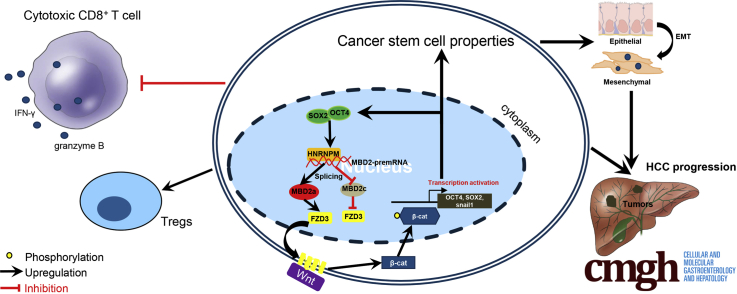
Abstract
Background & Aims
Cancer stemness and immune evasion are closely associated and play critical roles in tumor development and resistance to immunotherapy. However, little is known about the underlying molecular mechanisms that coordinate this association.
Methods
The expressions of heterogeneous nuclear ribonucleoprotein M (HNRNPM) in 240 hepatocellular carcinoma (HCC) samples, public databases, and liver development databases were analyzed. Chromatin immunoprecipitation assays were performed to explore the associations between stem-cell transcription factors and HNRNPM. HNRNPM-regulated alternative splicing (AS) and its binding motif were identified by RNA-seq and RIP-seq. HNRNPM-specific antisense oligonucleotides were developed to explore potential therapeutic targets in HCC. CD8+ T cells that were co-cultured with tumor cells were sorted by flow cytometry assays.
Results
We identified an elevated oncofetal splicing factor in HCC, HNRNPM, that unifies and regulates the positive association between cancer stemness and immune evasion. HNRNPM knockdown abolished HCC tumorigenesis and diminished cancer stem cell properties in vitro and in vivo. Mechanistically, HNRNPM regulated the AS of MBD2 by binding its flanking introns, whose isoforms played opposing roles. Although MBD2a and MBD2c competitively bound to CpG islands in the FZD3 promoter, MBD2a preferentially increased FZD3 expression and then activated the WNT/β-catenin pathway. Interestingly, FZD3 and β-catenin further provided additional regulation by targeting OCT4 and SOX2. We found that HNRNPM inhibition significantly promoted CD8+ T cell activation and that HNRNPM- antisense oligonucleotides effectively inhibited WNT/β-catenin to enhance anti-programmed cell death protein-1 immunotherapy by promoting CD8+ T cell infiltration.
Conclusions
HNRNPM has a tumor-intrinsic function in generating an immunosuppressive HCC environment through an AS-dependent mechanism and demonstrates proof of the concept of targeting HNRNPM in tailoring HCC immunotherapeutic approaches.
Keywords: Cancer Stem Cell, Hepatocellular Carcinoma, Immune Escape, Immunotherapy, RNA Splicing
Abbreviations used in this paper: AS, alternative splicing; ASO, antisense oligonucleotides; CCK-8, Cell Counting Kit-8; ChIP, chromatin immunoprecipitation assays; CLIP, UV crosslinking and immunoprecipitation; CSC, cancer stem cell; DMEM, Dulbecco’s modified Eagle’s medium; EN, endoderm; ES, embryonic stem cell; FBS, fetal bovine serum; HCC, hepatocellular carcinoma; HNRNPM, heterogeneous nuclear ribonucleoprotein M; IHC, immunohistochemistry; LV, lentivirus; MSP, methylation-specific PCR; OS, overall survival; PBS, phosphate-buffered saline; PD-1, programmed cell death protein 1; qRT-PCR, quantitative reverse transcription polymerase chain reaction; RIP, RNA immunoprecipitation; RNA-seq, high-throughput sequencing of RNA; shRNA, short hairpin RNA; TCGA, The Cancer Genome Atlas
Graphical abstract
Summary.
The study shows that heterogeneous nuclear ribonucleoprotein M has a tumor-intrinsic function in generating an immunosuppressive hepatocellular carcinoma environment by activating WNT/β-catenin pathway through alternative splicing-dependent mechanism and demonstrates the proof of the concept of targeting heterogeneous nuclear ribonucleoprotein M in tailoring hepatocellular carcinoma immunotherapeutic approaches.
Hepatocellular carcinoma (HCC) represents the second most common cause of cancer-related deaths worldwide and is increasing worldwide.1,2 HCC is well known for having an immunosuppressive tumor microenvironment with low tumor-infiltrating lymphocytes.3 Recent studies have demonstrated that tumor progression and metastasis may be linked to the emergence of cancer stem cells (CSCs).4, 5, 6 Recently, programmed cell death protein-1 (PD-1) blockade-based immunotherapy combined with targeted therapies has been approved as a first-line treatment for advanced HCC. Unfortunately, the objective response rate remains low in the 20% to 30% range, and the median response duration is relatively short, suggesting that HCC is resistant to PD-1 blockade.7
Cancer stemness and immune evasion have emerged as important features of HCC initiation, development, and metastasis. Mounting evidence indicates that CSCs are associated with the initiation, growth, metastasis, relapse, and drug resistance of HCC.8 To develop effective strategies for targeting CSCs in HCC, we need a better understanding of the molecular and epigenetic mechanisms that control CSC properties. A number of studies have focused on the inhibition of regulatory pathways that are critical for the stemness and tumorigenic potential of CSCs. Interestingly, cancer stemness has been found to be strongly associated with tumor cell-intrinsic immunosuppressive features.8 It is well-known that immunity plays a critical role in surveillance against emerging malignant cells from developing into tumors and the inhibition of tumor progression and metastasis.8,9 CSCs have to develop intrinsic mechanisms to escape immune surveillance during tumor development and growth. However, little is known about how cancer stemness and immune evasion are molecularly and epigenetically regulated.
The transcription factors OCT4 and SOX2 are master regulators of pluripotency in CSCs.10,11 Recently, functional genomics and molecular profiling approaches have been used to explore the broader role of core pluripotent factors in CSC biology and cancer development,4,10, 11, 12 Specifically, as one of the posttranscriptional gene regulatory mechanisms, alternative splicing (AS) enables a single gene to produce multiple mRNA variants and distinct protein isoforms, which may have different or even opposing roles in tumor biologic behaviors, such as proliferation, angiogenesis, drug resistance, and metastasis.13, 14, 15, 16 The muscleblind-like family of RNA binding proteins was found to repress pluripotency by mediating the expression of several somatic cell-specific protein isoforms, including FOXP1.17 However, the specific splicing factors and mechanistic links to cancer cell stemness and immune escape in HCC, which work in concert to reinforce a ground state of self-renewal and carcinogenesis, remain unresolved. The splicing factor heterogeneous nuclear ribonucleoprotein M (HNRNPM) has been reported to act as a crucial player in several cancer metastases and to regulate transcription.13,14,16,18 Although some splicing substrates have been identified,13,14,16,18 no stem cell-like property-specific and immune escape role in HCC has been established for HNRNPM.
In this study, we established mechanistic links between OCT4, SOX2, and HNRNPM and demonstrated that these factors work in concert to regulate the AS of MBD2. Specifically, MBD2a promotes while MBD2c suppresses FZD3, a Wnt/β-catenin signaling pathway receptor19 by competitively binding to its CpG islands, and then, β-catenin targets the promoters of OCT4 and SOX2 in HCC. In this regard, we developed a new therapeutic strategy, HNRNPM-specific antisense oligonucleotides (ASOs), to inhibit cancer stemness and potentiate antitumor immunity, providing important insights into the immune evasion of CSCs in HCC.
Results
HNRNPM Expression in HCC and Its Association With Prognosis
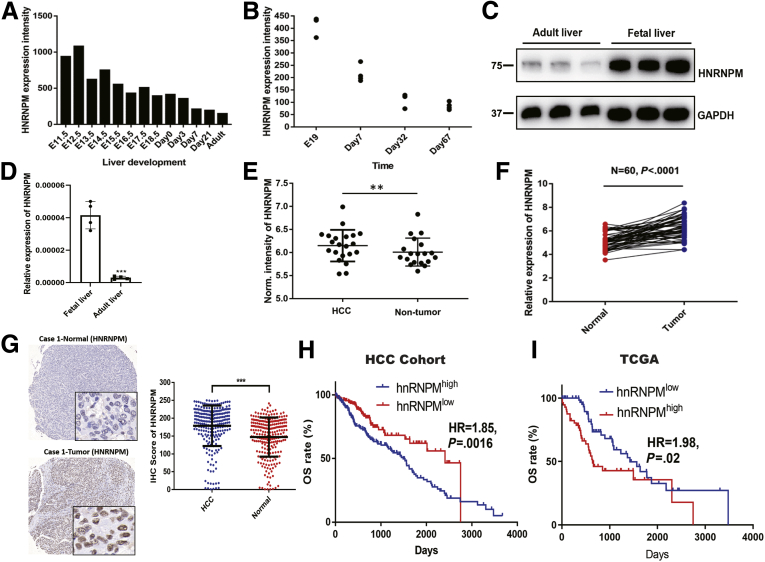
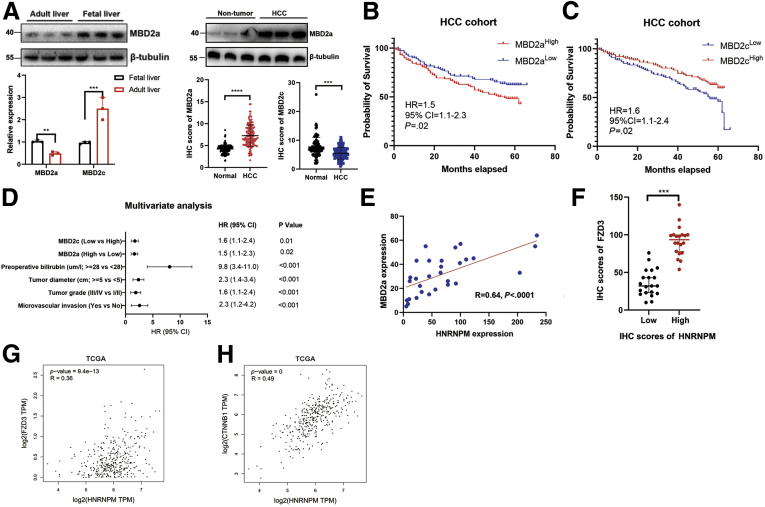
In previous microarray results of mouse liver development, HNRNPM was upregulated in mouse fetal livers compared with in adult livers20 (Figure 1, A). A publicly available database also revealed that HNRNPM was increased in mouse fetal livers from other cohorts21 (Figure 1, A, B). Measuring HNRNPM expression in human tissues revealed that HNRNPM was significantly higher in human fetal liver tissues than in adult liver tissues (Figure 1, C, D). These results demonstrated that HNRNPM is an embryonic gene.
Figure 1.
HNRNPM expression is increased in HCC and fetal liver tissues and is associated with prognosis.A, Normalized (Norm) HNRNPM expression levels during mouse liver development from GSE57824 data. B, HNRNPM expression levels during mouse liver development from GSE13149 data. C, Western blot analysis of HNRNPM protein levels in human fetal liver and adult liver tissues. D, Real-time qPCR analysis of HNRNPM mRNA levels in human fetal liver and adult liver tissues. Data are mean ± standard deviation of n = 3 independent samples. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 by the Student t test. E, Norm HNRNPM expression in HCC and normal liver tissues. ∗∗P < .01 by the Student t test. F, Real-time qPCR analysis of HNRNPM mRNA levels in 60 paired HCC and normal liver tissues. G, Representative images of HNRNPM by IHC in HCC and normal tissues. H, Kaplan-Meier analysis of HNRNPM in HCC cohort. I, Kaplan-Meier analysis of HNRNPM in TCGA cohort.
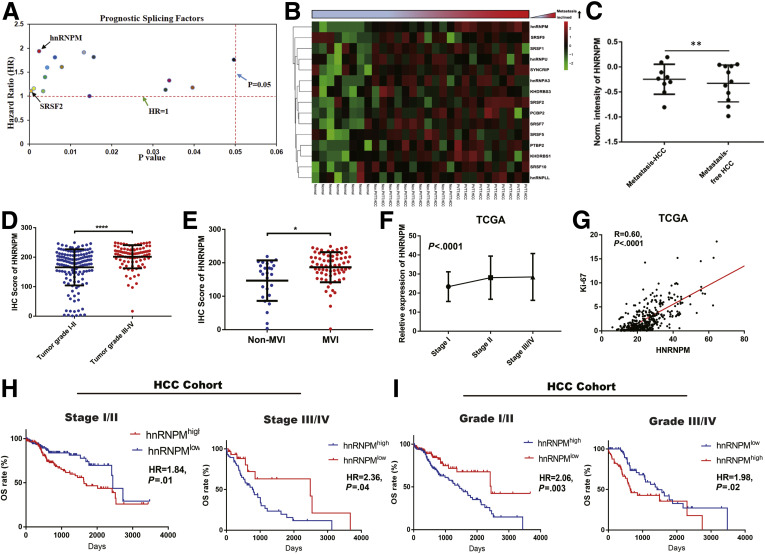
We then investigated HNRNPM expression in human HCC tissues. The relative expression of HNRNPM by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis in 60 paired HCC tissues showed significantly higher expression than that in noncancerous tissues. Similarly, tissue microarrays detecting HNRNPM protein levels in 240 HCC tissues and in 240 noncancerous hepatic tissues showed a stronger staining density of HNRNPM in HCC tissues (Figure 1, E-G). Kaplan-Meier analysis revealed that high HNRNPM protein levels in HCC tissues correlated with reduced overall survival (OS) (Figure 1, H-I). Oncomine Cox proportional hazards analysis revealed that HNRNPM ranked the highest in the SF database, contributing to detrimental effects on patient survival (Figure 2, A). In addition, the relative expression of HNRNPM in portal vein tumor thrombosis-HCC tissues was higher than that in non- portal vein tumor thrombosis HCC tissues and noncancerous hepatic tissues (Figure 2, B). Importantly, correlation regression analysis showed that a high HNRNPM protein level was significantly correlated with high tumor grade, the presence of microvascular invasion, poor tumor stage, high serum α-fetoprotein, multiple tumor numbers, high expression of the proliferation index (Ki-67), and large tumor size (Figure 2, C-G; Table 1, Table 2).
Figure 2.
HNRNPM was associated with clinopathological characteristics and poor prognosis in patients with HCC.A, Oncomine analysis showed the prognostic splicing factors from TCGA datasets. B, The selected prognostic splicing factors validated by real-time PCR in portal vein tumor thrombosis (PVTT) HCC, non-PVTT HCC, and normal liver tissues. C, The HNRNPM protein expression in metastasis and metastasis-free HCC tissues. D, The HNRNPM protein expression in tumor grade I/II and III/IV. E, The HNRNPM protein expression in no-microvascular invasion and microvascular invasion HCC tissues. F, The relative HNRNPM expression in tumor stage I/II/III/IV. G, The correlation analysis between Ki-67 and HNRNPM in TCGA database. H, Kaplan-Meier analyses of the correlations between HNRNPM level and overall survival in HCC tumor stage I/II and III/IV from our HCC cohort. I, Kaplan-Meier analyses of the correlations between HNRNPM level and OS in HCC tumor grade I/II and III/IV from our HCC cohort.
Table 1.
The Association of HNRNPM Expression With Clinical Characteristics in 240 Patients With HCC
| Characteristics | HNRNPM |
P-value | |
|---|---|---|---|
| Low | High | ||
| Number of patients | 120 | 120 | |
| Age, y | 54.6 ± 10.8 | 51.6 ± 11.4 | .030 |
| Sex | .406 | ||
| Female | 15 (12.5) | 11 (9.2) | |
| Male | 105 (87.5) | 109 (90.8) | |
| HBV | .432 | ||
| Negative | 15 (13.6) | 11 (10.2) | |
| Positive | 95 (86.4) | 97 (89.8) | |
| Anti-HCV | .566 | ||
| Negative | 114 (98.3) | 114 (99.1) | |
| Positive | 2 (1.7) | 1 (0.9) | |
| Postoperative TACE | 1.000 | ||
| No | 71 (59.2) | 71 (59.2) | |
| Yes | 49 (40.8) | 49 (40.8) | |
| Postoperative antivirus | .801 | ||
| No | 112 (93.3) | 111 (92.5) | |
| Yes | 8 (6.7) | 9 (7.5) | |
| Ascites | .582 | ||
| No | 112 (93.3) | 114 (95.0) | |
| Yes | 8 (6.7) | 6 (5.0) | |
| Macrovascular invasion | .811 | ||
| No | 111 (92.5) | 110 (91.7) | |
| Yes | 9 (7.5) | 10 (8.3) | |
| Microvascular invasion | .025 | ||
| No | 20 (36.4) | 6 (15.4) | |
| Yes | 35 (63.6) | 33 (84.6) | |
| Lymph node involved | 1.000 | ||
| No | 118 (98.3) | 118 (98.3) | |
| Yes | 2 (1.7) | 2 (1.7) | |
| Tumor number | < .001 | ||
| Single | 113 (94.2) | 76 (63.3) | |
| Multiple | 7 (5.8) | 44 (36.7) | |
| Tumor grade | < .001 | ||
| I/II | 92 (78.0) | 53 (44.9) | |
| III/IV | 26 (22.0) | 65 (55.1) | |
| Tumor diameter, cm | < .001 | ||
| <5 | 86 (71.7) | 56 (46.7) | |
| ≥5 | 34 (28.3) | 64 (53.3) | |
| Preoperative ALT, U/L | .206 | ||
| <45 | 69 (67.6) | 72 (75.8) | |
| ≥45 | 33 (32.4) | 23 (24.2) | |
| Preoperative AST, U/L | .591 | ||
| <45 | 75 (73.5) | 73 (76.8) | |
| ≥45 | 27 (26.5) | 22 (23.2) | |
| Preoperative bilirubin, um/L | .283 | ||
| <28 | 99 (99.0) | 91 (96.8) | |
| ≥28 | 1 (1.0) | 3 (3.2) | |
| Preoperative AFP, ng/mL | .007 | ||
| <400 | 84 (73.7) | 53 (55.8) | |
| ≥400 | 30 (26.3%) | 42 (44.2%) | |
Note: Data are presented as number (%) or mean ± standard deviation.
Note: Boldface P values indicate statistical significance.
AFP, α-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HNRNPM, heterogeneous nuclear ribonucleoprotein M; TACE, transhepatic arterial chemotherapy and embolization.
Table 2.
The Association of HNRNPM Expression With Clinical Characteristics in 371 Patients With HCC
| Parameters | HNRNPM Low expression | HNRNPM High expression | P-value |
|---|---|---|---|
| Number of patients | 185 | 186 | |
| BMI, kg/m2 | 26.4 ± 6.1 | 25.9 ± 10.4 | .066 |
| Albumin, g/dL | 4.3 ± 4.7 | 4.4 ± 5.2 | .929 |
| Creatinine, mg/dL | 1.1 ± 1.2 | 1.4 ± 1.8 | .008 |
| Platelet count, ×104 | 213.0 ± 114.0 | 232.1 ± 99.6 | .071 |
| Prothrombin time, s | 3.8 ± 4.4 | 3.7 ± 4.3 | .279 |
| Sex | .160 | ||
| Female | 54 (29.2) | 67 (36.0) | |
| Male | 131 (70.8) | 119 (64.0) | |
| AJCC stage | .009 | ||
| I/II | 138 (80.2) | 119 (68.0) | |
| III/IV | 34 (19.8) | 56 (32.0) | |
| Tumor grade | .017 | ||
| I | 32 (17.5) | 23 (12.6) | |
| II | 97 (53.0) | 80 (43.7) | |
| III/IV | 54 (29.5) | 80 (43.7) | |
| Embolization performed | .343 | ||
| No | 5 (21.7) | 6 (35.3) | |
| Yes | 18 (78.3) | 11 (64.7) | |
| Child-Pugh grade | .481 | ||
| A | 111 (93.3) | 77 (90.6) | |
| B | 8 (6.7) | 8 (9.4) | |
| Virus status | .782 | ||
| HBV | 39 (86.7) | 50 (84.7) | |
| HCV | 6 (13.3) | 9 (15.3) | |
| Ki-67 | < .001 | ||
| Low expression | 137 (74.1) | 48 (25.8) | |
| High expression | 48 (25.9) | 138 (74.2) | |
| Recurrence sites | .059 | ||
| Liver | 63 (82.9) | 53 (66.2) | |
| Lung | 6 (7.9) | 12 (15.0) | |
| Other | 7 (9.2) | 15 (18.8) |
Note: Data are presented as number (%) or mean ± standard deviation.
Note: Boldface P values indicate statistical significance.
AJCC, American Joint Committee on Cancer; BMI, body mass index; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HNRNPM, heterogeneous nuclear ribonucleoprotein M.
Furthermore, even within the cohort of patients with HCC with tumor stage I/II or III/IV or tumor grade I/II or III/IV, the association between high HNRNPM expression and poor prognosis remained obvious (Figure 2, H-I). Cox proportional hazards regression analysis further demonstrated that high HNRNPM expression in HCC tissues was an independent prognostic factor for reduced OS (Table 3). The prognostic value of HNRNPM was further verified by The Cancer Genome Atlas (TCGA) data analysis (Table 4). Analysis of HNRNPM expression in HCC cell lines (HCCLM3, MHCC97H, and Huh7) revealed a significantly higher HNRNPM expression level in HCC cells than in liver cancer cells with low metastatic characteristics, including MHCC97L and a hepatoblastoma cell line (HepG2) (Figure 3, A). Taken together, these data demonstrate that HNRNPM is an oncofetal protein whose expression is associated with serum α-fetoprotein, tumor differentiation, tumor size, and prognosis of patients with HCC.
Table 3.
Univariate and Multivariate Cox Regression Analysis of Overall Survival for HNRNPM (n = 240)
| Characteristics | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Sex | ||||
| Female | 1.0 | |||
| Male | 1.0 (0.5–2.0) | .999 | ||
| Age | 1.0 (1.0–1.0) | .659 | ||
| HBsAg | ||||
| Negative | 1.0 | |||
| Positive | 0.9 (0.4–1.7) | .645 | ||
| Anti-HCV | ||||
| No | 1.0 | |||
| Yes | 1.8 (0.4–7.5) | .398 | ||
| Postoperative TACE | ||||
| No | 1.0 | |||
| Yes | 2.6 (1.7–4.1) | < .001 | ||
| Preoperative antivirus | ||||
| No | 1.0 | |||
| Yes | 0.8 (0.3–1.9) | .566 | ||
| Ascites | ||||
| No | 1.0 | |||
| Yes | 1.2 (0.5–2.9) | .742 | ||
| Macrovascular invasion | ||||
| No | 1.0 | |||
| Yes | 3.3 (1.8–6.2) | < .001 | ||
| Microvascular invasion | ||||
| No | 1.0 | 1.0 | ||
| Yes | 2.7 (0.8–8.7) | .099 | 2.2 (1.2–4.1) | .009 |
| Lymph node involved | ||||
| No | 1.0 | |||
| Yes | 0.7 (0.1–4.9) | .698 | ||
| Tumor number | ||||
| Single | 1.0 | |||
| Multiple | 1.0 (0.5–1.8) | .976 | ||
| Tumor grade | ||||
| I/II | 1.0 | 1.0 | ||
| III/IV | 1.8 (1.2–2.8) | .009 | 1.5 (1.0–2.3) | .031 |
| Preoperative AFP, ng/mL | ||||
| <400 | 1.0 | 1.0 | ||
| ≥400 | 1.3 (0.8–2.2) | .270 | 1.7 (1.1–2.7) | .014 |
| Tumor diameter, cm | ||||
| <5 | 1.0 | 1.0 | ||
| ≥5 | 2.5 (1.6–3.9) | < .001 | 2.1 (1.4–3.2) | < .001 |
| Preoperative AST, U/L | ||||
| <45 | 1.0 | 1.0 | ||
| ≥45 | 2.5 (1.5–4.0) | < .001 | 2.1 (1.4–3.4) | < .001 |
| Preoperative ALT, U/L | ||||
| <45 | 1.0 | |||
| ≥45 | 1.2 (0.7–2.0) | .426 | ||
| Preoperative bilirubin, um/L | ||||
| <28 | 1.0 | 1.0 | ||
| ≥28 | 14.8 (5.0–43.4) | < .001 | 10.4 (3.6–29.9) | < .001 |
| HNRNPM | ||||
| Low expression | 1.0 | 1.0 | ||
| High expression | 2.1 (1.3–3.8) | < .001 | 2.4 (1.6–3.6) | < .001 |
Note: Boldface P values indicate statistical significance.
AFP, α-fetoprotein; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CI, confidence interval; HBsAg, surface antigen of the hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HNRNPM, heterogeneous nuclear ribonucleoprotein M; HR, hazard ratio; TACE, transhepatic arterial chemotherapy and embolization.
Table 4.
Univariate and Multivariate Cox Regression Analysis of Overall and Disease-free Survival for HNRNPM (n = 370) From TCGA Database
| Exposure |
Multivariate analysis |
|||
|---|---|---|---|---|
| Overall survival |
Disease-free survival |
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| HNRNPM | ||||
| Low expression | 1.0 | 1.0 | ||
| High expression | 1.8 (1.2–2.5) | .002 | 1.5 (1.1–1.9) | .005 |
| Sex | ||||
| Female | 1.0 | 1.0 | ||
| Male | 0.8 (0.6–1.2) | .252 | 0.9 (0.7–1.2) | .453 |
| Recurrence sites | ||||
| Liver | 1.0 | 1.0 | ||
| Lung | 2.9 (1.5–5.8) | .002 | 1.4 (0.8–2.3) | .194 |
| Other | 0.9 (0.4–1.7) | .674 | 0.5 (0.3–0.9) | .015 |
| AJCC stage | ||||
| I/II | 1.0 | 1.0 | ||
| III/IV | 2.5 (1.7–3.6) | < .001 | 2.1 (1.5–2.7) | < .001 |
| Tumor grade | ||||
| I | 1.0 | 1.0 | ||
| II | 1.2 (0.7–2.0) | .541 | 1.2 (0.8–1.8) | .293 |
| III/IV | 1.3 (0.7–2.2) | .400 | 1.3 (0.8–1.9) | .248 |
| Embolization performed | ||||
| No | 1.0 | 1.0 | ||
| Yes | 2.9 (0.8–9.8) | .094 | 2.7 (1.2–5.9) | .012 |
| Albumin, g/dL | 1.0 (0.9–1.0) | .547 | 1.0 (1.0–1.0) | .736 |
| Bilirubin, mg/dL | 1.0 (1.0–1.0) | .951 | 1.0 (1.0–1.0) | .176 |
| Child-Pugh grade | ||||
| A | 1.0 | 1.0 | ||
| B | 1.8 (0.7–4.5) | .238 | 1.3 (0.6–2.6) | .520 |
| Creatinine, mg/dL | 0.8 (0.5–1.2) | .263 | 1.0 (0.9–1.2) | .735 |
| Platelet count, ×104 | 1.0 (1.0–1.0) | .080 | 1.0 (1.0–1.0) | .383 |
| Prothrombin time, s | 1.0 (1.0–1.1) | .152 | 1.0 (1.0–1.1) | .548 |
| Vital status | ||||
| HBV | 1.0 | 1.0 | ||
| HCV | 0.7 (0.3–1.6) | .372 | 0.7 (0.3–1.4) | .310 |
| Ki-67 | ||||
| Low expression | 1.0 | 1.0 | ||
| High expression | 1.8 (1.2–2.5) | .001 | 1.7 (1.3–2.2) | < .001 |
Note: Boldface P values indicate statistical significance.
AJCC, American Joint Committee on Cancer; CI, confidence interval; HBV, hepatitis B virus; HCV, hepatitis C virus; HNRNPM, heterogeneous nuclear ribonucleoprotein M; HR, hazard ratio; TCGA, The Cancer Genome Atlas.
Figure 3.
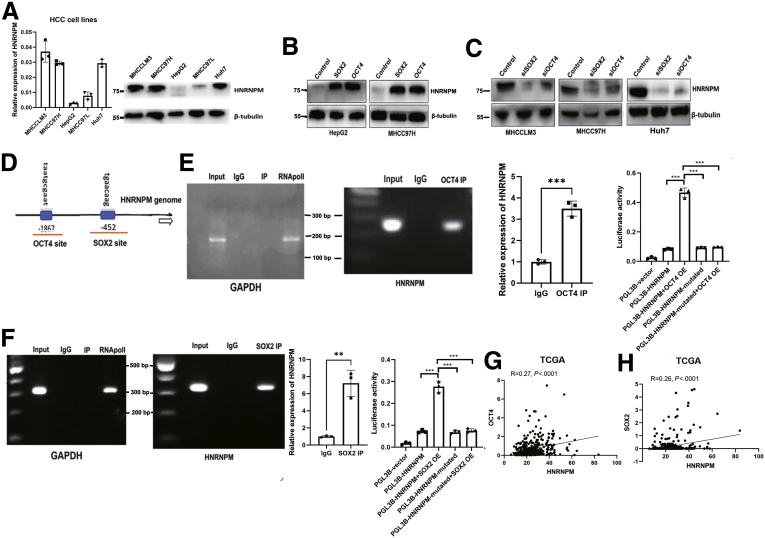
Cell stem cell transcriptional factors SOX2 and OCT4 bind with promoter and upregulate the expression of HNRNPM.A, The basic expression of HNRNPM in different HCC cell lines. B-C, Western blot analysis of HNRNPM expression when overexpressing (B) or depletion of (C) OCT4 and SOX2. D, The predicted binding site for OCT4 and SOX2 with HNRNPM promoter. E, OCT4 directly binds with HNRNPM promoter by ChIP assays and luciferase assays. F, SOX2 directly binds with HNRNPM promoter by ChIP assays and luciferase assays. G-H, Correlation analysis between OCT4 (G), SOX2 (H), and HNRNPM from TCGA database.
OCT4 and SOX2 Upregulate HNRNPM Expression
To elucidate the mechanisms regulating HNRNPM expression in HCC, we first focused on OCT4 and SOX2, which are well-known transcription factors maintaining stem cell-like properties and tumor initiation of HCC cells.22, 23, 24, 25 Ectopic expression of OCT4 or SOX2 upregulated HNRNPM expression (Figure 3, B). Reciprocally, knockdown of OCT4 or SOX2 decreased HNRNPM expression (Figure 3, C). Bioinformatic analysis of the binding sites for OCT4 and SOX2 in the HNRNPM promoter predicted one OCT4- and SOX2-binding site (Figure 3, D). Chromatin immunoprecipitation assays confirmed the binding of OCT4 and SOX2 to the HNRNPM promoter (Figure 3, E-F). In addition, luciferase assays confirmed the binding of OCT4 and SOX2 to the HNRNPM promoter (Figure 3, E-F). Correlation analysis also confirmed that HNRNPM was significantly associated with OCT4 and SOX2 expression from the TCGA database (Figure 3, G-H). These data demonstrated that OCT4 and SOX2 upregulate HNRNPM expression by directly binding to the HNRNPM promoter.
Oncogenic and Stem Cell-like Role of HNRNPM in HCC Cells
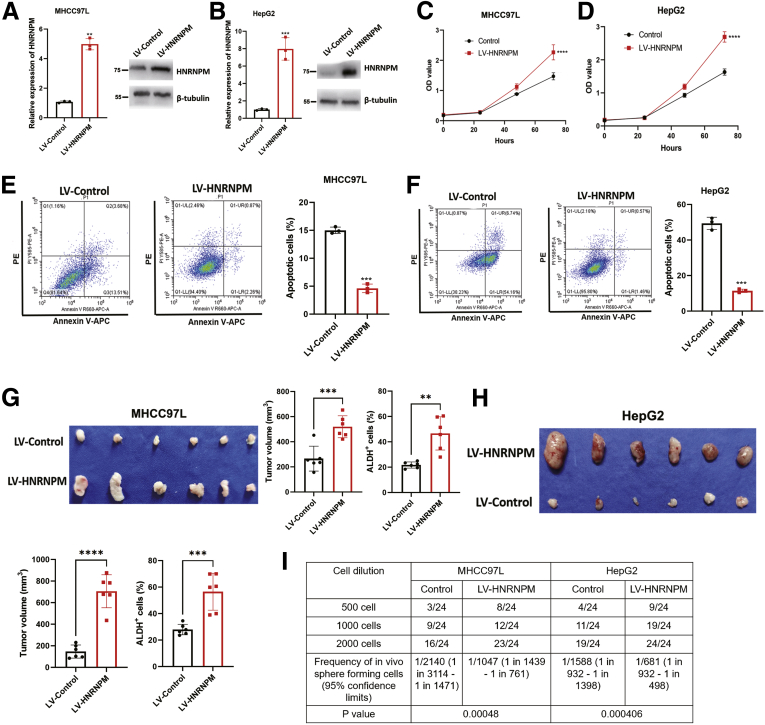
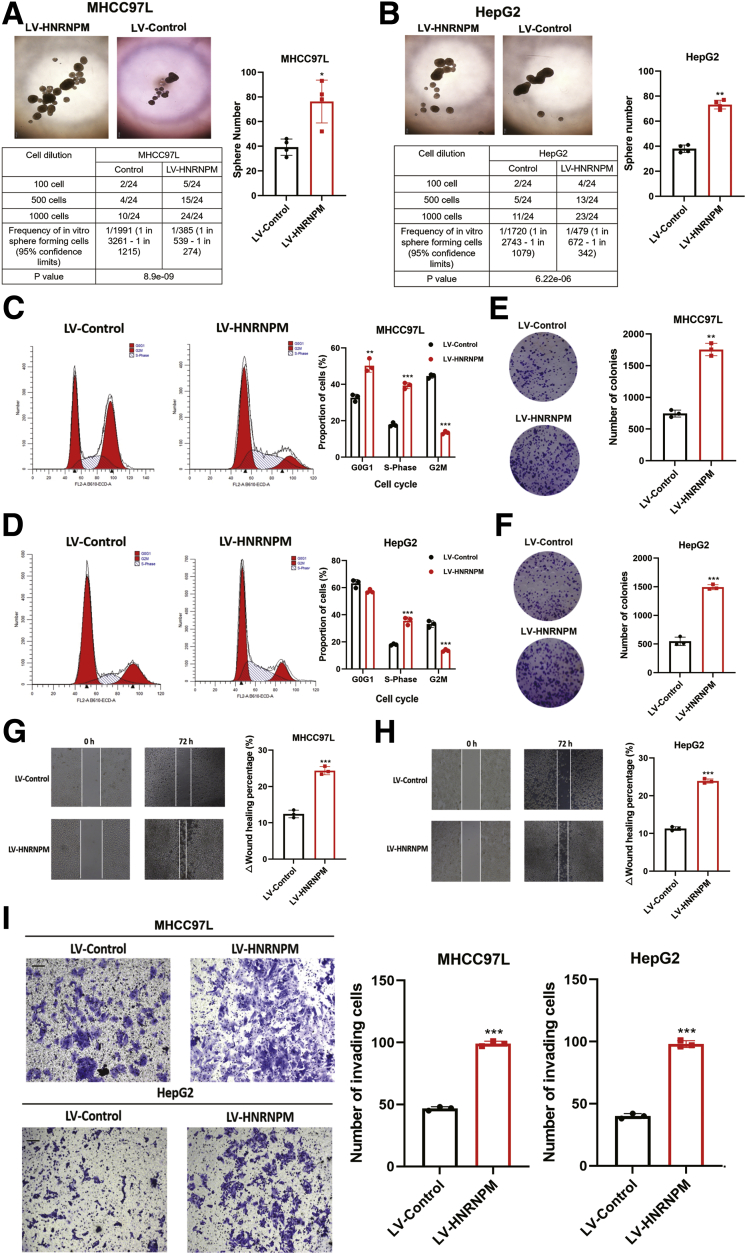
To examine the roles of HNRNPM in maintaining stem cell-like properties and hepatocarcinogenesis, we stably overexpressed HNRNPM in MHCC97L and HepG2 cells (Figure 4, A-B). First, we studied the changes in sphere formation of HNRNPM-overexpressing HCC cells. Our results showed that sphere formation was significantly higher in lentivirus (LV)-HNRNPM cells than in LV-Control cells (Figure 5, A-B). Additionally, limiting dilution assays showed that HNRNPM overexpression resulted in a significant increase in the frequency of CSCs compared with the control groups (Figure 5, A-B). Furthermore, ectopic expression of HNRNPM promoted cell growth, as determined by flow cytometry analysis, Cell Counting Kit-8 (CCK-8) assays, and colony formation assays (Figure 5, C-F; Figure 4, C-D). In addition, HNRNPM inhibited cell apoptosis, as determined by flow cytometry analysis (Figure 4, E-F), and significantly promoted cell migration and invasion (Figure 5, G-I). We next investigated the role of HNRNPM in HCC tumorigenesis in vivo. HepG2 and MHCC97L cells stably overexpressing HNRNPM or control cells were injected subcutaneously into athymic nude mice. HepG2 and MHCC97L cells overexpressing HNRNPM developed larger tumors than control cells, and in terms of the stem cell-like role, the proportion of ALDH+ subpopulation cells in the LV-HNRNPM group was significantly higher than that in the control group (Figure 4, G-H). Furthermore, limiting dilution spheroid formation assay in vivo showed the self-renewal ability was dramatically increased upon the third transplantation, we determined the CSC frequencies in the control and HNRNPM overexpressed HCC cells of third recipient mice (Figure 4, I).
Figure 4.
The tumorigenesis effects of HNRNPM overexpression in MHCC97L and HepG2 cells.A-B, The mRNA and protein levels of HNRNPM in MHCC97L (A) and HepG2 cells (B) stably overexpressing HNRNPM. C-D, The cell proliferation by CCK-8 assays stably MHCC97L (C) and HepG2 cells (D) stably overexpressing HNRNPM. ∗∗∗∗P < .0001 as compared with control. E-F, The cell apotosis by flow cytometry stably MHCC97L (E) and HepG2 cells (F) stably overexpressing HNRNPM. Data were from 3 independent experiments. ∗∗P < .01 by the Student t test. G-H, The in vivo effects in BALB/c nude mice in MHCC97L (G; n = 6) and HepG2 cells (H; n = 6) stably overexpressing HNRNPM. ∗∗P < .01 by the Student t test. I, The CSC frequency was determined from a limiting dilution assay performed with HCC cells depleting HNRNPM from the third transplant recipient mice (n = 6). The ELDA web tool was used to calculate the frequency of CSCs.
Figure 5.
HNRNPM drives HCC tumorigenesis and maintains CSC properties.A-B, Sphere formation and limiting dilution assays when overexpressed HNRNPM in MHCC97L and HepG2 cells. ∗P < .05; ∗∗P < .01 by the Student t test. The number of spheroids formed as a fraction of the number of cells seeded per well is given. Data are from 3 independent experiments. C-D, Cell cycle detected by flow cytometry when overexpressed HNRNPM in MHCC97L and HepG2 cells. ∗P < .05; ∗∗P < .01 by the Student t test. E-F, Colony formation assay when overexpressed HNRNPM in MHCC97L and HepG2 cells. ∗P < .05; ∗∗P < .01 by the Student t test. G-H, Cell migration assay when overexpressed HNRNPM in MHCC97L and HepG2 cells. ∗P < .05 by the Student t test. I, Cell invasion assays when overexpressed HNRNPM in MHCC97L and HepG2 cells. Results are presented as mean ± standard error of the mean, n = 3. ∗P < .05 by the Student t test.
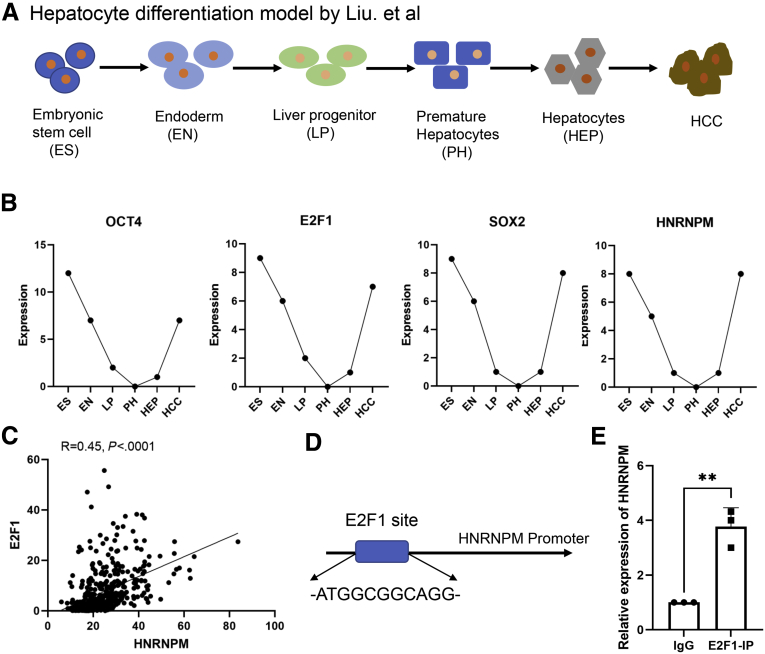
Furthermore, Liu et al established an in vitro hepatocyte differentiation model,26 which was defined by 6 stages, including embryonic stem cell (ES), endoderm (EN), liver progenitor cell, premature hepatocytes, hepatocytes, and HCC (Figure 6, A). Genes in the pluripotency and stem cell self-renewal signaling pathways, including OCT4, SOX2, and E2F1, were mainly restricted to the ES, EN, and HCC stages. To explore the oncogenic and stem cell-like role of HNRNPM, we also established this hepatocyte differentiation model and found that HNRNPM was first highly expressed in ES and was downregulated in the EN stage and liver progenitor cell, premature hepatocyte, and hepatocyte stages and finally upregulated significantly in the HCC stage, which was consistent with the results of OCT4 and E2F1 in this model (Figure 6, B). Additionally, Liu et al identified E2F1 as the most important upstream activator of ES+ tumors with oncofetal properties in a hepatocyte differentiation model. We then performed a correlation analysis between E2F1 and HNRNPM expression, which showed that HNRNPM was significantly correlated with E2F1 in HCC from TCGA databases (R = 0.45; P < .0001) (Figure 6, C). Furthermore, bioinformatic analysis of the binding site for E2F1 in the HNRNPM promoter predicted one E2F1-binding site (Figure 6, D). Chromatin immunoprecipitation assays confirmed the binding of E2F1 to the HNRNPM promoter (Figure 6, E). Collectively, these data suggested that HNRNPM drives HCC tumorigenesis and manipulates stem cell-like properties in HCC cells.
Figure 6.
The oncofetal properties of HNRNPM in hepatocyte differentiation model.A, The model scheme in hepatocyte differentiation model. B, The expression of OCT4, E2F1, SOX2, and HNRNPM in different stages from hepatocyte differentiation model. C, The correlation analysis between HNRNPM and E2F1 from TCGA databases. D, The potential binding site for E2F1 to HNRNPM promoter. E, E2F1 directly bind with HNRNPM promoter by ChIP assays. Data were from 3 independent experiments. ∗∗P < .01 by the Student t test.
HNRNPM is Required for Maintaining Stem Cell-like Properties and Tumorigenesis of HCC Cells
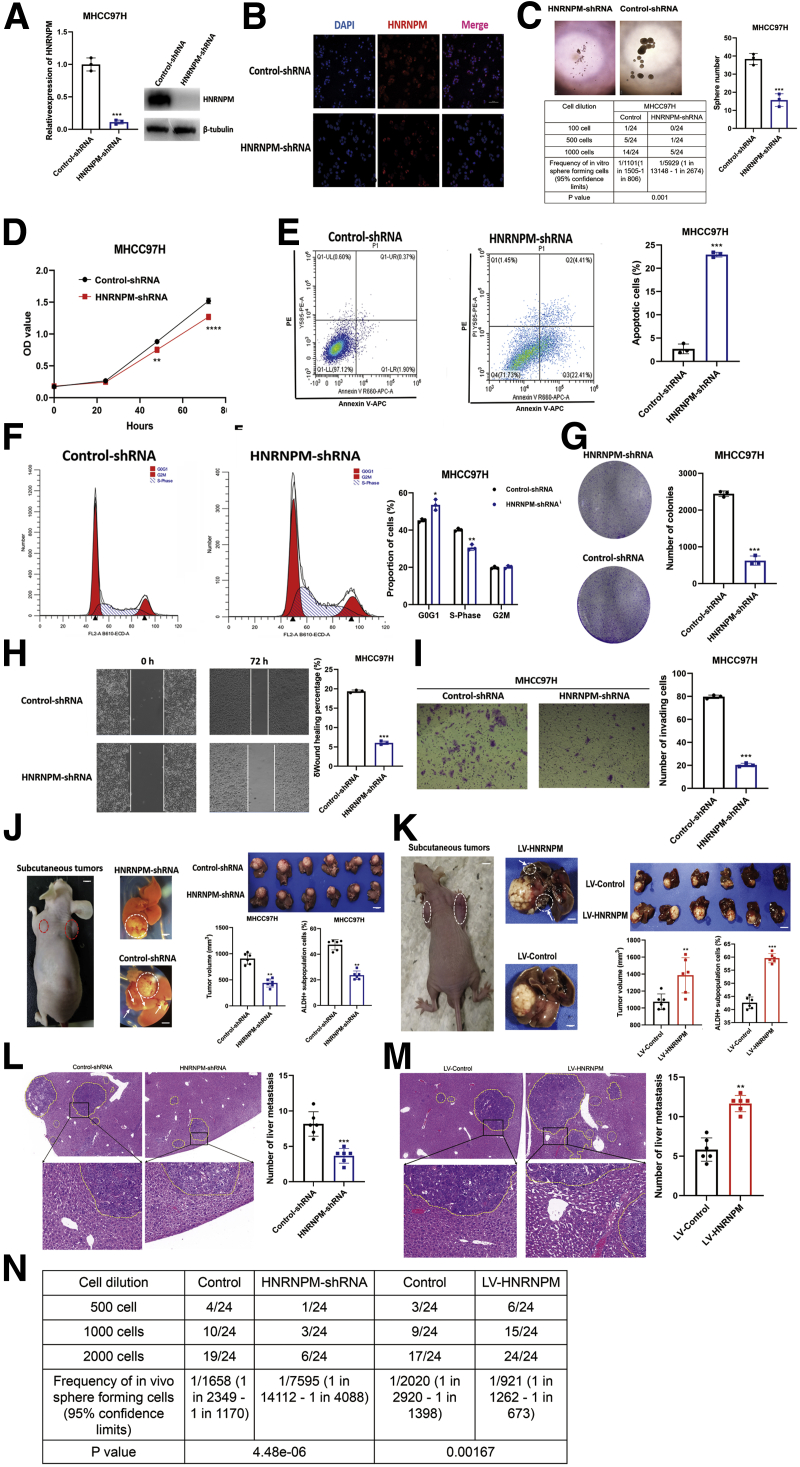
To further elucidate the role of HNRNPM in maintaining stem cell-like properties and hepatocarcinogenesis, we stably knocked down HNRNPM using effective independent short hairpin RNAs (shRNAs) in MHCC97H cells (Figure 7, A). Depletion of HNRNPM significantly inhibited MHCC97H sphere formation, cell growth and survival, and induced apoptosis (Figure 7, B-G). Furthermore, limiting dilution assays showed that HNRNPM depletion resulted in a significant decrease in the frequency of CSCs compared with the control groups (Figure 7, C). In addition, depletion of HNRNPM also inhibited cell migration and invasion (Figure 7, H-I). Next, MHCC97H cells stably depleting HNRNPM or control cells were injected subcutaneously into nude mice, and then, we established an orthotopic liver tumor model in nude mice. Strikingly, depletion of HNRNPM significantly inhibited tumorigenesis and ALDH+ subpopulation cells of MHCC97H in vivo (Figure 7, J). Inoculating MHCC97H cells into the liver of nude mice showed that depletion of HNRNPM significantly inhibited liver colonization and metastasis of MHCC97H cells (Figure 7, J, L). To further confirm the dramatic roles of overexpressing HNRNPM in liver colonization of HCC in vivo, we stably overexpressed HNRNPM in MHCC97H cells (Figure 7, K). The results showed that overexpression of HNRNPM significantly promoted liver colonization and metastasis of MHCC97H cells in vivo (Figure 7, K, M).
Figure 7.
HNRNPM is required for tumorigenesis of HCC cells.A, The mRNA and protein levels of HNRNPM in MHCC97H cells stably depleting HNRNPM. B, The protein levels of HNRNPM by immunofluorence stably depleting HNRNPM. C, Sphere formation and limiting dilution assays when depleting HNRNPM in MHCC97H cells. The number of spheroids formed as a fraction of the number of cells seeded per well is given. Data are from 3 independent experiments. ∗∗P < .01 by the Student t test. D, The cell proliferation by CCK-8 assays stably depleting HNRNPM in MHCC97H cells. Results are presented as mean ± standard error of the mean, n = 3. ∗P < .05; ∗∗P < .01 by the Student t test. E, The cell apotosis by flow cytometry stably depleting HNRNPM in MHCC97H cells. Results are presented as mean ± standard error of the mean, n = 3. ∗P < .05; ∗∗P < .01 by the Student t test. F, Cell cycle detected by flow cytometry when depleting HNRNPM in MHCC97H cells. G, Colony formation assay when depleting HNRNPM in MHCC97H cells. Results are presented as mean ± standard error of the mean, n = 3. ∗P < .05; ∗∗P < .01 by the Student t test. H, Cell migration assay when depleting HNRNPM in MHCC97H cells. Results are presented as mean ± standard error of the mean, n = 3. ∗∗∗P < .001 by the Student t test. I, Cell invasion assays when depleting HNRNPM in MHCC97H cells. J-K, The in vivo effects in BALB/c nude mice (n = 6 per group) when overexpressed and depleted HNRNPM. Results are presented as mean ± standard error of the mean, n = 6. ∗P < .05; ∗∗P < .01 by the Student t test. L-M, The number of liver metastasis in BALB/c nude mice when overexpressed and depleted HNRNPM. Results are presented as mean ± standard error of the mean, n = 6. ∗P < .05; ∗∗P < .01 by the Student t test. N, The CSC frequency was determined from a limiting dilution assay performed with HCC cells from the third transplant recipient mice. The ELDA web tool was used to calculate the frequency of CSCs.
As we observed that the self-renewal ability was dramatically reduced upon the third transplantation, we determined the CSC frequencies in the control and HNRNPM depletion or overexpression of HCC cells of third recipient mice by limiting dilution analysis. We found that the deletion or overexpression of HNRNPM resulted in a decrease (1/7595 vs 1/1658) or increase (1/921 vs 1/2020) in the frequency of CSCs compared with the control groups (Figure 7, N). These results identified HNRNPM as playing a vital role in maintaining CSCs and promoting tumorigenesis in vivo or in vitro, and HNRNPM acted as a potential therapeutic target for HCC.
Global Landscape of HNRNPM-affected AS and Gene Expression in HCC Cells
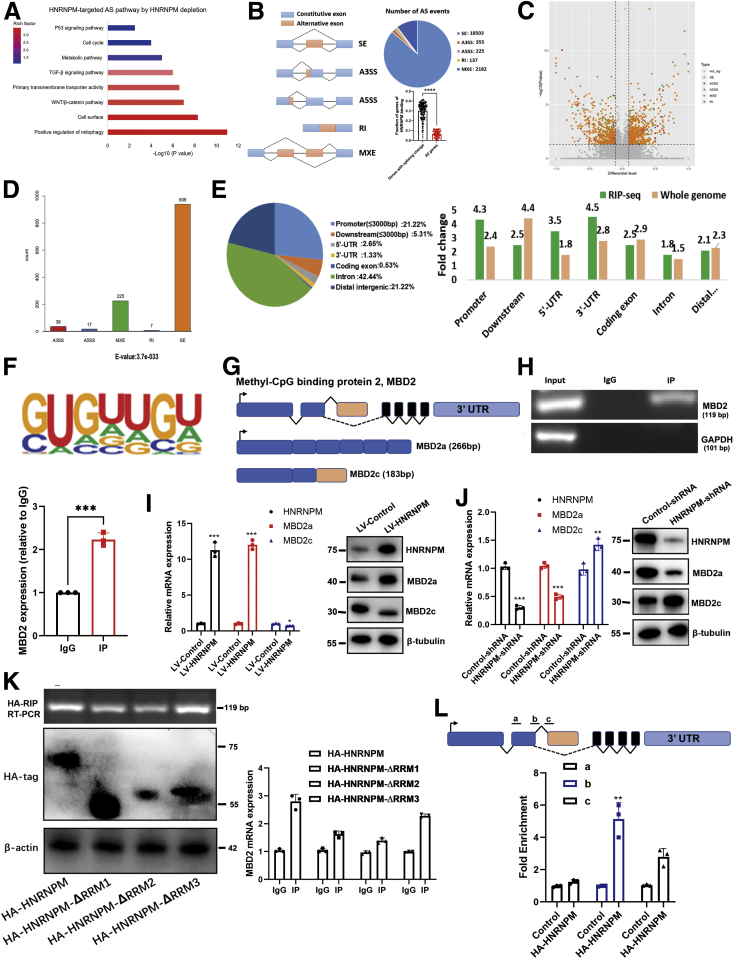
To screen HNRNPM-regulated AS events involved in hepatocarcinogenesis, we conducted high-throughput sequencing of RNA (RNA-seq) on the wild-type and knockdown cell lines of MHCC97H. Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that HNRNPM-targeted splicing control was significantly associated with the WNT/β-catenin pathway and positive regulation of mitophagy and the TGF-β pathway (Figure 8, A), supporting the role of HNRNPM in HCC growth and metastasis. With ∼100 million 150-nt paired-end reads, we identified a total of 21402 HNRNPM-regulated AS events in MHCC97H cells, which could be classified into 5 AS categories (Figure 8, B). HNRNPM binding is significantly associated with these splicing changes (Figure 8, B), supporting the direct effect of HNRNPM-RNA interaction on AS. The majority of these AS events belonged to skipped exons. Next, we identified 1224 AS events (Table 5) with significant change of percentage spliced-in values (percentage spliced-in ≥0.15; P < .05) (Figure 8, C-D), of which skipped exon was the majority among all 5 types of AS events (76.7%).
Figure 8.
The genome-wide landscape and global alternative splicing of HNRNPM.A, Kyoto Encyclopedia of Genes and Genomes analysis of HNRNPM-targeted splicing events. B, Quantification of the different AS events regulated by HNRNPM. A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site; MXE, mutually exclusive exon; RI, retained intron; SE, skipped exon. ∗∗∗P < .001 by the Student t test. C-D, The quantification of significant AS events regulated by HNRNPM (P < .05). E, HNRNPM-RIP-seq peaks were enriched in 5′UTR, promoter and 3′ UTR. All RIP-seq peaks were categorized according to the distribution on different genomic elements andcompared with the genomic background. F, De novo motif analysis identifying GU-repeat motif as the only enriched motif within the top HNRNPM RIP-seqpeaks. G, Schematic diagram of MBD2 molecular model. H, The RIP experiment showed HNRNPM directly binded with MBD2. I, The shift of MBD2a and MBD2c between HNRNPM overexpressed stably transduced and control MHCC97H cells. J, The shift of MBD2a and MBD2c between HNRNPM shRNA stably transduced and control MHCC97H cells. K, The RMMs of HNRNPM bind to MBD2 by RIP experiments. L, The potential binding of HNRNPM to MBD2 pre-mRNA by CLIP assay.
Table 5.
The Significant Alternative Splicing Events by Comparing Depletion of HNRNPM With Wild-type HCC Cells
| Gene name | AS type | P value | FDR | Group 1 value | Group 2 value | Group difference |
|---|---|---|---|---|---|---|
| APLP2 | SE | 0 | 0 | 0.691–0.613 | 0.239–0.243 | 0.411 |
| DNMT3B | SE | 1.54E−12 | 7.13E−09 | 1.0–1.0 | 0.116–0.0 | 0.942 |
| PBRM1 | A3SS | 8.74E−11 | 3.10E−08 | 0.0–0.0 | 1.0–0.817 | −0.908 |
| THNSL2 | SE | 2.15E−10 | 7.96E−07 | 1.0–1.0 | 0.0–0.111 | 0.945 |
| PRR7-AS1 | SE | 5.01E−10 | 1.54E−06 | 0.0–0.0 | 1.0–1.0 | −1 |
| RP5-1185I7.1 | SE | 1.14E−09 | 3.00E−06 | 1.0–1.0 | 0.333–0.13 | 0.768 |
| LINC00958 | SE | 1.55E−09 | 3.19E−06 | 1.0–1.0 | 0.0–0.0 | 1 |
| CCDC18 | SE | 1.47E−09 | 3.19E−06 | 0.0–0.0 | 1.0–1.0 | −1 |
| MBD2 | A3SS | 1.71E−05 | 0.00372 | 0.771–1.0 | 0.259–0.278 | 0.617 |
| WDSUB1 | MXE | 1.71E−09 | 3.72E−06 | 1.0–1.0 | 0.0–0.0 | 1 |
| CYP2C18 | SE | 2.59E−09 | 4.36E−06 | 0.0–0.0 | 1.0–1.0 | −1 |
| GMPR2 | SE | 2.40E−09 | 4.36E−06 | 1.0–0.6 | 0.0–0.0 | 0.8 |
| ZNF107 | SE | 4.09E−09 | 6.31E−06 | 0.0–0.111 | 1.0–0.714 | −0.801 |
| TEX41 | MXE | 1.11E−08 | 8.07E−06 | 0.0–0.0 | 1.0–1.0 | −1 |
| U2AF1L5 | MXE | 1.11E−08 | 8.07E−06 | 0.0–0.0 | 1.0–1.0 | −1 |
| YWHAZ | MXE | 2.62E−08 | 1.15E−05 | 0.47–0.516 | 0.662–0.653 | −0.164 |
| FAM171A1 | MXE | 2.60E−08 | 1.15E−05 | 0.0–0.0 | 0.778–0.583 | −0.68 |
| RP11-762I7.5 | SE | 8.79E−09 | 1.25E−05 | 1.0–1.0 | 0.0–0.0 | 1 |
| PI4KB | SE | 1.38E−08 | 1.77E−05 | 1.0–0.933 | 0.565–0.467 | 0.451 |
| PATZ1 | SE | 1.44E−08 | 1.77E−05 | 1.0–1.0 | 0.133–0.176 | 0.846 |
| SMG1P3 | SE | 2.47E−08 | 2.86E−05 | 1.0–1.0 | 0.162–0.279 | 0.779 |
| BID | SE | 2.66E−08 | 2.90E−05 | 0.443–0.511 | 0.88–0.82 | −0.373 |
| TNFSF13 | SE | 3.93E−08 | 4.04E−05 | 0.269–0.0 | 1.0–1.0 | −0.865 |
| BTN2A2 | MXE | 1.62E−07 | 5.90E−05 | 1.0–1.0 | 0.5–0.286 | 0.607 |
| GLDN | MXE | 2.28E−07 | 7.10E−05 | 1.0–1.0 | 0.504–0.245 | 0.625 |
| SMARCAD1 | A3SS | 4.78E−07 | 8.48E−05 | 0.771–1.0 | 0.259–0.278 | 0.617 |
| CTTN | SE | 1.16E−07 | 0.000107 | 0.921–0.88 | 0.752–0.78 | 0.135 |
| POGZ | SE | 1.12E−07 | 0.000107 | 0.963–1.0 | 0.802–0.675 | 0.243 |
| MARCH7 | SE | 1.24E−07 | 0.000109 | 1.0–1.0 | 0.0–0.476 | 0.762 |
A3SS, Alternative 3′ splicing site; AS, alternative splicing; HCC, hepatocellular carcinoma; HNRNPM, heterogeneous nuclear ribonucleoprotein M; MXE, mutually exclusive exon; SE, skipped exon.
Additionally, HNRNPM- RNA immunoprecipitation (RIP) sequencing analysis was performed to detect HNRNPM-associated RNA peaks. We categorized the distribution of the binding sites across different genomic elements and found that HNRNPM-associated RNA peaks were mostly enriched in introns, promoters, 5′ UTRs and 3′ UTRs (Figure 8, E; Table 6). De novo motif analysis showed that the most enriched binding motif in MHCC97H cells is a GU-enriched pattern (Figure 8, F), consistent with previous studies of HNRNPM-RNA interactions in other cell types.13,15,18
Table 6.
The Results of HNRNPM-RIP Analysis
| Gene name | Reference | FPKM experiment | FPKM control | Fold change |
|---|---|---|---|---|
| NME1-NME2 | 17 | 248.5939 | 0 | 8.751501665 |
| MIR3658 | 1 | 102.4301 | 0.577572 | 6.351095673 |
| TMEM189-UBE2V1 | 20 | 22.45659 | 0 | 5.316242172 |
| MBD2 | 18 | 10.60723 | 5.953727 | 2.132356326 |
| SNORD58B | 18 | 24.18816 | 0.423759 | 4.519596825 |
| AC090617.4 | 17 | 10.12713 | 0 | 4.2107537 |
| SNORA50B | 22 | 9.741061 | 0.013225 | 4.119218916 |
| RNA5SP298 | 10 | 8.234012 | 0 | 3.92999811 |
| OVCA2 | 17 | 17.40486 | 0.558641 | 3.858893033 |
| RPS10-NUDT3 | 6 | 6.868194 | 0.000195 | 3.686458733 |
| HOXA11-AS1_2 | 7 | 12.31942 | 0.392512 | 3.628603535 |
| SNORA50A | 16 | 9.722084 | 0.244401 | 3.569755575 |
| AL662899.3 | 6 | 13.52493 | 0.54494 | 3.527270187 |
| SLX1B | 16 | 5.94223 | 0 | 3.495457891 |
| LSM4 | 19 | 68.49814 | 4.875267 | 3.443554269 |
| PET100 | 19 | 19.23275 | 1.06697 | 3.427615848 |
| SNX3 | 6 | 137.8464 | 10.6208 | 3.395929081 |
| AC026464.4 | 16 | 5.462482 | 0 | 3.385201292 |
| BCL2L2-PABPN1 | 14 | 50.60795 | 3.760887 | 3.347114379 |
HNRNPM, Heterogeneous nuclear ribonucleoprotein M; RIP, RNA immunoprecipitation.
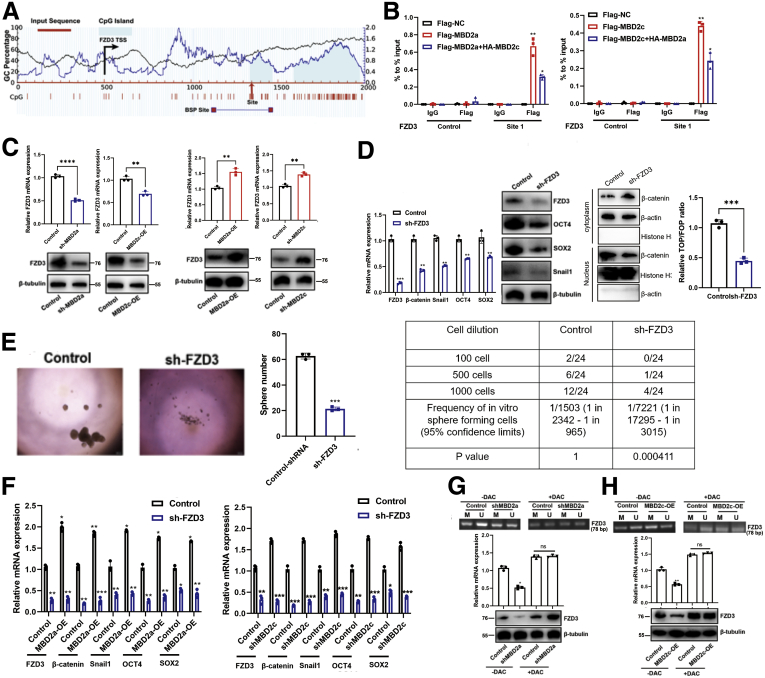
HNRNPM Promoted MBD2 Alternative Splicing in HCC Cells
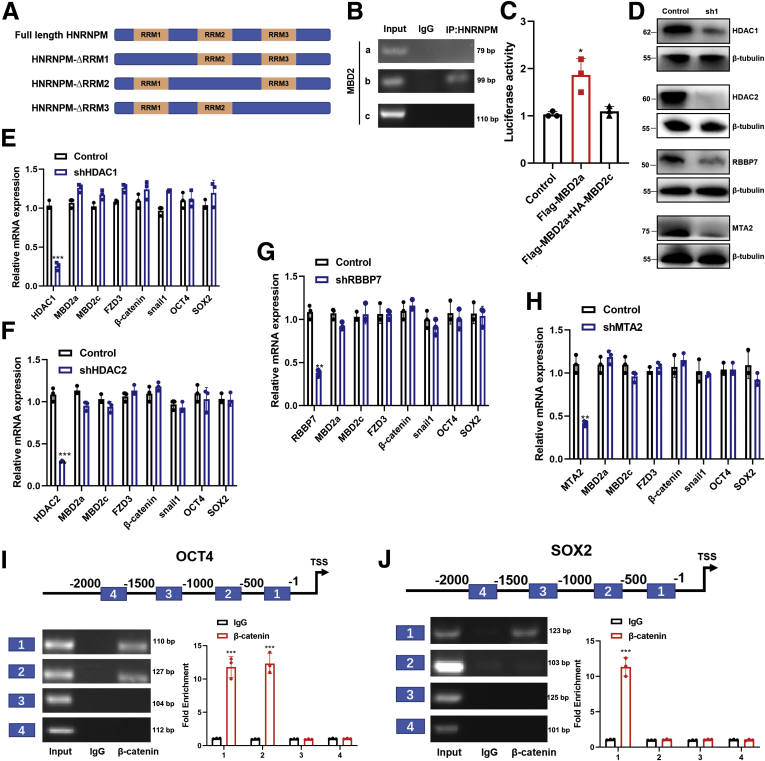
To gain insights into the molecular mechanism underlying the protumorigenic and stem cell-like role of HNRNPM, we intersected the results of HNRNPM-RIP analysis and transcriptomic sequencing. Among the intersection results (Table 7), we noted MBD2 (gene ID ENSG00000134046; Figure 8, G). The long MBD2 isoform has 6 exons and 411 amino acids, namely, MBD2a, whereas its splicing short isoform, MBD2c, has 3 exons and 302 amino acids. To further validate the RIP-seq results, we performed RIP-qPCR analysis and showed that HNRNPM interacted with MBD2 directly (Figure 8, H). Additionally, our qPCR and immunoblotting assays further demonstrated that overexpression of HNRNPM increased MBD2a and decreased MBD2c (Figure 8, I). The opposite results were observed by specific shRNAs targeting NRNPM in MHCC97H cells (Figure 8, J). Furthermore, when introducing different domain deletion mutants of HNRNPM (HNRNPM-ΔRRM1, -ΔRRM2, -ΔRRM3), RIP experiments showed that MBD2 mRNA bound to all domains of HNRNPM (Figure 8, K; Figure 9, A). To examine the specific binding sites of HNRNPM with MBD2, in vivo crosslinking followed by immunoprecipitation (CLIP) and the following RT–PCR results showed that HNRNPM-wt was bound to intron 2 with a high affinity (Figure 8, L; Figure 9, B), which is consistent with that in IPS cells.27 Taken together, our data revealed that HNRNPM promoted MBD2 splicing, inducing more MBD2a and lower MBD2c.
Table 7.
The Intersection Results of HNRNPM-RIP Analysis and Transcriptomic Sequencing
| Gene name | RIP fold change | AS type | FDR | Group difference |
|---|---|---|---|---|
| PPDPF | 2.737252418 | SE | 0.140194 | 0.28 |
| DNAAF4-CCPG1 | 2.310591698 | SE | 0.091385 | −0.161 |
| MBD2 | 2.132356326 | A3SS | 0.00372 | 0.617 |
| TRIM7-AS | 2.308789616 | SE | 0.007814 | −0.581 |
| TRIM7-AS | 2.308789616 | SE | 0.063631 | 0.248 |
| TRAPPC6A | 2.139013812 | SE | 0.006874 | −0.517 |
| APOC1P1 | 1.995491031 | SE | 0.028979 | −0.834 |
| PMVK | 1.698755769 | MXE | 0.077641 | −0.132 |
| UBE2C | 1.606328987 | MXE | 0.027552 | −0.131 |
| UBE2C | 1.606328987 | SE | 0.036757 | 0.619 |
A3SS, Alternative 3′ splicing site; AS, alternative splicing; HNRNPM, heterogeneous nuclear ribonucleoprotein M; MXE, mutually exclusive exon; RIP, RNA immunoprecipitation; SE, skipped exon.
Figure 9.
DNA methylation controls MBD2-mediated FZD3 transcription.A, The shematic diagram of HNRNPM domains. B, The specific binding site for MBD2 with HNRNPM by CLIP assay. C, The luciferase assay for FZD3 transcription activity when overexpressing MBD2a or MBD2a and MBD2c. Data were from three independent experiments. ∗P < .05. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. D-H, qPCR analysis of MBD2a, MBD2c, FZD3, β-catenin, and Snail1 mRNA transcripts in MHCC97H cells stably expressing NC, shRNAs targeting HDAC1, HDAC2, RBBP7, or MTA2. Immunoblot analysis showed the knockdown efficiency of shRNAs targeting HDAC1, HDAC2, RBBP7, or MTA2 in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. I-J, β-catenin promotes the expression of OCT4 (I) and SOX2 (J) by binding its promoter. Data were from 3 independent experiments. ∗P < .05. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test.
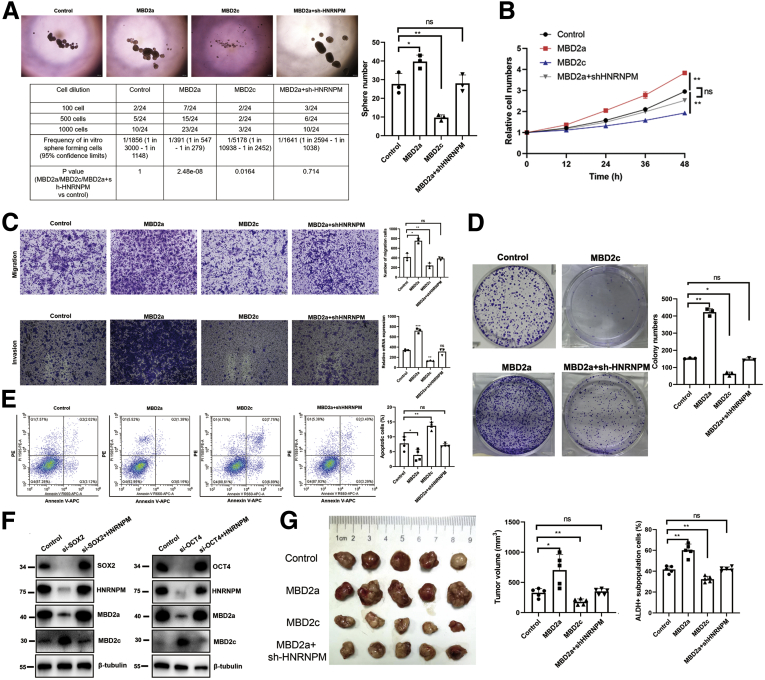
MBD2a Induces, Whereas MBD2c Represses, HCC Progression, and CSC Properties
We next investigated the functional roles of MBD2 isoforms in maintaining CSC properties and HCC tumorigenesis. First, we performed in vitro and in vivo experiments, and interestingly, compared with the control group, MHCC97H cells overexpressing MBD2a showed a significant increase in sphere formation, CSC frequency, cell growth and survival, and reduced apoptosis, cell migration, and invasion (Figure 10, A-E), whereas these protumorigenic effects were rescued by HNRNPM depletion. However, HCC cells with MBD2c overexpression inhibited sphere formation, CSC frequency, cell growth and survival, and induced apoptosis, cell migration and invasion (Figure 10, A-E). Furthermore, we established MHCC97H cells stably expressing OCT4- and SOX2-specific siRNAs and found downregulation of HNRNPM and MBD2a and upregulation of MBD2c. Furthermore, the overexpression of HNRNPM blocked the knockdown of OCT4- and SOX2-induced switching from MBD2c to MBD2a (Figure 10, F). Next, MHCC97H cells overexpressing MBD2a, MBD2c, or control were injected subcutaneously into nude mice, and strikingly, the MBD2a group significantly promoted tumorigenesis and ALDH+ subpopulation cells (Figure 10, G). These results were rescued by HNRNPM depletion, whereas MBD2c inhibited tumorigenesis and ALDH+ subpopulation cells in vivo compared with the control group (Figure 10, G).
Figure 10.
MBD2a induces, whereas MBD2c represses, HCC tumorigenesis and CSC properties.A, Sphere formation and limiting dilution assays when overexpressing MBD2a or with HNRNPM depletion, MBD2c in MHCC97H cells. B, The cell proliferation by CCK-8 assays when overexpressing MBD2a or with HNRNPM depletion, MBD2c in MHCC97H cells. C, Cell migration and migration assay when overexpressing MBD2a or with HNRNPM depletion, MBD2c in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05. D, Colony formation assay when overexpressing MBD2a or with HNRNPM depletion, MBD2c in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05. E, The cell apotosis by flow cytometry when overexpressing MBD2a or with HNRNPM depletion, MBD2c in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05. F, The protein expression of HNRNPM, MBD2a, MBD2c when downregulating SOX2, OCT4, and together with overexpressing HNRNPM by Western blot experiments. G, The in vivo effects in BALB/c nude mice when overexpressing MBD2a (n = 5) or with HNRNPM depletion (n = 5), MBD2c (n = 5). ns, Non-significant. ∗P < .05, ∗∗P < .01. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test.
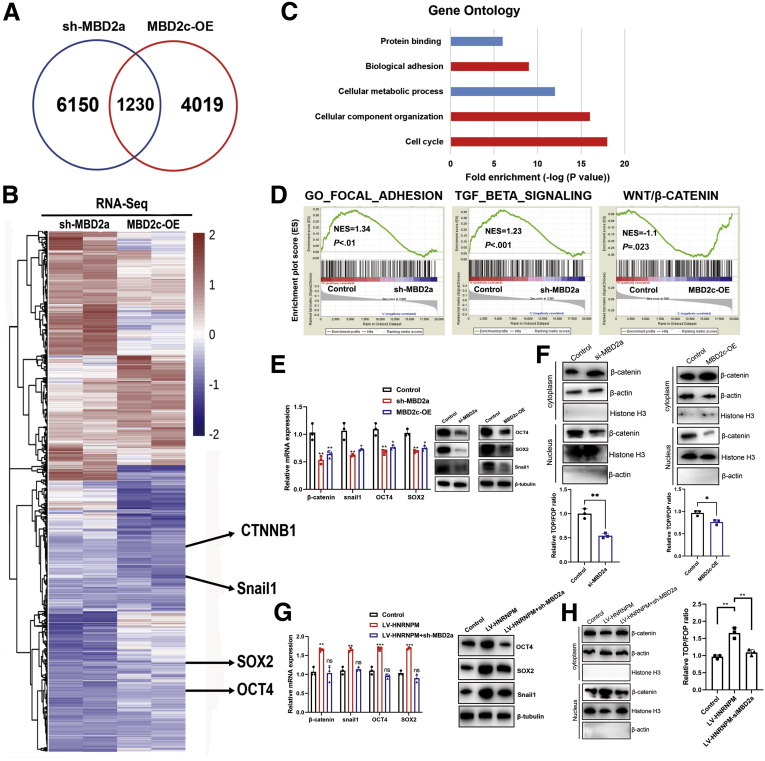
RNA-seq was performed in MHCC97H cells with MBD2a knockdown or MBD2c overexpression. RNA-seq data showed that 1230 genes were similarly regulated by MBD2a knockdown and MBD2c overexpression, which accounted for 20.1% of MBD2a-knockdown and 30.6% of MBD2c-overexpression regulated genes, respectively (Figure 11, A). A heat map showed all genes regulated by shMBD2a or MBD2c overexpression (up- and down-regulated genes were defined as having a log2-fold change greater than 0.5 or less than −0.5, respectively) (Figure 11, B). Gene ontology term enrichment analysis (DAVID: https://david.ncifcrf.gov/) showed that genes similarly affected by MBD2a knockdown and MBD2c overexpression play prominent roles in multiple biological processes involved in tumor proliferation and metastasis, including the cell cycle, cellular component organization, and biological adhesion (Figure 11, C). Gene enrichment analysis of RNA-seq data revealed that MBD2c overexpression or shMBD2a was positively associated with the GO_FOCAL_ADHESION and TGF_BETA_SIGNALING gene signatures but was negatively correlated with the Wnt/β-Catenin pathway (Figure 11, D). Therefore, these analyses confirmed the negative enrichment of metastasis- or stem cell-like-related gene signatures in MBD2c-overexpressing or MBD2a-knockdown cells compared with control cells.
Figure 11.
The coregulated genes by MBD2a and MBD2c in HCC cells.A-B, Venn diagram of the RNA-seq data showing the genes commonly regulated by MBD2a and MBD2c. C-D, Gene Ontology (GO) enrichment analysis. The top 5 GO terms in the indicated categories with the lowest P values are shown. E, The expression of Snail1, OCT4, SOX2 mRNA, and proteins was measured by qPCR and Western blot in MHCC97H cells expressing shRNAs targeting MBD2a, and in MHCC97H cells stably expressing MBD2c. Data were from 3 independent experiments. ∗P < .05 as compared with controls. F, The expression of β-catenin by nuclear/cytoplasmic protein fractionation and TOP/FOP-flash reporter assays when silencing MBD2a or overexpressing MBD2c. Data were from 3 independent experiments. ∗P < .05; ∗∗P < .01 by the Student t test. G, The expression of Snail1, OCT4, SOX2 mRNA, and proteins was measured by qPCR and Western blot in MHCC97H cells expressing HNRNPM and shRNA targeting MBD2a. Data were from 3 independent experiments. ∗∗∗P < .001 as compared with controls; ns, Not significant; P > .05. H, The expression of β-catenin by nuclear/cytoplasmic protein fractionation and TOP/FOP-flash reporter assays when overexpressing HNRNPM and silencing MBD2a. Data were from 3 independent experiments. ∗∗P < .01. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test.
The top 20 genes with altered expression according to RNA-seq data (Table 8) involved in stem cell-like properties and metastasis were studied in MHCC97H cells expressing shMBD2a or MBD2c (Figure 11, E). Notably, MBD2a enhanced while MBD2c suppressed the expression of β-catenin, snail1, OCT4, and SOX2 at both the mRNA and protein levels (Figure 11, B, E). Specifically, Western blot analysis of the TOP/FOP luciferase reporter assay and translocation of β-catenin from the cytoplasm to the nucleus showed that Wnt/β-catenin pathway activity could be significantly suppressed or activated by MBD2a/MBD2c overexpression, respectively (Figure 11, F). Further experiments showed that HNRNPM-induced snail1, OCT4, and SOX2 accumulation and Wnt/β-catenin pathway activity was abolished by MBD2a knockdown (Figure 11, G-H). Collectively, these data prove that MBD2a and MBD2c have competitively antagonistic roles in regulating β-catenin, snail1, OCT4, and SOX2 expression, which provides a molecular correlation to our findings that MBD2a and MBD2c play opposite roles in HCC tumorigenesis and maintaining CSC properties.
Table 8.
Genes With Altered Expression According to RNA-seq Data Involved in HCC Cell Tumorigenesis and CSC Properties
| Gene | NC vs shMBD2a |
NC vs MBD2c OE |
||||
|---|---|---|---|---|---|---|
| NC | sh2a | log2 (fold change) | NC | MBD2c | log2(fold change) | |
| FZD3 | 6.7542 | 2.13453 | −3.4521583 | 5.5828213 | 1.2648953 | −2.921362 |
| BCL6 | 10.5128 | 5.66543 | −1.19144 | 11.5697 | 7.97015 | −0.453377 |
| FN1 | 97.165 | 59.348 | −1.0632 | 101.797 | 78.634518 | −1.033386 |
| CXXC4 | 2.9779 | 0.056784 | −4.5743 | 1.24439 | 0.60213 | −1.111473 |
| NRP1 | 64.612 | 35.478 | −0.694769 | 72.787 | 96.0794 | −0.564434 |
| POU5F1 | 4.643 | 1.8953 | −1.10862 | 5.3391 | 2.1555 | −0.533557 |
| PTPRJ | 21.597 | 12.274 | −0.627586 | 39.665 | 25.8322 | −0.664897 |
| CTNNB1 | 210.5 | 86.614 | −0.626239 | 230.6745 | 155.633 | −0.565406 |
| SNX9 | 13.236 | 12.969 | −0.607511 | 29.64685 | 14.438397 | −0.678946 |
| FBP1 | 19.12 | 43.029 | 0.909213 | 19.32576 | 39.1543 | 1.02807 |
| JAK2 | 5.2499 | 0.9521 | −3.83621 | 4.62095 | 1.94382 | −0.574803 |
| SOX2 | 11.33 | 2.467552 | −2.38735 | 11.45719 | 2.6532866 | −1.21284 |
| HMGB1 | 371.36 | 57.828 | −2.33299 | 335.593 | 102.706 | −0.675133 |
| SATB1 | 7.97543 | 2.462 | −2.0718 | 6.7554 | 3.56073 | −0.555625 |
| LRP6 | 7.7443 | 3.63432 | −1.9576 | 8.9765 | 6.3201424 | −0.576317 |
| CYP1B1 | 39.53379 | 11.43783 | −1.84371 | 37.1218 | 14.8415 | −1.21455 |
| SNAI1 | 8.3776 | 2.03224 | −1.78365 | 7.7558 | 0.8656907 | −1.876549 |
| FZD1 | 19.5336 | 7.5432299 | −1.51276 | 18.0155 | 10.5791 | −0.685639 |
| RRAD | 2.15321 | 0.4356 | −1.42891 | 4.6547 | 2.03482 | −0.350023 |
| NET1 | 39.532 | 15.63453 | −1.13324 | 42.0497 | 23.6004 | −0.473372 |
CSC, Cancer stem cell; HCC, hepatocellular carcinoma.
MBD2a and MBD2c Competitive Binding to CpG Islands in the FZD3 Promoter
A previous study28 reported that MBD2a and MBD2c could competitively bind to the promoter of FZD1 in breast cancer. However, in HCC, among our top 10 genes with altered expression according to RNA-seq data, FZD3, rather than FZD1, an important upstream regulator of the Wnt/β-catenin pathway, is on our top list of interest. FZD3 has been revealed to not only activate the Wnt/β-catenin pathway but also upregulate core cell cycle protein components in melanomas with a hyperactive BRAF oncogene.29 In contrast to FZD1, the HNRNPM/MBD2a/FZD3 axis may explain our results in HCC, such as increased tumor growth, invasion, and CSC phenotype. As reported, hypermethylation of frizzled family proteins was associated with inactivation of Wnt/β-catenin signaling.19,29 Moreover, we selected the potential MBD2 binding site on CpG islands of FZD3 (Figure 12, A). Thus, we hypothesized that MBD2a and MBD2c could also competitively bind to the promoter of FZD3. Next, chromatin immunoprecipitation assays (ChIP)-PCR was performed in MHCC97H cells with Flag antibody against Flag-tagged MBD2a without or with overexpression of HA-MBD2c. As a result, ChIP-PCR data revealed that both MBD2a and MBD2c could bind to this site in the promoter of FZD3 (Figure 12, B). To explore whether MBD2a and MBD2c bind to the FZD3 promoter in vitro in a competitive way, we constructed GAL4-DBD-FZD3 plasmids, and luciferase reporter assays showed that the transcriptional activity of FZD3 in cells overexpressing both MBD2c and MBD2a was significantly decreased compared with cells overexpressing only MBD2a without MBD2c overexpression (Figure 12, B). These data suggested that both MBD2a and MBD2c bound to FZD3, and the binding exhibited a mutual inhibitory competition pattern between MBD2a and MBD2c, confirming that MBD2a and MBD2c colocalized on chromatin and competitively bound to the same sequences (Figure 12, B).
Figure 12.
MBD2a and MBD2c competitive binding to the CpG islands in the FZD3 promoter.A, Bioinformatic analysis of FZD3 gene. Location of CpG islands (light blue), TSS (black), qPCR sites (site 1 with red arrow), and BSP-amplified regions were indicated in this graph. Indicated positions were relative to the TSS (+1). BSP, Bisulfite sequencing PCR; TSS, transcription start site. B, ChIP experiments were performed using IgG or Flag antibody in indicated MHCC97H cells. The occupancy of the predicted DNA binding site for MBD2a or MBD2c within CpG islands of the FZD2 promoter assessed using qPCR. Data are presented as the mean (± standard deviation) of 3 independent experiments. ∗P < .05 as compared with the corresponding Flag-EV+HA-EV samples. C, qPCR and immunoblot analyses of FZD3 expression in MHCC97H cells stably expressing shRNAs targeting MBD2a, or overexpression of MBD2c. ∗P < .05; ∗∗P < .01 as compared with the corresponding control or EV control samples. D, qPCR and immunoblot analyses of β-catenin, FZD3, OCT4, SOX2, and Snail1 expression in MHCC97H cells stably expressing NC or FZD3 shRNAs. ∗∗P < .01; ∗∗∗P < .001 as compared with control samples. E, Sphere formation and limiting dilution assays for shRNA targeting FZD3 in MHCC97H cells. Data were from 3 independent experiments, ∗∗∗P < .001 by the Student t test. F, MHCC97H cells stably expressing EV, MBD2a (left panel), control NTC, MBD2c shRNAs (right panel) were further transfected with viruses expressing control NC, or FZD3 shRNAs, then the expression of FZD3, OCT4, SOX2, β-catenin, and Snail1 were measured by qPCR. ∗P < .05 as compared with the corresponding controls. G, MHCC97H cells stably expressing empty virus (EV), MBD2c (H), or control, MBD2a shRNAs (G) were further treated without or with DAC (20 μM) for 48 hours, followed by MSP detection of the methylation level of CpG island in FZD3 promoter and qPCR and immunoblot analyses (bottom panel) of the mRNA and protein expression of FZD3. PCR products are amplified with primers that recognize the methylated (M) or unmethylated sequences (U). ∗P < .05 as compared with the corresponding EV or control samples. ns, Not significant. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test.
Next, qPCR and immunoblotting assays showed that FZD3 expression was repressed by MBD2a depletion or MBD2c overexpression and enhanced by MBD2c depletion and MBD2a overexpression in MHCC97H cells (Figure 12, C-D). FZD3 inhibition markedly suppressed the activity of the WNT/β-catenin pathway and the expression of snail1, OCT4, and SOX2 at both the mRNA and protein levels in HCC cells, sphere formation, and CSC frequency (Figure 12, E). More interestingly, suppression of FZD3 dramatically abolished the elevated expression of β-catenin, snail1, OCT4, and SOX2 induced by MBD2a overexpression and MBD2c repressed in HCC cells (Figure 12, F), suggesting that FZD3 is involved in MBD2a- and MBD2c-regulated EMT and cancer cell self-renewal. MBD2a has long been considered a transcriptional repressor by recognizing and binding to hypermethylated DNA promoters together with other complexes.30 However, our results showed that knockdown of histone deacetylase complexes HDAC1/2, metastasis-associated gene 1 (MTA2) or retinoblastoma-binding protein 7 (RBBP7), the partners of MBD2a that repress gene transcription, exhibited no effect on the mRNA levels of MBD2a, MBD2c, FZD3, and β-catenin (Figure 9, D-H), indicating that MBD2a- and MBD2c-regulated FZD3 transcription is independent of this mechanism. Previous studies reported that MBD2a positively regulates gene transcription by acting as a DNA demethylase by removing repressive methyl residues.30,31 Thus, we performed methylation-specific PCR (MSP) assays in MHCC97H cells. The results of MSP (Figure 12, G, H) assays showed that methylation of the FZD3 promoter was enhanced by shMBD2a or MBD2c overexpression, which resulted in a significant reduction in the mRNA transcript and protein levels of FZD3 (Figure 12, G, H). In the presence of 5-aza-2'-deoxycytidine (decitabine; DAC), an inhibitor of DNA methyltransferase activity, the hypermethylated status of the FZD3 promoter was repressed, and FZD3 mRNA levels increased (Figure 12, G, H). These results suggest that MBD2a promotes FZD3 transcription by reducing the methylation levels of the CpG islands of FZD3, whereas MBD2c functions oppositely. Taken together, MBD2a and MBD2c inversely regulate the expression of β-catenin by competitively binding to FZD3.
SOX2, OCT4, HNRNPM, MBD2a, and FZD3 Comprise a Positive Feedback Loop
Consistent with HNRNPM expression, the expression of MBD2a was higher in fetal liver tissues, decreased in adult liver tissues, and re-increased in HCC tissues (Figure 13, A), which indicated that MBD2a may also be involved in the stem cell-like phenotype-driven axis during HCC tumorigenesis. However, MBD2c showed the opposite results, expressing higher in normal vs HCC tissues. Furthermore, Kaplan-Meier analysis showed that high expression of MBD2a and low expression of MBD2c indicated poorer prognosis in patients with HCC (Figure 13, B-C), and multivariate analysis revealed that high expression of MBD2a and low expression of MBD2c were independent prognostic factors in patients with HCC (Figure 13, D). As previously reported, the Wnt/β-catenin pathway can regulate the expression of stem cell markers, such as CD44 and CD133.32 Targeting β-catenin can inhibit the tumor proliferation, invasion, and chemoresistance of CSCs in vivo and in vitro.32 Hence, we hypothesized that OCT4, SOX2, HNRNPM, MBD2a, and FZD3 may comprise a positive feedback loop during HCC tumorigenesis. To further confirm this hypothesis, our ChIP experiments showed that β-catenin could bind to the promoters of OCT4 and SOX2 in MHCC97H cells at -1 to 1000 bp and -1 to 500 bp, respectively (Figure 9, I-J). In addition, the expression of HNRNPM was highly correlated with MBD2a, FZD3, and β-catenin (Figure 13, E-H). These interesting results may provide several potential therapeutic targets in HCC treatment.
Figure 13.
A, The relative expression of MBD2a, MBD2c in fetal liver, adult liver, HCC, and adjacent noncancerous liver tissues. ∗∗∗P < .001 by the Student t test. B-C, The Kaplan-Meier analyses of the correlations between MBD2a (B), MBD2c (C) level and overall survival of n = 100 patients with HCC. The median MBD2a or MBD2c level was used as the cutoff. D, The multivariate analysis for MBD2a and MBD2c in patients with HCC. E, The correlation analysis between HNRNPM and MBD2a in patients with HCC (n = 30) by IHC experiments. F, FZD3 and HNRNPM expression in protein levels in HCC tissues with strong or weak HNRNPM staining intensity. The median HNRNPM staining intensity was used as the cutoff (n = 60 HCC tissues). ∗∗∗P < .001. G-H, The correlation between the expression of HNRNPM and FZD3 (G), β-catenin (H) from TCGA datasets.
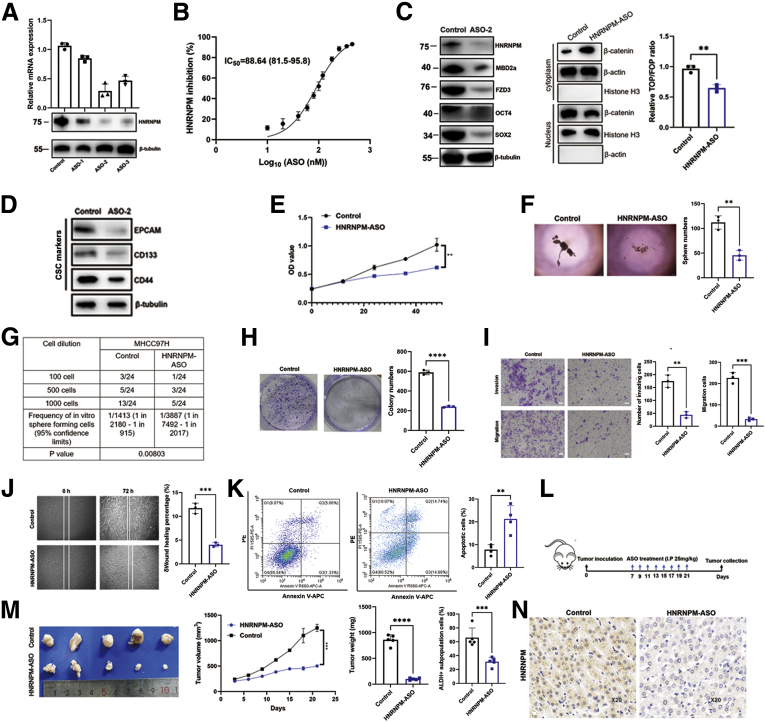
LNA-modified ASOs Targeting HNRNPM Inhibit HCC Progression
The tumor-promoting role of HNRNPM in HCC in the current study, as well as results from other cancer models,13,18 established it as a potentially novel target for cancer therapy. To evaluate the therapeutic potential of targeting HNRNPM, we applied a set of 3 LNA-modified ASOs with potent HNRNPM-targeting activity in vitro and in vivo. We therefore validated it for feasibility and tolerability in vitro and in vivo for HCC. qRT-PCR and Western blot assays showed that ASO-2 was the most effective in targeting HNRNPM mRNA and protein expression (Figure 14, A), and it significantly decreased both HNRNPM mRNA and protein expression in a dose-dependent manner with IC50 values of approximately 88.64 nM (Figure 14, B). Furthermore, HNRNPM ASOs also inhibited the activity of the WNT/β-catenin pathway and the expression of MBD2a, FZD3, OCT4, SOX2, and CSC markers, including CD44, EpCAM, and CD133, indicating that this ASO was a potent inhibitor of HNRNPM and CSCs in HCC (Figure 14, C-D). To determine whether HNRNPM ASOs exert antitumor effects similar to those of HNRNPM knockdown, we initially employed in vitro experimental assays. Compared with the control group, MHCC97H treated with HNRNPM ASO showed a significant decrease in sphere formation, CSC frequency, cell growth, and survival (Figure 14, E-H). In addition, treatment with HNRNPM ASO also inhibited cell migration and invasion and induced apoptosis (Figure 14, I-K). Next, we started the therapeutic evaluation in an HCC model (Figure 14, L). BALB/c nude mice bearing MHCC97H primary tumors ∼2 mm in diameter were randomized into 2 groups and treated with the negative control oligonucleotide or HNRNPM-specific ASO. HNRNPM-specific ASO treatment significantly inhibited tumorigenesis and ALDH+ subpopulation cells in vivo (Figure 14, M-N). Collectively, these results indicated that the HNRNPM ASO may serve as an effective potential therapeutic target in HCC.
Figure 14.
The effects of HNRNPM-specific ASO for HCC in vivo and in vitro.A, The expression of HNRNPM was significantly correlated with MBD2a. B, The IC50 of ASO-2 for MHCC97H cells. C, The protein expression of HNRNPM, MBD2a, FZD3, OCT4, SOX2, and β-catenin related assays when treated with ASO-2 in HCC cells. Data were from 3 independent experiments. ∗P < .05 by the Student t test. D, The CSC markers expression by ASO treatment. E, The CCK-8 experiment when treated with HNRNPM-specific ASO in MHCC97H cells. F, Sphere formation and limiting dilution assays when treated with HNRNPM-specific ASO in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05 by the Student t test. G, Limiting dilution assays when treated with HNRNPM-specific ASO in MHCC97H cells. H, Colony formation assay when treated with HNRNPM-specific ASO in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05. I-J, Invasion assay (I) and cell migration (J) and when treated with HNRNPM-specific ASO in MHCC97H cells. Data were from 3 independent experiments. ∗P < .05 by the Student t test. K, The HCC cell apoptosis changes by ASO treatment. Data were from 3 experiments. ∗∗P < .01 by the Student t test. L, The schematic diagram of ASO-2 treating nude mice when inoculating the tumor cells. M, The effects of HNRNPM-specific ASO when treated ASO I.P by 25 mg/kg (n = 5). ∗∗∗P < .001 by the Student t test. N,. The HNRNPM expression in tumors when treating HNRNPM-ASO by IHC experiments.
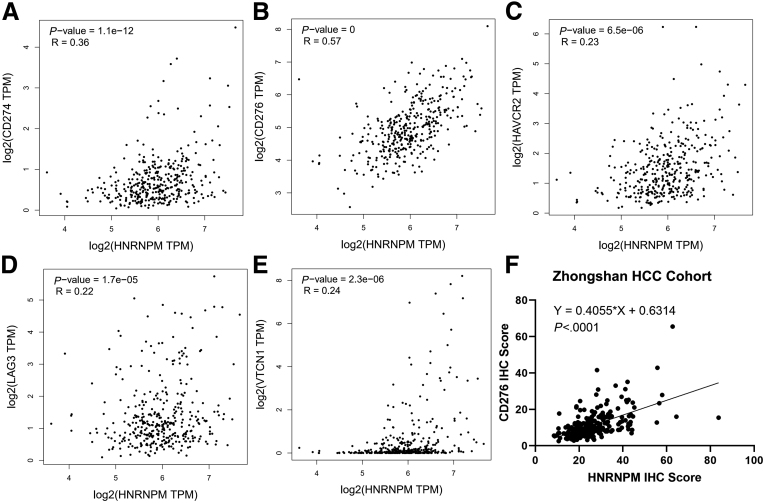
Targeting HNRNPM Enhances PD-1 Blockade Immunotherapy in WNT-activated HCC
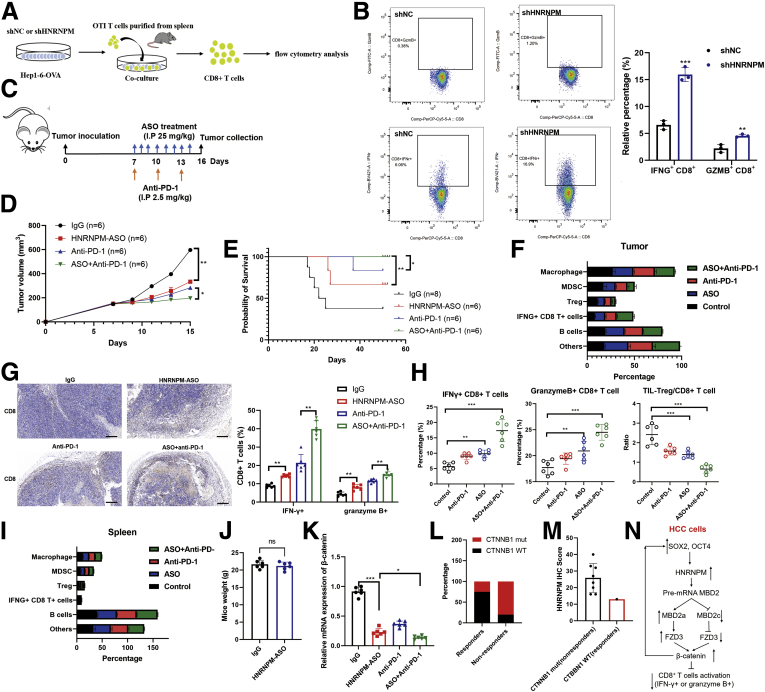
Recently, several studies in patients with HCC have shown that β-catenin activation correlates with T cell exclusion and resistance to anti-PD-1 therapy, which has also been validated in other human cancers.9,33, 34, 35 Hence, theoretically, inhibiting the activity of β-catenin could restore immune surveillance and enable resistant tumor immunotherapy efficacy. Therefore, to determine whether HNRNPM plays a role in T cell tumor immunity in HCC, we first analyzed its correlation with the RNA expression of several immune checkpoint molecules in TCGA HCC datasets. HNRNPM expression was positively associated with the expression of these genes, especially B7-H3 (CD276) (Figure 15, A-E), which was also validated in HCC tissues by quantitative immunohistochemistry (IHC) results (Figure 15, F). These results may suggest that HNRNPM may act as an immune-inhibitory molecule for T cell functions. Second, to test whether the attenuation of HNRNPM expression affects T cell-mediated antitumor function, we used sh-HNRNPM in ovalbumin (OVA)-expressing Hep1-6 HCC (Hep1-6-OVA) cells and cocultured them with OTI CD8+ T cells isolated from the spleens of OTI mice (Figure 16, A). A significantly increased proportion of IFN-γ+ or granzyme B+ CD8+ T cells was observed when Hep1-6-OVA cells and OTI CD8+ T cells were cocultured (whose TCR is known to react with the OVA peptide 257-264) (Figure 16, B), which indicated that tumor-intrinsic HNRNPM functions as a suppressive molecule that restricts T cell activation.
Figure 15.
Expression of HNRNPM correlated with immune checkpoint in human HCC.A-E, Expression correlation between HNRNPM and immune checkpoint gene RNA amounts in the TCGA HCC database, n = 370, HNRNPM (HNRNPM), PD-L1 (CD274), B7-H3 (CD276), B7-H4 (VTCN1), LAG-3 (LAG3), and TIM-3 (HAVCR2). B, Pearson correlation analysis of HNRNPM and CD276 immune checkpoint expressions in human HCC tissue microarray based on the IHC results, n = 240.
Figure 16.
HNRNPM inhibition curbs immune escape and enhances PD-1 blockade by promoting CD8+ T cells activation phenotype.A, Schematic diagram of Hep1-6-OVA cells co-cultured with OTI cells. B, The flow cytometry analysis of IFN-γ+ or granzyme B+ CD8+ T cells between control and shHNRNPM groups. Data were from 3 independent experiments. ∗∗∗P < .001. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. C, Schematic diagram of ASO and anti-PD-1 therapy in C57/BJ6 mice. D, Tumor inhibition by IgG (n = 6), HNRNPM-ASO (n = 6), anti-PD-1 (n = 6), or combination therapy (n = 6) in C57/BJ6 mice. ∗P < .05. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. E, Survival analysis of IgG (n = 6), HNRNPM-ASO (n = 6), anti-PD-1 (n = 6), or combination therapy (n = 6) in C57/BJ6 mice. ∗P < .05. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. F, The profiles of immune cells in tumors by HNRNPM-ASO, anti-PD-1 or combination therapy. G, CD8+ T cells infiltration in HNRNPM-ASO, anti-PD-1 or combination therapy groups. ∗∗P < .01. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. H, The changes of Treg, IFNG+, GMZB+ CD8+ T cells in control, HNRNPM-ASO, anti-PD-1 or combination therapy groups in tumor-bearing C57/BJ6 mice. ∗∗∗P < .001. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. I, The immune cells infiltration landscape of spleen in control, HNRNPM-ASO, anti-PD-1, or combination therapy groups in tumor-bearing C57/BJ6 mice. J, The mice weight between controls and HNRNPM-ASO group. ns, Non-significant. K, The relative expression of β-catenin in HNRNPM-ASO, anti-PD-1, or combination therapy groups. ∗∗∗P < .001. P values were calculated using 1-way analysis of variance and Dunnett’s multiple comparison test. L-M, The distribution of CTNNB1 mutation in PD-1 responders or non-responders. N, The study model diagram.
Given that HNRNPM plays an immunosuppressive role in vitro, we then subcutaneously inoculated Hep1-6-OVA cells into C57BL/6 mice. At 7 days after inoculation, we intraperitoneally injected HNRNPM-specific ASO (25 mg/kg) every day for 10 days and PD-1 blockade (2.5 mg/kg) every 2 days 3 times (Figure 16, C). Interestingly, combination therapy exhibited marked antitumor efficacy (Figure 16, D-E) and altered the immune landscape toward antitumor immunity with decreased proportions of Tregs and MDSCs, a decreased TIL-Treg/CD8 ratio, and increased proportions of IFN-γ+ and granzyme B+ CD8+ T cells (Figure 16, F-H). Furthermore, no difference was observed in any parameter examined in spleen tissues (Figure 16, I), indicating that the combination therapy played a specific role in affecting TILs without influencing systemic immunity. Further examination revealed no differences in body weight among the different groups, suggesting that HNRNPM-ASO may have limited general toxicity in mouse models (Figure 16, J). Additionally, β-catenin expression was significantly downregulated in the combinational therapy group and HNRNPM-ASO group (Figure 16, K). The association between β-catenin activation and resistance to anti-PD-1 therapy has been observed in patients with HCC:34 only 1 of the 9 patients with HCC activating mutations in CTNNB1 responded to anti-PD-1 or anti-PD-L1 therapy, whereas 60% of CTNNB1 WT patients responded. To test this further, we collected tumor specimens from 14 patients with HCC treated with anti-PD-1 therapy at Zhongshan Hospital. Overall, 4 patients (25%) responded to anti-PD-1 therapy, with a median survival time of 20.1 months, and 10 (75%) patients did not respond, with a median survival of 5.4 months (P = .023) (Figure 16, L). Of note, 9 patients harbored CTNNB1 mutations, 8 were non-responders with relatively high expression of HNRNPM, and 1 was a responder with relatively low expression of HNRNPM (Figure 16, M).
Discussion
Growing evidence demonstrates that cancer stemness and immune evasion play a critical role in tumor development, progression, and metastasis.8 In this study, we identified an oncofetal splicing factor, HNRNPM, whose expression was activated in fetal liver tissues, silenced in adult liver tissues, and significantly increased in HCC tissues. High HNRPM expression is a robust predictor of poor prognosis in patients with HCC. The core pluripotent factors OCT4 and SOX2 activate HNRNPM expression in HCC. Functional studies showed that HNRNPM significantly drives and is necessary for HCC CSC properties and tumorigenesis. Through transcriptome sequencing and RIP sequencing, we identified an important HNRNPM-modulated AS event of MBD2, which predominantly contains MBD2a and MBD2c. HNRNPM shifts the MBD2c isoform to MBD2a. MBD2a and MBD2c competitively bind to CpG islands in the FZD3 promoter, and FZD3 expression was repressed by MBD2a depletion or MBD2c overexpression. Additionally, targeting HNRNPM could inhibit cancer stemness and potentiate antitumor immunity by inhibiting FZD3, providing important insights into the immune evasion of CSCs in HCC.
These observations suggest that the core pluripotent factors OCT4 and SOX2 must balance a stochastic transcriptional ground state and respond rapidly to exogenous cues to properly orchestrate the cancer cell lineages required for tumor growth, all from a relatively modest number of protein-coding genes. Alternative splicing represents a likely pathway whereby core pluripotency factors can dynamically regulate proteome diversity to support high-fidelity lineage commitment. Although several examples of alternatively spliced gene products have been functionally validated in pluripotent cells,11,17,27 a general framework that mechanistically links OCT4 or SOX2 with specific splicing factors, pre-mRNA substrates, and canonical regulators of gene transcription has yet to be described.
We found a mechanistic link between an oncofetal splicing factor, a splicing event, and HCC tumorigenesis (Figure 16, N). The HNRNPM protein family comprises a type of conserved splicing factor that has important roles in the development of multiple tissues.36, 37, 38, 39, 40 HNRNPM was recently proposed to induce the epithelial to mesenchymal transition and maintain a mesenchymal phenotype in cancers.13,15,16 Consistently, we found that HNRNPM was expressed at high levels in fetal liver tissues, silenced in adult liver tissues, and reactivated in human HCC tissues and cell lines, particularly in Huh7 cells, which have relatively high expression of CD133 and EpCAM, markers of CSCs.6,11 These data establish interdependent genetic and functional links between OCT4, SOX2, and HNRNPM in HCC cells. We confirmed a different expression pattern for MBD2 isoforms and found that HNRNPM biochemically targets the pre-mRNA of this methyl-DNA binding protein. We also observed a reciprocal link between SOX2, OCT4, and MBD2a, manifested at the level of gene expression and self-renewal phenotype in HCC. Interestingly, HCC cells displayed a distinct phenotype in response to overexpression of HNRNPM or overexpression of MBD2a, suggesting that the splicing factor likely targets additional gene products. Notwithstanding a comprehensive analysis of HNRNPM gene targets, our current results provide compelling mechanistic evidence that the functional role of OCT4 and SOX2 in HCC cells extends to the pathways that regulate gene splicing.
Although HNRNPM promotes cell proliferation, maintains CSC properties, and inhibits cell apoptosis in vitro, the in vivo function of HNRNPM is more intensive, as inhibiting HNRNPM almost completely abolished tumorigenesis in mice, and overexpression of HNRNPM was sufficient to drive tumorigenesis. These results highlight the attractive therapeutic potential of targeting HNRNPM. Significantly, we observed that MBD2a and MBD2c competitively bound to the CpG islands of FZD3 and inversely regulated the expression of downstream β-catenin, an EMT marker.41 MBD2 has been considered a transcriptional repressor; however, our results showed that MBD2a positively regulates FZD3 and β-catenin expression at the transcriptional level and that knockdown of HDAC components did not influence their mRNA transcripts. Previous reports documented that MBD2a, as a DNA demethylase, upregulates uPA and Foxp3 expression by removing repressive methyl residues and thereby giving rise to promoter-specific gene transcriptional activation.30,31,42 Moreover, it was speculated that MBD2 recruits “activators” to turn on gene expression. The complexes formed by MBD2 with other proteins, such as cAMP-responsive factor (CEBPA), MBD2-interacting protein, transforming-acid-coiled–coil (TACC3), focal adhesion kinase (FAK/PYK2) and nerve growth factor-inducible protein A (NGFI-A), in most cases, are mutually exclusive with HDAC-containing complexes, thus relieving the repression potential of MBD2 even prior to eventual demethylation.30,31,42, 43, 44, 45 In addition, it is not yet clear whether MBD2a directly promotes DNA demethylation or performs this function by recruiting or regulating other DNA demethylation enzymes, such as ten eleven translocation DNA demethylases or ten eleven translocation DNA demethylases partner MLL, observed in Treg cells.46 Due to the structural difference at the C-terminus of MBD2a and MBD2c, it is worthwhile to study whether MBD2c binds to HAT, TACC3, HTLV-1 TAX1, or other unknown MBD2a-binding transcriptional activators.43,47 Here, we found that the methylation level of FZD3 was increased when MBD2a was depleted or MBD2c was overexpressed by MSP assays (Figure 12, G, H). Intriguingly, after the addition of DAC, an inhibitor of DNA methyltransferase, to the culture of these cells, the increased methylation level of FZD3 was reversed, which resulted in an elevated FZD3 mRNA level (Figure 12, G, H). Thus, our results indicate that altered the methylation status of the FZD3 promoter when manipulating the expression of MBD2 splice variants could be responsible for the change in FZD3 mRNA expression. Our results significantly extended the distinct functions of MBD2 splice variants from self-renewal of human pluripotent stem cells and somatic reprogramming27 to tumorigenesis and metastasis.
Moreover, growing evidence suggests that tumor resistance to PD-1 blockade immunotherapy is probably due to a lack of CD8+ T cell infiltration into the tumor microenvironment after treatment.8,33, 34, 35 The response rate of HCC to PD-1 blockade is approximately 20% to 30%, and the majority of HCC is irresponsive to PD-1 blockade,9 suggesting that HCC might have an intrinsic mechanism against immunotherapy. However, the underlying mechanisms of immunotherapy resistance are poorly understood. Several molecular pathways involving WNT/β-catenin have been found to inhibit antitumor immunity.8,33, 34, 35 Interestingly, these pathways also play an important role in maintaining cancer stemness. As shown in our study, compared with the control group, PD-1 monotherapy showed little effect in our spontaneous HCC murine models, which validated the results that β-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in HCC.34 Underlying this biology, we proposed a new potential combinational therapy with curbing β-catenin activation and PD-1 blockade to restore immune surveillance in β-catenin-driven HCC tumors.
However, we found that HNRNPM inhibition significantly improved PD-1 blockade treatment by inhibiting the WNT/β-catenin pathway and recruiting more CD8+ T cells. Our work showed that increasing HNRNPM in HCC is an important molecular mechanism that mediates the tumor immunosuppressive environment in HCC. Taken together, our results have important implications in developing a new combination treatment for advanced cancer by targeting HNRNPM to eliminate CSCs and to activate tumor cell-intrinsic immune responses in HCC.
Methods
Tissue Samples and Clinicopathological Data
Surgical specimens of 240 tumor tissues and paired adjacent nontumor cirrhotic liver tissues were obtained from patients undergoing curative resection in 2010 at the Liver Cancer Institute, Zhongshan Hospital, Fudan University, with written consent. After surgical excision, 480 specimens were fixed immediately in 3.7% buffered formaldehyde solution and embedded in 480 paraffin. Then, 5-μm continuous sections were prepared for IHC of HNRNPM. All patients were postsurgically monitored until June 20, 2015. The histopathologic diagnosis was based on the World Health Organization criteria. The tumor grade was determined by the classification proposed by Edmondson and Steiner. The Child-Pugh scoring system was used to assess liver function. Tumor stage was determined according to the Tumor-Node-Metastasis classification system established by the 2010 International Union Against Cancer. The Research Ethics Committee of Zhongshan Hospital approved the ethical use of human subjects for this study, and informed consent was obtained from each patient. Fetal liver tissues were obtained with informed consent from patients who underwent pregnancy termination in the Zhongshan Hospital of Fudan University. Ethical consent was granted from the Committee on Ethics of Zhongshan Hospital, Fudan University. OS was defined as the interval between surgery and death or between surgery and the last observation point. For surviving patients, the data were censored at the last follow-up. Disease-free survival was defined as the interval between surgery and the date of any diagnosed relapse (intrahepatic recurrence and extrahepatic metastasis). Additionally, RNA-seq data from a total of 372 human HCC samples and corresponding clinical information were downloaded from TCGA (https://cancergenome.nih.gov/). A total of 372 patients with HCC and HNRNPM gene expression data were extracted, and prognostic analysis was performed with clinical information.
RNA Sequencing Analysis
The RNAs from MHCC97H cells stably silencing HNRNPM or control were isolated, quantified, and purified prior to generation of the cDNA library. The sequencing libraries were generated using the TruSeq Stranded Total RNA Library Prep Kit from Illumina according to the manufacturer’s instructions. The samples were paired-end sequenced with a read length of 100 bp on an Illumina HiSeq 2500 sequencer. The process of sequencing was controlled by Illumina Data collection software. The library construction and sequencing was performed at Shanghai Biotechnology Corporation. After pre-treatment of the raw reads (filtering and QC), directional sequencing reads were mapped against the human reference genome version 19 using the TopHat algorithm. After alignment to the genome, the expression level of genes was determined on the basis of the value of reads per kilobase per million reads, which was calculated by cuffdiff. Gene ontology analysis was conducted on differentially expressed genes using the DAVID GO database to search for enriched pathways. MISO package was used to analyze the differentially regulated AS events.
Immunohistochemistry
IHC was performed with rabbit anti-human HNRNPM (1:50; sc-20002, 1D8, SANTA CRUZ). Phosphate-buffered saline (PBS) was used to substitute the primary antibody as a negative control (only with bio-IgG and ABC complex). IHC images were acquired under a DM6000B microscope (Leica, Germany).
Immunofluorescence Staining
Tumor tissues were collected from tumor-bearing mice on day 16. Tumor tissues were embedded in OCT (Sakura 4583) and frozen at −80 °C. Tissues were cut into 8-mm pieces transversally and adhered to microscope slides (ZSGB-BIO ZLI-9506). Sections were then blocked with 5% goat serum (ZSGB-BIO) for 1 hour and incubated with antibodies directly against CD8a (KT15) at 4°C overnight in the dark. The slides were washed 3 times with PBS. 1 mg/ml DAPI (Life technology) were added and incubated for 5 minutes. After a final wash step, sections were mounted using the Fluoromount-G (SouthernBiotech 0100-01). Immunofluorescence was visualized utilizing a confocal microscope (ZEISS LSM880). Antibodies and staining details are listed in Table 9.
Table 9.
List of Antibodies Used in This Research
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-HNRNPM antibody | Santa Cruz | Cat# sc-134360 |
| Anti-OCT4 antibody | Cell Signaling Technology | Cat# 2750 |
| Anti-SOX2 antibody | Cell Signaling Technology | Cat# 3579 |
| Anti-MBD2 antibody | Santa Cruz | Cat# sc-514062 |
| Anti-MBD2a antibody | Abcam (For IHC) | Cat# ab133196 |
| Anti-MBD2c antibody | Abclonal | Cat# Q2688 |
| Anti-β-catenin antibody | Abcam (For ChIP) | Cat# ab32572 |
| Anti-Snail1 antibody | Proteintech | Cat# 13099-1-AP |
| Goat Anti-Rabbit IgG (H + L)- | Bio-Rad | AB-11042881 Cat#1706515 |
| Goat Anti-Mouse IgG (H + L)- | Bio-Rad | Cat#1706516; RRID: AB_11125547 |
| HDAC1 Polyclonal Antibody | Proteintech | Cat#10197-1-AP; RRID: AB_2118062 |
| HDAC2 Polyclonal Antibody | Proteintech | Cat#12922-3-AP; RRID: AB_2118516 |
| MTA2 Polyclonal Antibody | Proteintech | Cat#66195-1-Ig; RRID: AB_2877118 |
| RBBP7 (RbAp46) Polyclonal | Proteintech | Cat#20365-1-AP |
| Anti-HA-tag antibody | Cell Signaling Technology | Cat# 3724 |
| Anti-Flag-tag antibody | Cell Signaling Technology | Cat#14793 |
| Anti-E2F-1 Antibody | Cell Signaling Technology | Cat#3742 |
| Anti-CD44 antibody | Cell Signaling Technology | Cat#96848 |
| Anti-EpCAM antibody | Cell Signaling Technology | Cat#93790 |
| Anti-CD133 antibody | Cell Signaling Technology | Cat#64326 |
ChIP, Chromatin immunoprecipitation assays; IHC, immunohistochemistry.
Cell Preparation
Six cell lines were utilized in this research. MHCC97H, MHCC97L, and MHCCLM3 (highly metastatic human HCC cell lines) were established at our institute. HepG2 and PLC/PRF/5 cells (low-metastatic human HCC cell lines) were obtained from the Chinese Academy of Sciences Cell Bank. All cells were cultured at 37 °C in an atmosphere containing 5% CO2 and in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum.
Western Blot Analysis
Total cell and tissues lysates were prepared in a 1× sodium dodecyl sulfate buffer. Identical quantities of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose filter membranes. After incubation with antibodies specific for HNRNPM (1:200; sc-20002, 1D8, SANTA CRUZ), OCT4 (1:1,000; ab109183, EPR2054, Abcam), SOX2 (1:1,000; ab92494, EPR3131, Abcam), Flag (1: 5000, F1804, Sigma-Aldrich), GAPDH (1:1000, 6004-1-Ig, Proteintech), or β-actin (1:5,000; 66009-1-Ig, Proteintech), the blots were incubated with IRdye 800-conjugated goat anti-rabbit IgG or IRdye 700-conjugated goat anti-mouse IgG and were detected using an Odyssey infrared scanner (Li-Cor). β-actin or GAPDH was used as a loading control for Western blots. The list of antibodies was showed in Table 9.
Cell Proliferation, Colony Formation, and Flow Cytometry Assays
For cell proliferation assays, a total of 3000 cells were seeded into 96-well plates. After 12 hours of culture, cell proliferation was assessed using the Cell Counting Kit-8 (Dojindo Laboratories) according to the manufacturer’s protocol. The cell proliferation curves were plotted using the absorbance at each time point. For colony formation assays, 3000 cells were seeded in the 6-well plates or 3.5-cm dishes and incubated with normal medium for 10 days. Clones were fixed and stained with 0.5% crystal violet, and the number of colonies was counted. We applied flow cytometry to detect cell cycle and cell apotosis according to the manufacturer’s protocol.
Flow Cytometry
Briefly, 4 different group tumor tissues were digested at 37 °C for 30 minutes with 1 mg/mL Collagenase D and 0.1 mg/mL DNase I (Roche). Digestion was stopped by EDTA, and cells were filtrated through 70-mm cell strainers and washed twice with PBS containing 1 mM EDTA and 2% fetal bovine serum (FBS) (staining buffer). Cells were re-suspended in the staining buffer and stained with following antibodies on ice for 30 minutes: anti-CD45, anti-CD8, anti-IFNg, anti-Granzyme B, anti-CD11b, anti-CD11c, anti-FOXP3, anti-CD25, anti-F4/80, Ly6C were purchased from BioLegend. For intracellular staining, cells were fixed with fixation buffer (Biolegend) on ice for 15 minutes, and then washed twice with Intracellular Staining Permeabilization Wash Buffer (Biolegend). Antibodies against IFN-g (Clone XMG1.2) and Granzyme B (Clone: QA16A02) were added and incubated for 1 hour on ice. The cytokine producing cells were determined by flow cytometry. The flow cytometry data were collected on Fortessa (BD) and analyzed by FlowJo (Tree Star). For cell sorting, CD8+ T cells that were co-cultured with tumor cells for 6 hours were collected and washed with culture medium. Re-suspended cells were stained with anti-CD8a antibodies (Clone: 53-6.7) for 30 min on ice. After a washing step, cells were sorted on a BD FACS AriaIII (BD) and lysed in the buffer RLT plus (QIAGEN).
Animal Studies
The animal studies were approved by the Committee on Ethics of Zhongshan hospital, Fudan University. Male athymic BALB/c nude mice (5 weeks old) were used for animal studies. Cells (3 × 106) were injected subcutaneously into the left or the right flanks of mice. Tumors were allowed to grow for 3 to 5 weeks. Tumors growth was recorded weekly with a calliper and tumor volume was calculated as a × b2 × 0.5 (a, longest diameter; b, shortest diameter). Liver colonization assays were performed with 2 × 106 cells (orthotopic implantation). Tumors were allowed to grow for 4 weeks, and then the mice were euthanized, and the liver colonization number was counted. No statistical method was used to determine sample size. The experiments were not randomized. The investigators recording tumor growth and colonization were blinded to mouse allocation.
RNA Extraction and Real-time qPCR Analysis
Total RNA was isolated using FineProtect Universal RNA Extraction Kit (Genfine Biotech Co., Ltd; Beijing, China; Catalog No: R203). First-strand cDNA was generated using the GenFQ III Reverse Transcriptase (Genfine Biotech Co., Ltd; Beijing, China; Catalog No.: A107) according to the manufacturer’s protocol. PCR and real-time qPCR was performed in the StepOne Real-Time PCR System (Applied Biosystems) using was using 2 × Taq Master Mix (Vazyme, Nanjing, China; Catalog No: P112-01) and GenFQ SYBR qPCR Master Mix (Genfine Biotech Co., Ltd; Beijing, China; Catalog No: A104), respectively. The gene-specific primers are shown in Table 9. GAPDH was employed as an endogenous control for mRNAs. The relative expression of RNAs was calculated using the comparative Ct method. The primer sequences used for splicing assays are shown in Table 10.
Table 10.
List of Primers Sequences and shRNA Sequences Used in this Research
| Real-time qPCR primers sequences | Sequence (5'→3') |
|---|---|
| HNRNPM sense | ctaacgcgagtgtatctcgag |
| HNRNPM anti-sense | cgctcccggctgcctccttgg |
| β-actin sense | GGGAAATCGTGCGTGACATTAAG |
| β-actin anti-sense | TGTGTTGGCGTACAGGTCTTTG |
| GAPDH sense | GGTCTCCTCTGACTTCAACA |
| GAPDH anti-sense | GTGAGGGTCTCTCTCTTCCT |
| MBD2a-Fwd | AGCAAGCCTCAGTTGGCAAGGT |
| MBD2a-Rev | TGTTCATTCATTCGTTGTGGGTCTG |
| MBD2c-Fwd | AGCAAGCCTCAGTTGGCAAGGT |
| MBD2c-Rev | TGAAAGCGCATGCCATGGTGCA |
| CTNNB1-Fwd | CAGAAGCTATTGAAGCTGAGG |
| CTNNB1-Rev | TTCCATCATGGGGTCCATAC |
| SNAI1-Fwd | TCGGAAGCCTAACTACAGCGA |
| SNAI1-Rev | AGATGAGCATTGGCAGCGAG |
| FZD3-Fwd | ATGGCTTTGAGATGGATTGTC |
| FZD3-Rev | GGCACATCCTCAAGGTTATAGGT |
| RBBP7-Fwd | TTGAGTGGACATCTCCTAAGTG |
| RBBP7-Rev | CCTGGTGTCCCATATCATAAGT |
| HDAC1-Fwd | CTTTAACCTGCCTATGCTGATG |
| HDAC1-Rev | CTCATTCGTGTTCTGGTTAGTC |
| HDAC2-Fwd | GCCCCATAAAGCCACTGCCGAAG |
| HDAC2-Rev | GCTCCAGCAACTGAACCGCCAG |
| MTA2-Fwd | GCCAAACCCTAACCAGATCA |
| MTA2-Rev | CAGGCATACCACTGAGCAGA |
| Flag-MBD2a-Fwd | GCTAGCATCGATACGCGTATGCGCGCGCACCCGG |
| Flag-MBD2a-Rev | TGCGGATCCTTCGAACTAGTTTAGGCTTCATCTCCACTGTCCATT |
| HA-MBD2c-Fwd | GCTAGCATCGATACGCGTATGCGCGCGCACCCGG |
| HA-MBD2c-Rev | TGCGGATCCTTCGAACTAGTTTATGGAGGAAAGGATTGGTTCTGCC |
| shRNAs | |
| MBD2a shRNA | CCGGGTAGCAATGATGAGACCCTTTCTCGAGAAAGGGTCTCATCATTGCTACT |
| HDAC1 shRNA | CCGGGCCGGTCATGTCCAAAGTAATCTCGAGATTACTTTGGACATGACCGGC |
| HDAC2 shRNA | CCGGCAGTCTCACCAATTTCAGAAACTCGAGTTTCTGAAATTGGTGAGACTGT |
| RBBP7 shRNA | CCGGCCTCCAGAACTCCTGTTTATTCTCGAGAATAAACAGGAGTTCTGGAGGT |
| MTA2 shRNA | GCCGGCGGACTTTCCTAATTGGAGTTCTCGAGAACTCCAATTAGGAAAGTCCGT |
| Control siRNA | UUCUCCGAACGUGUCACGUTT |
| OCT4 siRNA | AACAUGUGUAAGCUGCGGCCCdTdT |
| SOX2 siRNA | CUGCAGUACAACUCCAUGATT |
| MSP primers | |
| FZD3 MSP M-Fwd | GGAGAGGAGAAATATTTTTTAAGGAGTA |
| FZD3 MSP M-Rev | ACTCTAACCTACTACAAAACTCCAC |
| FZD3 MSP U-Fwd | AATATTTTTTAAGGAGTAAAATTGGG |
| FZD3 MSP U-Rev | ACTCTAACCTACTACTACAAAACTCCAC |
| MBD2_RIP_a_FWD | AAGCAAGCCTCAGTTGGCAAGGT |
| MBD2_RIP_a_REV | GAGAGGATCGTTTCGCAGTCTCTGT |
| MBD2_RIP_b_FWD | AAACAGAGACTGCGAAACGATCCTC |
| MBD2_RIP_b_REV | GGGTATGGGGACATGCACGGG |
| MBD2_RIP_c_FWD | TGCTGGGTACCTATAAAAGGGGCT |
| MBD2_RIP_c_REV | GCATGCCATGGTGCAGGACGA |
| ChIP assays primers sequences | Sequence (5'→3') |
| OCT4-HNRNPM Site sense | gatgcctcctctatcggactc |
| OCT4-HNRNPM Site anti-sense | cgtctgggctttgtgtgagcg |
| SOX2-HNRNPM Site sense | GTGATCCGCCGCCTCGGCCTCCCAT |
| SOX2-HNRNPM Site anti-sense | CTTGTTCAACCCGCGGTCCGGCACA |
| CTNNB1-OCT4 Site sense1 | ACTGGTTCATGTGGGGAAGGT |
| CTNNB1-OCT4 Site anti-sense1 | GGCTGGGGCAGCCCT |
| CTNNB1-OCT4 Site sense1 | AGGGATGGGCTGCCCCA |
| CTNNB1-OCT4 Site anti-sense1 | GGAGGAGGCCGGGAGCG |
| CTNNB1-SOX2 Site sense | ACCGTATGGCGTGTACCA |
| CTNNB1-SOX2 Site anti-sense | GAAGATGGCCGCCAGAT |
| E2F1-HNRNPM Site sense | GCCCAGACGCGGAGAAAA |
| E2F1-HNRNPM Site anti-sense | GCCGCTCTCTTCCTCCATTT |
| Antisense oligonucleotides sequences | |
| ASO ID | Sequence |
| ASO-1 | #T∗#C∗# G∗A∗T∗A∗C∗G∗A∗G∗A∗C∗C∗T∗C∗T∗G∗A∗A∗T∗T∗T∗ #T∗#C∗#T |
| ASO-2 | #T∗#C∗# G∗C∗C∗G∗A∗C∗A∗T∗C∗A∗A∗G∗A∗T∗G∗G∗A∗G∗A∗A∗ #T∗#C∗#T |
| ASO-3 | #T∗#C∗# C∗A∗T∗G∗G∗A∗G∗C∗G∗C∗A∗T∗T∗G∗G∗C∗T∗C∗T∗G∗ #T∗#C∗#T |
| Negative control | #T∗#C∗# T∗A∗T∗C∗G∗T∗G∗A∗T∗G∗T∗T∗ #T∗#C∗#T |
Note: # indicates LNA-modified nucleotides and ∗ indicates PTO linkages.
ASO, Antisense oligonucleotides; MSP, methylation-specific polymerase chain reaction; qPCR, quantitative polymerase chain reaction; shRNA, short hairpin RNA.
Lentivirus Production and Construction of Stable Cell Lines
For construction of lentiviral vector expressing human HNRNPM, the CDS of HNRNPM was subcloned into the NotI and BamHI sites of the lentiviral vector pLV8/EF-1a/RFP/Puro (GenePharma). To produce LV overexpressing HNRNPM, HEK-293T cells were co-transfected with the resulting vector described above, pGag/Pol, pRev, and pVSV-G (GenePharma) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s guidelines. Infectious LV was harvested at 72 hours post transfection and filtered through 0.45-μm PVDF filters and then concentrated using Lenti-X Concentrator (Clontech). The virus-containing pellet was dissolved in DMEM, aliquoted and stored at −80 ºC. Recombinant LV was designated as LV-HNRNPM. We used empty vector as a negative control and designated this as LVControl. MHCC97H, HepG2, and huh7 cells were infected with LV-HNRNPM or LVControl in the presence of 8 μg ml−1 Polybrene (Sigma-Aldrich) and selected with puromycin (2 μg ml−1).
Two pairs of cDNA oligonucleotides to suppress HNRNPM expression were designed and synthesized. After annealing, double-strand oligonucleotides were inserted into the SuperSilencing shRNA expression vector pGPU6/GFP/Neo (GenePharma). The resulting vectors were designated as HNRNPM shRNA-1 and HNRNPM shRNA-2. We used a scrambled shRNA as a negative control and designated this as control shRNA. The shRNA sequences are shown in Table 10. MHCC97H cells were transfected with the resulting vectors and selected with neomycin (800 μg ml−1). The same cDNA oligonucleotides with HNRNPM shRNA were synthesized. After annealing, double-strand oligonucleotides were inserted into the linear lentiviral vector pGLV10/U6/RFP/Puro (GenePharma). To produce lentivirus suppressing HNRNPM expression, HEK-293T cells were co-transfected with the resulting vector described above, pGag/Pol, pRev and pVSV-G (GenePharma) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s guidelines. Infectious LV was harvested at 72 hours post transfection and filtered through 0.45-μm PVDF filters and then concentrated using Lenti-X Concentrator (Clontech). The virus-containing pellet was dissolved in DMEM, aliquoted and stored at −80 ºC. Recombinant LV was designated as LV-shHNRNPM. We used a scrambled shRNA as a negative control and designated this as LV-shControl. MHCC97H, MHCCLM3, HepG2, and PLC/PRF/5 cells were infected with LV-shHNRNPM or LV-shControl in the presence of 8 μg ml−1 Polybrene (Sigma-Aldrich) and selected with puromycin (2 μg ml−1).
To obtain cell lines stably expressing MBD2c, or stably depleting MBD2a, MHCC97H cells were transfected with the plasmid pcDNA3.1- FLAG- MBD2a, or shMBD2c and selected with neomycin (800 μ g ml−1).
ChIP assays
ChIP assays were performed using an OCT4 antibody (5 μg per reaction; 2750, Cell Signaling Technology), or a SOX2 antibody (5 μg per reaction; 5024, D6D9, Cell Signaling Technology), and the EZMagna ChIP A/G (17-10086, Millipore) according to the manufacturer’s protocols. ChIP-derived DNA was quantified using real-time qPCR analysis. The primers specific for the HNRNPM and OCT4, SOX2 promoters are shown in Table 10.
UV CLIP Assays
After cell cultured 48 hours, UV crosslinking was performed at 400 mJ cm2 with XLE-1000 UV crosslinker (Spectroline). Immunoprecipitation was performed with anti-HNRNPM antibody (5 μg per reaction; catalog No. sc-33652, Santa Cruz) and the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Primer sequences used for CLIP assays are shown in Table 9.
RNA Immunoprecipitation Assays
pcDNA3.1-HA-HNRNPM, pcDNA3.1-HA-ΔRRM1-HNRNPM, pcDNA3.1-HA-ΔRRM2-HNRNPM, pcDNA3.1-HA-ΔRRM3-HNRNPM was co-transfected into MHCC97H cells. After 48 hours, cells were used to perform RIP experiments using a HA antibody (5 μg per reaction; HA-Tag [C29F4] Rabbit mAb #3724) and the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. In brief, cells were cross-linked in 0.1% formaldehyde prior to lysis. Cell lysates were sonicated and immunoprecipitated, and the eluates were reverse-crosslinked. Relative occupancy values were calculated by determining the IP efficiency and normalized to the level observed by immunoprecipitation using non-specific IgG.
Luciferase Reporter Assay
Full length promoter of HNRNPM, or the HNRNPM promoter harboring wild-type or potential SOX2, OCT4 site mutants were inserted into the pSI-CHECK-2 dual-luciferase reporter vector. HEK293 cells were co-transfected with the reporter plasmids along with control, OCT4, or SOX2 using Lipofectamine 2000 (Invitrogen) in a 48-well plate. Luciferase activity was measured after transfection for 48 hours using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase was normalized to firefly luciferase activity.
Methylation-specific PCR
Genomic DNA from cells was extracted and then underwent bisulfite modification utilizing the Bisulfite Conversion Kit (Active Motif). Modified DNA was amplified by PCR. Sequences of the primers designed to detect the methylation status of CpG sites were listed as in Table 10.
In Vivo and Vitro Limiting Dilution Assays
Non-mouse cells from each xenograft were sorted on the basis of the indicated surface phenotypes and injected subcutaneously in NSG mice. Each mouse received the same cell dose of indicated fractions from the same xenograft and was either harvested when tumor generated from any fraction reached 1.5 cm diameter (usually 6–10 weeks), or 20 weeks later. Mice were considered negative for tumor formation when there was no palpable tumor. For functional assays of HNRNPM and MBD2a overexpression or depletion, cells were sorted for the respective population and injected into mice at indicated doses. The frequency of cancer-initiating cells was calculated using the Web-based tool Extreme Limiting Dilution Assay (ELDA; Walter and Eliza Hall Institute, Parkville VIC, Australia).
TOP/FOP-flash Reporter Assays
Briefly, tumor cells (5 × 104) were seeded into a 24-well plate, and TOP-Flash reporter plasmids and pTK-RL plasmids were transiently co-transfected into the cells using Lipofectamine 2000 (Invitrogen). After transfection for 48 hours, the Dual-Luciferase Reporter Assay (Promega) was applied according to the manufacturer’s instructions. For the TOP/FOP-Flash assay, TOP/FOP-Flash (Genechem) was co-transfected into cells along with HNRNPM depletion or overexpression, HNRNPM-ASO, MBD2a silence, FZD3 silence, or MBD2c overexpression, and/or control. The TOP/FOP-Flash values were normalized to the Renilla reniformis (Promega) reading and the TOP/FOP ratio was measured. Experiments were performed in triplicate.
Nuclear/Cytoplasmic Protein Fractionation
Nuclear and cytoplasmic fractionation was performed according to The Kit protocols from Abcam (ab113474). After been centrifuged, the nuclear and the cytosolic fraction were collected respectively. Equal volumes of the nuclear and cytoplasmic lysates were tested by Western blot.
Antisense Oligonucleotides
Three sets of ASOs were designed based on the sequence of the human HNRNPM gene. LNA-modified nucleotides were inserted into the flanks of ASOs. Main criterion for sequence selection was selectivity to avoid undesired off-target effects. LNA-modified ASOs were ordered from Microsynth or Axolabs. For in vitro testing, ASOs were resuspended in H2O; for in vivo experiments, ASOs were resuspended in PBS. ASOs were added to cells in vitro without the use of a transfection reagent and used for in vivo studies without a delivery system. Sequences of selected HNRNPM ASOs and control oligonucleotide used in this study are listed in Table 10. Control oligonucleotide was derived from a previous study. In order to investigate in vitro and vivo efficacy of HNRNPM-specific LNA-modified ASOs, target knockdown efficacy of HNRNPM-specific LNA-modified ASOs was transfected into HCC cells and injected BALB/c nude mice intraperitoneally by 20 mg/kg every 2 days.
Generation of Hepatocyte-like Cells
In brief, hESCs were passaged onto a feeder free system until a confluence of 50% to 70% was attained. Then cells cultured in RPMI-1640 (Life Technologies) supplemented with 100 ng/mL activin A (R&D Systems) and 25 ng/mL Wnt3 a (R&D Systems) for 3 days. To induce hepatic EN, cells were grown in KO/DMEM (Life Technologies) supplemented with 25 nm/mL keratinocyte growth factor (R&D Systems) and 2% FBS (Gibco) for 2 days and then further cultured in the KO/DMEM containing 20% SR, 1 mM glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, and 1% dimethyl sulfoxide (DMSO) for 4 to 7 days. The final maturation step to obtain hepatocyte-like cells involved culturing the cells in mature medium containing 10% FBS, 10 ng/mL hepatocyte growth factor (R&D Systems), 20 ng/mL oncostatin M (R&D Systems), and 0.5 μM dexamethasone (R&D Systems) for 7 more days.
Statistical Analyses
All statistical analyses were performed using GraphPad Prism Software. For comparisons, the Student t test (2-sided), the nonparametric Mann-Whitney test, the Wilcoxon signed-rank test, the Pearson χ2 test, the log-rank test, the Fisher exact test, and Pearson correlation analysis were performed as indicated. A P value < .05 was considered significant. When representative figures are shown, these are representative of 3 independent repeats.
Acknowledgments
CRediT Authorship Contributions
Guiqi Zhu (Formal analysis: Lead; Validation: Lead; Writing – original draft: Lead)
Yi Wang (Formal analysis: Equal; Writing – original draft: Equal)
Biao Wang (Data curation: Equal; Resources: Equal)
Wei-Ren Liu (Conceptualization: Equal; Data curation: Lead)
Shuang-Shuang Dong (Methodology: Equal; Project administration: Equal)
Er-Bao Chen (Investigation: Lead; Project administration: Lead)
Jia-Liang Cai (Methodology: Equal; Project administration: Equal)
Jing-Lei Wan (Methodology: Lead; Software: Lead)
Jun-Xian Du (Software: Equal; Supervision: Equal)
Li-Na Song (Project administration: Lead; Resources: Equal; Software: Supporting)
Shi-Ping Chen (Resources: Supporting; Software: Supporting)
Lei Yu (Methodology: Equal; Resources: Equal; Software: Supporting)
Zheng-Jun Zhou (Formal analysis: Equal; Investigation: Supporting)
Zheng Wang (Formal analysis: Supporting; Investigation: Supporting; Methodology: Equal)
Jian Zhou (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Ying-Hong Shi (Investigation: Equal; Methodology: Equal; Project administration: Equal)
Jia Fan (Investigation: Equal; Resources: Equal; Validation: Equal)
Zhi Dai, PhD (Funding acquisition: Lead; Investigation: Equal; Validation: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This project was supported by grants from the National Natural Science Foundation of China (Grant numbers: 82072670, 81871916), the Leading Project of the Science and Technology Commission of Shanghai Municipality (No. 21Y21900100), and the Project of Shanghai Municipal Health Commission (No. 202140269).
Contributor Information
Ying-Hong Shi, Email: shi.yinghong@zs-hospital.sh.cn.
Jia Fan, Email: fan.jia@zs-hospital.sh.cn.
Zhi Dai, Email: dai.zhi@zs-hospital.sh.cn.
References
- 1.Gordan J.D., Kennedy E.B., Abou-Alfa G.K., Beg M.S., Brower S.T., Gade T.P., Goff L., Gupta S., Guy J., Harris W.P., Iyer R., Jaiyesimi I., Jhawer M., Karippot A., Kaseb A.O., Kelley R.K., Knox J.J., Kortmansky J., Leaf A., Remak W.M., Shroff R.T., Sohal D.P.S., Taddei T.H., Venepalli N.K., Wilson A., Zhu A.X., Rose M.G. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J Clin Oncol. 2020;38:4317–4345. doi: 10.1200/JCO.20.02672. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.D., Heimbach J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371 doi: 10.1136/bmj.m3544. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich B., Gertz E.M., Schäffer A.A., et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut. 2021 doi: 10.1136/gutjnl-2021-325288. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Rogoff H.A., Keates S., Gao Y., Murikipudi S., Mikule K., Leggett D., Li W., Pardee A.B., Li C.J. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci U S A. 2015;112:1839–1844. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang T., Guo C., Xia T., Zhang R., Zen K., Pan Y., Jin L. LncCCAT1 promotes breast cancer stem cell function through activating WNT/β-catenin signaling. Theranostics. 2019;9:7384–7402. doi: 10.7150/thno.37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou L., Wang D., Sheng D., Xu J., Chen W., Qin Y., Du R., Yang X., He X., Xie N., Liu S., Zhang L. NOTCH4 maintains quiescent mesenchymal-like breast cancer stem cells via transcriptionally activating SLUG and GAS1 in triple-negative breast cancer. Theranostics. 2020;10:2405–2421. doi: 10.7150/thno.38875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma L., Wang L., Khatib S.A., Chang C.W., Heinrich S., Dominguez D.A., Forgues M., Candia J., Hernandez M.O., Kelly M., Zhao Y., Tran B., Hernandez J.M., Davis J.L., Kleiner D.E., Wood B.J., Greten T.F., Wang X.W. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Hepatol. 2021;75:1397–1408. doi: 10.1016/j.jhep.2021.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda A., Hamilton P.T., Zhang A.W., Pattnaik S., Becht E., Mezheyeuski A., Bruun J., Micke P., de Reynies A., Nelson B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116:9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding J.J., Nandakumar S., Armenia J., Khalil D.N., Albano M., Ly M., Shia J., Hechtman J.F., Kundra R., El Dika I., Do R.K., Sun Y., Kingham T.P., D'Angelica M.I., Berger M.F., Hyman D.M., Jarnagin W., Klimstra D.S., Janjigian Y.Y., Solit D.B., Schultz N., Abou-Alfa G.K. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019;25:2116–2126. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chovanec P., Collier A.J., Krueger C., Várnai C., Semprich C.I., Schoenfelder S., Corcoran A.E., Rugg-Gunn P.J. Widespread reorganisation of pluripotent factor binding and gene regulatory interactions between human pluripotent states. Nat Commun. 2021;12:2098. doi: 10.1038/s41467-021-22201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michael A.K., Grand R.S., Isbel L., Cavadini S., Kozicka Z., Kempf G., Bunker R.D., Schenk A.D., Graff-Meyer A., Pathare G.R., Weiss J., Matsumoto S., Burger L., Schübeler D., Thomä N.H. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science. 2020;368:1460–1465. doi: 10.1126/science.abb0074. [DOI] [PubMed] [Google Scholar]
- 12.Ling S., Shan Q., Zhan Q., Ye Q., Liu P., Xu S., He X., Ma J., Xiang J., Jiang G., Wen X., Feng Z., Wu Y., Feng T., Xu L., Chen K., Zhang X., Wei R., Zhang C., Cen B., Xie H., Song P., Liu J., Zheng S., Xu X. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–1334. doi: 10.1136/gutjnl-2019-319616. [DOI] [PubMed] [Google Scholar]
- 13.Chen T.M., Lai M.C., Li Y.H., Chan Y.L., Wu C.H., Wang Y.M., Chien C.W., Huang S.Y., Sun H.S., Tsai S.J. hnRNPM induces translation switch under hypoxia to promote colon cancer development. EBioMedicine. 2019;41:299–309. doi: 10.1016/j.ebiom.2019.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llères D., Denegri M., Biggiogera M., Ajuh P., Lamond A.I. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 2010;11:445–451. doi: 10.1038/embor.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passacantilli I., Frisone P., De Paola E., Fidaleo M., Paronetto M.P. hnRNPM guides an alternative splicing program in response to inhibition of the PI3K/AKT/mTOR pathway in Ewing sarcoma cells. Nucleic Acids Res. 2017;45:12270–12284. doi: 10.1093/nar/gkx831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang T., An Z., Zhang C., Wang Z., Wang X., Liu Y., Du E., Liu R., Zhang Z., Xu Y. hnRNPM, a potential mediator of YY1 in promoting the epithelial-mesenchymal transition of prostate cancer cells. Prostate. 2019;79:1199–1210. doi: 10.1002/pros.23790. [DOI] [PubMed] [Google Scholar]
- 17.Han H., Irimia M., Ross P.J., Sung H.K., Alipanahi B., David L., Golipour A., Gabut M., Michael I.P., Nachman E.N., Wang E., Trcka D., Thompson T., O’Hanlon D., Slobodeniuc V., Barbosa-Morais N.L., Burge C.B., Moffat J., Frey B.J., Nagy A., Ellis J., Wrana J.L., Blencowe B.J. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun H., Liu T., Zhu D., Dong X., Liu F., Liang X., Chen C., Shao B., Wang M., Wang Y. HnRNPM and CD44s expression affects tumor aggressiveness and predicts poor prognosis in breast cancer with axillary lymph node metastases. Genes Chromosomes Cancer. 2017;56:598–607. doi: 10.1002/gcc.22463. [DOI] [PubMed] [Google Scholar]
- 19.Mo D., Jiang P., Yang Y., Mao X., Tan X., Tang X., Wei D., Li B., Wang X., Tang L., Yan F. A tRNA fragment, 5'-tiRNA(Val), suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019;457:60–73. doi: 10.1016/j.canlet.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Wang F., Yuan J.H., Wang S.B., Yang F., Yuan S.X., Ye C., Yang N., Zhou W.P., Li W.L., Li W., Sun S.H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 21.Li T., Huang J., Jiang Y., Zeng Y., He F., Zhang M.Q., Han Z., Zhang X. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics. 2009;93:235–242. doi: 10.1016/j.ygeno.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.S., Heo J., Libbrecht L., Chu I.S., Kaposi-Novak P., Calvisi D.F., Mikaelyan A., Roberts L.R., Demetris A.J., Sun Z., Nevens F., Roskams T., Thorgeirsson S.S. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 23.Lee T.K., Castilho A., Cheung V.C., Tang K.H., Ma S., Ng I.O. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Shan J., Shen J., Liu L., Xia F., Xu C., Duan G., Xu Y., Ma Q., Yang Z., Zhang Q., Ma L., Liu J., Xu S., Yan X., Bie P., Cui Y., Bian X.W., Qian C. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004–1014. doi: 10.1002/hep.25745. [DOI] [PubMed] [Google Scholar]
- 25.Zhu P., Wang Y., He L., Huang G., Du Y., Zhang G., Yan X., Xia P., Ye B., Wang S., Hao L., Wu J., Fan Z. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J Clin Invest. 2015;125:3795–3808. doi: 10.1172/JCI81979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M., Yan Q., Sun Y., Nam Y., Hu L., Loong J.H., Ouyang Q., Zhang Y., Li H.L., Kong F.E., Li L., Li Y., Li M.M., Cheng W., Jiang L.X., Fang S., Yang X.D., Mo J.Q., Gong Y.F., Tang Y.Q., Li Y., Yuan Y.F., Ma N.F., Lin G., Ma S., Wang J.G., Guan X.Y. A hepatocyte differentiation model reveals two subtypes of liver cancer with different oncofetal properties and therapeutic targets. Proc Natl Acad Sci U S A. 2020;117:6103–6113. doi: 10.1073/pnas.1912146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y., Loh Y.H., Li H., Cesana M., Ficarro S.B., Parikh J.R., Salomonis N., Toh C.X., Andreadis S.T., Luckey C.J., Collins J.J., Daley G.Q., Marto J.A. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Sun L., Cai Y., Shen S., Zhang T., Wang N., Wu G., Ma W., Li S.T., Suo C., Hao Y., Jia W.D., Semenza G.L., Gao P., Zhang H. Hypoxia-induced suppression of alternative splicing of MBD2 promotes breast cancer metastasis via activation of FZD1. Cancer Res. 2021;81:1265–1278. doi: 10.1158/0008-5472.CAN-20-2876. [DOI] [PubMed] [Google Scholar]
- 29.Li C., Nguyen V., Clark K.N., Zahed T., Sharkas S., Filipp F.V., Boiko A.D. Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc Natl Acad Sci U S A. 2019;116:4548–4557. doi: 10.1073/pnas.1813802116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita H., Fujii R., Aratani S., Amano T., Fukamizu A., Nakajima T. Antithetic effects of MBD2a on gene regulation. Mol Cell Biol. 2003;23:2645–2657. doi: 10.1128/MCB.23.8.2645-2657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lembo F., Pero R., Angrisano T., Vitiello C., Iuliano R., Bruni C.B., Chiariotti L. MBDin, a novel MBD2-interacting protein, relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol Cell Biol. 2003;23:1656–1665. doi: 10.1128/MCB.23.5.1656-1665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Jossin Y., Chai G., Lien W.H., Tissir F., Goffinet A.M. Feedback regulation of apical progenitor fate by immature neurons through Wnt7-Celsr3-Fzd3 signalling. Nat Commun. 2016;7 doi: 10.1038/ncomms10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anson M., Crain-Denoyelle A.M., Baud V., Chereau F., Gougelet A., Terris B., Yamagoe S., Colnot S., Viguier M., Perret C., Couty J.P. Oncogenic β-catenin triggers an inflammatory response that determines the aggressiveness of hepatocellular carcinoma in mice. J Clin Invest. 2012;122:586–599. doi: 10.1172/JCI43937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz de Galarreta M., Bresnahan E., Molina-Sánchez P., Lindblad K.E., Maier B., Sia D., Puigvehi M., Miguela V., Casanova-Acebes M., Dhainaut M., Villacorta-Martin C., Singhi A.D., Moghe A., von Felden J., Tal Grinspan L., Wang S., Kamphorst A.O., Monga S.P., Brown B.D., Villanueva A., Llovet J.M., Merad M., Lujambio A. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–1141. doi: 10.1158/2159-8290.CD-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 36.Li H., Liu J., Shen S., Dai D., Cheng S., Dong X., Sun L., Guo X. Pan-cancer analysis of alternative splicing regulator heterogeneous nuclear ribonucleoproteins (hnRNPs) family and their prognostic potential. J Cell Mol Med. 2020;24:11111–11119. doi: 10.1111/jcmm.15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Low Y.H., Asi Y., Foti S.C., Lashley T. Heterogeneous nuclear ribonucleoproteins: implications in neurological diseases. Mol Neurobiol. 2021;58:631–646. doi: 10.1007/s12035-020-02137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu D., Osuka S., Zhang Z., Reichert Z.R., Yang L., Kanemura Y., Jiang Y., You S., Zhang H., Devi N.S., Bhattacharya D., Takano S., Gillespie G.Y., Macdonald T., Tan C., Nishikawa R., Nelson W.G., Olson J.J., Van Meir E.G. BAI1 suppresses medulloblastoma formation by protecting p53 from Mdm2-mediated degradation. Cancer Cell. 2018;33:1004–1016.e5. doi: 10.1016/j.ccell.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X., Li X., Cheng Y., Wu W., Xie Z., Xi Q., Han J., Wu G., Fang J., Feng Y. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat Commun. 2014;5:4581. doi: 10.1038/ncomms5581. [DOI] [PubMed] [Google Scholar]
- 40.Zhu W., Ding J., Sun L., Wu J., Xu X., Wang W., Li H., Shen H., Li X., Yu Z., Chen G. Heterogeneous nuclear ribonucleoprotein A1 exerts protective role in intracerebral hemorrhage-induced secondary brain injury in rats. Brain Res Bull. 2020;165:169–177. doi: 10.1016/j.brainresbull.2020.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Flahaut M., Meier R., Coulon A., Nardou K.A., Niggli F.K., Martinet D., Beckmann J.S., Joseph J.M., Mühlethaler-Mottet A., Gross N. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta-catenin pathway. Oncogene. 2009;28:2245–2256. doi: 10.1038/onc.2009.80. [DOI] [PubMed] [Google Scholar]
- 42.Mei L., Xiong W.C. FAK interaction with MBD2: a link from cell adhesion to nuclear chromatin remodeling? Cell Adh Migr. 2010;4:77–80. doi: 10.4161/cam.4.1.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angrisano T., Lembo F., Pero R., Natale F., Fusco A., Avvedimento V.E., Bruni C.B., Chiariotti L. TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res. 2006;34:364–372. doi: 10.1093/nar/gkj400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya S.K., Ramchandani S., Cervoni N., Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 45.Detich N., Theberge J., Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- 46.Wang L., Liu Y., Han R., Beier U.H., Thomas R.M., Wells A.D., Hancock W.W. Mbd2 promotes foxp3 demethylation and T-regulatory-cell function. Mol Cell Biol. 2013;33:4106–4115. doi: 10.1128/MCB.00144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ego T., Tanaka Y., Shimotohno K. Interaction of HTLV-1 Tax and methyl-CpG-binding domain 2 positively regulates the gene expression from the hypermethylated LTR. Oncogene. 2005;24:1914–1923. doi: 10.1038/sj.onc.1208394. [DOI] [PubMed] [Google Scholar]