Abstract
Most genetic studies concerning risk genes in Alzheimer’s disease (AD) are from Caucasian populations, whereas the data remain limited in the Chinese population. In this study, we systematically explored the relationship between AD and risk genes in mainland China. We sequenced 33 risk genes previously reported to be associated with AD in a total of 3604 individuals in the mainland Chinese population. Common variant (MAF ≥ 0.01) based association analysis and gene-based (MAF < 0.01) association test were performed by PLINK 1.9 and Sequence Kernel Association Test-Optimal, respectively. Polygenic risk score (PRS) was calculated, and receiver operating characteristic curve (AUC) was computed. Plasma Aβ42, Aβ40, total tau (T-tau), and neurofilament light chain (NFL) were tested in a subgroup, and their associations with PRS were conducted using the Spearman correlation test. Six common variants varied significantly between AD patients and cognitively normal controls after the adjustment of age, gender, and APOE ε4 status, including variants in ABCA7 (n = 5) and APOE (n = 1). Among them, four variants were novel and two were reported previously. The AUC of PRS was 0.71. The high PRS was significantly associated with an earlier age at onset (P = 4.30 × 10−4). PRS was correlated with plasma Aβ42, Aβ42/Aβ40 ratio, T-tau, and NFL levels. Gene-based association test revealed that ABCA7 and UNC5C reached statistical significance. The common variants in APOE and ABCA7, as well as rare variants in ABCA7 and UNC5C, may contribute to the etiology of AD. Moreover, the PRS, to some extent, could predict the risk, onset age, and biological changes of AD.
Subject terms: Diseases, Medical research
Introduction
Being the most prevalent dementia type in the elderly worldwide, Alzheimer’s disease (AD) accounts for an estimated 60–80% of all dementia cases [1]. AD is characterized by cognitive impairments, such as memory loss, disorientation. Amyloid-β (Aβ) accumulation and tau neurofibrillary tangles are the main pathological hallmarks in the AD brain [2]. Amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) are the causative genes of AD [3]. It is estimated that AD has a heritability of 70%, suggesting AD is a highly heritable disease [4].
Thanks to the rapid development of high-throughput sequencing technologies, over 30 AD risk genes have been identified by genome-wide association studies (GWASs) [4]. Apolipoprotein E (APOE) remains the strongest genetic risk factor in AD [5]. In 2009, the first two GWAS studies in AD demonstrated that CLU, CR1, and PICALM were risk genes of AD [6, 7]. In the second year, another GWAS identified that BIN1 was associated with AD [8]. In the subsequent GWAS studies, ABCA7, MS4A gene cluster (MS4A6A, MS4A6E), EPHA1, CD33, and CD2AP reached genome-wide statistical significance [9, 10]. Furthermore, 19 genes were related to AD, of which 11 genes were novel, including HLA-DRB5/HLA-DRB1, PTK2B, SORL1, FERMT2, etc. [11]. Besides, rare variants were identified by next-generation sequencings, such as PLD3 and ABCA7 [12, 13]. Interestingly, some rare AD-associated variants are located in AD risk genes with common variants related to AD, such as ABCA7 and SORL1, indicating that these genes are involved in the etiology of AD through multiple pathways [3].
Most genetic studies of AD are from Caucasian populations, whereas the genetic data of the Chinese population are limited. Genetic heterogeneity existed among different populations. Even for APOE ε4, the most prominent genetic risk factor in AD, its contribution to AD varied among different ethnic groups [14]. To investigate the roles of risk genes in the Chinese population systematically, we genotyped 33 AD risk genes in a large-scale Chinese population.
Materials and methods
Subjects
For targeted panel sequencing, we recruited 1192 AD patients from Xiangya Hospital and 2412 controls from a community in Changsha. The AD patients were diagnosed by two neurologists specializing in neurodegenerative disease. The AD patients met the National Institute on Aging-Alzheimer’s Association criteria for probable AD [15]. A battery of neuropsychological tests was performed in the AD patients by an experienced clinical neuropsychologist, including Mini-Mental State Exam (MMSE), Montreal Cognitive Assessment (MoCA), Clinical Dementia Rating (CDR), the activity of daily living (ADL), and Neuropsychiatric Inventory (NPI). The MMSE was also administered to the controls. The clinical data were collected by PhD students guided by experienced neurologists. Participants with causative mutations for AD (APP, PSEN1, and PSEN2) had been excluded by Sanger sequencing. This study was approved by the Ethics Committee of Xiangya Hospital, Central South University, China. Written informed consent was obtained from each participant or guardian.
Targeted sequencing
We designed a targeted sequencing panel composed of 33 AD risk genes, including APOE, BIN1, CD2AP, EPHA1, CLU, MS4A6A, MS4A6E, CD33, TTR, TMEM106B, PTK2B, SLC24A4, RIN3, DSG2, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2, CASS4, CR1, ABCA7, SORL1, TREM2, ADAM10, PLD3, PICALM, UNC5C, AKAP9, TTC3, PLCG2, and ABI3. These risk genes were identified by GWAS approaches or next-generation sequencing studies in AD cohorts. Our panel used biotinylated RNA probes to capture known DNA sequences from the human reference GRCh37. The designed probes and genomic locations for the 33 AD risk genes are shown in Supplementary Files 1 and 2. The panel’s design workflow involves five steps: (1) probe design, (2) oligo pool synthesis, (3) probe production, (4) wet-lab testing, and (5) data quality control and analysis. Genomic DNA was extracted from peripheral blood leukocytes using a QIAGEN kit. All DNA samples were normalized to 100 ng/μL. The genomic DNA was fragmented into 150–200 bp length fragments by Biorupter Pico. End-repairing, A-tailing, adaptor ligation, and an 11-cycle pre-capture PCR amplification were conducted in fragmented DNA. The fragmented DNA was captured by the targeted panel and sequenced on Illumina NovaSeq 6000 platform. The low-quality reads of fastq data were filtered out by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, the paired-end sequence reads were aligned to the human reference genome (UCSC hg19/GRCH37) using the BWA software (version 0.7.15, http://bio-bwa.sourceforge.net) [16]. Picard (version 2.18.7, http://broadinstitute.github.io/picard/) was used to remove duplicate sequence reads and index the sequencing data. The quality-score recalibration, local realignments, variant calling, and filtering were conducted by the Genome Analysis Toolkit (version 3.2, https://software.broadinstitute.org/gatk/) [17]. The variants were annotated using ANNOVAR (https://hpc.nih.gov/apps/ANNOVAR.html) [18]. Common variants and rare variants were classified on the basis of minor allele frequencies (MAF) at a threshold of 0.01 (common variants: MAF ≥ 0.01; rare variants: MAF < 0.01). Besides, ReVe, an algorithm developed by our team, was used to predict the pathogenicity of missense variants [19]. In our study, we defined the damaging variants as loss-of-function (LoF) variants or missense variants with ReVe > 0.7. LoF variants were considered as the variants resulting in stop, frameshift, or splice-site disruption. The variants were named based on the guidelines of the Human Genome Variation Society [20].
Blood sampling and analyses
In our study, a subgroup of 333 AD patients and 130 controls underwent plasma biomarkers testing. Specifically, the venous blood was collected and stored frozen at −80 °C before analysis. Plasma Aβ42, Aβ40, t-tau, and neurofilament light chain (NFL) levels were determined using the single-molecule array (Simoa)-HD1 platform (Simoa; Quanterix, USA). Aβ42, Aβ40, and t-tau levels were determined using a multiplex array (Neurology 3-Plex A Advantage Kit, N3PA), and NFL levels were measured via a single-analyte array (NF-light). Specifically, the calibrators were kept at room temperature. The N3PA assay or NF-Light assay definition was imported under Custom Assay. Plasma samples were manually diluted 4× with sample diluent. The beads were vortexed for at least 30 s and the prepared reagents (Bead Reagent, Detector Reagent, SBG Reagent, Sample Diluent) were added into the reagent bay. Meanwhile, the resorufin ß-D-galactopyranoside was loaded into the sample bay. Finally, the concentration of each sample was assessed by Simoa software with Neat or the standard 4× dilution protocol for AD patients and controls. All samples were measured with the two-step immunoassay. All measurements were conducted by well-trained technicians who were blinded to the clinical information.
Statistical analysis
The variants with genotyping rate <95%, Hardy–Weinberg equilibrium P value < 1 × 10−6 in the controls, genotype quality ≤ 20, allelic balance out of 25%/75% ratio of referent and alternate allele reads in the heterozygote, and allelic balance out of 95% ratio of in the homozygote were filtered out with the use of PLINK 1.9. We performed the common variant (MAF ≥ 0.01) based association analysis between 1192 AD patients and 2412 controls using PLINK 1.9 [21]. Age, gender, and APOE ε4 status (APOE ε4+, APOE ε4–) were adjusted by PLINK 1.9 for each common variant. Linkage disequilibrium (LD) patterns of significant variants were reconstructed using Haploview version 4.2 [22].
PRS was generated using PRSice-2 [23], and the receiver operating characteristic curve (ROC) was drawn by the R software (version 4.0.3, R Project for Statistical Computing). The area under the ROC (AUC) was calculated. Moreover, the participants were divided into four groups based on the PRS quartile. Using the Cox proportional hazard model, we investigated the associations of PRS scores with the cumulative incidence rate of AD. The associations of PRS and plasma biomarkers were performed using the Spearman correlation test.
In addition, using the Sequence Kernel Association Test-Optimal (SKAT-O test) [24], we performed the gene-based association test by combining rare variants between AD patients and controls. Rare variants were further classified as followings: rare damaging variants (MAF < 0.01, LoF or ReVe > 0.7), rare damaging missense variants (MAF < 0.01, ReVe > 0.7), rare LoF variants (MAF < 0.01, LoF), rare missense variants (MAF < 0.01, missense), and rare synonymous variants (MAF < 0.01, synonymous). Age, gender, and APOE ε4 status were adjusted by SKAT-O. A cutoff P value * n < 0.05 was considered statistically significant (n is defined by the number of common variants or genes).
Results
Demographic and clinical information
On average, the sequencing coverage (or sequencing depth) was 621.81× and the percentage of base sequences ≥20× was 98.36%. A total of 1194 AD patients and 2412 controls were enrolled. The average onset age of AD patients was 63.93 years, and the average age of controls was 64.76 years. With regard to age, no significant difference was observed between AD patients and controls (P = 0.06). The MMSE scores of AD patients were significantly higher than those of controls (P = 4.84 × 10−6). Furthermore, in the AD patients, the average MoCA, CDR, ADL, and NPI scores were 8.46, 1.29, 34.41, and 18.05, respectively.
Plasma Aβ42 levels and Aβ42/Aβ40 ratio in AD were lower than those than in controls (Aβ42: P = 5.32 × 10−3, Aβ42/Aβ40: P = 9.11 × 10−12). The levels of plasma T-tau and NFL were higher than those in controls (T-tau: P = 9.72 × 10−16, NFL: P = 2.20 × 10−16) (Table 1).
Table 1.
Characteristics of AD patients and controls.
| AD | Control | P value | |
|---|---|---|---|
| Number | 1192 | 2412 | – |
| Age (years), median (IQR) | 64 (18) | 65 (10) | 0.06 |
| Gender (M/F) | 475/717 | 1157/1255 | 4.84 × 10−6 |
| MMSE, median (IQR) | 12 (10) | 27 (3) | 1.20 × 10−12 |
| MoCA, median (IQR) | 8 (8) | – | – |
| CDR, median (IQR) | 1 (1) | – | – |
| ADL, median (IQR) | 31 (15) | – | – |
| NPI, median (IQR) | 12 (19) | – | – |
| Plasma Aβ42, pg/mL, median (IQR) | 13.42 (4.33) | 14.22 (3.69) | 5.32 × 10−3 |
| Plasma Aβ40, pg/mL, median (IQR) | 266.91 (68.97) | 256.03 (59.74) | 2.03 × 10−2 |
| Plasma Aβ42/Aβ40, median (IQR) | 0.0013 (0.0002) | 0.0014 (0.0003) | 9.11 × 10−12 |
| Plasma T-tau, pg/mL, median (IQR) | 3.96 (1.44) | 3.08 (0.94) | 9.72 × 10−16 |
| Plasma NFL, pg/mL, median (IQR) | 21.38 (13.12) | 11.38 (7.36) | 2.20 × 10−16 |
MMSE Mini-mental State Examination, MoCA Montreal Cognitive Assessment, CDR Clinical Dementia Rating, ADL activities of daily living, NPI Neuropsychiatric Inventory, IQR inter-quartile range, Aβ amyloid β, T-tau total tau, NFL neurofilament light chain.
Common variant association test
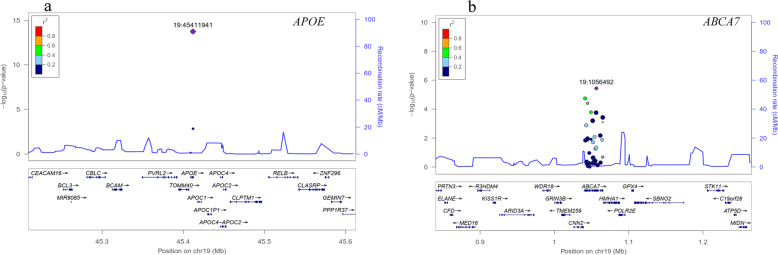
After quality control, 217 common variants were identified in AD patients and controls. After adjusting for age, gender, and APOE ε4 status, 34 variants were nominally associated with AD risk, including variants in ABCA7 (n = 18), NME8 (n = 3), APOE (n = 2), BIN1 (n = 2), SORL1 (n = 2), INPP5D (n = 2), UNC5C (n = 1), CLU (n = 1), MS4A6E (n = 1), PICALM (n = 1), and TEME106B (n = 1) (adjusted P < 0.05). Based on Bonferroni corrected P value (P < 2.30 × 10−4, 0.05/217), six variants differed significantly between AD patients and controls, involving APOE rs429358 (adjusted P = 1.82 × 10−14), ABCA7 rs3752246 (adjusted P = 3.66 × 10−6), ABCA7 rs3752229 (adjusted P = 1.83 × 10−5), ABCA7 rs3764648 (adjusted P = 3.98 × 10−5), ABCA7 rs4147914 (adjusted P = 1.64 × 10−4), and ABCA7 rs150594667 (adjusted P = 1.77 × 10−4) (Table 2 and Fig. 1). The LD patterns of variants in the ABCA7 (rs3752246-rs3752229-rs3764648-rs4147914-rs150594667) were similar between AD patients and controls (Supplementary Fig. 1). In our study, the nominal common variants with adjusted P < 0.05 are listed in Supplementary File 3. Given that nominal variants may also play important roles in AD, we performed the network biology approach using the Network Assisted Genomic Association (NAGA) [25]. NAGA study revealed that several genes may be implicated in the AD etiology, including APOE, APOC2, APOC1, APOC4, CLPTM1, TOMM40, etc. (Supplementary Fig. 2).
Table 2.
The significant common variants between AD patients and controls.
| Gene | Position | Rs ID | Effect allele | MAF | OR (95% CI) | P | Adjusted P | |
|---|---|---|---|---|---|---|---|---|
| AD | NC | |||||||
| APOE | 19:45411941 | rs429358 | C | 0.276 | 0.099 | 5.718 (3.661–8.932) | 1.07 × 10−83 | 1.82 × 10−14 |
| ABCA7 | 19:1056492 | rs3752246 | G | 0.371 | 0.311 | 1.291 (1.159–1.438) | 5.15 × 10−7 | 3.66 × 10−6 |
| ABCA7 | 19:1041352 | rs3752229 | G | 0.376 | 0.322 | 1.259 (1.133–1.399) | 5.81 × 10−6 | 1.83 × 10−5 |
| ABCA7 | 19:1044753 | rs3764648 | T | 0.345 | 0.292 | 1.256 (1.126–1.400) | 6.80 × 10−6 | 3.98 × 10−5 |
| ABCA7 | 19:1049269 | rs4147914 | A | 0.398 | 0.346 | 1.221 (1.100–1.354) | 1.65 × 10−5 | 1.64 × 10−4 |
| ABCA7 | 19:1056149 | rs150594667 | T | 0.030 | 0.013 | 1.983 (1.386–2.835) | 5.03 × 10−7 | 1.77 × 10−4 |
Effect allele represents the minor allele.
MAF minor allele frequency, AD Alzheimer’s disease, NC controls, OR odds ratio, CI confidence interval; Adjusted P adjusted by age, gender, and APOE ε4 status.
Fig. 1. Regional association plots.
Regional association plots of the APOE (a) and ABCA7 loci (b). Purple diamonds represent the sentinel variant in the corresponding locus. Colors show the LD measured as R2 between the sentinel variant and its neighboring variants. cMMb centimorgans per megabase.
Discriminative and predictive performance of PRS
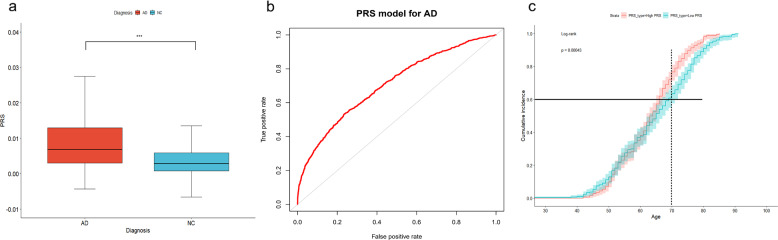
PRS was generated using PRSice-2. As expected, the PRS values in AD patients were significantly higher than those in controls (P < 2.2 × 10−16) (Fig. 2a). The AUC of the model was 0.71 (95% confidence interval: 0.69–0.72) (Fig. 2b). The effects on AD occurrence were evaluated using a Cox proportional hazards model. Based on their individual PRS, all AD cases were separated into quartiles. Using the Cox proportional hazards model, we found that the highest PRS quartile was significantly associated with an earlier onset age compared to those in the lowest quartile (high PRS vs low PRS, OR = 1.36, P = 4.30 × 10−4, 95% CI: 1.15–1.60). For instance, the expected onset age for 60% to develop AD was around 70 years in the low PRS group, later that that in the high PRS group (the expected age of onset was about 66 years). Meanwhile, at the age of 70, the cumulative incidence rates of AD patients in the high PRS group were higher than that of the low PRS group, which were approximately 70% and 60%, respectively (Fig. 2c).
Fig. 2. Discriminative and predictive performance of PRS.
PRS between AD patients and controls (a) (***P < 0.0001, PRS polygenic risk score). The discriminative ability of PRS model (b). The cumulative incidence of AD in high and low PRS groups (c).
Correlations between PRS and AD plasma biomarkers
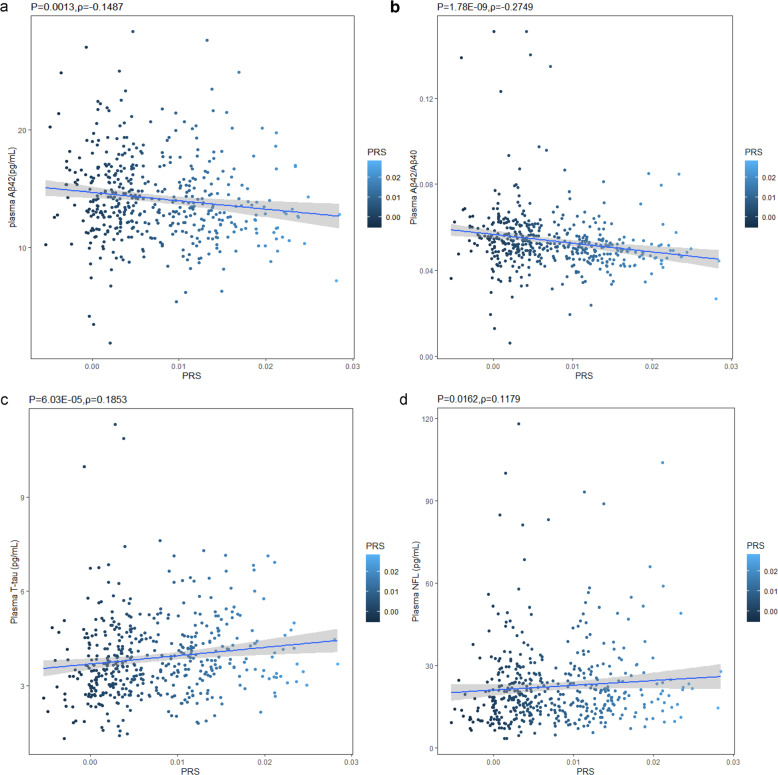
PRS was inversely associated with plasma Aβ42 (P = 0.0013, Spearman ρ = −0.1487) and the ratio of Aβ42/Aβ40 (P = 1.78 × 10−9, Spearman ρ = −0.2749). No significant correlation between PRS and plasma Aβ40 was observed (P = 0.9170, Spearman ρ = 0.0049). Meanwhile, PRS was positively correlated with plasma T-tau (P = 6.03 × 10−5, Spearman ρ = 0.1853) as well as plasma NFL (P = 0.0162, Spearman ρ = 0.1179) (Fig. 3). Furthermore, using general linear regression, the associations of plasma Aβ42, Aβ42/Aβ40 ratio, and T-tau with PRS remained significant even after adjusting for age and gender (Aβ42, β = −2.941, adjusted P = 0.0034; Aβ42/Aβ40 ratio, β = −4.496, adjusted P = 8.77 × 10−6; T-tau, β = 2.877, adjusted P = 0.0042). Also, plasma NFL was nominally associated with PRS after the adjustment of age and gender (β = 1.777, adjusted P = 0.0762).
Fig. 3. Correlations between PRSs and plasma biomarkers (Spearman correlation coefficients (ρ) and P values were used to evaluate the correlations).
PRS and Aβ42 (a); PRS and Aβ42/Aβ42 ratio (b); PRS and T-tau (c); PRS and NFL (d).
Gene-level aggregation testing
After quality control, 4277 rare variants were identified in our study. The rare variants were collapsed together within genes and their joint effects were investigated. P value less than 1.52 × 10−3 was considered significant based on Bonferroni correction (0.05/33). When analyzing rare damaging variants and rare damaging missense variants, ABCA7 all reached statistical significance (adjusted P = 1.32 × 10−3 and adjusted P = 7.48 × 10−4, respectively). Gene-based association analysis on rare missense variants revealed that UNC5C and ABCA7 were significantly associated with AD (adjusted P = 1.14 × 10−3 and adjusted P = 1.20 × 10−3, respectively) (Table 3).
Table 3.
Significant genes between AD and controls in the SKAT-O test.
| Classification | Gene | No. of variants | SKAT-O test | |
|---|---|---|---|---|
| P | Adjusted P | |||
| Rare damaging variants (MAF < 0.01, LoF or ReVe > 0.7) | ABCA7 | 55 | 1.08 × 10−4 | 1.32 × 10−3 |
| Rare damaging missense variants (MAF < 0.01, ReVe > 0.7) | ABCA7 | 47 | 5.23 × 10−5 | 7.48 × 10−4 |
| Rare missense variants (MAF < 0.01, missense) | UNC5C | 34 | 2.41 × 10−3 | 1.14 × 10−3 |
| Rare missense variants (MAF < 0.01, missense) | ABCA7 | 138 | 2.96 × 10−4 | 1.20 × 10−3 |
No. of variants number of the variants in the test, SKAT-O Sequence Kernel Association Test-Optimal, Adjusted P adjusted by age, gender, and APOE ε4 status.
Discussion
A number of risk genes contribute to the development of AD. However, the vast majority of studies were performed in the Caucasian population. Most studies focused on reported variants based on array-based SNP genotyping. In this study, we systematically screened 33 AD risk genes in the mainland Chinese population. In the common variant association test, six variants located within APOE and ABCA7 differed significantly between AD patients and controls. PRS was associated with onset age and plasma biomarkers of AD. Pathway enrichment analysis revealed that several processes were associated with AD. Furthermore, gene-based association analyses demonstrated that UNC5C and ABCA7 were associated with AD risk.
APOE, located in chromosome 19q13.2, is the most important risk gene for AD. We found that the ε4 allele of APOE (rs429358) conferred susceptibility to AD, which was similar to the finding in the Caucasian population [26]. The recent large-scale GWAS also revealed that APOE ε4 remains the strongest genetic risk factor [27]. Generally, one APOE ε4 allele enhanced the risk of developing AD by about 3.7 times in the Caucasian population [28], while our study demonstrated that one APOE ε4 allele increased the risk of AD by 5.7 times. Similarly, in the Japanese population, the APOE ε4 allele also exhibited a higher risk effect on AD compared to the Caucasian population [26]. These findings underscored that APOE ε4 may be more harmful in the Asian population than in the Caucasian population.
We identified that five ABCA7 common risk variants were correlated to AD risk, including rs3752246, rs3752229, rs3764648, rs4147914, and rs150594667. Among them, rs3752246 was described previously while the remaining four variants were novel [29]. ATP-binding cassette, sub-family A, member 7 (ABCA7) is composed of 47 exons and encodes a 220-kDa protein. ABCA7 is expressed in brain tissue and linked to lipid metabolism, regulation of phagocytosis as well as amyloid-β metabolism [19]. In 2011, the SNP rs3764650 of ABCA7 obtained genome-wide significance in the Caucasian population, firstly suggesting that ABCA7 is a risk gene of AD [9]. The subsequent large Caucasian-based GWAS studies revealed that rs3752246 and rs4147929 were significantly associated with AD [10, 11]. A meta-analysis revealed that three variants increased the risk of developing AD, namely rs3764650, rs3752246, and rs4147929 [29]. Therefore, the association between ABCA7 and AD is well established. In our study, we confirmed that rs3752246 was associated with AD, supporting its risk role in the pathogenesis of AD. In addition, the remaining four significant variants were in strong LD with rs3752246 in our sample (rs3752229 vs rs3752246: D’ = 0.78, R2 = 0.57; rs3764648 vs rs3752246: D’ = 0.87, R2 = 0.69; rs4147914 vs rs3752246: D’ = 0.78, R2 = 0.52; rs150594667 vs rs3752246: D’ = 1.00, R2 = 0.01). These findings indicated that they may tag the same functional variant [30]. Further larger sample studies and functional experiments are warranted to validate their roles in AD. In addition, using NAGA, we found that several genes, including APOE, APOC2, APOC1, APOC4, CLPTM1, TOMM40, etc., were implicated in AD pathogenesis. These genes involve APOE itself or genes located near the APOE gene [31, 32], indicating the important role of APOE in the etiology of AD.
Common variants contribute to AD; however, their effects are relatively limited. PRS has been widely applied in predicting individuals at high risk for common diseases [33]. Using the genotype data from the International Genomics of Alzheimer’s Project, the PRS exhibited 0.75–0.84 prediction accuracy of AD risk [34]. In the Chinese population, Li et al. genotyped 35 SNPs and PRS models were built, demonstrating 0.61–0.66 prediction accuracy of AD risk [35]. In addition, in a recent large Chinese GWAS study, the top AUC was 0.73 when combining the significant variants and APOE status [36]. Similarly, in our study, we found that the AUC of the RPS model in AD was 0.71 (ranging from 0.69 to 0.72), indicating the PRS model could predict AD risk to some extent in the Chinese population. Furthermore, we revealed that high PRS was associated with an earlier onset age, and the cumulative incidence rate of the high PRS group was higher than that of the low PRS group in the same age. Leonenko et al. identified that PRS could predict the age-specific risk for developing AD [37]. Meanwhile, another study also revealed that PRS was correlated with onset age and AD risk in the Chinese population [35]. Accordingly, the high PRS might help clinicians to prioritize the individuals who most likely to develop AD and benefit from early prevention as well as treatment.
We observed that PRS was associated with decreased plasma Aβ42 levels and Aβ42/Aβ40 ratio as well as increased plasma T-tau and NFL levels. In 2018, the ATN classification system was issued, composed of β amyloid deposition (“A”), pathologic tau (“T”), and neurodegeneration (“N”). Although the ATN classification system greatly facilitates the diagnosis of AD, the invasive cerebrospinal fluid sampling and expensive PET scan constrain their widespread use [38]. Plasma Aβ42, Aβ42/Aβ40 ratio, total tau, and NFL are accessible and potentially useful biomarkers in AD [39–41]. A subgroup of our sample demonstrated that plasma biomarkers were significantly altered in AD patients, further supporting their utility for screening and diagnosing of AD. Interestingly, our study revealed a significant relationship between PRS and plasma biomarkers. The effects of genetic risk on AD biomarkers have been studied previously. CSF Aβ42, Aβ42/Aβ40 ratio, T-tau, and P-tau were correlated with PRS in AD patients and controls [35]. In cognitively healthy elders, PRS was associated with CSF NFL levels in individuals without Aβ42 pathology [42]. In the Hong Kong Chinese AD cohort, PRS was associated with plasma Aβ42 level and Aβ42/Aβ40 ratio [43]. Our investigation firstly identified that PRS was associated with plasma NFL, T-tau, and confirmed that PRS was related to plasma Aβ42 level and the ratio Aβ42/Aβ40. We highlighted that the aggregate genetic risk may modulate the individual pathogenic and biological alterations. Besides, given that the pathological changes occurred over two decades before clinical symptoms onset [44], the utility of PRS may be promising in identifying the subjects with abnormal plasma AD biomarkers.
Gene-based analysis observed two genes, ABCA7 and UNC5C, were significantly associated with AD by the SKAT-O test. Intriguingly, in our study, ABCA7 modulated the risk of AD both in common variant association tests and gene-based analysis. Accumulating evidence showed that ABCA7 is a significant risk gene harboring both common and rare risk variants in the development of AD [30]. The high burden of ABCA7 LoF variants and missense variants was observed previously in AD [13, 45]. We found that ABCA7 rare damaging variants were enriched in AD cases, which was in line with a study conducted in the French cohort and further underscored the damaging role of ABCA7 rare variants in AD across different populations [46]. UNC5C localizes on 4q22.3 and encodes UNC5C mediating neuronal apoptosis [47]. A rare coding mutation, UNC5C T835M, segregated with AD in two families and associated with AD in large case–control cohorts [48]. Our group previously revealed that several rare coding variants may confer a certain risk of AD [49]. A rare missense variant, UNC5C D353N, existed in five affected individuals in the AD family and may be involved in AD [50]. Our study determined that the burden of rare missense variants in UNC5C was significantly associated with AD, further indicating that UNC5C was implicated in AD via the modulation of rare variants.
Conclusions
The common variant association test indicated that APOE and ABCA7 were associated with AD in the mainland Chinese population. PRS is of potential use in assessing the risk and onset age of AD as well as plasma AD biomarkers. Gene-level aggregation testing indicated that ABCA7 and UNC5C may contribute to the etiology of AD in the mainland Chinese population.
Supplementary information
Acknowledgements
The authors are grateful to all subjects for participation in our study.
Author contributions
BJ, XWX, JCL, and LS designed the experiment and analyzed the data. ZHY, LG, XXL, YFZ, LZ, XW, XXL, HL, YLJ, ZJL, YZ, QJY, and WWZ collected the data. BJ and XWX wrote this manuscript. LS edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key R&D Program of China (No. 2020YFC2008500, 2017YFC0840100, 2017YFC0840104, and 2018YFC1312003), National Natural Science Foundation of China (No. 81671075, 81971029, 81701134, 82071216, and 81901171), Innovation platform and talent plan of Hunan Province (2019SK2335), and the Youth Science Foundation of Xiangya Hospital (2018Q020).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bin Jiao, Xuewen Xiao.
Supplementary information
The online version contains supplementary material available at 10.1038/s41386-021-01258-1.
References
- 1.2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 2020;16:391–460. [DOI] [PubMed]
- 2.van der Kant R, Goldstein LSB, Ossenkoppele R. Amyloid-β-independent regulators of tau pathology in Alzheimer disease. Nat Rev Neurosci. 2020;21:21–35. doi: 10.1038/s41583-019-0240-3. [DOI] [PubMed] [Google Scholar]
- 3.Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci. 2020;23:311–22. doi: 10.1038/s41593-020-0599-5. [DOI] [PubMed] [Google Scholar]
- 4.Bellenguez C, Grenier-Boley B, Lambert JC. Genetics of Alzheimer’s disease: where we are, and where we are going. Curr Opin Neurobiol. 2020;61:40–8. doi: 10.1016/j.conb.2019.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 8.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–4. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg S, Stefansson H, Jonsson T, Johannsdottir H, Ingason A, Helgason H, et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat Genet. 2015;47:445–7. doi: 10.1038/ng.3246. [DOI] [PubMed] [Google Scholar]
- 14.Hallman DM, Boerwinkle E, Saha N, Sandholzer C, Menzel HJ, Csázár A, et al. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–49. [PMC free article] [PubMed] [Google Scholar]
- 15.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr., Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Zhao T, Zhang Y, Zhang K, Shi L, Chen Y, et al. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46:7793–804. doi: 10.1093/nar/gky678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–9. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:giz082. [DOI] [PMC free article] [PubMed]
- 24.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlin DE, Fong SH, Qin Y, Jia T, Huang JK, Bao B, et al. A fast and flexible framework for network-assisted genomic association. iScience. 2019;28:155–61. doi: 10.1016/j.isci.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–56. [PubMed] [Google Scholar]
- 27.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021;20:68–80. doi: 10.1016/S1474-4422(20)30412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma FC, Wang HF, Cao XP, Tan CC, Tan L, Yu JT. Meta-analysis of the association between variants in ABCA7 and Alzheimer’s disease. J Alzheimers Dis. 2018;63:1261–7. doi: 10.3233/JAD-180107. [DOI] [PubMed] [Google Scholar]
- 30.De Roeck A, Van Broeckhoven C, Sleegers K. The role of ABCA7 in Alzheimer’s disease: evidence from genomics, transcriptomics and methylomics. Acta Neuropathol. 2019;138:201–20. doi: 10.1007/s00401-019-01994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak GA, Zhou Z, Silzer TK, Barber RC, Phillips NR, Alzheimer’s Disease Neuroimaging Initiative, Breast and Prostate Cancer Cohort Consortium, and Alzheimer’s Disease Genetics Consortium Two-stage Bayesian GWAS of 9576 individuals identifies SNP regions that are targeted by miRNAs inversely expressed in Alzheimer’s and cancer. Alzheimers Dement. 2020;16:162–77. doi: 10.1002/alz.12003. [DOI] [PubMed] [Google Scholar]
- 32.Shao Y, Shaw M, Todd K, Khrestian M, D’Aleo G, Barnard PJ, et al. DNA methylation of TOMM40-APOE-APOC2 in Alzheimer’s disease. J Hum Genet. 2018;63:459–71. doi: 10.1038/s10038-017-0393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–84. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li WW, Wang Z, Fan DY, Shen YY, Chen DW, Li HY, et al. Association of polygenic risk score with age at onset and cerebrospinal fluid biomarkers of Alzheimer’s disease in a Chinese cohort. Neurosci Bull. 2020;36:696–704. doi: 10.1007/s12264-020-00469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia L, Li F, Wei C, Zhu M, Qu Q, Qin W, et al. Prediction of Alzheimer’s disease using multi-variants from a Chinese genome-wide association study. Brain. 2020;144:924–37. [DOI] [PMC free article] [PubMed]
- 37.Leonenko G, Sims R, Shoai M, Frizzati A, Bossù P, Spalletta G, et al. Polygenic risk and hazard scores for Alzheimer’s disease prediction. Ann Clin Transl Neurol. 2019;6:456–65. doi: 10.1002/acn3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jack CR, Jr., Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–62. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simrén J, Leuzy A, Karikari TK, Hye A, Benedet AL, Lantero-Rodriguez J, et al. The diagnostic and prognostic capabilities of plasma biomarkers in Alzheimer’s disease. Alzheimers Dement. 2021;17:1145–56. [DOI] [PMC free article] [PubMed]
- 40.Nakamura A, Kaneko N, Villemagne VL, Kato T, Doecke J, Doré V, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554:249–54. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 41.Preische O, Schultz SA, Apel A, Kuhle J, Kaeser SA, Barro C, et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat Med. 2019;25:277–83. doi: 10.1038/s41591-018-0304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoog I, Kern S, Najar J, Guerreiro R, Bras J, Waern M, et al. A non-APOE polygenic risk score for Alzheimer’s disease is associated with cerebrospinal fluid neurofilament light in a representative sample of cognitively unimpaired 70-year olds. J Gerontol A Biol Sci Med Sci. 2021;76:983–90. doi: 10.1093/gerona/glab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Chen Y, Ip FCF, Lai NCH, Li YYT, Jiang Y, et al. Genetic and polygenic risk score analysis for Alzheimer’s disease in the Chinese population. Alzheimers Dement (Amst). 2020;12:e12074. doi: 10.1002/dad2.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, Vermeulen S, et al. Mutations in ABCA7 in a Belgian cohort of Alzheimer’s disease patients: a targeted resequencing study. Lancet Neurol. 2015;14:814–22. doi: 10.1016/S1474-4422(15)00133-7. [DOI] [PubMed] [Google Scholar]
- 46.Le Guennec K, Nicolas G, Quenez O, Charbonnier C, Wallon D, Bellenguez C, et al. ABCA7 rare variants and Alzheimer disease risk. Neurology. 2016;86:2134–7. doi: 10.1212/WNL.0000000000002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Wang BL, Sun FR, Li JQ, Cao XP, Tan L. The role of UNC5C in Alzheimer’s disease. Ann Transl Med. 2018;6:178. doi: 10.21037/atm.2018.04.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wetzel-Smith MK, Hunkapiller J, Bhangale TR, Srinivasan K, Maloney JA, Atwal JK, et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer’s disease and increases neuronal cell death. Nat Med. 2014;20:1452–7. doi: 10.1038/nm.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao B, Liu X, Tang B, Hou L, Zhou L, Zhang F, et al. Investigation of TREM2, PLD3, and UNC5C variants in patients with Alzheimer’s disease from mainland China. Neurobiol Aging. 2014;35:2422.e9–2422.e11. doi: 10.1016/j.neurobiolaging.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Korvatska O, Leverenz JB, Jayadev S, McMillan P, Kurtz I, Guo X, et al. R47H variant of TREM2 associated With Alzheimer disease in a large late-onset family: clinical, genetic, and neuropathological study. JAMA Neurol. 2015;72:920–7. doi: 10.1001/jamaneurol.2015.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.