Abstract
Neuroimaging and genetic biomarkers have been widely studied from discriminative perspectives towards Alzheimer’s disease (AD) classification, since neuroanatomical patterns and genetic variants are jointly critical indicators for AD diagnosis. Generative methods, designed to model common occurring patterns, could potentially advance the understanding of this disease, but have not been fully explored for AD characterization. Moreover, the introduction of a supervised component into the generative process can constrain the model for more discriminative characterization. In this study, we propose an original method based on supervised topic modeling to characterize AD from a generative perspective, yet maintaining discriminative power at differentiating disease populations. Our topic modeling jointly exploits discretized image features and categorical genetic features. Diagnostic information - cognitively normal (CN), mild cognitive impairment (MCI) and AD - is introduced as a supervision variable. Experimental results on the ADNI cohort demonstrate that our model, while achieving competitive discriminative performance, can discover topics revealing both well-known and novel neuroanatomical patterns including temporal, parietal and frontal regions; as well as associations between genetic factors and neuroanatomical patterns.
Keywords: Alzheimer’s disease, MRI, genetics, topic modeling, mixed membership model, generative method
I. Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease with rising frequency in the global aging population. In AD, cell death can result in regional atrophy in brain regions (e.g. entorhinal cortex, hippocampus). The patterns of atrophy revealed from structural MRI (sMRI) may correlate with different symptoms or progression routes. The volumetric measures of regions including entorhinal cortex and hippocampus [1], [2] are common image biomarkers characterizing AD pathology. AD also has substantial genetic heritability [3], such that genetic variants play an important role in AD progression. The basic unit for a genetic variant is a single nucleotide polymorphism (SNP), among which rs7412 and rs429358 are well-known for AD [4]. While imaging and genetics both provide important biomarkers to understand and diagnose AD, they are very different in their representation. Inter-subject and disease heterogeneity make this task even more challenging. Topic modeling based on mixed membership models provides a powerful methodological framework to capture homogeneity and heterogeneity of different types of features and is well-suited for high-dimensional data representations [5], [6].
Interactions between genetics and imaging, such as associations between certain SNPs and regional volumetric measures calculated from sMRI, is gaining attention in the neuroimaging community both in general population studies [7], [8] and in AD studies [9], [10], and has been utilized for AD classification [11]. Discriminative methods have been widely studied for AD classification using sMRI data [12], [13], [2], [14], and have been exploited for AD characterization using sMRI and genetic data independently [15]. Generative topic modeling opens up new ways to explicitly characterize the generative process and discover common occurring patterns in AD populations, and could potentially advance the understanding and early diagnosis of AD. A recent study [16] identified atrophy patterns in AD patients using voxel-based morphometry (VBM) and latent Dirichlet allocation (LDA) [17], which is a classic unsupervised generative topic model. Here they found some latent brain atrophy factors that overlap with AD subtypes defined in a retrospective neuropathology cohort [18]. However, such fully unsupervised generative methods may lack the power of discovering patterns of interest. Alternatively, one can introduce supervised components into the model to exploit the advantages of both generative and discriminative methods. A recent study [19] applied such strategy using a diagnostic label to guide a joint projection and sparse regression model to study brain-wide and genome-wide associations for AD.
In AD characterization, evidences and features are exploited for subject stratification at different scales. At the coarsest scale, subjects are assigned to diagnosis groups, i.e. whether having AD, mild cognitive impairment (MCI) or being cognitively normal (CN). Clinicians provide such diagnosis by exploiting a comprehensive set of evidences such as cognitive tests (e.g. MMSE, Mini-Mental State Exam [20]) image-based markers, and genetic variants. This grouping is most commonly regarded as a “hard” assignment and can be exploited as a supervision for discriminative characterization using supervised learning methods. At a finer scale, for subjects with AD, it is desirable to further define AD subtypes associated with different mechanisms or at different stages, which would likely benefit from different monitoring or intervention options. In addition, within CN and MCI groups, certain subjects will remain CN (or MCI), while others will progress to AD [4]. This heterogeneity could potentially be captured by biomarkers embedded in “micro”-genetics and “macro”-image biomarkers [21]. However, this concept still lacks consensus and is not included in routine clinical practice. Such latent subtypes can be modeled via “soft” assignment or mixed-membership. Supervised topic modeling [22], [23] is a constructive extension of the unsupervised versions, and can combine subject stratification at different scales for a more robust and discriminative characterization.
In this study, we infer grouping of subjects from image and genetic biomarkers and analyze the prevalent patterns that emerge in the AD and MCI populations. We first investigate supervised topic modeling by encoding “co-atrophy” patterns of brain regions as “topics” and characterize AD with latent neuroanatomical patterns using image features. We then jointly model image and genetic features to discover jointly occurring image and genetic AD biomarkers. We apply the proposed model for three types of population stratification: 1) CN vs.AD, 2) CN vs. MCI vs. AD and 3) MCI stable vs. MCI progressing to AD subjects. Experimental results on the ADNI cohort demonstrate that our model can discover topics that reveal both well-known and novel neuroanatomical patterns as well as associations between neuroanatomical and genetic factors implicated in AD.
II. Method
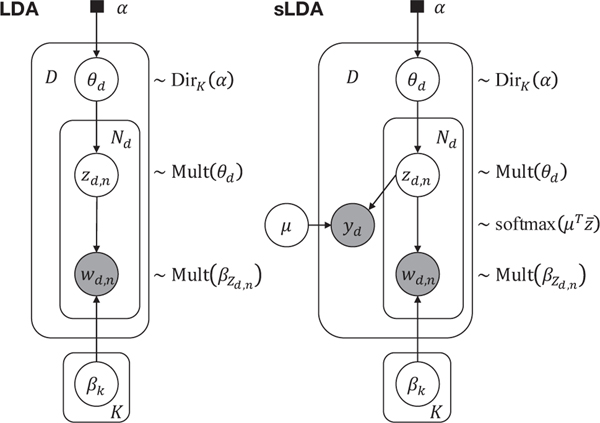
We first introduce the problem setting of AD characterization using the classic LDA topic model formalism. Then we introduce our supervised LDA (sLDA) model where the diagnosis group information (e.g. CN vs. MCI vs. AD) is integrated as a supervision variable to enable more discriminative characterization. Graphical representations of these two models are provided in Fig. 1.
Fig. 1:
Graphical representations of the classic (unsupervised) LDA model and the supervised LDA (sLDA) model. Hollow circles denote latent variables, shaded circles denote observed variables, solid squares denote hyper-parameters, arrows denote dependence relationship, and plates denote replicated structures.
A. Problem Setting in LDA
The LDA model is built on a bag-of-words representation of the input observations. It analyzes data from a set of documents d ∈ [1,...,D] known as corpus, and characterizes each document as a mixture of K topics β1:K, whereas topics are defined via learning the probabilistic distribution of possible words wd,n with n ∈ [1,...,Nd] for each document d. A Dirichlet prior is assumed for the topic proportions θd in each document. And the topic assignment zd,n of each word wd,n is assumed to be drawn from a multinomial distribution parameterized by θd. The posterior distribution is modeled as follows:
| (1) |
In our LDA setting for AD diagnosis, we use both image features and genetic variants of individuals in the topic modeling. For image features, we use subcortical and cortical volume and cortical thickness. The feature values are discretized via binning, and are normalized by the maximum value in the total training population to ensure equivalent contributions in the model among different features. The genetic features encode the number of alleles (A, T, C and G) for certain AD-related SNPs (details in Section III-A) and take three values 0, 1 and 2, which represent the occurrence of the reference allele on two homologous chromosomes [24].
We use an analogy to LDA previously applied for survey analysis [25], where observations are answers to individual questions. Each “document” is the collection of answers by a single respondent. Topics encode common occurring patterns of likely answers for each question (“word”), and topic proportions represent how much each individual exhibits those patterns. In our case, each subject is viewed as a survey “document”. Brain regions and SNPs are the shared survey questions, and the feature values (e.g. cortical thickness of a certain brain region) are the answers. Hence, SNPs are regarded as survey questions with three possible answers, while image features are regarded as questions with preset possible responses, where the variety depends on the binning size. When using image features alone, atrophy patterns across the brain regions are the “topics” to discover. When using both image and SNP features, the “topics” are the integrated patterns of brain region atrophy and jointly occurring genetic variants. Each individual expresses the discovered topics at variable levels.
B. Supervised LDA
The expressive power of the basic LDA to discover topics comes at a price. The posterior distributions can exhibit many local modes, resulting in very unstable solutions. Furthermore,
Algorithm 1:
Generative process for the sLDA model.
| For each individual d ∈ [1,...,D]: |
| a. Draw topic mixture proportions: θd ~ DirichletK(α), where α is a hyper-parameter. |
| b. For each “word” n ∈ [1,...,Nd] within individual d: i. Draw a topic assignment: zd,n ~ Multinomial(θd). ii. Draw an image / genetic feature: wd,n ~ Multinomial(βzd,n). |
| c. Draw the discrete response variable: where is the empirical frequencyvector of different topics in subject d, and μ is the classification weighting matrix to be estimated. |
unsupervised topic modeling lacks constraints to differentiate disease populations, and is thus likely to find topics in the data that are not reflective of the structures of interests [22]. In our preliminary experiments, we found the distribution of topics learned with a classic unsupervised LDA model to be nearly uniform. Therefore, we propose to introduce diagnostic information into our model to generate topics with patterns more discriminative of a disease state.
One simple way to incorporate diagnostic information is to screen features with a univariate association model. A more sophisticated approach is to integrate the diagnostic variable into the graphical model, with a categorical distribution from pre-defined categories (e.g. CN vs. MCI vs. AD), like other variables. However, since diagnosis groups are inferred from multiple biomarkers, this variable should be treated hierarchically differently. It could be treated as a response (at the end of an arrow), or it could be used to inform the model (at the head of an arrow). In this study, we treat diagnosis groups as a response. Such a model is referred to as supervised LDA model [22], [23].
The generative process using the proposed sLDA model (Fig. 1) is detailed in Algorithm 1, where K topics β1:K are discovered on D individuals, and each βk is a probability vector over the image or genetic features. In the bag-of-words representation, the count of each “word” is the feature value, and Nd is the sum of all counts (feature values) per subject. The hyper-parameter α is set as the inverse of the number of topics 1/K [22]. Different from the original LDA model [17], we treat β1:K as unknown constants to be estimated, rather than random variables, as suggested in [22].
The model is estimated using a variational expectation-maximization (EM) algorithm, where the E-step is the mean-field variational inference of the posterior distribution of latent variables z,θ, and the M-step is maximum likelihood estimation of the parameters β and μ. In variational inference, we first define a family of distributions over the latent variables indexed by the variational parameters, and then estimate the variational parameters to minimize the Kullback-Leibler divergence between the variational distribution q and the exact posterior distribution. This way, the inference problem is turned into an optimization problem. The variational distribution q with the optimal parameter is then an approximate of the posterior distribution. For mean-field family, each latent variable is independent and associated with its own variational parameter. In our setting, the variational distribution of latent variables z,θ is:
| (2) |
where ϕ is a multinomial variational parameter, γ is the Dirichlet variational parameter. The estimated q(θd|γd) is used to generate θ in the inference.
III. Experimental Results
We tested our proposed sLDA topic modeling using either image features only or combining image and genetic features, for three supervised tasks. 1) Post hoc 2-class classification task of CN vs. AD. Since these two groups are relatively separable, this is often used as a proof-of-concept classification task. 2) 3-class classification task (CN vs. MCI vs. AD), which is more realistic but also more challenging. 3) 2-class classification task of MCI stable (MCI-s) subjects vs. MCI progressing to AD (MCI-p) subjects. This is a challenging task but is important for understanding disease progression and facilitating early diagnosis.
Our supervised sLDA estimates not only topics but also classification labels. The classification accuracy is used to select the optimal value for the number of topics K*, and results on topic proportions and compositions are reported using this value.
A. Data and Preprocessing
Imaging and genetic data used for training and evaluation are derived from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu/).
The imaging data used in this study consists of the T1-weighted structural MRI scans from N = 820 individuals enrolled in ADNI-1 study. We included scans for all the 820 subjects at baseline visit, and the subset of N = 740 scans available from the first (month-6) follow-up visit. The number of subjects in each diagnosis group at month-6 is different from baseline due to conversion, reversion, or drop-out. The MRI images were acquired using 1.5 T MRI scanners1. The diagnosis labels are the cross-sectional diagnosis results at scan time. Eighty-four neuroanatomical measures (cortical thickness, cortical volume, subcortical volume), generated from the FreeSurfer cross-sectional pipeline [26]2,3 are used as image-derived features, after quantification into 10 regular bins [16]. Since atrophy patterns are of interest for our tasks, we use the opposite of the discretized values and then add a constant to convert the features into positive values.
Genetic data is available for N = 757 of the 820 individuals. We selected 1) two SNPs (rs429358, rs7412) that define APOE genotype, a major genetic risk factor for AD; and 2) all SNPs provided in the dataset “ADNI_cluster_01_forward_757LONI”. We preprocessed this data following the steps carried out in [7] using PLINK [24]. Specifically, subjects with discordant sex information, large missingness (> 10%), highly-related subjects in the study (identity-by-state, IBS similarity > 0.2) were excluded. SNPs with low occurrence, high missingness, deviating from Hardy-Weinberg equilibrium were excluded. The level of similarity of individual subjects was visualized using multi-dimensional scaling (MDS) and only those with most similar genetic ancestry were included [7].
The final set after preprocessing consists of N = 697 subjects (out of the 757) with quality controlled (QC) genetic data including 556,165 SNPs. The SNPs were screened with logistic regression using PLINK under additive assumption with sex and baseline age as covariates and the AD versus CN label as the discriminatory response. A total of twenty-nine SNPs with p-value below 1e-4 were identified as AD-related SNPs, among which, the most significant (p < 1e-6) SNPs are rs429358 (one defining SNP of APOE genotype), rs11857713 and rs4778636. For each SNP, the feature is the number of reference alleles (risky alleles returned by the logistic regression).
The data used in each diagnosis group is summarized in TABLE I and TABLE II. The subjects in ADNI were diagnosed based on comprehensive clinical dementia and MCI evaluations. The detailed procedure can be found in the manual provided by ADNI4. The N = 697 baseline subjects with both genetic and image data are used to evaluate the sLDA models with supervisions from diagnostic labels (CN, MCI and AD).
TABLE I:
Number of subjects extracted from ADNI with respect to data type and diagnostic categories
| Diagnosis label╲Data | CN | MCI | AD | Total |
|---|---|---|---|---|
| QC Genetics | 193 | 339 | 165 | 697 |
| MRI scans at baseline | 230 | 399 | 191 | 820 |
| MRI scans at month-6 | 219 | 332 | 189 | 740 |
| MRI and QC genetics at baseline | 193 | 339 | 165 | 697 |
| MRI and QC genetics at month-6 | 185 | 291 | 161 | 637 |
TABLE II:
Demographic information for the 820 baseline subjects
| Diagnosis | CN | MCI | AD | Total |
|---|---|---|---|---|
| # Subjects | 230 | 399 | 191 | 820 |
| Age ± std | 75.85 ± 5.02 | 74.69 ± 7.45 | 75.27 ± 7.46 | 75.15 ± 6.87 |
| MMSE ± std | 29.11 ± 1.00 | 27.03 ± 1.77 | 23.31 ± 2.04 | 26.75 ± 2.67 |
| Gender M/F | 120/110 | 257/142 | 100/91 | 477/343 |
The cohort is split into training and test sets with a ratio of 4:1. K-medoids undersampling is used during training to account for group imbalance. The follow-up data (month-6) is used to study the reproducibility of the discovered discriminative topics. Finally, among the baseline subjects, we used the N = 96 MCI-s subjects who stayed MCI during a follow-up period of at least two years, and the N = 124 MCI-p subjects who progressed to AD within two years, to train the sLDA for characterizing MCI progression (training/test split ratio = 4:1).
B. Optimal Numbers of Topics
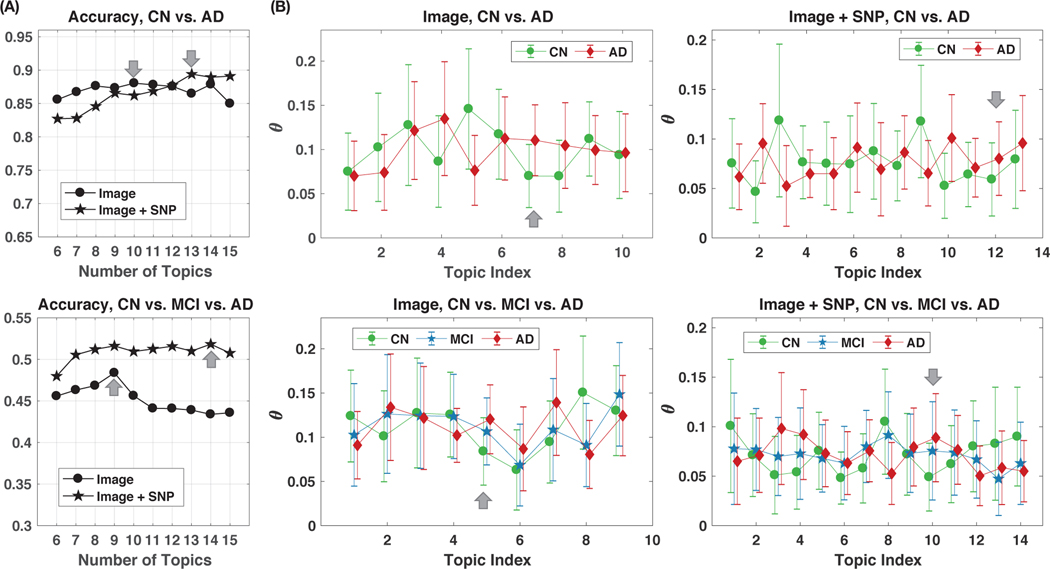
We report results in Fig. 2 (A) using a total of 113 features (84 image-based and 29 SNP-based). The number of topics K is set per feature type and classification task, as the value K* maximizing the average test sLDA classification accuracy. Average accuracy is computed over 20 runs with different random initializations of β1:K. We limited the range of tested values for K to [6,15] to limit under-fitting (K too low), overfitting (K too large), while maintaining interpretability [22].
Fig. 2:
Illustrations of topic proportions θ estimated from sLDA models, and the classification accuracies. (A) Classification accuracy of diagnosis groups versus number of topics: (top) CN vs. AD classification task, (bottom) CN vs. MCI vs. AD classification task. Grey arrows indicate the selected optimal numbers of topics K* in each setting. (B) Estimated topic proportions θ (mean ± standard deviation) in different diagnosis groups when modeling with image features only or with both image and genetic features. Numbers of topics are determined by the highest classification accuracy in Fig. 2 (A). Grey arrows point to the discriminative topics that are visualized and interpreted later (as in Fig. 3 (A)).
Results in Fig. 2 (A) lead to the following parameterization: with image features only, K*=10 for the 2-class classification of CN vs. AD, and K*=9 for the 3-class classification of CN vs. MCI vs. AD; with image and genetic features, K*=13 for the 2-class classification of CN vs. AD, and K*=14 for the 3-class classification of CN vs. MCI vs. AD. More topics are needed when combining image and genetic features, which was expected. Using image feature alone achieves an accuracy of 88.0% for AD vs. CN classification. Adding genetic biomarkers increases the classification accuracies from 88.0% to 89.4% in the CN vs. AD classification task, and from 48.4% to 52.2% in the CN vs. MCI vs. AD classification task.
We emphasize here that the goal of our sLDA learning is not to beat fully-discriminative methods for disease classification, but rather to investigate the possibility to discover AD-related imaging and genetic patterns inside certain topics, while optimizing the classification accuracy metric to set the number of topics and ensure discriminancy.
C. Discriminative Topics and Features for Characterizing CN vs. MCI vs. AD Populations
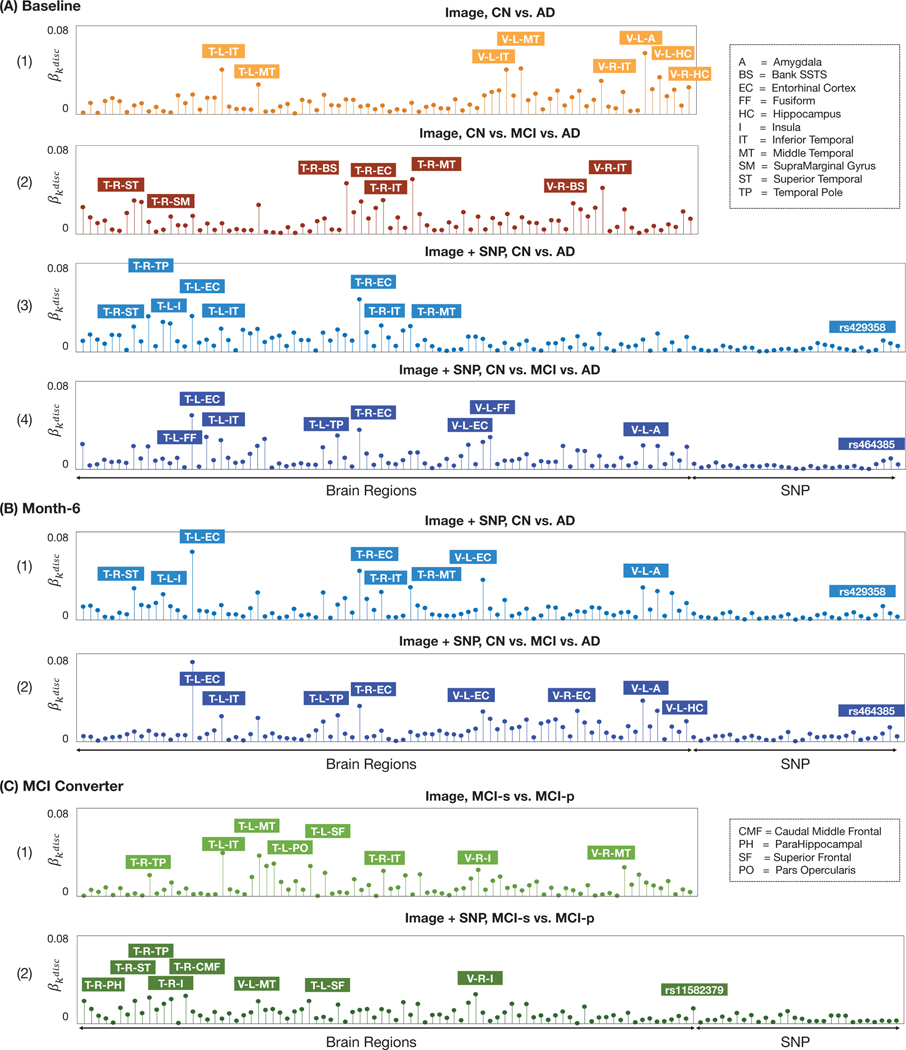
Topics learned with optimal discriminative power are now investigated for interpretation. In Fig. 2 (B), we show the average topic proportions θ within all baseline subjects in different diagnosis groups, where the topics were learned using the optimal number of topics K* associated with the highest average test classification accuracy. For the four classification setups, the most discriminative topics are shown in Fig. 3 (A), encoded with their proportion values of brain regions and SNP features. For a classification setup, the most discriminative topic kdisc is the topic that appears more frequently in the disease than in the normal subjects and is identified as the one exhibiting the most significant AD vs. CN distribution differences, when evaluated via t-tests. The features (brain regions or SNPs) that have the largest proportion values in the selected discriminative topics are annotated in each plot.
Fig. 3:
Visualization of the most discriminative topics (exhibiting most significant differences in AD vs. CN or MCI-s vs. MCI-p) as feature proportion vectors returned by our sLDA models. In each plot, features with highest proportion values are annotated. (A) Results with ADNI baseline data: (1) CN vs. AD task using image features; (2) CN vs. MCI vs. AD task using image features; (3) CN vs. AD task using image and genetic features; (4) CN vs. MCI vs. AD task using image and genetic features. (B) Results with ADNI month-6 (follow-up) data: (1) CN vs. AD task using image and genetic features; (2) CN vs. MCI vs. AD task using image and genetic features. (C) Results for modeling MCI converters: (1) MCI-s vs. MCI-p using image features; (2) MCI-s vs. MCI-p using image and genetic features. T-=Thickness, V-=Volume; L-=Left, R-=Right.
Our results reveal a well-known neuro-anatomical circuit implicated in AD [1], constituted of medial temporal lobe structures including the hippocampus, entorhinal cortex (a gateway between the hippocampus and neocortex), temporal pole, and fusiform cortex. But in the CN vs. AD classification tasks (Fig. 3 (A-1, A-3)), other temporal lobe regions are revealed, including the inferior-, middle- and superior-temporal cortices. When using both image and genetic features, Fig. 3 (A-3) reveals a pattern with temporal lobe structures and SNP rs429358, a well-known genetic risk factor of AD [4]. In the CN vs. MCI vs. AD classification tasks, a heterogeneous pattern involving parietal (supramarginal gyrus) and temporal lobes is revealed in Fig. 3 (A-2); in Fig. 3 (A-4), the anatomical pattern involves temporal lobe regions, while the SNP with the largest proportion in is rs464385, associated with gene PEX26 (peroxisomal biogenesis factor 26).
D. Reproducibility over Training Data
We further re-train the proposed sLDA model using both image and genetic data in ADNI follow-up (month-6) visit, to evaluate the consistency of the discovered discriminative topics and features. We again exploited the stratification of 1) CN vs. AD and 2) CN vs. MCI vs. AD diagnosis groups.
We set the number of topics K to the optimal value identified with the baseline data as in Section III-C. The discovered discriminative topics are shown in Fig. 3 (B). Again, medial temporal structures including hippocampus, entorhinal cortex, temporal pole, and temporal cortex regions including inferior-, middle-, superior-temporal cortices appear in the discriminative topics. As with the vast majority of generative topic modeling, our sLDA model has a non-convex objective function, leading to some variability in discovered topics and feature proportions when altering the training data. Nonetheless, we observe that the dominant brain regions and SNPs in maintain a high-level of consistency after re-training with the follow-up data. When training for the CN vs. AD classification task, Fig. 3 (B-1) reveals again a joint pattern of temporal lobe structures and SNP rs429358. When training for the CN vs. MCI vs. AD classification task, Fig. 3 (B-2) reveals a pattern with temporal lobe structures and again SNP rs464385.
E. Results on Characterizing MCI Conversion
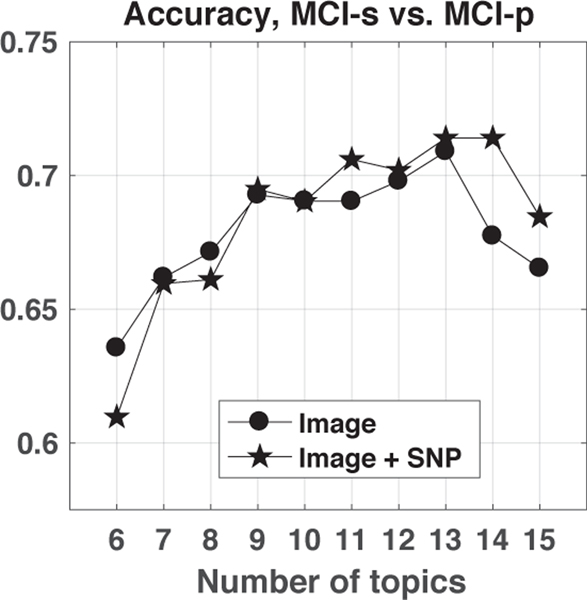
The sLDA model was also tested to characterize the conversion of MCI to AD within two years, which is important for understanding disease progression. We compare modeling with image features only versus incorporating genetic variants. The optimal number of topics K* is again determined via optimal classification accuracy. As shown in Fig. 4, when modeling with image features only, the highest test classification accuracy is 70.9%, corresponding to K* = 13 topics. Incorporating genetic variants slightly increased the test classification accuracy from 70.9% to 71.4%, with K* = 14 topics.
Fig. 4:
Classification accuracy of diagnosis groups MCI-stable (MCI-s) vs. MCI-progressing to AD (MCI-p) versus number of topics.
The discriminative topic with the most significant difference in MCI-s vs. MCI-p is shown in Fig. 3 (C). Besides temporal lobe structures including parahippocampal gyrus, temporal pole, middle temporal cortex, and inferior temporal cortex, we also observe the inclusion of frontal regions including caudal middle frontal, superior frontal cortex, and SNP rs11582379 associated with gene GFI1 (growth factor independent 1 transcriptional repressor).
Collectively, the classification accuracy values with the sLDA model and the corresponding optimal numbers of topics K* are summarized in Table III.
TABLE III:
Classification accuracy values and the corresponding optimal numbers of topics (ACC%/K*)
| Task╲Features | Image | Image+genetics |
|---|---|---|
| CN vs. AD | 88.0% / 10 | 89.4% / 13 |
| CN vs. MCI vs. AD | 48.4% / 9 | 52.2% / 14 |
| MCIs vs. MCIp | 70.9% / 13 | 71.4% / 14 |
IV. Discussions and Conclusions
In this study, we formulate an original sLDA generative model to combine image and genetic features into discriminative topics. The proposed model aims to identify, in topics more prevalent in AD / MCI / MCI-p, neuroanatomical and genetic patterns implicated in AD and in MCI progression. Our results reveal, with confirmation on a reproducibility study, associations between specific SNPs and image features from the temporal lobe on the AD vs. CN classification task, and the medial temporal lobe in the CN vs. MCI vs. AD classification task. Our results on the MCI progression prediction identify discriminative features in both frontal and temporal lobes, which reflects the heterogeneity of MCI anatomical patterns [27]. Regarding genetic variants, the SNPs rs429358 (APOE) and rs464385 (PEX26) were identified as informative for characterizing AD vs. CN in our results from both baseline and follow-up data. The former is a well known AD-associated risk factor [4]. While the latter is not well-documented in the AD literature, a study [28] has suggested that peroxisomal deficit is linked to Alzheimer’s disease. The SNP rs11582379 associated with gene GFI1, was found to be informative for characterizing MCI-s vs. MCI-p, whose link to AD, however, is not previously reported. Similar to the discussion in Section III D about reproducibility over training data, genetic biomarkers also result in changes to the rank order of the image features because of the non-deterministic nature of the algorithm, the inherent association of features, and the non-linear feature integration in the modeling process. However, the anatomical patterns are all coherent with current knowledge on AD.
Supervised by diagnosis labels, our model also maintains discriminative power. For CN vs. AD classification task, using image features alone, our model reaches state-of-the-art accuracy, compared with the 86% accuracy reported in [12] using a multivariate approach based on sMRI features, and is further improved when combining image-genetics features. The increase is particularly obvious in the CN vs. MCI vs. AD classification, but only modest in CN vs. AD classification. Because of the relatively larger neuroanatomical difference between CN and AD, the neuroanatomical biomarkers suffice to capture the class differences and the additive predictive value of genetic biomarkers becomes less important. But we want to emphasize that the genetic features contribute to the discovered topics with non-zero coefficients. For the MCI stable vs. progression to AD classification, our accuracy of around 71% is higher than a previous study [13] which reported an optimal accuracy of 64.4% using multiple methods, but on a slightly different sample in ADNI. Therefore, our proposed model, primarily designed for generative modeling of interpretable feature patterns, can achieve competitive discriminative performance in predicting MCI conversion.
Compared to discriminative models, the topics discovered by our generative modeling open up new ways for disease understanding. Compared to basic (unsupervised) LDA, our sLDA model provides a more robust and discriminative characterization, benefiting from the guidance of supervised population stratification. Our sLDA model graphical representation is flexible and can easily accommodate and test additional interactions and features, which could potentially further boost classification performance, and provide better characterization. We could introduce functional MRI, cognitive tests or vascular measures [29], to investigate structure-function, structure-cognition or structure-vasculature relationships. Association of individual expressions of patterns from the discriminative topics with disease state is under investigation. As future work, the proposed model will be extended to a longitudinal design with more timepoints which could help discover patterns that are associated with intra-subject longitudinal disease progression.
Acknowledgments:
This work was supported by NIH/NHLBI R01-HL121270. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Contributor Information
Jie Yang, Department of Biomedical Engineering, Columbia University, New York, NY, USA.
Xinyang Feng, Department of Biomedical Engineering, Columbia University, New York, NY, USA.
Andrew F. Laine, Departments of Biomedical Engineering, Radiology and Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, USA..
Elsa D. Angelini, Departments of Biomedical Engineering and Radiology, Columbia University, New York, NY, USA, and NIHR Imperial Biomedical Research Centre, ITMAT Data Science Group, Imperial College London, UK.
REFERENCES
- [1].Leandrou S, Petroudi S, Kyriacou PA, Reyes-Aldasoro CC, and Pattichis CS, “Quantitative MRI brain studies in mild cognitive impairment and Alzheimer’s disease: A methodological review,” IEEE Rev Biomed Eng, vol. 11, pp. 97–111, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Feng X, Yang J, Laine AF, and Angelini ED, “Alzheimer’s disease diagnosis based on anatomically stratified texture analysis of the hippocampus in structural MRI,” in IEEE Int Symp Biomed Imaging (ISBI), 2018, pp. 1546–1549.
- [3].Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW et al. , “Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease,” Nat Genet, vol. 45, no. 12, p. 1452, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, and Van der Flier WM, “Alzheimer’s disease,” Lancet, vol. 388, no. 10043, pp. 505–17, 2016. [DOI] [PubMed] [Google Scholar]
- [5].Airoldi EM, Blei D, Erosheva EA, and Fienberg SE, Handbook of Mixed Membership Models and Their Applications. CRC Press, 2014. [Google Scholar]
- [6].Song J, Yang J, Smith B, Balte P, Hoffman EA, Barr RG, Laine AF, and Angelini ED, “Generative method to discover emphysema subtypes with unsupervised learning using lung macroscopic patterns (LMPs): The MESA COPD study,” in IEEE Int Symp Biomed Imaging (ISBI), 2017, pp. 375–378. [DOI] [PMC free article] [PubMed]
- [7].Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivierès S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M. et al. , “Common genetic variants influence human subcortical brain structures,” Nature, vol. 520, no. 7546, pp. 224–229, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, and Smith SM, “Genome-wide association studies of brain imaging phenotypes in UK Biobank,” Nature, vol. 562, no. 7726, pp. 210–216, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moon SW, Dinov ID, Kim J, Zamanyan A, Hobel S, Thompson PM, and Toga AW, “Structural neuroimaging genetics interactions in Alzheimer’s disease,” J Alzheimers Dis, vol. 48, no. 4, pp. 1051–1063, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD et al. , “Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort,” Neuroimage, vol. 53, no. 3, pp. 1051–1063, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ning K, Chen B, Sun F, Hobel Z, Zhao L, Matloff W, and Toga AW, “Classifying Alzheimer’s disease with brain imaging and genetic data using a neural network framework,” Neurobiol Aging, vol. 68, pp. 151–158, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sabuncu MR and Konukoglu E, “Clinical prediction from structural brain MRI scans: a large-scale empirical study,” Neuroinformatics, vol. 13, no. 1, pp. 31–46, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M-O, Chupin M, Benali H, Colliot O, Initiative ADN et al. , “Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database,” Neuroimage, vol. 56, no. 2, pp. 766–781, 2011. [DOI] [PubMed] [Google Scholar]
- [14].Feng X, Yang J, Lipton ZC, Small SA, and Provenzano FA, “Deep learning on MRI affirms the prominence of the hippocampal formation in Alzheimer’s disease classification,” bioRxiv, 2018.
- [15].Varol E, Sotiras A, Davatzikos C, and Initiative ADN, “HYDRA: revealing heterogeneity of imaging and genetic patterns through a multiple max-margin discriminative analysis framework,” Neuroimage, vol. 145, pp. 346–364, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, and Yeo BT, “Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer’s disease,” Proc Natl Acad Sci USA, vol. 113, no. 42, pp. E6535–E6544, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Blei DM, Ng AY, and Jordan MI, “Latent dirichlet allocation,” J Mach Learn Res, vol. 3, no. Jan, pp. 993–1022, 2003. [Google Scholar]
- [18].Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, and Dickson DW, “Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study,” Lancet Neurol, vol. 10, no. 9, pp. 785–796, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou T, Thung K-H, Liu M, and Shen D, “Brain-wide genome-wide association study for Alzheimer’s disease via joint projection learning and sparse regression model,” IEEE Trans Biomed Eng, vol. 66, no. 1, pp. 165–175, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Folstein MF, Folstein SE, and McHugh PR, “mini-mental state: a practical method for grading the cognitive state of patients for the clinician,” Journal of psychiatric research, vol. 12, no. 3, pp. 189–198, 1975. [DOI] [PubMed] [Google Scholar]
- [21].Batmanghelich NK, Dalca A, Quon G, Sabuncu M, and Golland P, “Probabilistic modeling of imaging, genetics and diagnosis,” IEEE Trans Med Imaging, vol. 35, no. 7, pp. 1765–1779, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mcauliffe JD and Blei DM, “Supervised topic models,” in Neural Inf Process Syst (NIPS), 2008, pp. 121–128.
- [23].Chong W, Blei D, and Li F-F, “Simultaneous image classification and annotation,” in IEEE Comput Soc Conf Comput Vis Pattern Recognit (CVPR), 2009, pp. 1903–1910. [Google Scholar]
- [24].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, and Sham PC, “PLINK: a tool set for whole-genome association and population-based linkage analyses,” Am J Hum Genet, vol. 81, no. 3, pp. 559–575, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Erosheva EA, Fienberg SE, and Joutard C, “Describing disability through individual-level mixture models for multivariate binary data,” Ann Appl Stat, vol. 1, no. 2, p. 346, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fischl B, “FreeSurfer,” NeuroImage, vol. 62, no. 2, pp. 774–781, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bell-McGinty S, Lopez OL, Meltzer CC, Scanlon JM, Whyte EM, DeKosky ST, and Becker JT, “Differential cortical atrophy in subgroups of mild cognitive impairment,” Arch Neurol, vol. 62, no. 9, pp. 1393–1397, 2005. [DOI] [PubMed] [Google Scholar]
- [28].Berger J, Dorninger F, Forss-Petter S, and Kunze M, “Peroxisomes in brain development and function,” Biochim Biophys Acta Mol Cell Res, vol. 1863, no. 5, pp. 934–955, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-Prez JM, Evans AC, and The I. Alzheimers Disease Neuroimaging, “Early role of vascular dysregulation on late-onset alzheimers disease based on multifactorial data-driven analysis,” Nature Communications, vol. 7, p. 11934, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]