Abstract
Determining cell death mechanisms occurring in patient and animal tissues is a longstanding goal that requires suitable biomarkers and accurate quantification. However, effective methods remain elusive. To develop more powerful and unbiased analytic frameworks, we developed a machine learning approach for automated cell death classification. Image sets were collected of HT-1080 fibrosarcoma cells undergoing ferroptosis or apoptosis and stained with an anti-transferrin receptor 1 (TfR1) antibody, together with nuclear and F-actin staining. Features were extracted using high-content-analysis software, and a classifier was constructed by fitting a multinomial logistic lasso regression model to the data. The prediction accuracy of the classifier within three classes (control, ferroptosis, apoptosis) was 93%. Thus, TfR1 staining, combined with nuclear and F-actin staining, can reliably detect both apoptotic and ferroptotis cells when cell features are analyzed in an unbiased manner using machine learning, providing a method for unbiased analysis of modes of cell death.
Introduction
Regulated cell death is a complex and tightly regulated phenomenon, involving intricate molecular mechanisms. For numerous cell death processes, molecular markers have been developed that identify cells undergoing apoptosis1 or necroptosis2 through immunolabeling. Such markers may be used in cell culture and tissue histopathological applications to examine the prevalence of cell death processes, which may improve the treatment and diagnosis of diseases in which these processes are implicated.
Ferroptosis is a form of regulated cell death characterized by the iron-dependent accumulation of lipid peroxides, as well as the loss of cellular antioxidant repair capabilities.3 The enzyme glutathione peroxidase 4 (GPX4) is a cellular regulator of lipid peroxidation levels, and several ferroptosis inducers have been developed that specifically target the activity of this enzyme through direct inhibition (e.g., RSL3).4 A second class of ferroptosis inducers (e.g., IKE and erastin) causes inactivation of GPX4 through depletion of glutathione via inhibition of the antiporter system xc–.5 Ferroptosis has been implicated in several disease pathologies, such as degenerative diseases and organ injury.6,7 Furthermore, ferroptosis induction may have potential as a cancer treatment strategy.8,9
Toward the goal of specific identification of ferroptosis in tissue samples, we previously discovered an effective ferroptosis-staining reagent, 3F3 anti-Ferroptotic Membrane Antibody (3F3-FMA), that can be used to stain cells and tissue samples directly.10 The antigenic target of 3F3-FMA is transferrin receptor 1 (TfR1), a membrane receptor that internalizes iron-bound transferrin through receptor-mediated endocytosis.11 This iron uptake activity of TfR1 contributes to intracellular iron levels necessary for ferroptosis.12 3F3-FMA, as well as other anti-TfR1 antibodies, exhibits an increase in total and membrane-localized fluorescence when used to stain cells undergoing ferroptosis in culture (compared to vehicle-treated control cells). TfR1 has been used to identify the occurrence of ferroptosis in traumatic brain injury13 and myocardial ischemia/reperfusion injury,14 among other uses. Thus, TfR1 serves as a biomarker to facilitate the identification of ferroptosis in cell and tissue contexts.
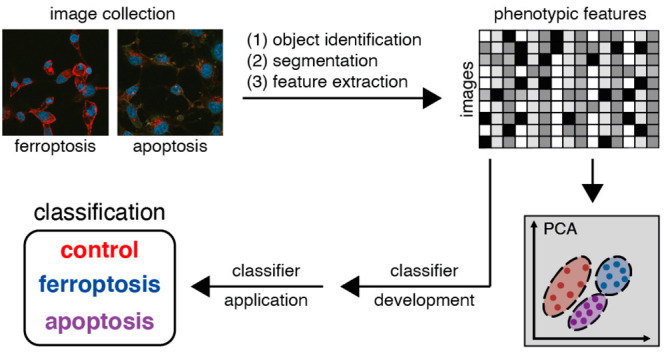
The identification of plasma membrane fluorescence as a distinguishing feature between ferroptosis and other cell death processes upon staining with anti-TfR1 antibodies was discovered using visual inspection; here, we sought instead to evaluate the use of machine learning as an unbiased tool to detect ferroptotic cells. Machine learning methods facilitate the high-throughput analysis of cell image sets versus tedious and subjective manual processes; in cell biology applications, machine learning can increase processing capabilities and objectivity. The supervised machine learning pipeline involves image collection and preprocessing, object detection, and feature extraction and prioritization.15 Our goals were to assess the machine learning potential in discriminating ferroptosis, apoptosis, and control-treated samples as well as to provide a pipeline for identification of features that best distinguish those cell death modalities in our setting.
Therefore, after collecting images of fluorescently stained cells treated with vehicle only or undergoing ferroptosis or apoptosis, images were analyzed via high-content-image analysis, and a classifier was trained on the extracted data. The trained classifier corresponds to a nonexclusive list of informative features with assigned coefficients, which was validated with a second data set by successfully predicting the same classes. These results expand and strengthen the applicability of biomarkers, such as 3F3-FMA/TfR1, for differentiating cell death mechanisms in an objective and high-throughput manner.
Results and Discussion
To explore the application of machine learning to the classification of different cell death modalities, we collected large numbers of images of cells fixed and immunofluorescently stained with 3F3 anti-Ferroptotic Membrane Antibody (3F3-FMA), a ferroptosis-specific antibody with TfR1 as its target antigen. Specifically, HT-1080 cells were treated with ferroptosis inducers (RSL3, a GPX4 inhibitor, or IKE, a system xc– inhibitor), an apoptosis inducer (staurosporine, STS),16 or DMSO vehicle control. In addition to being stained with anti-TfR1 3F3-FMA (labeled with AlexaFluor 594), cells were stained with DAPI as a nuclear marker and FITC-phalloidin as a cytoplasmic (F-actin) marker to assist identification of cellular features for machine learning classification (see below).
Machine learning tools are designed to adapt to any data pattern associated with the task to learn. There were several important aspects to consider in collecting images for machine learning classification. First, all treatments within a day (i.e., using the same microscope settings) were balanced. Moreover, we collected all images of the discovery data on day 1 and the validation data later on a different day. Second, the extent of cell death was standardized across different conditions to analyze cells in an early stage of cell death induction. Specifically, we fixed cells under each treatment condition when they reached 10–20% cell death, so that cell death had been initiated, but not to the extent of excessive end-stage necrosis. At this point, the cells should still have intact cell membrane integrity and not have detached from the surface. The CellTiter-Glo (CTG) viability assay, which measures intracellular ATP levels as an indicator of viability, was used to monitor the extent of cell death. We performed a pilot study and established optimal concentration and time point ranges for each treatment (Figure S1).
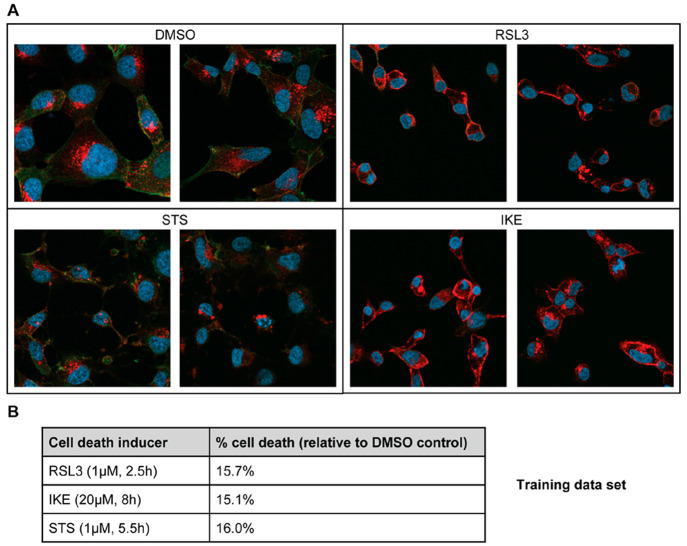
Guided by the results of the pilot study, the first image set for training and discovery of classifiers was collected, and immunofluorescence experiments were performed when the extent of cell death reached 10–20% compared to DMSO control treatment in parallel CTG assays (Figure 1). Viewing the images, the characteristic membrane localization of the 3F3-FMA signal can be seen in ferroptotic cells compared to the DMSO control,10 and characteristic membrane blebbing can be observed in apoptotic cells.17
Figure 1.
Images undergoing different cell death modalities for machine learning analysis. (A) HT-1080 cells were incubated with ferroptosis inducers RSL3 (1 μM) or IKE (20 μM), apoptosis inducer STS (1 μM), or DMSO control. Nuclei were stained with DAPI (blue). TfR1 was labeled with 3F3-FMA and Alexa Fluor 594 secondary antibody (red). F-actin was labeled with FITC-phalloidin (green). Images were captured using a Zeiss LSM800 confocal microscope at 63×/1.40 oil DIC objective. For each treatment, representative images from the training data set are depicted. (B) In parallel with the immunofluorescence experiments, CellTiter-Glo viability assays were used to monitor the percentage cell death for each treatment, and cells were fixed when percentage cell death reached 10–20%. The concentrations and time points that resulted in this extent of cell death in each set are listed for each treatment.
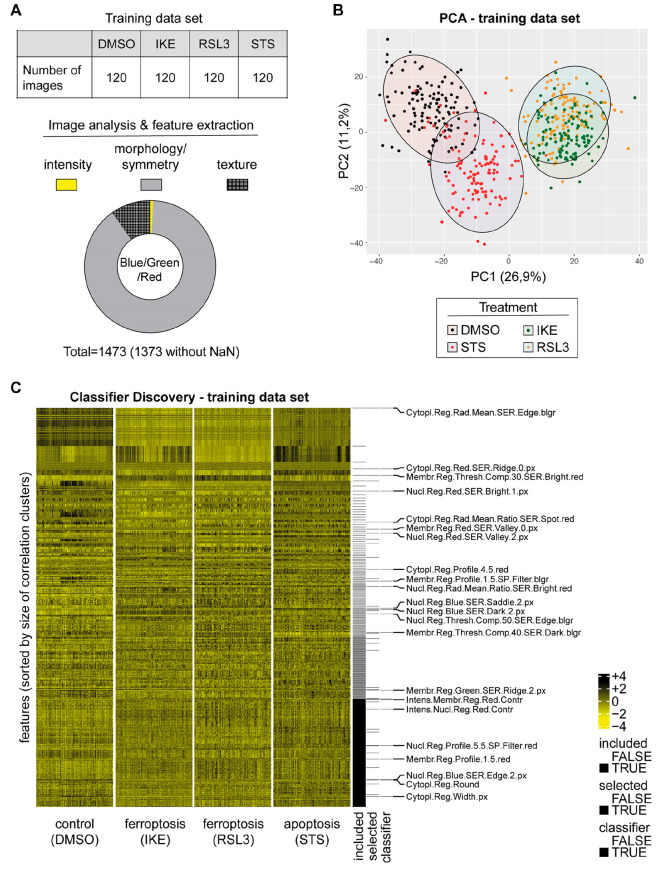
For the training set, once the cells were fixed and stained with DAPI, FITC-phalloidin, and anti-TfR1 3F3-FMA, 120 images were collected per treatment condition (DMSO control, RSL3, IKE, STS) with an average of 10 cells per image (Figure 2A), which corresponds to a cell density of approximately 80% for DMSO-treated cells. Subsequently, we analyzed images with the PerkinElmer Columbus high-content-analysis software. For this purpose, nuclei were identified using the DAPI signal, and based on this, the cytoplasm and the membrane regions were segmented using the F-actin signal (Figure S2). The intensity, the morphology, and the symmetry of the objects, as well as the texture and structure of the fluorescence signal, were determined within these cell segments for the blue, green, and red channels, respectively. Consequently, we were able to extract a large number of features for each image. Importantly, during the analysis, the features for single cells were averaged for each image (median). This gave rise to 120 observations per treatment for each feature. The blue (DAPI) and green (FITC-phalloidin) channel provided together 738 features, while the red (TfR1) channel provided 735 features (Figure S2). Among these features, there were frequently used features such as “Number of Nuclei”, “Nucleus Intensity”, and “Nucleus Roundness”. As expected, different effects are visible for basic features after treatment, but no reasonable classification could be made (Figure S3A–C). In order to validate the quality of the data, we analyzed the membrane fluorescence intensity for the TfR1 signal. As expected, we found a significant increase in TfR1 fluorescence intensity after treatment with RSL3 and IKE but not upon treatment with DMSO or STS (Figure S3D).
Figure 2.
Feature extraction and classifier discovery. (A) The experiment consisted of 120 images per condition (DMSO, IKE, RSL3, STS). The image analysis software extracted 1473 features for the blue, green, and red fluorescence signals. The features can roughly be grouped in intensity, morphology/symmetry, and texture features. Undefined values (NaN, “Not a Number”). (B) Principal component analysis of 1373 features extracted from the images. Individual images are visualized as points on the scatter plot of the first two principal components. The color code is according to the treatment label (black = DMSO, yellow = RSL3, green = IKE, and red = STS) and was added after the PCA was conducted. (C) Feature matrix of the training data set (scaled for visualization purposes) is cleared for highly correlated features (“included”) and informative features are isolated by pairwise logistic lasso regressions (“selected”). Finally, a multinomial logistic lasso regression model is fitted to the reduced feature matrix, and a classifier is identified (“classifier”: 23 features with corresponding regression model coefficients). blgr = bluegreen
We then removed all features that contained undefined values (NaN, “Not a-Number”) and reduced the number of features from 1473 to 1373. We performed a principal component analysis (PCA) with the data matrix of 1373 features and a total of 480 observations (= 120 images per condition; DMSO, IKE, RSL3, and STS) and visualized principal components 1 and 2 (Figure 2B). The cells treated with RSL3 and IKE separated well from the other samples in the first principal component (Figure 2B). As expected, the RSL3-treated and IKE-treated samples overlapped in the first two principal components, as both induce the same type of cell death modality, namely ferroptosis. Cells treated with STS also separated from the DMSO population, although to a lesser extent compared to ferroptosis inducers. STS differs not only from the vehicle DMSO but also from RSL3 and IKE, although cell death in the CTG viability assay performed in parallel was almost identical. This indicated that the staining and analysis strategy was able to distinguish vehicle-treated from ferroptosis, and from apoptosis.
This data set was then used for supervised machine learning to build a classifier that would allow the determination of whether treatments of cells with certain substances trigger ferroptosis or apoptosis (Figure 2C).
A classifier is a mathematical function or procedure that assigns a sample to one or several classes, usually by calculating class scores for each sample (i.e., image) from its feature values. With respect to the type of mathematical procedure, classifiers vary in terms of interpretability and transferability to new data sets. Multinomial logistic regression models using the lasso (least absolute shrinkage and selection operator) inherently provide a feature selection and return a vector of coefficients for the selected features, called signature, which is directly interpretable and transferable.
For numerical stability of a treatment classifier, all non-normally distributed features (Shapiro–Wilk test of normality in discovery data, alpha = 0.05) were Box-Cox transformed (parameters lambda1 = 0 and lambda2 = 1 if the p value of this test was increased by transformation). Reduction of dimensionality was carried out by removal of redundancies (according to feature-pairwise Pearson correlation of |r| > 0.9 in discovery data) and by preselection of informative features through treatment-pairwise logistic lasso regression analysis. Notably, only informative features of limited correlation among each other were used for signature discovery. The CRAN package glmnet was used to perform multinomial logistic lasso regression.19 For classification of three groups (DMSO; IKE/RSL3; STS), a signature of 23 features was identified (Table S1). These features have biological meanings and can be interpreted as such: for instance, the feature “Membrane.Region.Red.SER.Valley.0.px” is based on texture changes (= SER.Valley.0.px; SER = Spots, Edges and Ridges) of the TfR1 staining (= Red) within the cell membrane (= Membrane.Region). We have previously shown that TfR1 plasma membrane intensity staining changes under ferroptotic conditions.10 Thus, it is plausible that this feature should be represented in a classifier signature. Interestingly, the signature also consists of features that are not TfR1 related. For example, the feature “Nucleus.Region.Blue.SER.Saddle.2.px” describes a texture (SER.Saddle.2.px) in the nucleus that is determined using the blue channel (DNA staining). Importantly, this particular texture changes upon treatment with apoptosis inducers, which is expected as apoptosis induces alterations to DNA and chromatin structure. Similar to these two examples, the biological context of features can be interpreted.
Together, this unbiased approach to classifier identification offers the possibility of discovering features that previously have not been considered in cell death. Hence, this strategy allows the development of a signature using features whose changes human eyes would not necessarily perceive and helps to more accurately classify cell death states. Notably, there are highly correlated features in the full data set (Table S2), which are potentially replaceable in the classifier (after refitting the coefficients). Features that were not included in the classifier are not necessarily uninformative—they were not selected, because they do not contribute additional information to improve the classifier.
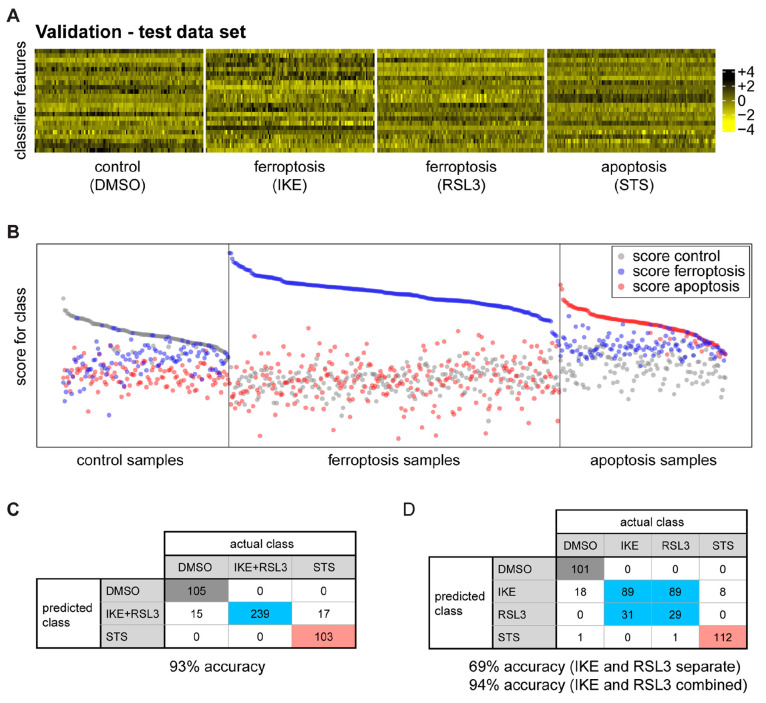
We then collected an independent second image set—using the same conditions with viabilities in the 80–90% range (Figure S4A)—in order to generate biological replicates for model validation (Figure S4B). For this experiment, termed the “validation experiment”, we ran an identical analysis to extract image data and generated the same set of features as was used in the “training experiment”. For model validation, the data from the validation experiment was used to challenge the identified classifier. The coefficients of the 23 features in the classifier were used to predict the class of the samples in the validation experiment, i.e., control, ferroptosis, or apoptosis (Figure 3A,B). The accuracy of prediction for the three classes of control (DMSO), ferroptosis (RSL3+IKE), or apoptosis (STS) was 93% (447 out of 479 cases correct; Figure 3C).
Figure 3.
Model validation. (A) The classifier was applied to the independent test data set for model validation. (B) Comparison of the known class with the predicted class measures classifier performance. Each class is enriched in the corresponding samples, thereby validating the model. (C and D) Confusion tables for the multiclass prediction. (C) DMSO, IKE+RSL3, and STS classes are predicted with an accuracy of 93%. (D) DMSO, IKE, RSL3, and STS are predicted with 94% accuracy, when IKE and RSL3 are combined.
A four-class classifier trained to distinguish the three inducers (IKE, RSL3, and STS), as well as the DMSO control, did not differentiate between IKE and RSL3, as expected. Both classes were assigned identically to IKE (89 cases each) or RSL3 (31 and 29 cases) and minimally to STS (0 or 1 case). Combining IKE and RSL3 resulted in an accuracy of 94% (Figure 3D). Consistently, even when excluded from model discovery, IKE validation set images were constantly identified as RSL3-like by two-class logistic lasso regression classifiers trained to discriminate DMSO control from RSL3 or STS from RSL3 (120 of 120 and 113 of 120 images, respectively–see supplementary PDF file “MachineLearning_Ferroptosis_SI.pdf”: “Binary Prediction”). Importantly, this suggests that both ferroptosis inducers induce a similar phenomenology with respect to the features extracted from the images.
The classifier performed well for detecting ferroptosis, as TfR1 is a known ferroptosis marker, and features from this channel are prominently represented in the signature. However, we were intrigued that apoptosis was also readily distinguished from the control group using the developed signature.
This classifier is based on images of cells treated with ferroptosis or apoptosis inducers and stained with anti-TfR1 3F3-FMA, DAPI, and FITC-Phalloidin. It is important to consider that for any new (unknown) small molecule that is desired to be tested with this classifier, the concentration and incubation times reducing the viability to 80–90% have to be identified in advance. Standardized microscopy image acquisition of treated cells in combination with this classifier could provide the information on whether the substances induce ferroptosis or apoptosis. As with any analysis tool, some refinement might be needed.
Further, this work may have important implications for tissue analysis and allow for a high-throughput, objective procedure to identify ferroptosis and other cell death modalities in a tissue context, whether with animal disease models or patient samples. One such application may involve assessing the response of cancer patients to therapy.9
This classifier cannot directly be applied to images taken under entirely different conditions (treatments, staining, etc.). However, we present a workflow on how researchers can develop a classifier based on a training image set for various cell death processes with the help of standardization of experiments and corresponding analysis tools. Hence, this strategy may serve as a blueprint to be employed for the detection of other cell death pathways, including necroptosis and pyroptosis, and ultimately a universal classifier that detects and classifies all of the major types of cell death.
Methods
Cell Culture
HT-1080 (ATCC Cat# CRL-7951, RRID:CVCL 0317) cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% nonessential amino acids. Cells were grown in a humidified incubator at 37 °C and 5% CO2.
CellTiter-Glo Assay
HT-1080 cells were plated in technical triplicates in white opaque 96-well plates at 15 000 cells/100 μL media per well. For the pilot experiment, the cells were treated with 1 μM RSL3, 20 μM IKE, or 1 μM staurosporine (STS) at different time points. For the immunofluorescence experiments, the cells were treated at the time points determined in the pilot experiment and several time points before and after. A total of 100 μL of 50% CellTiter-Glo (Promega) and 50% cell culture medium was added to each well, and the cells were incubated and shaken for 2 min at RT. Luminescence was measured using a Victor X5 plate reader (PerkinElmer).
Immunofluorescence (IF)
HT-1080 cells were treated with 1 μM RSL3, 20 μM IKE, or 1 μM STS on poly lysine-coated coverslips (Sigma-Aldrich P4832) in 24-well plates. When the cell death percentage reached around 10–20% (determined using the CellTiter-Glo assay), media were removed, and the cells were gently washed with PBS2+ (PBS with 1 mM CaCl2 and 0.5 mM MgCl2) twice, ensuring the cells did not dry out. The cells were fixed and permeabilized with 4% PFA in PBS with 0.1% Triton X-100 (PBT), with 200 μL per well. The plates were covered with foil, and the cells were incubated and shaken at RT for 15–20 min. The PFA was disposed of safely, and the cells were washed with PBT three times. The cells were blocked with 5% normal goat serum (NGS; ThermoFisher 50197Z) in PBT for 1 h at RT. The cells were then incubated with mouse 3F3 anti-Ferroptotic Membrane Antibody (3F3-FMA) at a 1:500 dilution in PBT with 1% bovine serum albumin (BSA) and 5% NGS at 4 °C overnight. The cells were washed with PBT for 5 min three times. The cells were then incubated with goat antimouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Thermo Fisher Scientific Cat# A-11032, RRID:AB_2534091) at 1:200 dilution, and FITC-phalloidin at 1:1000 dilution in PBT with 1% BSA for 1 h at RT. The cells were washed with PBT for 5 min three times. The cells were placed on slides using Prolong Diamond antifade mountant with DAPI (ThermoFisher P36962). All images were collected on a Zeiss LSM 800 confocal microscope using a Plan-Apochromat 63×/1.40 oil DIC objective with constant laser intensity for all images.
Automated Image Analysis
Image analysis was performed using Columbus software version 2.8.0 (PerkinElmer). In the following, the analysis steps in Columbus are described: the DAPI and FITC signals were smoothened for the cell segmentation process using Median filters to reduce noise signals. Nuclei were detected via the DAPI signal. The FITC channel was used to define the cytoplasm and membrane region. In a next step, morphology/symmetry features, texture (SER features), and intensity properties of the DAPI, FITC, and red channel were calculated for each cell region (nuclei, cytoplasm, and membrane). Moreover, we applied a filter to remove border objects (nuclei that cross image borders). For the detailed analysis pipeline in Columbus, please see Figure S2 and the analysis sequences.
Statistical Data Analysis: Transformation and Feature Selection
From two data sets containing 480 samples each (120 DMSO, 120 IKE, 120 RSL3, 120 STS) 1473 features were generated and exported by the Columbus imaging software. The data sets were filtered for completeness, i.e., all features containing “not-a-number” (NaN) were excluded from analysis, resulting in 1373 features. The data set generated first was assigned to model discovery, the second data set to model validation. Features that were non-normally distributed in the discovery data according to the Shapiro test for normality (p < 0.05) were log-transformed (i.e., log(1 + x) also known as two-parameter Box–Cox transformation with lambda1 = 0 and lambda2 = 1), if the transformed data were closer to normality in terms of the Shapiro-test p value. Of all pairs of features that were highly correlated in the discovery data (i.e., absolute Pearson correlation coefficient of larger than 0.9), one member was excluded from analysis iteratively; starting with the feature participating in the largest number of correlations in the training data set for classifier discovery, which was preserved, all highly correlated features were removed from both data sets.
Classifier Discovery
Further feature preselection was conducted on the discovery data by logistic regression for pairwise classification among control, ferroptosis, and apoptosis using the lasso (least absolute shrinkage and selection operator).18 All features that were selected at least once in the pairwise logistic regressions were preserved in the training data set for classifier discovery, on which the classifier was trained. For classification, a multinomial logistic regression model with the lasso was used, resulting in a signature for sample classification. Lambda.1se was used as a criterion for selection of the optimal penalty parameter. The quality of this signature was determined in terms of accuracy of classification of the validation data, where true class membership is known. The importance of signature features was estimated by the product of the standard deviation of the transformed feature in the discovery data and the coefficient in the regression model. All statistical calculations were conducted using R version 4.0.3; for lasso regression, the glmnet package was used.19
Data Availability Statement
The data underlying this study (raw data as txt files, R code Rmd file, and complete and intermediate Rdata files) are openly available in Columbia University Academic Commons at 10.7916/3hdp-9j07.
Acknowledgments
B.R.S. is supported by NCI grants P01CA87497 and R35CA209896, National Institute of Neurological Disorders and Stroke (NINDS) grants R61NS109407 and R33NS109407, and NIH grant UG3CA256962. J.J. was supported by the Columbia College Science Scholars program.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.1c00953.
Description of concepts, data transformation, binary prediction as proof of principle, multiclass prediction using multinomial logistic regression lasso model (PDF)
Pilot study results using CellTiter-Glo viability assay to determine optimal cell death treatment concentration and time point, workflow of automated image analysis, segmentation, and feature extraction, boxplots of representative features, viability data and principal component analysis of validation experiment, analysis sequences (Figures S1–S4) (PDF)
Signature of features in cell death classification (Table S1) (XLSX)
List of features highly correlated to features in signature (Table S2) (XLSX)
Author Contributions
∇ These authors contributed equally
Author Contributions
J.J., K.S., K.H., and B.R.S. conceptualized experiments. J.J. performed imaging collection and viability assays. K.S. performed image analysis, feature extraction, and PCA. D.S. and K.U. formulated the machine learning model and performed classification. J.J., K.S., D.S., K.U., K.H., and B.R.S. performed data analysis. All authors contributed to the writing and editing of the manuscript.
The authors declare the following competing financial interest(s): B.R.S. is an inventor on patents and patent applications involving small molecule drug discovery, ferroptosis, and 3F3-FMA; cofounded and serves as a consultant to Inzen Therapeutics, Nevrox Limited, Exarta Therapeutics, and ProJenX Inc.; and serves as a consultant to Weatherwax Biotechnologies Corporation and Akin Gump Strauss Hauer & Feld LLP. All other authors declare no competing financial interest.
Supplementary Material
References
- Denton D.; Kumar S. Immunostaining Using an Antibody against Active Caspase-3 to Detect Apoptotic Cells in Drosophila. Cold Spring Harb Protoc 2015, 2015, 576–579. 10.1101/pdb.prot086215. [DOI] [PubMed] [Google Scholar]
- He P.; Ai T.; Yang Z.-H.; Wu J.; Han J. Detection of necroptosis by phospho-MLKL immunohistochemical labeling. STAR Protoc. 2021, 2, 100251. 10.1016/j.xpro.2020.100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B. R.; Friedmann Angeli J. P.; Bayir H.; Bush A. I.; Conrad M.; Dixon S. J.; Fulda S.; Gascon S.; Hatzios S. K.; Kagan V. E.; Noel K.; Jiang X.; Linkermann A.; Murphy M. E.; Overholtzer M.; Oyagi A.; Pagnussat G. C.; Park J.; Ran Q.; Rosenfeld C. S.; Salnikow K.; Tang D.; Torti F. M.; Torti S. V.; Toyokuni S.; Woerpel K.A.; Zhang D. D. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S.; SriRamaratnam R.; Welsch M. E.; Shimada K.; Skouta R.; Viswanathan V. S.; Cheah J. H.; Clemons P. A.; Shamji A. F.; Clish C. B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larraufie M.-H.; Yang W. S.; Jiang E.; Thomas A. G.; Slusher B. S.; Stockwell B. R. Incorporation of metabolically stable ketones into a small molecule probe to increase potency and water solubility. Bioorg. Med. Chem. Lett. 2015, 25, 4787–4792. 10.1016/j.bmcl.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.; Stockwell B. R.; Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland A.; Wang Y.; Wu W.; Lan X.; Han X.; Li Q.; Wang J. Ferroptosis and Its Role in Diverse Brain Diseases. Molecular Neurobiology 2019, 56, 4880–4893. 10.1007/s12035-018-1403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli J. P.; Krysko D. V.; Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 2019, 19, 405–414. 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- Chen X.; Kang R.; Kroemer G.; Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin Oncol. 2021, 18, 280–296. 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- Feng H.; Schorpp K.; Jin J.; Yozwiak C. E.; Hoffstrom B. G.; Decker A. M.; Rajbhandari P.; Stokes M. E.; Bender H. G.; Csuka J. M.; et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020, 30, 3411–3423. 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Zak O.; Aisen P.; Harrison S. C.; Walz T. Structure of the Human Transferrin Receptor-Transferrin Complex. Cell 2004, 116, 565–576. 10.1016/S0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Yang W. S.; Stockwell B. R. Synthetic lethal screening identifies compounds activating iron-dependent, non-apoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008, 15, 234–245. 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Gao C.; Yan Y.; Cheng Z.; Chen G.; Rui T.; Luo C.; Gao Y.; Wang T.; Chen X.; et al. Ruxolitinib exerts neuroprotection via repressing ferroptosis in a mouse model of traumatic brain injury. Exp. Neurol. 2021, 342, 113762. 10.1016/j.expneurol.2021.113762. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Cai L.; Wang S.; Wang J.; Chen B. Baicalin Prevents Myocardial Ischemia/Reperfusion Injury Through Inhibiting ACSL4Mediated Ferroptosis. Front Pharmacol. 2021, 12, 628988. 10.3389/fphar.2021.628988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.; Gerlich D. W. Machine learning in cell biology – teaching computers to recognize phenotypes. J. Cell Sci. 2013, 126, 5529–5539. 10.1242/jcs.123604. [DOI] [PubMed] [Google Scholar]
- Bertrand R.; Solary E.; O’Connor P.; Kohn K. W.; Pommier Y. Induction of a Common Pathway of Apoptosis by Staurosporine. Exp. Cell Res. 1994, 211, 314–321. 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H.; Kerr J. F. R.; Currie A. R. Cell Death: The Significance of Apoptosis. Int. Rev. Cytol. 1980, 68, 251–306. 10.1016/S0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J. R Stat Soc. Series B Stat Methodol. 2011, 73, 273–282. 10.1111/j.1467-9868.2011.00771.x. [DOI] [Google Scholar]
- Friedman J.; Hastie T.; Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 2010, 33, 1–22. 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this study (raw data as txt files, R code Rmd file, and complete and intermediate Rdata files) are openly available in Columbia University Academic Commons at 10.7916/3hdp-9j07.