Abstract
RNA molecules can show high levels of cooperativity in their global folding and interactions with divalent ions. However, cooperativity at individual ligand-RNA interaction sites remains poorly understood. Here we investigated binding of thiamine and methylene diphosphonic acid (MDP, a soluble structural analog of pyrophosphate) to the thiamine pyrophosphate (TPP) riboswitch. These ligands each bind weakly at proximal sub-sites, with 10 μM and 1 mM affinities, respectively. Affinity of MDP moderately improves when thiamine or thiamine-like fragments are pre-bound to the RNA. Covalent linking of thiamine and MDP substantially increases riboswitch binding to a notable high affinity of 20 nM. Crystal structures and single-molecule correlated chemical probing revealed favorable induced fit effects upon binding of individual ligands and, unexpectedly, a substantial thermodynamically unfavorable RNA structural rearrangement upon binding of the linked thiamine-MDP ligand. Thus, linking of two ligands of modest affinity, accompanied by an unfavorable structural rearrangement, yields a potent linked RNA-binding compound. Since complex ligands often bind riboswitches and other RNAs at proximal sub-sites, principles derived from this work inform and support fragment-linking strategies for identifying small molecules that interact with RNA specifically and with high affinity.
Graphical Abstract
Introduction
Cooperativity and induced fit are critical in molecular recognition and biological function1–4. Ligand cooperativity has been extensively studied in protein-small molecule ligand systems1,5,6, and principles derived from these studies have been used to guide fragment-based ligand design for multiple protein targets1,5–8. The field of RNA-targeted drug discovery is undergoing a notable shift to emphasize low-molecular-weight, drug-like molecules9–12, and fragment-based approaches represent a promising strategy for discovering molecules with favorable properties that bind RNA13–22. RNA molecules clearly experience significant cooperativity at the level of their global folding and interaction with divalent ions23,24, in binding by large oligonucleotide ligands25,26, and for interactions between dimer and multivalent ligands with duplex RNAs27. However, structure-function relationships that underlie cooperativity at individual RNA-ligand interaction sites are poorly understood. For example, it is currently unknown what initial affinity and binding cooperativity support obtaining potent ligands by an RNA-targeted fragment strategy.
Cooperativity occurs when the binding of a ligand to one binding site in a macromolecule affects the macromolecule in such a way that binding of a second ligand occurs more readily than it would without the first ligand present. Core tenets of fragment-based ligand development are that a high-affinity ligand can be developed by linking two ligands that individually bind with low affinity and that the binding affinity of the linked compound can be approximated by summing the affinities of the two ligands5,28. Generally, there is an entropic advantage to linking two fragments; in addition, there are potential contributions from induced fit. In principle, it is possible to achieve “super-additivity” upon fragment linking, if the entropic advantage of linking two fragments were sufficiently large1.
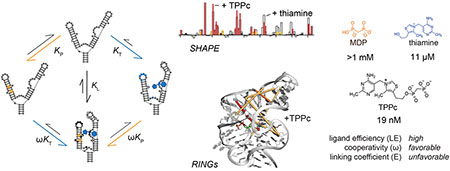
Riboswitches are useful test cases for investigating cooperativity relationships in RNA systems, as substituents of complex riboswitch ligands often bind at defined sub-sites in the ligand binding pocket29. The thiamine pyrophosphate (TPP) riboswitch is an excellent model system for understanding the ligandability of RNA because, first, the TPP riboswitch is largely unstructured and “floppy” in the absence of ligand16,30 and, second, the thiamine and the pyrophosphate moieties interact with the RNA at distinct sub-sites (Fig. 1A)31–33. RNAs that recognize a ligand via two or more sub-sites or that fold such that multiple ligandable sites lie close in three-dimensional space are ideal models for understanding the effects of ligand linking on RNA-ligand interactions.
Figure 1.
Ligand binding to the thiamine and MDP sub-sites in TPP riboswitch RNA. (A) Structure of the TPP-bound binding site31,32. TPP ligand is shown as sticks, with regions corresponding to thiamine and MDP in TPPc colored orange and blue. Nucleotides that become more or less constrained upon TPPc binding, judged by SHAPE reactivity determined in this study, are colored black and white, respectively; other nucleotides are shown in gray. Magnesium ions are shown as spheres. (B) Thermodynamic cycle for binding by thiamine (blue, KT) and MDP (orange, KP) fragments. Nucleotides that become more or less constrained upon ligand binding as indicated by decreased or increased SHAPE reactivity are denoted by closed and open circles, respectively. Flexibility changes upon binding by the linked fragments, TPPc (KL), shown in bottom quadrant, reveal similar, but more widespread, flexibility changes than observed upon binding by thiamine or thiamine-like fragments alone.
Here we analyze binding of thiamine, methylene diphosphonic acid (MDP, a structural analog of pyrophosphate), and the thiamine-MDP conjugate to the TPP riboswitch to gain insight into how this RNA binds to small-molecule ligands, and to understand the potency that can be realized by linking low molecular mass, weakly binding ligands. Our study of the thermodynamic and structural effects of fragment-like ligand cooperativity support an optimistic assessment of fragment-based ligand discovery directed toward RNA targets.
Results
Cooperative binding of constituent fragments in the TPP riboswitch
To investigate potential cooperativity in the TPP riboswitch RNA-TPP ligand system, we analyzed constituents of the native TPP ligand: thiamine (and related thiamine analogues) and methylene diphosphonic acid (MDP) (Fig. 1A). We used MDP, which is soluble in the presence of millimolar concentrations Mg2+ (an ion essential for RNA folding), whereas pyrophosphate is not. We measured binding affinities of the riboswitch for the TPP fragments and analogs using isothermal calorimetry (ITC). ITC directly measures the enthalpy (ΔH) of binding, and the resulting data can be fit to a global binding model to obtain dissociation constants (Kd), the Gibbs free energy (ΔG), and entropy (–TΔS). We determined binding affinities of the TPP riboswitch for multiple ligands at 1 mM Mg2+, where MDP is soluble. Thiamine bound to the RNA with a KT of 11 μM under our conditions (Fig. 2A), consistent with prior work34,35. MDP bound much more weakly with a KP of 1.2 mM (Fig. 2B).
Figure 2.
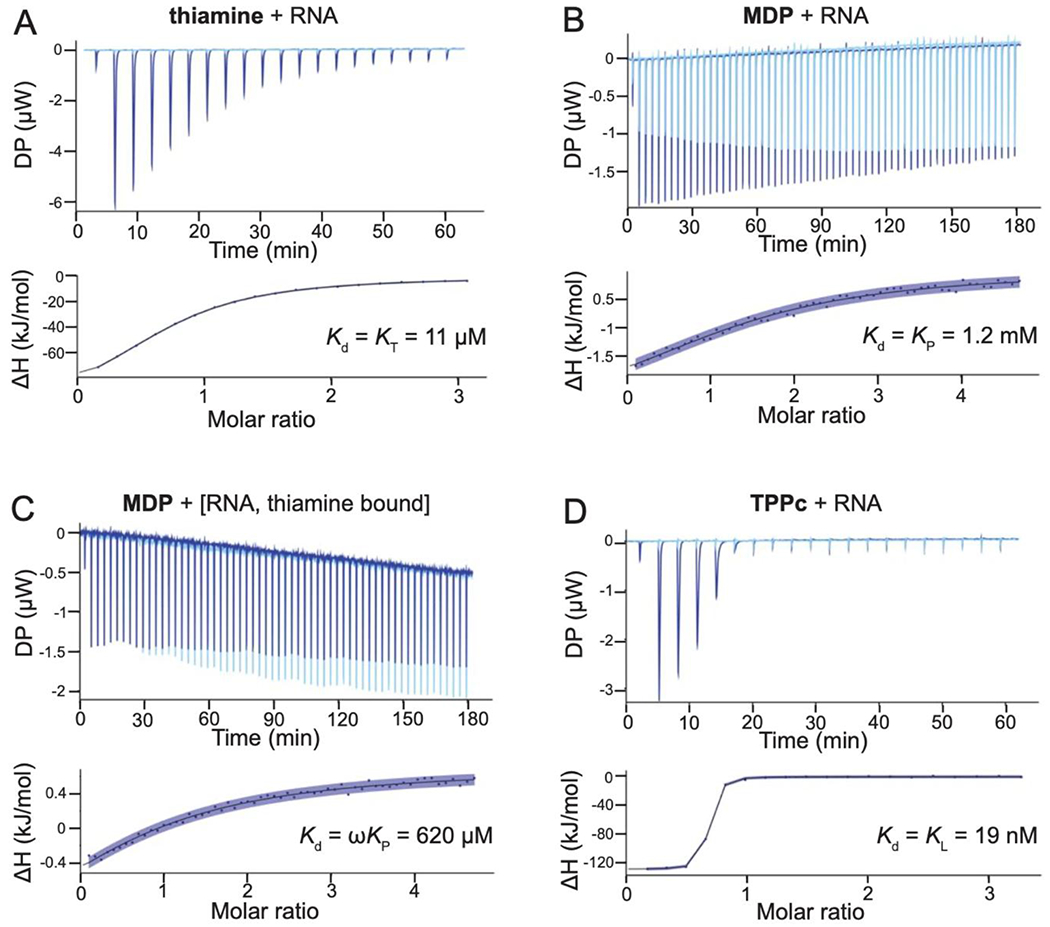
RNA ligand affinities determined by isothermal titration calorimetry. ITC traces obtained upon titration of (A) thiamine into the riboswitch, (B) MDP into the riboswitch, (C) MDP into the thiamine-bound riboswitch, and (D) TPPc into the riboswitch. Background traces (ligand titrated into buffer) are shown as light blue, experimental traces in dark blue. Curve fits are shown with the 95% confidence intervals in blue shading. Experimental steps taken to obtain accurate data for weak binding ligands are detailed in the Methods.
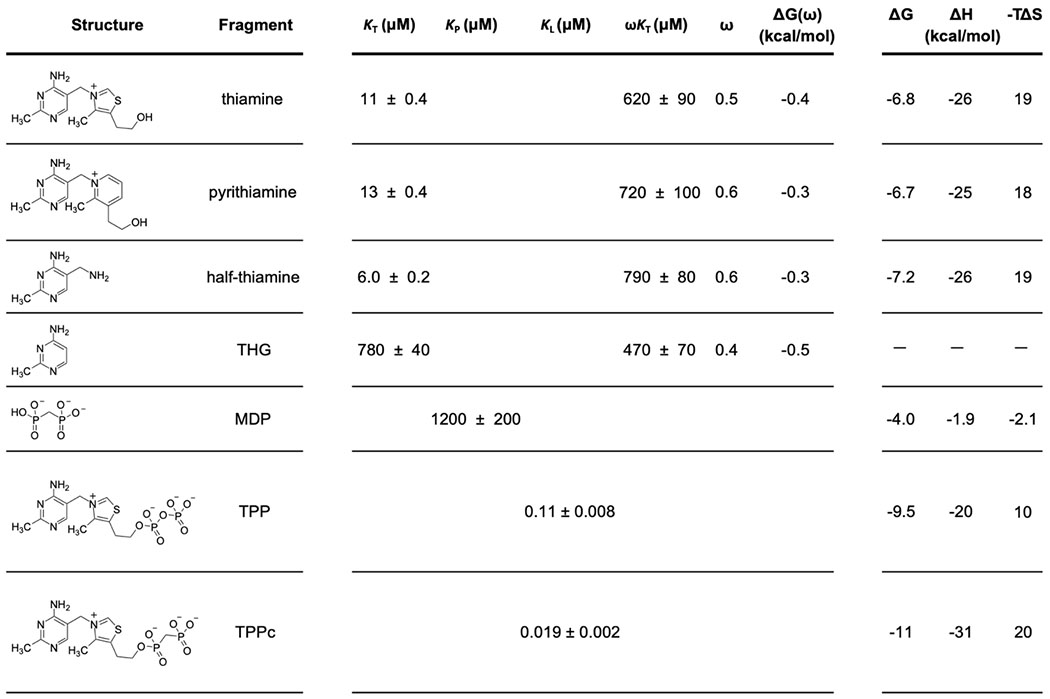
We then measured cooperativity between these groups by pre-binding the riboswitch RNA with a saturating concentration of thiamine or thiamine analog and then titrating MDP into this RNA-fragment complex. The cooperativity factor, ω, is a measure of degree to which binding of one fragment is enhanced (values <1.0) or inhibited (values >1.0) by the presence of another bound fragment3. The cooperativity factor was calculated as the ratio of the equilibrium dissociation constant for MDP binding to the riboswitch, as pre-bound by thiamine (ωKP), to that in the absence of thiamine (KP). Cooperativity with MDP was investigated for thiamine (Fig. 2C) and thiamine derivatives (Fig. S1). The KT values for thiamine and its derivatives varied by 70-fold (Table 1). We observed consistent, modest cooperativity values for MDP for all thiamine derivatives with ω of approximately 0.5, corresponding to a Gibbs free energy change of –0.4 kcal/mol.
Table 1.
Structures, equilibrium dissociation constants, and thermodynamic values for thiamine, thiamine analogs, MDP, TPP, and TPPc. Binding data were obtained by ITC; error estimates for dissociation constants are based on 95% confidence intervals of curve fits. KT, KP, KL, and ωKT refer to binding constants illustrated in Fig. 1. Cooperativity values were calculated as ω = ωKP/KP; ΔG(ω) = −RT ln ω. ΔH values were measured directly from ITC experiments; ΔG and −TΔS values were calculated from ΔH and Kd. –, not determined due to poor ITC curve fit.
|
Effect of fragment linking on binding
To model binding of the native-like linked TPP ligand in our fragment system, we synthesized an analog of TPP containing the MDP moiety in place of pyrophosphate (TPPc, Table 1). TPPc bound with considerably higher affinity than either of the constituent fragments alone or for the fragments bound in combination: The dissociation constant of the TPPc ligand, KL, was 19 nM (Fig. 2D). Consistent with prior work35, TPPc bound 6-fold more tightly to the TPP riboswitch than did the native TPP ligand (Table 1).
As expected, both fragment components and TPPc have thermodynamically favorable binding interactions with the riboswitch RNA. Binding interactions for thiamine and its analogs were largely enthalpically driven (∆H values ranged from −20 to −26 kcal/mol). Binding by MDP contributed a small additional favorable entropy component (Table 1). The free energy change upon binding of the riboswitch by TPPc was roughly equal to the sum of free energy changes observed upon binding of the individual sub-site ligands.
Structures of the thiamine, TPP and TPPc-bound riboswitches
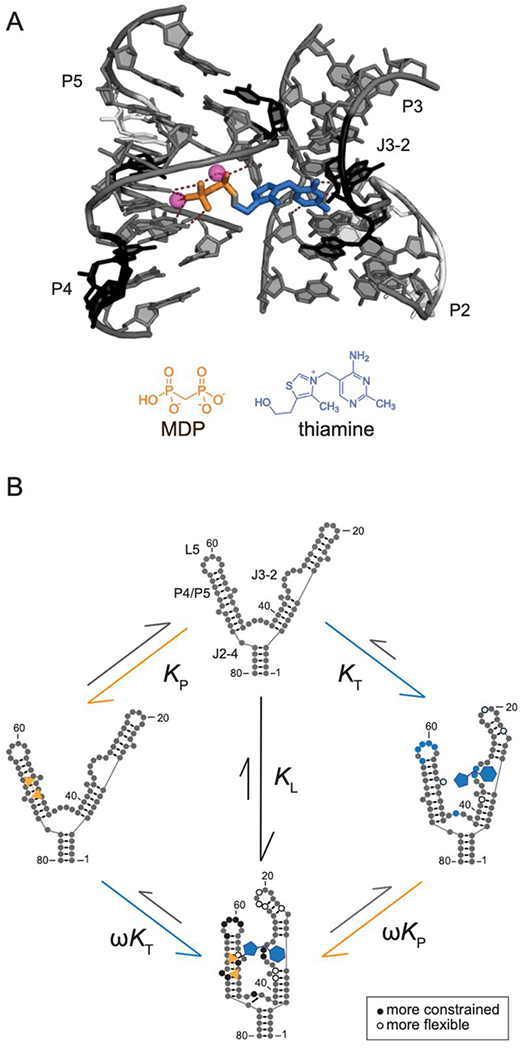
To understand the molecular bases for the cooperativity between thiamine and MDP binding and for the high-affinity binding of TPPc, we determined X-ray crystal structures of the thiamine-, TPPc-, and TPP-bound riboswitch RNAs (at 2.9, 2.5 and 2.2 Å resolution, respectively) (Fig. 3, Fig. S2, Table S1). In all cases, the thiamine head group (THG) bound similarly and formed hydrogen bonds in the J3-2 pocket (G40, G42 and A43) (Fig. 3). In the thiamine-bound structure, the quality of the electron density map for RNA nucleotides that comprise the thiamine-binding sub-site was higher than for the pyrophosphate sensor site, as observed previously16, consistent with the pyrophosphate pocket being partially dynamic (Fig. S2A). Density is also weak around the hydroxyethyl group, suggesting flexibility. No metal ion-mediated intermolecular interactions were observed in the thiamine-bound structure. G72 lies along and forms van der Waals interactions the hydroxyethyl moiety of thiamine and makes a hydrogen bond with G60 (Fig. 3A).
Figure 3.
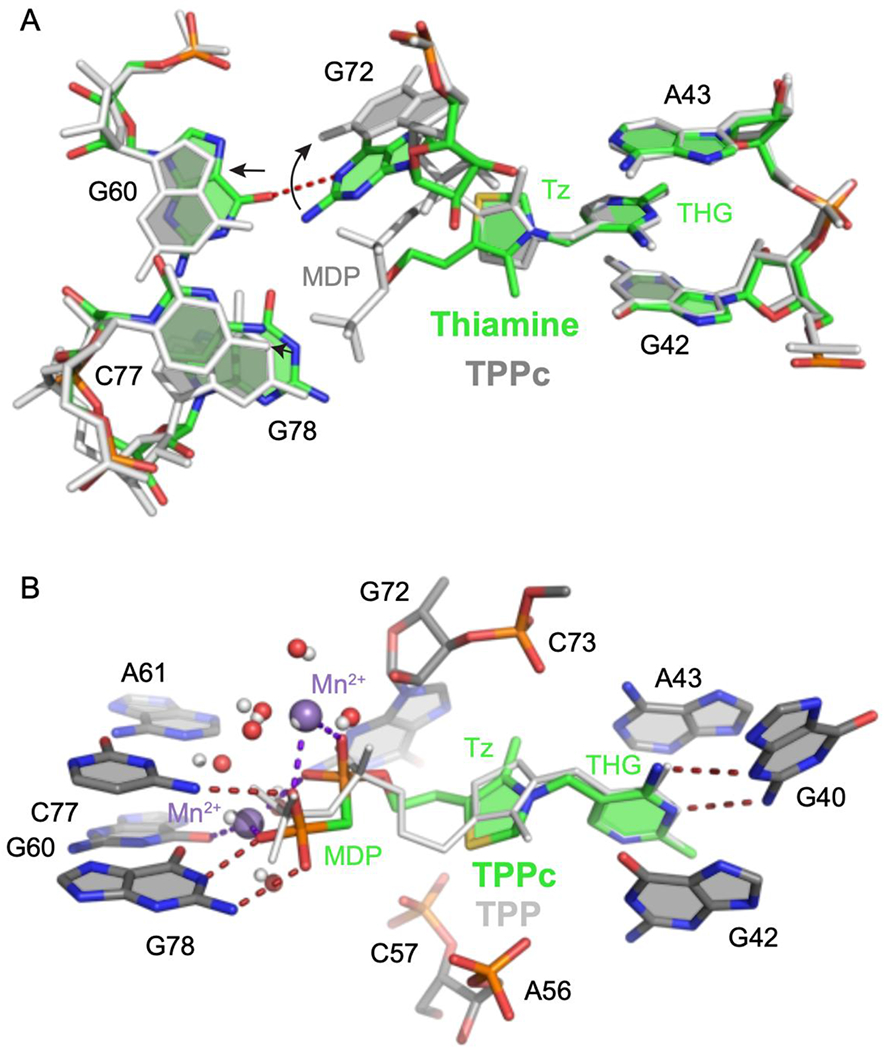
Comparison of TPP riboswitch structures, bound by thiamine, TPP and TPPc. (A) Comparison of thiamine- (colored) and TPPc-bound (light gray) riboswitch structures. Metal ions and water molecules were omitted from the TPPc structure for clarity. Arrows indicate shifts in positions of nucleotides near thiamine. THG, thiamine head group; Tz, thiazole. (B) Comparison of TPPc- (colored) and TPP-bound (light gray) structures. Ligand-bound Mn2+ ions and coordinated waters in the TPPc structure are depicted in violet and red spheres, respectively; ions and coordinated water molecules for the TPP-bound structure are shown with small spheres (in light gray). Hydrogen bonds and metal-ligand coordination are shown with dashed lines.
The TPP and TPPc structures reveal that the RNA undergoes significant conformational changes relative to the thiamine structure. As expected, and consistent with extensive prior studies31–33, TPP and TPPc bind the riboswitch in an extended conformation, with the MDP/pyrophosphate group bound to the stacked P4-P5 helices (involving residues G60, A61, C77, G78). These interactions are mediated by two divalent metal cations (Fig. 3B). As a result, nucleotides that create the MDP/pyrophosphate recognition pocket, G60 and G78, move away from the ligand, and G72 swings away from the thiamine-MDP linker.
The thiazolium ring does not specifically interact with the RNA in either structure, and its presence appears to hamper ligand binding slightly, as indicated by two-fold higher affinity for half-thiamine (Table 1). Strikingly, linking MDP to thiamine to form TPPc flips the thiazolium moiety relative to the thiamine- and TPP-bound structures. Thiamine and TPP bind such that the sulfur atom of the thiazolium ring points toward G72 (upwards, in Fig. 3 and Fig. S2C), a conformation unambiguously identified in a prior 2.05 Å structure31. In the TPPc-bound structure, the electron density best supports a model in which the sulfur atom points downwards (Fig. 3A, Fig. S2B). Thus, despite binding in the same sub-site, the thiamine moiety of TPPc binds RNA in a conformation different from both thiamine and the thiamine moiety of TPP.
Comparison of the TPPc- and TPP-bound structures reveals plausible sources of the 6-fold higher affinity of the TPPc complex (Fig. 3B). The substitution of the angular sp3-hybridized oxygen atom with the tetrahedral sp3-hybridized methylene moiety changes the geometry of MDP in TPPc relative to that of the pyrophosphate moiety in TPP, that apparently induces flipping of the thiazole moiety. This change also increases the intermolecular interface between the RNA and TPPc by ~10 Å2 relative to the TPP-RNA interface, possibly contributing stronger van der Waals interactions. Substituting the non-bridging oxygen atom by a carbon atom increases the electronegativity of terminal oxygen atoms36 and the methylene phosphonate should form stronger interactions with the metal cation co-ligands and stronger hydrogen bonds with G78 and C77.
Consequences of ligand binding on internucleotide structural communication
We used SHAPE chemical probing to reveal conformational adjustments in the riboswitch in solution, in the absence of ligand and in the presence of thiamine or TPPc. SHAPE measures local nucleotide flexibility and detects changes in local structure upon ligand binding37,38. Differences in SHAPE reactivity profiles for the riboswitch in the absence of ligand versus in the presence of thiamine were significant and localized in the L5 and J3-2 regions of the RNA; SHAPE reactivity was lower in these regions in the presence of the ligand (Fig. 4A, 4B, left; Fig. S3). The SHAPE reactivity profile in the presence of TPPc revealed additional protections in the L5 and J3-2 regions of the riboswitch, relative to that observed with thiamine, and a decrease in SHAPE reactivity in the P4-P5 stem (Fig. 4C, left). These data are fully consistent with our crystallographic analysis showing thiamine binds at J3-2 and the MDP group binds in the P4-P5 region (Fig. 3).
Figure 4.
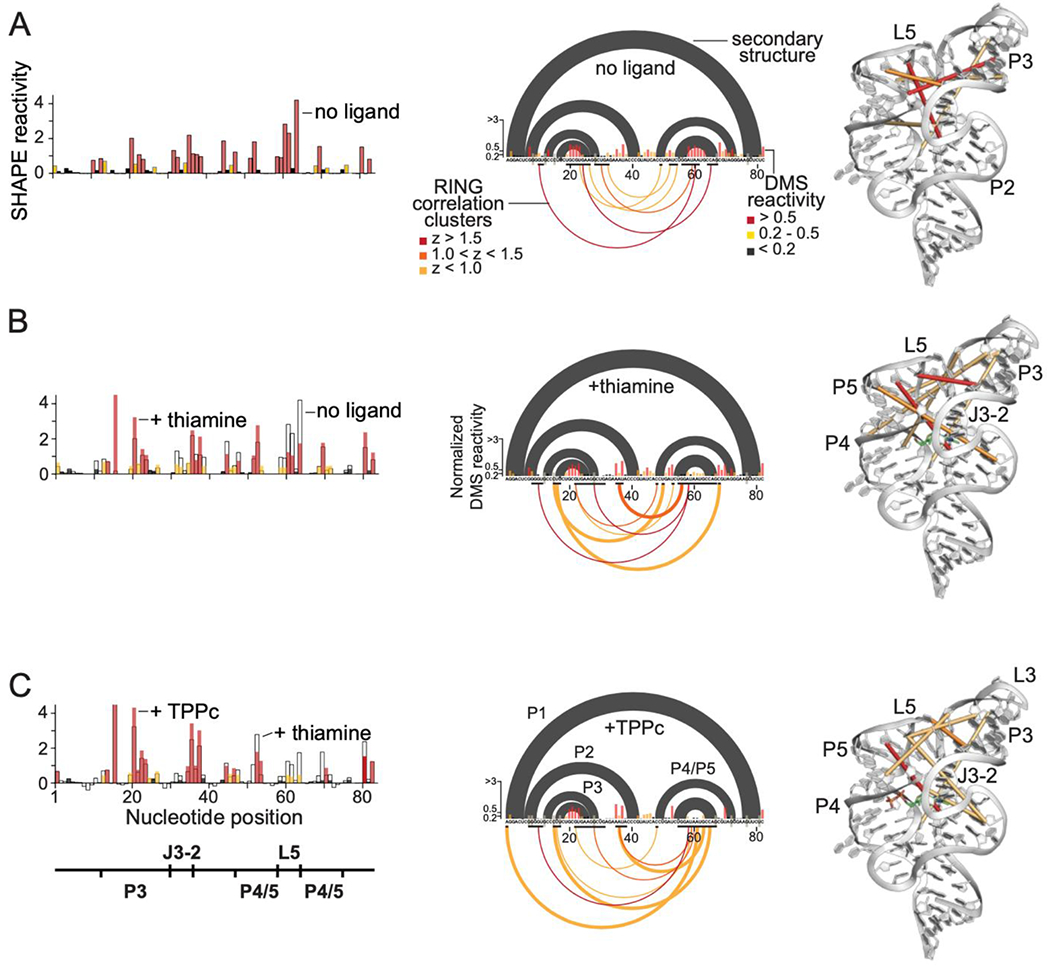
Solution probing of secondary and tertiary structures of the TPP riboswitch bound to thiamine and to TPPc. Left to right: SHAPE reactivity profiles, RING-MaP data, and RING correlation cluster centroids plotted on the three-dimensional TPP riboswitch structure for (A) TPP riboswitch RNA alone, (B) thiamine-bound riboswitch, and (C) TPPc-bound riboswitch. SHAPE reactivities for each complex are labeled. For RING-MaP data, accepted secondary structure (black arcs) and DMS reactivities (histograms) are shown above the x-axis. Below the axis, black bars denote nucleotides comprising a cluster; arcs link the centroid nucleotides of a given cluster, colored by z-score.
Changes in the through-space interactions in the riboswitch RNA upon binding thiamine or TPPc were further examined using single-molecule, correlated chemical probing (RING-MaP)39,40. We employed RING-MaP, using dimethyl sulfate (DMS), to measure correlations between nucleotides co-modified in the same RNA strand and thereby to evaluate through-space structural communication as mediated by each ligand. RING data were visualized as cluster centroids for nucleotides with correlated RING reactivities adjacent in the primary sequence. In the absence of ligand, there is pre-existing structural communication involving the L5, P2, and P3 structural elements of the riboswitch, consistent with partial formation of the long-range tertiary interaction involving L5 and P3 (Fig. 4A, center and right; Fig. S4). Upon addition of thiamine, both the number and complexity of through-space interactions increase. New interactions between the J3/2 and L5 regions form, and interactions between the P3 and P4/P5 helices increase (Fig. 4B, center and right). In the presence of the linked ligand, TPPc, a dense network of through-space interactions was maintained but, critically, a subset of the through-space interactions changed. When bound to TPPc, the L5 loop formed a nexus for many of the strongest interactions (Fig. 4C, center and right). Nearly all the correlations in the presence of TPPc were between L5 and other regions, especially J3/2 and P3. The RING-MaP studies thus suggest that the bisphosphonate moiety of the TPPc ligand anchors interactions that bring the two arms of the TPP riboswitch together (Fig. 1B).
Notably, there were not simply more RING interactions in the presence of TPPc than in the presence of thiamine. Instead, both single molecule RING data (Fig. 4) and crystallographic analysis (Fig. 3) reveal substantial differences in local tertiary structure, implying that a structural rearrangement is required to realize the additional binding energy afforded by MDP.
Discussion
The TPP riboswitch forms a simple and common RNA structure based on a three-helix junction, is relatively unstructured as a free RNA, binds its canonical ligand via two sub-sites, and undergoes a large structural change upon ligand binding. This riboswitch is thus representative of RNA motifs that might be targeted by small molecules generally. We examined the effects of fragment linking on affinity for the TPP riboswitch by evaluating binding by low molecular mass fragments, thiamine and MDP, and their linked conjugate, TPPc.
Thiamine and MDP bound with affinities of 11 and 1,200 μM, respectively. Linking these compounds yielded TPPc, which bound with 19 nM affinity, corresponding to a 600-fold increase in affinity over thiamine. Thiamine contributes −6.8 kcal/mol of favorable interactions, and MDP contributes −4.0 kcal/mol of interaction energy. Linking these compounds produced a compound with −11 kcal/mol of favorable binding interaction energy, almost exactly equal to the sum of the individual binding energies (Table 1). Thus, we observed only a small cooperative effect over that expected based on affinities of the individual moieties. Linking the thiamine and MDP fragments to form the TPPc ligand, which binds in an active site optimized by evolution for gene regulation in bacteria, is merely additive rather than super-additive.
Our findings are broadly supportive of fragment-linking strategies as applied to RNA targets. First, we created a high-affinity ligand by linking thiamine and MDP fragments, each of which has only modest affinity. The ligand efficiency for TPPc, a measure of the quality of interactions formed between a ligand and macromolecule (calculated as the ∆GL divided by the number of non-hydrogen atoms) is 0.40, substantially exceeding the value of 0.3 that is usually taken as the lower bound for an atom-efficient interaction41 (Fig. 5A).
Figure 5.
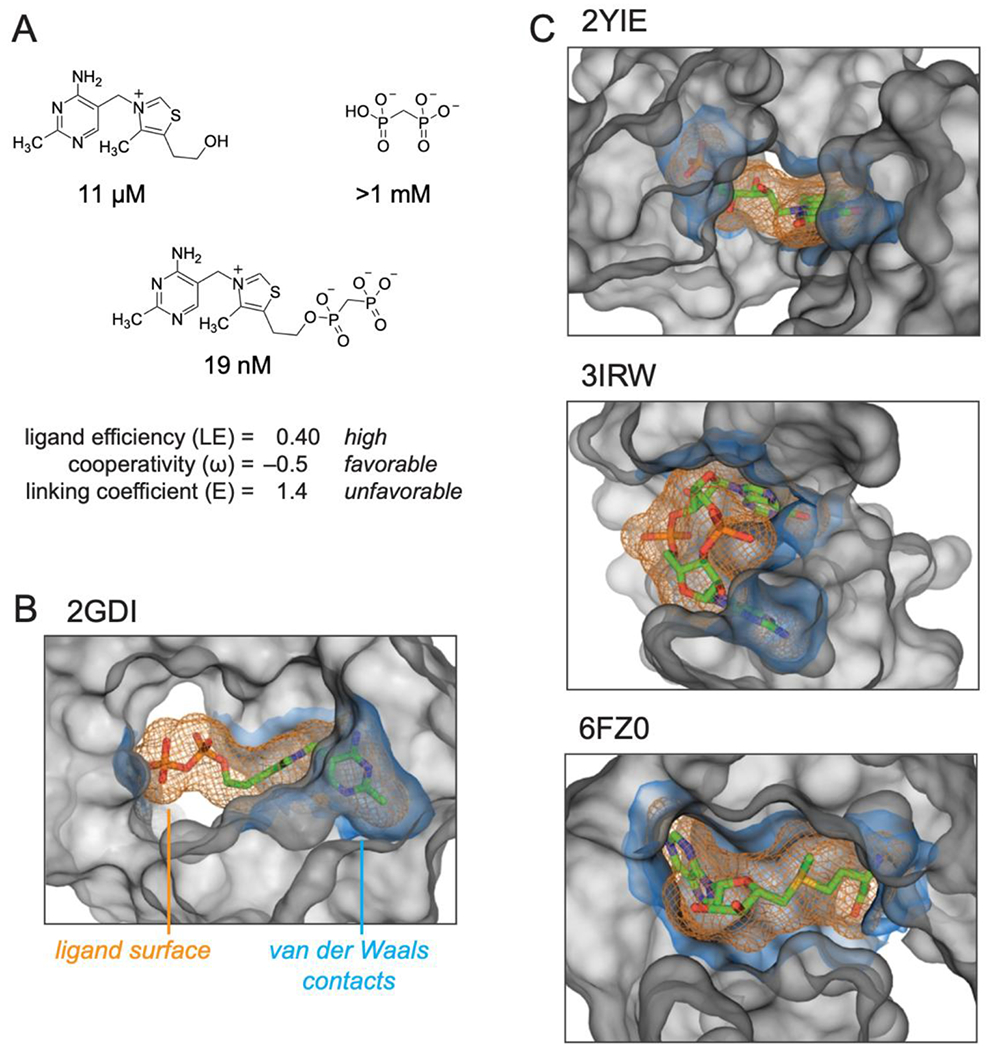
Summary of the structures and properties of thiamine and MDP fragments and the linked compound, TPPc, and comparison with representative RNA structures that interact with their ligands using well-defined sub-sites. (A) TPPc features. Ligand efficiency (LE) is equal to the binding energy per non-hydrogen ligand atom, LE = ΔG / N; ligand cooperativity, ω, is defined in Table 1; linking coefficient (E) was calculated as E = KL / (KT KP). (B) TPP riboswitch binding pocket. (C) Representative examples of riboswitch ligands that bind in defined sub-sties: FMN43, cyclic di-GMP44, and SAM-V riboswitches45. Ligands are shown as sticks, orange mesh shows ligand molecular envelope. RNA atoms within 6 Å of ligand are shown as a grey surface that delineates the topography of the binding pocket. The RNA surface within 2 Å of the ligand, equal to the van der-Waals contact distance57, is shown in blue. VDW surfaces were calculated using HOLLOW57 and were visualized in PyMOL (Schrodinger, LLC). PDB IDs for each structure are shown.
Second, the sub-site pockets are individually optimizable. A plurality of binding interactions originates from the thiamine moiety and variation of this group afforded derivatives with affinities ranging from 6 to 800 μM. Regardless of affinity, fragment-like groups as small as THG and as large as pyrithiamine bound to the TPP riboswitch with modest cooperativity with MDP (ω ≈ 0.5; Table 1). Binding in the pyrophosphate sub-site could be improved by the substitution of a single oxygen atom for a methylene group, based on this work and a prior study35. Thus, structure-activity relationships can be explored independently for ligands that bind to adjacent RNA sub-sites.
Third, a high-affinity molecule can be created by linking of two fragments even if linking itself does not provide a substantial binding enhancement. The linking coefficient, E, is a measure of the degree to which a multivalent ligand system exhibits cooperativity when covalently linked8. A linking coefficient of 1.0 implies that the covalent linker neither hinders nor helps the binding of the linked molecule, whereas values less and greater than 1.0 imply cooperativity through covalent linkage versus thermodynamic destabilization, respectively. Linking coefficients vary by orders of magnitude in protein systems3,8 and cooperative binding by relatively complex oligonucleotide25,26 and multivalent27 ligands is well established for RNA. The linking coefficient for TPPc is 1.4 (Fig. 5A), indicating that conjugation of thiamine and MDP is moderately detrimental and that the majority of the positive cooperativity (ω ≈ 0.5) originates from the binding of (and conformational changes induced by) the fragments themselves. Crystal structures (Fig. 3) and single-molecule RING data (Fig. 4) reveal that (energetically unfavorable) RNA structural rearrangements occur upon binding of TPPc relative to binding of the individual fragments. The thiazole and hydroxyethyl groups of the thiamine moiety bind the RNA with different orientations upon conjugation with MDP, likely contributing to the unfavorable linking coefficient. Indeed, favorable overall energetic effects are achieved with an apparently non-ideal covalent linkage.
Features of the TPP-binding ligand pocket are conserved in other RNA-ligand interactions. The thiamine and MDP RNA-interacting groups are joined by a linker region with no or few contacts between the linker atoms and the RNA (Fig. 5B). The linker lies in a solvent accessible hole, a feature consistent with the small effect of linking the fragments on overall binding affinity. Similarly, in the FMN riboswitch, the three-ring isoalloxazine moiety and the phosphate moiety bind to distinct sub-sites42,43. These sites are separated by a large solvent accessible channel (Fig. 5C, top). The cyclic-di GMP riboswitch also has a two sub-site architecture; in this case, the two guanosine-binding sites face the solvent accessible exterior of the RNA44. In the SAM-V riboswitch, RNA sub-sites interact with the adenosyl moiety and with the distal end of the methionine (Fig. 5C, center and bottom)45. The ribose linker between these groups is substantially exposed to solvent. In each of these four complexes, and presumably in yet-discovered examples, the ligand is comprised of two fragment-like entities connected by a short linker that spans a solvent exposed region of the RNA.
In sum, we have examined sub-site ligand binding in the TPP bacterial riboswitch system and found that high affinity binding to TPPc is achieved without highly cooperative or super-additive interactions between sub-sites. The region linking the two fragments of TPP lies in a solvent-accessible hole in the RNA, and the linker makes few contacts with the RNA. All of these features appear to bode well for fragment-based ligand discovery strategies for RNA.
Experimental
Compounds
Small-molecule compounds were obtained from Millipore-Sigma and were used without further purification. Thiamine bisphosphonate 5, denoted here as TPPc, was synthesized as reported46 (Fig. S5). Briefly, thiamine propyl disulfide 1 was coupled with mono-deprotected diphosphoric acid 247, followed by treatment with triphenylphosphine to obtain benzyl-protected thiamine bisphosphonate 4. The debenzylation of 4 with TMSBr afforded the desired thiamine bisphosphonate 5. The 1H and 13C NMR spectra of 5 matched reported values46.
RNA preparation
The single-stranded DNAs (Integrated DNA Technologies) encoding the T7 promoter and E. coli thiM TPP riboswitch used for ITC experiments had the following sequence: 5′-GAAAT TAATA CGACT CACTA TAGGC AGTA CTCG GGGTG CCCTT CTGCG TGAAG GCTGA GAAAT ACCCG TATCA CCTGA TCTGG ATAAT GCCAG CGTAG GGAAG TGCT G-3′; primer binding sites are underlined. For synthesis of the template for in vitro transcription for SHAPE and RING probing, the sequence included the T7 promoter, the TPP riboswitch, and flanking structure cassettes48: 5′-GAAAT TAATA CGACT CACTA TAGGC CTTCG GGCCA AGGAC TCGGG GTGCC CTTCT GCGTG AAGGC TGAGA AATAC CCGTA TCACC TGATC TGGAT AATGC CAGCG TAGGG AAGTT CTCGA TCCGG TTCGC CGGAT CCAAA TCGGG CTTCG GTCCG GTTC-3′; primer binding sites are underlined. DNA was amplified by PCR (Q5 hot-start high-fidelity polymerase; NEB) to create templates for in vitro transcription. In vitro transcription was carried out with 5 mM NTPs (New England Biolabs), 300-800 nM DNA template, 0.02 U/μL yeast inorganic pyrophosphatase (New England Biolabs), 0.05 mg/mL T7 polymerase in 25 mM MgCl2, 40 mM Tris-HCl, pH 8.0, 2.5 mM spermidine, 0.01% Triton, 10 mM DTT. Typical reaction volumes were 10 mL. Reactions were incubated at 37 °C for 4 h, then Turbo DNase (RNase-free, Invitrogen) was added to a final concentration of 0.04 U/μL and incubated at 37 °C for 30 min, followed by a second DNase addition to a total final concentration of 0.08 U/μL with an additional 30-min incubation; enzymatic reactions were halted by the addition of EDTA to a final concentration of 50 mM and placed on ice. RNA was extracted by phenol-chloroform-isoamyl alcohol extraction (buffered to pH 6.7 with 1 M Tris). The RNA was then exchanged into 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, via centrifugal concentration (Amicon Ultra centrifugal filters, 10K MWCO, Millipore-Sigma), and stored at −20 °C.
Isothermal titration calorimetry
ITC experiments were carried out (Microcal PEAQ-ITC automated instrument, Malvern Analytical) under RNase-free conditions49. In vitro transcribed RNA was exchanged into folding buffer containing 100 mM HEPES-Na, pH 8.0, 200 mM potassium acetate, and 1 mM MgCl2 using centrifugal concentration (Amicon Ultra centrifugal filters, 10K MWCO, Millipore-Sigma). Ligands were dissolved in folding buffer (to minimize heat of mixing upon addition of ligand to RNA) at a concentration equal to 10-20 times the experimental concentration of RNA. The RNA concentration was quantified (Nanodrop UV-VIS spectrometer, ThermoFisher Scientific), diluted to approximately 1-10 times the expected Kd in buffer, and the diluted RNA was quantified to establish the final experimental concentration. The RNA was heated at 65 °C for 5 min, placed on ice for 5 min, and allowed to fold at 37 °C for 15 min. For cooperative binding experiments, thiamine or a thiamine analogue was pre-bound to the RNA by adding 0.1 volume of 10 times the desired final concentration of the bound ligand, followed by incubation at room temperature for 10 min. RNA and ligand concentrations and c-values are provided in Table S2.
Each experiment involved two ITC runs; one in which the ligand was titrated into RNA (the experimental trace) and one in which the same ligand was titrated into buffer (the control trace). ITC experiments were performed using the following parameters: 25 °C cell temperature, 8 μCal/sec reference power, 750 RPM stirring speed, high feedback mode, 0.2 μL initial injection, and 180 s spacing between injections. The number of injections and volume pre-injection varied based on whether the ligand was a tight (<500 μM) or weak (>500 μM) binder. Tight-binding ligands were titrated using 20 injections of 2 μL each over 4 s; weak binding ligands were titrated using 60 injections of 0.6 μL, each over 1.2 s.
Extraction of RNA-ligand binding parameters from ITC data
ITC data were analyzed (MicroCal PEAQ-ITC Analysis Software; Malvern Analytical) by adjusting the baseline for each injection peak manually to resolve any incorrectly picked injection endpoints, subtracting the control trace from the experimental trace using point-to-point subtraction, and fitting a least-squares regression line to the data using the Levenberg-Marquardt algorithm.
Considerations for ITC analysis of weakly binding ligands
In the case of weakly binding ligands (>500 μM), the limitations of working at low c-values50 were specifically mitigated by the following: (i) curves were required to reach full receptor saturation, (ii) the control trace was subtracted from the experimental trace using point-to-point subtraction, (iii) N was manually fixed to 1.0, and (iv) experimental replicates were obtained to assess replicability. These procedures were sufficient to allow comparison between experimental conditions and to determine whether low-affinity binders in a series are increasing or decreasing in affinity due to cooperative effects.
Cooperativity and linking parameters
The ω value is a measure of the cooperativity observed for non-linked fragments, A and B, and quantifies the additional binding affinity conferred by having a primary fragment (A) pre-bound. ω is calculated as: ω = KA+B / KB. The corresponding Gibbs free energy of cooperativity (ΔGω) is calculated as ΔGω = −RT ln ω. E is the affinity of the linked compound (L) relative to coupled binding by the constituent ligands (A and B). If the binding energies of the two fragments are exactly additive (no cooperativity), E equals 1. E is calculated as: E = KL / KA • KB. Ligand efficiency (LE) is a measure of the binding energy per non-hydrogen atom of a ligand to its binding partner and is calculated as LE = ΔG / N. For thermodynamic parameters of ligand binding, enthalpy (ΔH) was obtained experimentally by ITC, Gibbs free energy (ΔG) was calculated as ΔG = −RT ln K, and entropy (−TΔS) was calculated as −TΔS = ΔG – ΔH.
SHAPE and RING chemical probing
For SHAPE chemical probing, 5 pmol of RNA were diluted to 19.6 μL in RNase-free water at 4 °C. The RNA was heated at 95 °C for 2 min, and immediately cooled at 4 °C for 5 min. To the RNA was added 19.6 μL of 2× SHAPE buffer (final concentrations 50 mM HEPES-Na, pH 8.0, 200 mM potassium acetate, and 10 mM MgCl2), and the sample was incubated at 37 °C for 30 min. For the cooperative binding experiments, 24.3 μL of folded RNA were added to 2.7 μL of primary binding ligand in 1× SHAPE buffer to a final concentration of 10× the Kd for the ligand, and incubated at 37 °C. After 10 min, 24.3 μL of folded RNA was added to 2.7 μL of 10× ligand (in 1× SHAPE buffer to yield a final ligand concentration of 10× the Kd of the ligand). Solutions were mixed by pipetting and incubated for 10 min at 37 °C. A 22.5-μL aliquot of this solution was added to 2.5 μL of 10× SHAPE reagent (5-nitroisatoic anhydride, final concentration 25 mM51 in DMSO at 37 °C) and rapidly mixed by pipetting to achieve homogenous distribution of the SHAPE reagent. The SHAPE reagent was allowed to react for 15 min, and then the sample was placed on ice. Excess ligand, solvent, and hydrolyzed SHAPE reagent were removed (G-50 columns, GE Healthcare Life Sciences).
For RING-MaP experiments, 5 pmol of RNA was diluted to 9 μL in RNase-free water at 4 °C. The RNA was heated at 95 °C for 2 min, followed by cooling at 4 °C for 5 min. To the RNA was added 9 μL of 2× RING buffer (final concentrations 200 mM bicine, pH 8.0, 200 mM KOAc, 10 mM MgCl2), and the sample was incubated at 37 °C for 30 min. For the cooperative binding experiments, 18 μL of folded RNA were added to 2 μL of primary binding ligand in 1× RING buffer to a final concentration of 10× the Kd of the ligand, and the sample was incubated at 37 °C for 10 min. An 18-μL aliquot of folded RNA was added to 2 μL of 10× ligand (in 1× RING buffer to yield a final ligand concentration of 10× the Kd of the ligand). An 18-μL aliquot of the RNA-ligand sample was then added to 2 μL of DMS solution (1.7 M DMS in EtOH). After 6 min at 37 °C, the reaction was quenched by addition of 20 μL of ice cold 20% 2-mercaptoethanol solution and incubation at 4 °C for 3 min. A no-reagent control RNA was prepared identically, substituting neat EtOH for the DMS solution. Reactions were precipitated with isopropanol, followed by magnetic bead purification (Mag-Bind TotalPure NGS beads, Omega). RNA concentrations were determined (Qubit RNA HS Assay, Invitrogen).
Library preparation for massively parallel sequencing
RNA from SHAPE and RING probing experiments was first subjected to reverse transcription. To 5 μL modified RNA and 2 μL dNTP mix (10 mM each, New England Biolabs) was added 1 μL reverse transcription primer for a final concentration of 333 nM primer and 2.5 mM dNTPs. After incubation at 68 °C for 5 min, the sample was and placed on ice for 2 min. To this solution, 2 μL 10× NTP minus buffer (500 mM Tris-HCl, pH 8.0, 750 mM KCl, 100 mM DTT), 4 μL 5 M Betaine (Millipore Sigma), and 3 μL 40 mM MnCl2 were added and incubated at 25 °C for 2 min before adding 1 μL SuperScript II Reverse Transcriptase (Invitrogen). The reaction was incubated at 25 °C for 10 min to equilibrate, followed by extension at 42 °C for 90 min, then 10 cycles of 50 °C for 2 min and 42 °C for 2 min, and finally a 70 °C heat inactivation for 10 min before being placed on ice. The resulting cDNA product was purified (Agencourt RNAclean magnetic beads; Beckman Coulter), eluted into RNase-free water, and stored at −20 °C. The sequence of the reverse transcription primer was 5′-CGGGC TTCGG TCCGG TTC-3′.
DNA libraries were prepared for sequencing using sequential PCR reactions to amplify the DNA and add the necessary TruSeq adapters52. Forward and reverse SHAPE-MaP amplicon-specific primers for library preparation were 5′-CCCTA CACGA CGCTC TTCCG ATCTN NNNNG GCCTT CGGGC CAAGG A-3′ and 5′-GACTG GAGTT CAGAC GTGTG CTCTT CCGAT CTNNN NNTTG AACCG GACCG AAGCC CGATT T-3′, respectively; sequences overlapping the TPP riboswitch sequences are underlined. DNA was amplified by PCR using 200 μM dNTP mix (New England Biolabs), 500 nM forward primer, 500 nM reverse primer, 1 ng cDNA or double-stranded DNA template, 20% (v/v) Q5 reaction buffer (New England Biolabs), and 0.02 U/μL Q5 hot-start high-fidelity polymerase (New England Biolabs). Excess unincorporated dNTPs and primers were removed by affinity purification (Agencourt AmpureXP magnetic beads; Beckman Coulter; at a 0.7:1 sample to bead ratio). DNA libraries were quantified (Qubit dsDNA High Sensitivity assay kit, Invitrogen), checked for quality (Bioanalyzer 2100 on-chip electrophoresis instrument, Agilent), and sequenced (Illumina MiSeq high-throughput sequencer) to an average depth of 100,000 reads per sample.
SHAPE-MaP and RING-MaP data analysis
Following massively parallel sequencing, data were analyzed using the most recent versions of ShapeMapper53 to obtain SHAPE and DMS reactivity profiles and RingMapper40 to obtain RING correlations. RING correlation clusters were calculated by (i) removing correlations within a 20-nucleotide contact distance to select for tertiary structure correlations, (ii) creating clusters of all correlations within 2 nucleotides of one another (in both 5′ and 3′ directions) and within the same z-score category (<1, 1-5, or >5), and (iii) computing the median 5′ and 3′ correlation start sites as well as the mean z-score of all correlations within a cluster. Clusters were visualized as the 5′ and 3′ centroid nucleotides of all correlations within that cluster and the mean z-score.
X-ray crystallography
All complexes were crystallized using an RNA construct described previously16. To form complexes, RNA (0.15 mM) was incubated in a buffer containing 5 mM Tris-HCl, pH 8.0, 3 mM MgCl2, 10 mM NaCl, 0.1 M KCl, and 0.5 mM spermine with 0.5 mM of TPP, 0.75 mM TPPc, or 1 mM thiamine at 37 °C for 30 min and at 4 °C for 60 min prior to crystallization. For crystallization, 1.5 μL of the RNA-ligand complex was mixed with 0.75 μL of reservoir solution. For TPP and thiamine, the reservoir solution was 50 mM Bis-Tris HCl, pH 6.5, 0.5 M NH4Cl, 10 mm MnCl2, and 30% (v/v) PEG2000. For TPPc with Mn2+, the reservoir solution was 50 mM sodium cacodylate, pH 6.0, 0.35 M NH4Cl, 10 mm MnCl2, and 30% (v/v) PEG2000. For TPPc with Ca2+, reservoir solution was 50 mM sodium cacodylate, pH 6.0, 0.3 M NH4Cl, 10 mm CaCl2, and 30% (v/v) PEG2000. Crystallization was performed at 291K by hanging drop vapor diffusion. Rod-shaped crystals grew in 1 week. The crystals were cryoprotected in the reservoir solution supplemented with 15% glycerol and ligands at the concentration used for preparing complexes. Crystals were flash frozen by dipping into liquid nitrogen. Data for TPP- and thiamine-bound structures were collected at the 17-ID-2 beamline at NSLS-II (Brookhaven National Laboratory, 0.9793 Å wavelength). Data for TPPc were collected at the 24-ID-C beamline at Advanced Photon Source (Argonne National Laboratory, 0.9791 Å wavelength). Data were processed with HKL2000 (HKL Research) or XDS54. The structures were solved by molecular replacement using Phenix55 and the 2HOJ riboswitch RNA structure31 as a search model. Structures were refined in Phenix. Organic ligands, water molecules and ions were added at the late stages of refinement based on Fo-Fc and 2Fo-Fc and simulated annealing omit electron density maps. We specifically validated our models for conformations of the thiazolium ring of TPPc and TPP bound to RNA. Since we were not able to collect data of sufficient quality to observe the anomalous signal for the sulfur atom of TPP or TPPc, we based our refinement on the observable, larger size of this atom. Simultaneous refinement of two conformations for the TPP complex produces high (63 vs 37%) occupancy for the upward conformer of the ligand. In the TPPc complex, the experimental density map is well defined for the linker and strongly supports a predominant downward conformation; refinement with both TPPc conformations yielded downward conformation as major (68 vs 32% occupancy). Molecular interfaces were calculated by the PISA (European Bioinformatics Institute) service56,57.
Supplementary Material
Acknowledgements
This work was supported by NIH grants to K.M.W. (R01 AI068462 and R01-EUREKA GM098662) and A.S. (R01 GM112940). K.L. was supported by the UNC Lineberger Comprehensive Cancer Center. ITC experiments were performed at the UNC Macromolecular Interactions Facility (NCI P30CA016086). This work used NE-CAT beamlines (GM124165), a Pilatus detector (RR029205), an Eiger detector (OD021527) at the Advanced Photon Source (DE-AC02-06CH11357) of the Argonne National Laboratory; and FMX (17ID-2) beamline at NSLS-II (Brookhaven National Laboratory), supported by the NIH NIGMS (1P30GM133893) and BER-BO 070. NSLS-II is supported by DOE, BES-FWP-PS001.
Footnotes
Accession codes
PDB IDs for the TPP riboswitch structures with bound thiamine (with Mn2+ ion), TPP (Mn2+), TPPc (Mn2+), and TPPc (Ca2+) are 7TD7, 7TDA, 7TDB and 7TDC, respectively.
Supporting Information Available. Tables summarizing crystallographic and ITC experiments; and five figures of data on ITC, crystallography, SHAPE, and single molecule chemical probing experiments, and on TPPc synthesis.
Disclosure
K.M.W. is an advisor to and holds equity in Ribometrix.
References
- (1).Jencks WP On the Attribution and Additivity of Binding Energies. Proc. Natl. Acad. Sci 1981, 78 (7), 4046–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Olejniczak ET; Hajduk PJ; Marcotte PA; Nettesheim DG; Meadows RP; Edalji R; Holzman TF; Fesik SW Stromelysin Inhibitors Designed from Weakly Bound Fragments: Effects of Linking and Cooperativity. J. Am. Chem. Soc 1997, 119 (25), 5828–5832. [Google Scholar]
- (3).Murray CW; Verdonk ML The Consequences of Translational and Rotational Entropy Lost by Small Molecules on Binding to Proteins. J. Comput. Aided. Mol. Des 2002, 16 (10), 741–753. [DOI] [PubMed] [Google Scholar]
- (4).Hunter CA; Anderson HL What Is Cooperativity? Angew. Chemie - Int. Ed 2009, 48 (41), 7488–7499. [DOI] [PubMed] [Google Scholar]
- (5).Velvadapu V; Farmer BT; Reitz AB Fragment-Based Drug Discovery. The Practice of Medicinal Chemistry; 2015; 161–180. [Google Scholar]
- (6).Doak BC; Norton RS; Scanlon MJ The Ways and Means of Fragment-Based Drug Design. Pharmacol. Ther 2016, 167, 28–37. [DOI] [PubMed] [Google Scholar]
- (7).Hajduk PJ; Sheppard G; Nettesheim DG; Olejniczak ET; Shuker SB; Meadows RP; Steinman DH; Carrera GM; Marcotte PA; Severin J; et al. Discovery of Potent Nonpeptide Inhibitors of Stromelysin Using SAR by NMR. J. Am. Chem. Soc 1997, 119 (25), 5818–5827. [Google Scholar]
- (8).Ichihara O; Barker J; Law RJ; Whittaker M Compound Design by Fragment-Linking. Mol. Inform 2011, 30 (4), 298–306. [DOI] [PubMed] [Google Scholar]
- (9).Hermann T Small Molecules Targeting Viral RNA. WIREs. RNA 2016, 7 (6), 726–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Morgan BS; Forte JE; Culver RN; Zhang Y; Hargrove AE Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-Targeted Bioactive Ligands. Angew. Chemie - Int. Ed 2017, 56 (43), 13498–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rizvi NF; Smith GF RNA as a Small Molecule Druggable Target. Bioorganic Med. Chem. Lett 2017, 27 (23), 5083–5088. [DOI] [PubMed] [Google Scholar]
- (12).Warner KD; Hajdin CE; Weeks KM Principles for Targeting RNA with Drug-like Small Molecules. Nat. Rev. Drug Discov 2018, 17 (8), 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen L; Cressina E; Leeper FJ; Smith AG; Abell C A Fragment-Based Approach to Identifying Ligands for Riboswitches. ACS Chem. Biol 2010, 5 (4), 355–358. [DOI] [PubMed] [Google Scholar]
- (14).Cressina E; Chen L; Abell C; Leeper FJ; Smith AG Fragment Screening against the Thiamine Pyrophosphate Riboswitch ThiM. Chem. Sci 2011, 2 (1), 157–165. [Google Scholar]
- (15).Moumné R; Catala M; Larue V; Micouin L; Tisné C Fragment-Based Design of Small RNA Binders: Promising Developments and Contribution of NMR. Biochimie 2012, [DOI] [PubMed] [Google Scholar]
- (16).Warner KD; Homan P; Weeks KM; Smith AG; Abell C; Ferré-D’Amaré AR Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically. Chem. Biol 2014, 21, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zeiger M; Stark S; Kalden E; Ackermann B; Ferner J; Scheffer U; Shoja-Bazargani F; Erdel V; Schwalbe H; Göbel MW Fragment Based Search for Small Molecule Inhibitors of HIV-1 Tat-TAR. Bioorganic Med. Chem. Lett 2014, 24 (24), 5576–5580. [DOI] [PubMed] [Google Scholar]
- (18).Garavís M; López-Méndez B; Somoza A; Oyarzabal J; Dalvit C; Villasante A; Campos-Olivas R; González C Discovery of Selective Ligands for Telomeric RNA G-Quadruplexes (TERRA) through 19F-NMR Based Fragment Screening. ACS Chem. Biol 2014, 9 (7), 1559–1566. [DOI] [PubMed] [Google Scholar]
- (19).Bottini A; De SK; Wu B; Tang C; Varani G; Pellecchia M Targeting Influenza A Virus RNA Promoter. Chem. Biol. Drug Des 2015, 86 (4), 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Tran B; Pichling P; Tenney L; Connelly CM; Moon MH; Ferré-D’Amaré AR; Schneekloth JS Jr; Jones CP Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design. Cell Chem Biol. 2020, 27 (10), 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Suresh BM; Li W; Zhang P; Wang KW; Yildirim I; Parker CG; Disney MD General Fragment-Based Approach to Identify and Optimize Bioactive Ligands Targeting RNA. Proc. Natl. Acad. Sci 2020, 117 (52), 33197–33203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sreeramulu S; Richter C; Berg H; Wirtz Martin MA; Ceylan B; Matzel T; Adam J; Altincekic N; Azzaoui K; Bains JK; et al. Exploring the Druggability of Conserved RNA Regulatory Elements in the SARS-CoV-2 Genome. Angew. Chemie-Int. Ed 2021, 60 (35), 19191–19200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Williamson JR Cooperativity in Macromolecular Assembly. Nat. Chem. Biol 2008, 4 (8), 458–465. [DOI] [PubMed] [Google Scholar]
- (24).Peselis A; Gao A; Serganov A Cooperativity, Allostery and Synergism in Ligand Binding to Riboswitches. Biochimie 2015, 117, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bevilacqua PC; Johnson KA; Turner DH Cooperative and Anticooperative Binding to a Ribozyme. Proc. Natl. Acad. Sci 1993, 90 (18), 8357–8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Karbstein K; Carroll KS; Herschlag D Probing the Tetrahymena Group I Ribozyme Reaction in Both Directions. Biochemistry 2002, 41 (37), 11171–11183. [DOI] [PubMed] [Google Scholar]
- (27).Ursu MA; Childs-Disney JL; Andrews RJ; O’Leary CA; Meyera SM; Angelbello AJ; Moss WN; Disney MD Design of Small Molecules Targeting RNA Structure from Sequence. Chem. Soc. Rev 2020, 49 (20), 7252–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Shuker SB; Hajduk PJ; Meadows RP; Fesik SW Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science 1996, 274 (5292), 1531–1534. [DOI] [PubMed] [Google Scholar]
- (29).Mccown PJ; Corbino KA; Stav S; Sherlock ME; Breaker RR Riboswitch Diversity and Distribution. RNA 2017, 23 (7), 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Steen K; Rice G; Weeks K Fingerprinting Noncanonical and Tertiary RNA Structures by Differential SHAPE Reactivity. J Am Chem Soc. 2012, 134 (32), 13160–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Serganov A; Polonskaia A; Phan AT; Breaker RR; Patel DJ Structural Basis for Gene Regulation by a Thiamine Pyrophosphate-Sensing Riboswitch. Nature 2006, 441, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Edwards TE; Ferré-D’Amaré AR Crystal Structures of the Thi-Box Riboswitch Bound to Thiamine Pyrophosphate Analogs Reveal Adaptive RNA-Small Molecule Recognition. Structure 2006, 14 (9), 1459–1468. [DOI] [PubMed] [Google Scholar]
- (33).Lang K; Rieder R; Micura R Ligand-Induced Folding of the ThiM TPP Riboswitch Investigated by a Structure-Based Fluorescence Spectroscopic Approach. Nucleic Acids Res. 2007, 35 (16), 5370–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kulshina N; Edwards TE; Ferré-D’Amaré AR Thermodynamic Analysis of Ligand Binding and Ligand Binding-Induced Tertiary Structure Formation by the Thiamine Pyrophosphate Riboswitch. RNA 2010, 16, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chen L; Cressina E; Dixon N; Erixon K; Agyei-Owusu K; Micklefield J; Smith AG; Abell C; Leeper FJ Probing Riboswitch–Ligand Interactions Using Thiamine Pyrophosphate Analogues. Org. Biomol. Chem 2012, 10 (30), 5924–5931. [DOI] [PubMed] [Google Scholar]
- (36).Blackburn GM; England DA; Kolkmann F Monofluoro- and Difluoro-Methylenebisphosphonic Acids: Isopolar Analogues of Pyrophosphoric Acid. J. Chem. Soc. Chem. Commun 1981, No. 17, 930–932. [Google Scholar]
- (37).Wang B; Wilkinson K; Weeks KM Complex Ligand-Induced Conformational Changes in TRNA. Biochemistry 2008, 3454–3461. [DOI] [PubMed] [Google Scholar]
- (38).Sztuba-Solinska J; Shenoy SR; Gareiss P; Krumpe LRH; Le Grice SFJ; O’Keefe BR; Schneekloth JS Identification of Biologically Active, HIV TAR RNA-Binding Small Molecules Using Small Molecule Microarrays. J. Am. Chem. Soc 2014, 136, 8402–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Homan PJ; Favorov OV; Lavender CA; Kursun O; Ge X; Busan S; Dokholyan NV; Weeks KM Single-Molecule Correlated Chemical Probing of RNA. Proc. Natl. Acad. Sci 2014, 111 (38), 13858–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mustoe AM; Lama N; Irving PS; Olson SW; Weeks KM RNA Base Pairing Complexity in Living Cells Visualized by Correlated Chemical Probing. Proc. Natl. Acad. Sci 2019, 116 (49), 24574–24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Schultes S; De Graaf C; Haaksma EEJ; De Esch IJP; Leurs R; Krämer O Ligand Efficiency as a Guide in Fragment Hit Selection and Optimization. Drug Discov. Today Technol 2010, 7 (3), 157–162. [DOI] [PubMed] [Google Scholar]
- (42).Serganov A; Huang L; Patel DJ Coenzyme Recognition and Gene Regulation by a Flavin Mononucleotide Riboswitch. Nature 2009, 458 (7235), 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Vicens Q; Mondragón E; Batey RT Molecular Sensing by the Aptamer Domain of the FMN Riboswitch: A General Model for Ligand Binding by Conformational Selection. Nucleic Acids Res. 2011, 39 (19), 8586–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Smith KD; Lipchock SV; Ames TD; Wang J; Breaker RR; Strobel SA Structural Basis of Ligand Binding by a C-Di-GMP Riboswitch. Nat. Struct. Mol. Biol 2009, 16 (12), 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Huang L; Lilley DMJ Structure and Ligand Binding of the SAM-V Riboswitch. Nucleic Acids Res. 2018, 46 (13), 6869–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Klein E; Nghiêm HO; Valleix A; Mioskowski C; Lebeau L Synthesis of Stable Analogues of Thiamine Di- and Triphosphate as Tools for Probing a New Phosphorylation Pathway. Chem. Eur. J 2002, 8 (20), 4649–4655. [DOI] [PubMed] [Google Scholar]
- (47).Saady M; Lebeau L; Mioskowski C Selective Monodeprotection of Phosphate, Phosphite, Phosphonate, and Phosphoramide Benzyl Esters. J. Org. Chem 1995, 60 (9), 2946–2947. [Google Scholar]
- (48).Merino EJ; Wilkinson KA; Coughlan JL; Weeks KM RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc 2005, 127, 4223–4231. [DOI] [PubMed] [Google Scholar]
- (49).Gilbert SD; Batey RT Monitoring RNA-Ligand Interactions Using Isothermal Titration Calorimetry. Methods Mol. Biol 2009, 540, 97–114. [DOI] [PubMed] [Google Scholar]
- (50).Turnbull WB Divided We Fall? Studying Low Affinity Fragments of Ligands by ITC. Microcal Application Notes. 2005. [Google Scholar]
- (51).Busan S; Weidmann CA; Sengupta A; Weeks KM Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry 2019, 58, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Smola MJ; Rice GM; Busan S; Siegfried NA; Weeks KM Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension and Mutational Profiling (SHAPE-MaP) for Direct, Versatile and Accurate RNA Structure Analysis. Nat. Protoc 2015, 10 (11), 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Busan S; Weeks KM Accurate Detection of Chemical Modifications in RNA by Mutational Profiling (MaP) with ShapeMapper 2. RNA 2018, 24 (2), 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kabsch W XDS. Acta Cryst. 2010, 66 (2), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Liebschner D; Afonine PV; Baker ML; Bunkoczi G; Chen VB; Croll TI; Hintze B; Hung LW; Jain S; McCoy AJ; et al. Macromolecular Structure Determination Using X-Rays, Neutrons and Electrons: Recent Developments in Phenix. Acta Cryst. 2019, 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Krissinel E; Henrick K Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol 2007, 372 (3), 774–797. [DOI] [PubMed] [Google Scholar]
- (57).Ho BK; Gruswitz F HOLLOW: Generating Accurate Representations of Channel and Interior Surfaces in Molecular Structures. BMC Struct. Biol 2008, 8 (49). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


