DOI] [PubMed] [Google Scholar]
- 30.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, Garcia-Cardena G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J. Clin. Investig. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boon RA, Horrevoets AJ. Key transcriptional regulators of the vasoprotective effects of shear stress. Hamostaseologie. 2009;29(39–40):41. [PubMed] [Google Scholar]
- 32.Andueza A, Kumar S, Kim J, Kang DW, Mumme HL, Perez JI, Villa-Roel N, Jo H. Endothelial reprogramming by disturbed flow revealed by single-cell RNA and chromatin accessibility study. Cell Rep. 2020;33:108491. doi: 10.1016/j.celrep.2020.108491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondareva O, Tsaryk R, Bojovic V, Odenthal-Schnittler M, Siekmann AF, Schnittler HJ. Identification of atheroprone shear stress responsive regulatory elements in endothelial cells. Cardiovasc. Res. 2019;115:1487–1499. doi: 10.1093/cvr/cvz027. [DOI] [PubMed] [Google Scholar]
- 34.He M, Huang TS, Li S, Hong HC, Chen Z, Martin M, Zhou X, Huang HY, Su SH, Zhang J, Wang WT, Kang J, Huang HD, Zhang J, Chien S, Shyy JY. Atheroprotective flow upregulates ITPR3 (inositol 1,4,5-trisphosphate receptor 3) in vascular endothelium via KLF4 (Kruppel-Like Factor 4)-mediated histone modifications. Arterioscler. Thromb. Vasc. Biol. 2019;39:902–914. doi: 10.1161/ATVBAHA.118.312301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khachigian LM, Anderson KR, Halnon NJ, Gimbrone MA, Jr, Resnick N, Collins T. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler. Thromb. Vasc. Biol. 1997;17:2280–2286. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Lee PL, Tsai CS, Lee CI, Yang TL, Chuang HS, Lin WW, Lin TE, Lim SH, Wei SY, Chen YL, Chien S, Chiu JJ. Force-specific activation of Smad1/5 regulates vascular endothelial cell cycle progression in response to disturbed flow. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7770–7775. doi: 10.1073/pnas.1205476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmoud MM, Serbanovic-Canic J, Feng S, Souilhol C, Xing R, Hsiao S, Mammoto A, Chen J, Ariaans M, Francis SE, Van der Heiden K, Ridger V, Evans PC. Shear stress induces endothelial-to-mesenchymal transition via the transcription factor Snail. Sci. Rep. 2017;7:3375. doi: 10.1038/s41598-017-03532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajami NE, Gupta S, Maurya MR, Nguyen P, Li JY, Shyy JY, Chen Z, Chien S, Subramaniam S. Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc. Natl. Acad. Sci. U.S.A. 2017;114:10990–10995. doi: 10.1073/pnas.1707517114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis. 2016;19:9–24. doi: 10.1007/s10456-015-9485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maimari N, Pedrigi RM, Russo A, Broda K, Krams R. Integration of flow studies for robust selection of mechanoresponsive genes. Thromb. Haemost. 2016;115:474–483. doi: 10.1160/TH15-09-0704. [DOI] [PubMed] [Google Scholar]
- 42.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, Trapnell C, Ahituv N, Shendure J. A Genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:377–390. doi: 10.1016/j.cell.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell. 2017;66:285–299. doi: 10.1016/j.molcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Quillien A, Abdalla M, Yu J, Ou J, Zhu LJ, Lawson ND. Robust identification of developmentally active endothelial enhancers in zebrafish using FANS-assisted ATAC-Seq. Cell Rep. 2017;20:709–720. doi: 10.1016/j.celrep.2017.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panigrahi A, O'Malley BW. Mechanisms of enhancer action: The known and the unknown. Genome Biol. 2021;22:108. doi: 10.1186/s13059-021-02322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novodvorsky P, Chico TJ. The role of the transcription factor KLF2 in vascular development and disease. Prog. Mol. Biol. Transl. Sci. 2014;124:155–188. doi: 10.1016/B978-0-12-386930-2.00007-0. [DOI] [PubMed] [Google Scholar]
- 51.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. J. Biol. Chem. 2008;283:3942–3950. doi: 10.1074/jbc.M707882200. [DOI] [PubMed] [Google Scholar]
- 53.Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, Mericko P, Stadtfeld M, Zhou D, Cheng L, Graf T, MacRae CA, Lepore JJ, Lo CW, Kahn ML. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev. Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Vermot J, Forouhar AS, Liebling M, Wu D, Plummer D, Gharib M, Fraser SE. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7:e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goddard LM, Duchemin AL, Ramalingan H, Wu B, Chen M, Bamezai S, Yang J, Li L, Morley MP, Wang T, Scherrer-Crosbie M, Frank DB, Engleka KA, Jameson SC, Morrisey EE, Carroll TJ, Zhou B, Vermot J, Kahn ML. Hemodynamic forces sculpt developing heart valves through a KLF2-WNT9B paracrine signaling axis. Dev. Cell. 2017;43:274–289. doi: 10.1016/j.devcel.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stratman AN, Burns MC, Farrelly OM, Davis AE, Li W, Pham VN, Castranova D, Yano JJ, Goddard LM, Nguyen O, Galanternik MV, Bolan TJ, Kahn ML, Mukouyama YS, Weinstein BM. Chemokine mediated signalling within arteries promotes vascular smooth muscle cell recruitment. Commun. Biol. 2020;3:734. doi: 10.1038/s42003-020-01462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dekker RJ, van Thienen JV, Rohlena J, de Jager SC, Elderkamp YW, Seppen J, de Vries CJ, Biessen EA, van Berkel TJ, Pannekoek H, Horrevoets AJ. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am. J. Pathol. 2005;167:609–618. doi: 10.1016/S0002-9440(10)63002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ. Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 61.Abdelilah-Seyfried S, Tournier-Lasserve E, Derry WB. Blocking signalopathic events to treat cerebral cavernous malformations. Trends Mol. Med. 2020;26:874–887. doi: 10.1016/j.molmed.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Hu S, Liu Y, You T, Heath J, Xu L, Zheng X, Wang A, Wang Y, Li F, Yang F, Cao Y, Zhang H, van Gils JM, van Zonneveld AJ, Jo H, Wu Q, Zhang Y, Tang C, Zhu L. Vascular semaphorin 7A upregulation by disturbed flow promotes atherosclerosis through endothelial beta1 integrin. Arterioscler. Thromb. Vasc. Biol. 2018;38:335–343. doi: 10.1161/ATVBAHA.117.310491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gehrig J, Reischl M, Kalmar E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Muller F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat. Methods. 2009;6:911–916. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, Dean A. Enhancers navigate the three-dimensional genome to direct cell fate decisions. Curr. Opin. Struct. Biol. 2021;71:101–109. doi: 10.1016/j.sbi.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bussmann J, Wolfe SA, Siekmann AF. Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138:1717–1726. doi: 10.1242/dev.059881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hasan SS, Tsaryk R, Lange M, Wisniewski L, Moore JC, Lawson ND, Wojciechowska K, Schnittler H, Siekmann AF. Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol. 2017;19:928–940. doi: 10.1038/ncb3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hogan BM, Herpers R, Witte M, Helotera H, Alitalo K, Duckers HJ, Schulte-Merker S. Vegfc/Flt4 signalling is suppressed by Dll4 in developing zebrafish intersegmental arteries. Development. 2009;136:4001–4009. doi: 10.1242/dev.039990. [DOI] [PubMed] [Google Scholar]
- 69.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 70.Hayashi M, Pluchinotta M, Momiyama A, Tanaka Y, Nishikawa S, Kataoka H. Endothelialization and altered hematopoiesis by persistent Etv2 expression in mice. Exp. Hematol. 2012;40:738–750. doi: 10.1016/j.exphem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 71.Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- 72.Koyano-Nakagawa N, Garry DJ. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc. Res. 2017;113:1294–1306. doi: 10.1093/cvr/cvx133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore JC, Sheppard-Tindell S, Shestopalov IA, Yamazoe S, Chen JK, Lawson ND. Post-transcriptional mechanisms contribute to Etv2 repression during vascular development. Dev. Biol. 2013;384:128–140. doi: 10.1016/j.ydbio.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sumanas S, Choi K. ETS Transcription factor ETV2/ER71/Etsrp in hematopoietic and vascular development. Curr. Top. Dev. Biol. 2016;118:77–111. doi: 10.1016/bs.ctdb.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 76.Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, Schachterle W, Pulijaal VR, Mathew S, Chasen ST, Xiang J, Rosenwaks Z, Shido K, Elemento O, Rabbany SY, Rafii S. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFbeta suppression. Cell. 2012;151:559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palikuqi B, Nguyen DT, Li G, Schreiner R, Pellegata AF, Liu Y, Redmond D, Geng F, Lin Y, Gomez-Salinero JM, Yokoyama M, Zumbo P, Zhang T, Kunar B, Witherspoon M, Han T, Tedeschi AM, Scottoni F, Lipkin SM, Dow L, Elemento O, Xiang JZ, Shido K, Spence JR, Zhou QJ, Schwartz RE, De Coppi P, Rabbany SY, Rafii S. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature. 2020;585:426–432. doi: 10.1038/s41586-020-2712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neal A, Nornes S, Payne S, Wallace MD, Fritzsche M, Louphrasitthiphol P, Wilkinson RN, Chouliaras KM, Liu K, Plant K, Sholapurkar R, Ratnayaka I, Herzog W, Bond G, Chico T, Bou-Gharios G, De Val S. Venous identity requires BMP signalling through ALK3. Nat. Commun. 2019;10:453. doi: 10.1038/s41467-019-08315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bowley G, Kugler E, Wilkinson R, Lawrie A, van Eeden F, Chico TJA, Evans PC, Noel ES, Serbanovic-Canic J. Zebrafish as a tractable model of human cardiovascular disease. Br. J. Pharmacol. 2021;179:900. doi: 10.1111/bph.15473. [DOI] [PubMed] [Google Scholar]
- 80.Cao J, Ehling M, Marz S, Seebach J, Tarbashevich K, Sixta T, Pitulescu ME, Werner AC, Flach B, Montanez E, Raz E, Adams RH, Schnittler H. Polarized actin and VE-cadherin dynamics regulate junctional remodelling and cell migration during sprouting angiogenesis. Nat. Commun. 2017;8:2210. doi: 10.1038/s41467-017-02373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schnittler HJ, Franke RP, Akbay U, Mrowietz C, Drenckhahn D. Improved in vitro rheological system for studying the effect of fluid shear stress on cultured cells. Am. J. Physiol. 1993;265:C289–298. doi: 10.1152/ajpcell.1993.265.1.C289. [DOI] [PubMed] [Google Scholar]
- 82.Alestrom P, D'Angelo L, Midtlyng PJ, Schorderet DF, Schulte-Merker S, Sohm F, Warner S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020;54:213–224. doi: 10.1177/0023677219869037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) M. Westerfield; 1993. [Google Scholar]
- 84.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, Cho SW, Mumbach MR, Carter AC, Kasowski M, Orloff LA, Risca VI, Kundaje A, Khavari PA, Montine TJ, Greenleaf WJ, Chang HY. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C, Carroll JS. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this study are available under GEO accession number GSE198221.