Abstract
The nematode Caenorhabditis elegans is a powerful model organism for studying cell development, apoptosis, neuronal circuits, and aging. The isolate N2 is recognized by the C. elegans community as the reference wild-type strain. Interestingly, the lifespan of presumably isogenic C. elegans N2 worms—even when grown under comparable conditions—varies significantly amongst distinct laboratories. This hinders the inter-laboratory comparability of C. elegans lifespan data and raises questions regarding data interpretation and reproducibility. Here, we hypothesized slight alterations in experimental design and worm handling could explain the observed discrepancies. To test this hypothesis, we collected and assessed data from over 1000 published C. elegans N2 lifespan assays as well as corresponding methodological meta-data. We find that mean N2 lifespans range from approximately 7 days to upwards of 35 days, despite laboratories disclosing seemingly comparable experimental conditions. We further demonstrate that, in addition to temperature, the use of the chemical sterilizer 5-fluoro-2′-deoxyuridine (FUDR) may change N2 lifespan. Additionally, we observed differences in average N2 lifespan from experiments originating from distinct geographic locations, indicating a potential effect of location-specific factors on experimental outcomes. Taken as a whole, our work indicates the sum of many small, rather than a few critical, differences in experimental conditions may account for the observed variance in N2 lifespan. We also find that the absence of standardized experimental methods and the insufficient disclosure of experiment details in the peer-reviewed literature limits the inter-lab comparability of published results. We thus propose the establishment of a succinct reporting standard for C. elegans lifespan experiments to increase the reliability and reproducibility, and thus scientific value, of these studies.
Keywords: N2, C. elegans, Lifespan, Aging, Variation, Reproducibility
1. Introduction
Caenorhabditis elegans was first isolated from mushroom compost in Bristol, England in 1951. The following decade, Dr. Sydney Brenner pioneered the use of this roundworm to model biological phenomena (Sterken et al., 2015). After culturing worms from his own garden (which he named N1) in the early 1960’s, Dr. Brenner obtained a “Bristol” C. elegans isolate from the laboratory of Dr. Ellsworth Doherty in 1963 (Sterken et al., 2015; WormBook, 2017). Following several passages of these worms on agar plates, Brenner selected and isolated a single hermaphroditic worm from Doherty’s strain for cohort expansion—thus the N2 (Bristol) strain was born (Sterken et al., 2015; Ankeny, 2001; Riddle et al., 1997; Brenner, 1974). Today, the N2 strain serves as the genuine wild-type reference strain in C. elegans research across the world for the investigation of various scientific questions.
Using C. elegans N2 as a model system offers several advantages over other invertebrate models: First, C. elegans’ self-mating behavior and short reproductive cycle allow for the rapid and technically simple establishment of large, isogenic, and age-synchronized cohorts (Porta-de-la-Riva et al., 2012). Second, the anatomy, developmental cell ancestry, and neuronal connectivity of C. elegans N2 is fully mapped (Altun and Hall, 2009). Third, approximately 80% of C. elegans N2 genes have orthologs in humans, indicating a high translational potential for findings made using worm models (Lai, 2000). Indeed, C. elegans-based research has led to several fundamental discoveries impacting human health and biomedical research, including the first description of apoptosis (awarded the 2002 Nobel prize in Medicine), seminal work on RNA interference (awarded the 2006 Nobel prize in Medicine), and ground-breaking findings on the genetics of aging (Barbour, 2002; Fire et al., 1998; Vanfleteren and Braeckman, 1999). Fourth, in laboratory conditions C. elegans N2 has a relatively short lifespan, making it an attractive model for aging research (Altun and Hall, 2009). Fifth, the genetic toolbox for C. elegans research, including RNA interference and CRISPR-Cas9 technology, is readily available and efficient (Fire et al., 1998; Dickinson and Goldstein, 2016). Altogether, these characteristics render C. elegans into a powerful model to investigate important questions relevant to human health, disease, and aging (Tissenbaum and Using, 2015; Zhang et al., 2020).
In the wake of the current efforts to increase rigor and reproducibility in research, inter-lab differences in C. elegans N2 physiology and behavior have come under scrutiny. This includes differences in baseline N2 lifespan (Gems and Riddle, 2000). Here, we analyze and compare more than 1000 peer-reviewed and published N2 lifespan experiments and methodological meta-data with the goal to identify factors explaining the observed variability in N2 survival. A matrix of collected meta-data (titled “N2 Lifespans Final”) is shared via GitHub (https://github.com/jpcav97/N2-Lifespans). We show that differences in assay temperature, the use of 5-fluoro-2′-deoxyuridine (FUDR), as well as the geographic location of the laboratory may contribute to the observed inter-lab variability. We also find that insufficient disclosure of experimental details in peer-reviewed C. elegans longevity studies hinders both reproducibility and comparability of lifespan data across different laboratories.
2. Results
2.1. Data collection and characteristics
We first collected N2 lifespan data from 1018 independent experiments disclosed in 817 separate publications. We sampled publications from distinct journals and publishers to minimize any potential journal-specific bias. From each lifespan experiment, we extracted the percentage of worms alive on day 3, 5, 10, 15, 20, 30, 40 and 50 of adulthood from published lifespan curves as well as reported mean lifespans (if disclosed). To exclude the known impact of C. elegans diet on longevity in our analysis, we only collected N2 lifespan data from worms kept on E.coli OP50 as the food source (Win et al., 2013; Stuhr and Curran, 2020). Subsequently, we recorded experimental conditions reported in the materials and methods section, main text, figure legend, and/or supplemental information. A matrix of collected metadata (titled “N2 Lifespans Final”) is shared via GitHub (See Material and Methods, Sections 4.1–4.2). Our final dataset consisted of experimental data acquired in 36 different countries published between 1993 and 2021, shared by 517 unique last authors.
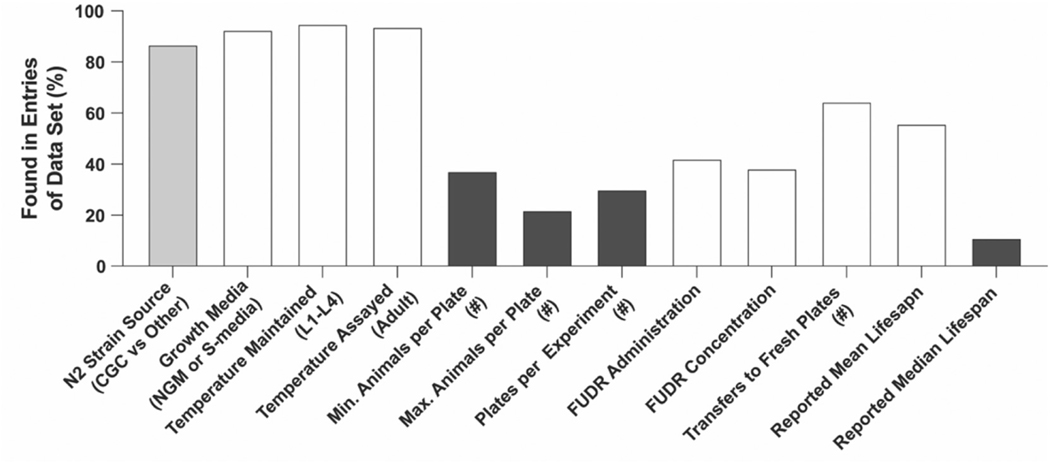
Of note, many of the experimental features for which we surveyed were underreported and hence could not be used for our analysis (Fig. 1). For example, only ~ 22% of experiments contained data on the number of animals per assay plate and ~ 30% of publications disclosed the number of replica plates used per experiment (Fig. 1). Consequently, we used only 7 of 12 features surveyed in our analysis. To account for major experimental differences, we categorized the data based on growth conditions (solid nematode growth media (NGM) or liquid S-media) and growth temperature in the larval stages and adulthood, as these were the most consistently reported and also known to impact lifespan (Vanfleteren and Braeckman, 1999; Win et al., 2013). Based on these features only, we identified 17 different experimental conditions used at least three times (Table 1). This exemplifies the diversity of experimental conditions used in C.elegans-based longevity research. Most experiments had worms maintained constantly at 20 °C on solid NGM plates (n = 702). Other frequently used conditions included worms maintained and assayed constantly at 15 °C on solid NGM plates (n = 18) and constantly at 25 °C on solid NGM plates (n = 66) (Table 1). We focused on these 3 conditions, as they were the most abundant, fully reported conditions that did not involve a temperature shift between the larval and assay periods. The curated dataset for analysis thus consisted of information from 786 N2 lifespan experiments reflecting 3 distinct and common assay conditions.
Fig. 1.
Levels of assay feature disclosed in analyzed data set.
White. Assay features used in analysis. Gray: assay feature disclosed in more than 50% of publications but not used for analysis. Dark gray: assay feature disclosed in less than 50% of experiments and not used in analysis.
Table 1.
Assay conditions identified in the data set based on growth media and temperature. Many experimental conditions were excluded due to insufficient reporting of conditions and/or insufficient number of entries.
| Assay Condition | Growth Media | Temperature Maintained (Through L4, °C) | Temperature Assayed (Adult, °C) | Count |
|---|---|---|---|---|
|
| ||||
| a1 | NGM | 20 | 20 | 702 |
| a2 | NGM | 25 | 25 | 66 |
| 3 | N/A | 20 | 20 | 51 |
| 4 | NGM | 20 | 25 | 27 |
| a5 | NGM | 15 | 15 | 18 |
| 6 | S-Medium | 20 | 20 | 17 |
| 7 | NGM | N/A | N/A | 17 |
| 8 | NGM | 25 | 20 | 16 |
| 9 | NGM | 15 | 20 | 11 |
| 10 | NGM | 22 | 22 | 10 |
| 11 | N/A | N/A | N/A | 10 |
| 12 | NGM | 15 | 25 | 9 |
| 13 | NGM | 20 | 15 | 7 |
| 14 | N/A | 25 | 25 | 6 |
| 15 | NGM | 25 | 15 | 5 |
| 16 | N/A | 15 | 15 | 3 |
| 17 | NGM | 21 | 21 | 3 |
| Other | 42 | |||
Assay condition used in analysis.
2.2. N2 lifespans are inconsistent across published experiments
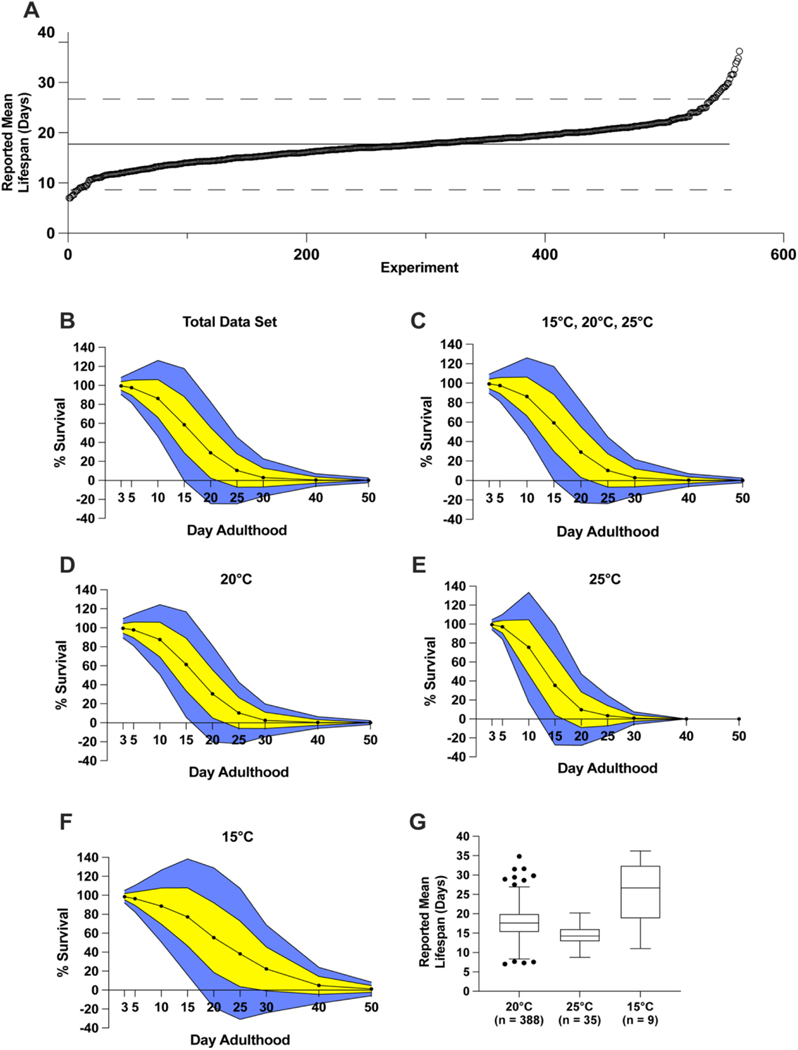
For our analysis, we focused on the percentage of worms alive on specific days of adulthood as well as reported mean lifespans, if reported in the manuscript or supplement. We found data for reported mean lifespan in approximately 55% (n = 432) of experiments for our 3 distinct groups and approximately 57% of all assessed publications (Fig. 1, Table S1). Reported mean lifespans for C. elegans N2 ranged from approximately 7 days to more than 35 days, with an average of 17.7 days (95% CI: 17.33–18.06) (Fig. 2A). Notably, 32 reported mean lifespans lay outside two standard deviations of the mean, with 9 outside the lower limit and 23 outside the upper limit (Fig. 2A). Assessing survival curves of the entire 1018 experiments in our dataset, we observed the largest deviations in survival amongst experiments on day 15 (± 29.54%) and day 20 (± 26.77%) of adulthood (Fig. 2B). We also calculated the average condition-specific reported mean lifespans and found large variations based on growth media and temperature (Fig. S1). We then collated our 3 distinct assay conditions and found the largest deviations still occurred on day 15 (± 29.34%) and day 20 (± 26.13%) (Fig. 2C). Deviations in the percentage of survival of more than ± 20% were consistently observed for all distinct temperatures at most time points (Fig. 2D–F). The least consistent results were observed for survival on day 20 of adulthood in experiments performed at 15 °C (± 36.87%) (Fig. 2F). As expected, we found a significant negative relationship (p < 0.0001) between assay temperature and mean N2 lifespan, as confirmed by an Analysis of Means (ANOM) test for unbalanced samples (95% confidence level) (Fig. S2A, Fig. 2G). We did not find a strong linear relationship between average lifespans at these temperatures after linear regression analysis (R2 0.09) (Fig. S12). The average of all reported mean lifespans for worms assayed at 15 °C was 25.2 days (95% CI: 18.69–31.64), at 20 °C was 17.7 days (95% CI: 17.29–18.08), and at 25 °C was 14.6 days (95% CI: 13.59–15.61) (Fig. 2G, Fig. S3A, Table S1). Together, these results demonstrate assay temperature is a key factor affecting C. elegans lifespan. Our results further indicate that factors other than assay temperature and growth media affect N2 physiology and longevity.
Fig. 2.
Variability of N2 lifespans in the published literature.
A. Reported mean lifespan of all entries in the data set. Average reported N2 lifespan was 17.7 days, as represented by the solid black line. Each circle represents an individual experiment. Dashed line indicates two standard deviations from the mean. B–F. Average lifespan on the respective day for the total data set (B), combination of assay conditions 1, 2, and 5 in Table 1 (C), and respective individual assay conditions (D–F). Yellow indicates 1 standard deviation from the mean; blue represents 2 standard deviations from the mean. Survival above 100% and below 0% is not physiologically possible. G. Tukey-method boxplot for experiments at 20 °C, 25 °C, and 15 °C (assay conditions 1, 2, and 5, respectively, from Table 1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. The usage of 5-fluoro-2′ -deoxyuridine (FUDR) may increase N2 lifespan
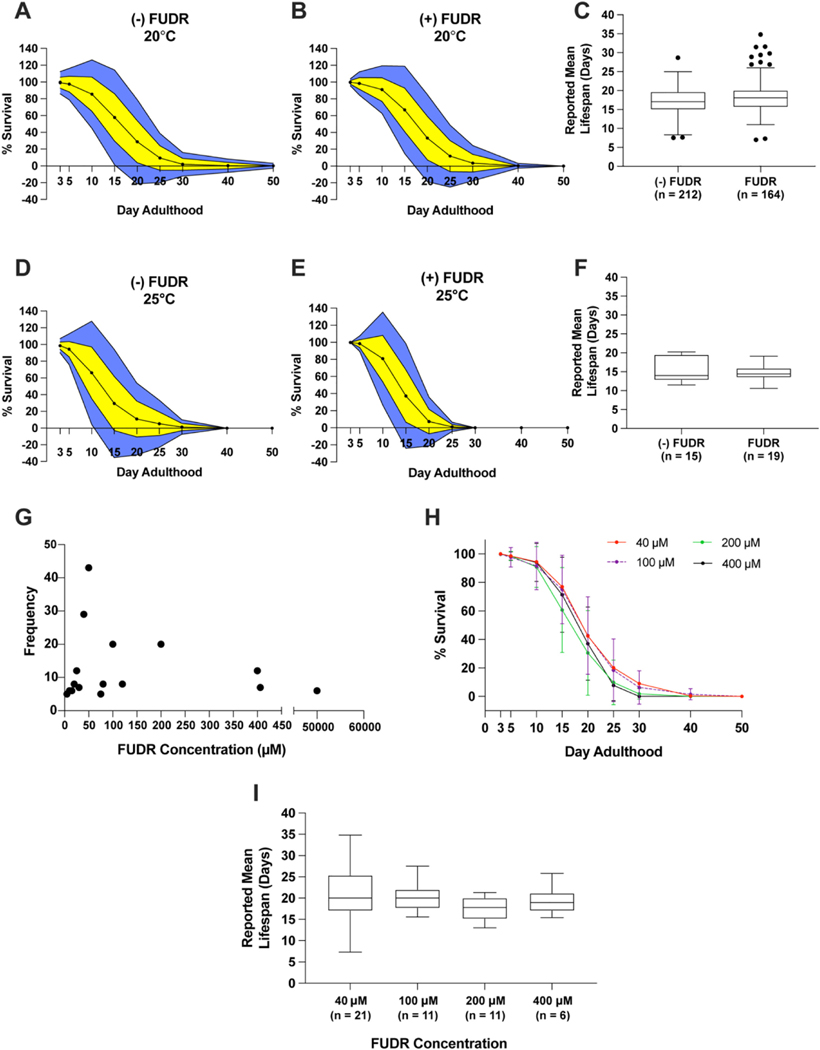
C. elegans lay eggs and produce viable progeny, hundreds per adult worm, beginning on day 1 of adulthood (Altun and Hall, 2009). The maturation of progeny into adults within approximately 72 h complicates the continuous identification and observation of parent assay worms. To circumvent this issue, chemical sterilizers, such as the thymidylate synthase inhibitor 5-fluoro-2′-deoxyuridine (FUDR), are often used to inhibit progeny maturation (Mitchell et al., 1979). We compared experimental data from N2 lifespans performed at 20 °C on solid NGM with or without FUDR. Sub-partitioning our datasets into ± FUDR was not sufficient to reduce or eliminate the previously observed inter-laboratory variance in N2 lifespan (Fig. 3A–B). Experiments which used FUDR had a reported mean N2 lifespan of 18.28 days (95% CI: 17.67–18.92) and those without FUDR showed a reported mean N2 lifespan of 17.18 days (95% CI: 16.73–17.72) at 20 °C (p = 0.0075) (Fig. 3C, Fig. S2B). However, given the variance in mean lifespans (FUDR SD = 3.7; (–) FUDR SD = 4.2) and an overlap of confidence intervals (Fig. S3B), the statistical significance of these differences has to be taken with caution. The use of FUDR resulted in no observable difference (p = 0.31) in N2 longevity at 25 °C (Fig. 3D–F, Fig. S2C). Amongst experiments using FUDR at 20 °C, reported mean N2 lifespans was not different with the usage of different FUDR concentrations (p = 0.34), which ranged from 5 μM-50 mM (Fig. 3G–I, Fig. S2D). Taken as a whole, our data indicate the use of FUDR in longevity experiments may extend N2 lifespan at 20 °C. Additionally, the presence or absence of FUDR alone is not sufficient to explain the observed inter-lab variance prevalent in published N2 survival experiments (Fig. 3A–B, D–E).
Fig. 3.
Analysis of the effect of 5-fluoro-2′-deoxyuridine on N2 lifespan.
A-B. Average lifespan on each respective day for experiments without (A) and with (B) FUDR at 20 °C. C. Tukey-method boxplot comparing experiments with and without FUDR at 20 °C. D-E. Average lifespan on the respective day for experiments without (D) and with (E) FUDR at 25 °C. F. Tukey-method boxplot comparing mean lifespan of experiments with and without FUDR at 25 °C. G. Scatter plot comparing the frequency of FUDR concentrations observed in the dataset. H. Average lifespan on each respective day for commonly used FUDR concentrations. Error bars represent 1 standard deviation from the mean at 20 °C. I. Tukey-method boxplot of reported mean lifespans of 4 commonly used FUDR concentrations at 20 °C. All experiments were on solid NGM agar. For A-B, D-E, yellow indicates 1 standard deviation from the mean; blue indicates 2 standard deviations from the mean. Survival above 100% and below 0% is not physiologically possible. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Worm manipulation has no significant effect on N2 longevity
In the absence of a chemical sterilizer for colony control, adult N2 worms in lifespan experiments must be transferred to fresh OP50 plates approximately every 48 h at least until they cease to reproduce. These transfers are a likely source of physical distress for worms. Thus, we wondered if repetitive worm transferring impacts longevity. We hypothesized increased manipulation (transfer)-dependent stress would decrease longevity. To test our hypothesis, we compared data from experiments in which worms were transferred to fresh assay plates either less (Group 1, “(–) manipulation”) or more (Group 2, “(+) manipulation”) than twice during the experiment. Our analysis showed no difference in mean reported N2 lifespan (p = 0.18) between these two groups, suggesting that worm handling-inflicted stress does not affect N2 longevity assays (Fig. 4A–C, Fig. S2E). Notably, the use of FUDR was similar between the two groups (Fig. S4). The large inter-laboratory variance in N2 survival remained and peaked at day 15 for Group 1 (± 25.65%) and Group 2 (± 29.51%) (Fig. 4A–B). A further sub-division of Group 2 based on worm transfer occurring more frequently during the reproductive phase confirmed that differences in worm handling did not affect N2 lifespans (Fig. 4D–F). Altogether, the data indicate worm manipulation/transfer does not influence N2 lifespan.
Fig. 4.
The effect of worm transfers on N2 lifespan.
A-C. Average survival curves for the respective day of adulthood (A-B) and Tukey-method boxplot of reported mean lifespans (C) for experiments which transformed worms less than (“(−) manipulation”) or more than (“(+) manipulation”) twice during the experiment. D-E. Average survival curves from the “manipulation” group which transferred worms everyday (D) or every 2–3 days (E) during the reproductive period. F. Overlay of the survival curves of D-E. Error bars represent 1 standard deviation from the mean. For A-B, D-E, yellow indicates 1 standard deviation from the mean and blue indicates 2 standard deviations from the mean. Survival above 100% and below 0% is not physiologically possible. All experiments were done on solid NGM at 20 °C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. Differences in N2 longevity are related to distinct geographic locations
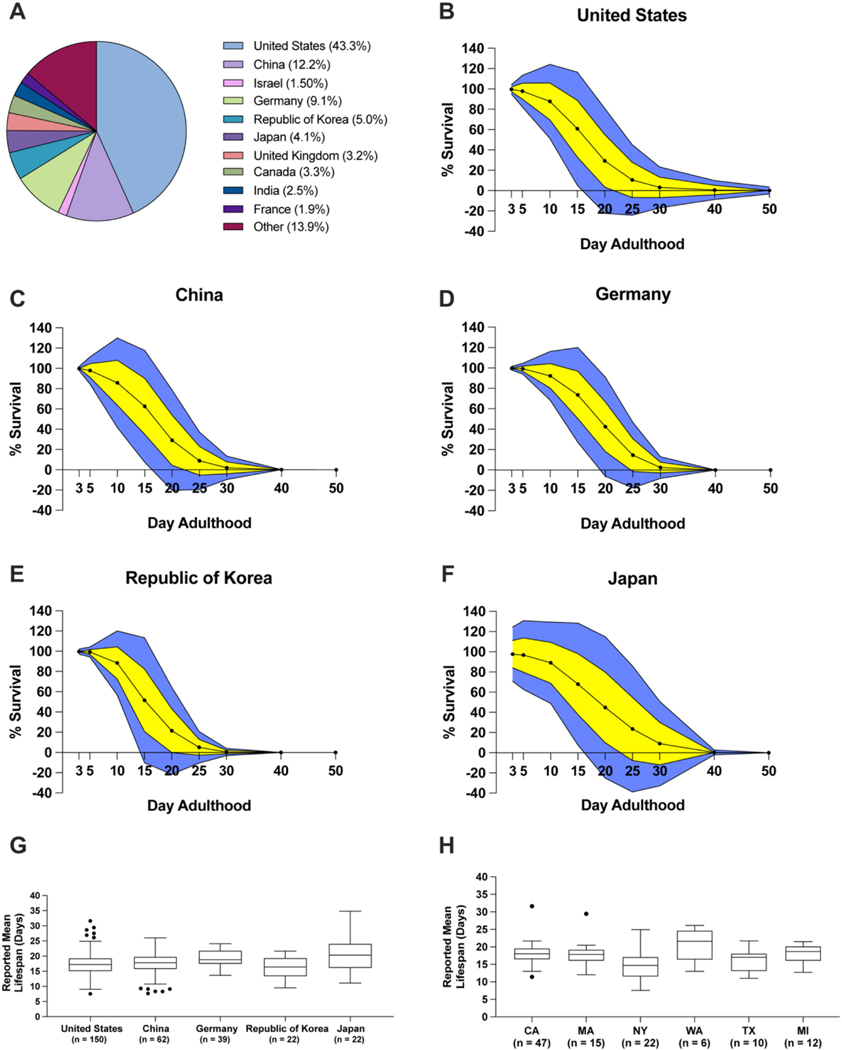
C. elegans lifespan experiments are potentially sensitive to subtle changes in environmental parameters. In addition to temperature, humidity, air pressure, and air quality are known to impact C. elegans health (Haghani et al., 2019). We thus wondered if differences in local climate influence N2 longevity. Our dataset contained information from experiments reported from 36 different countries. The three most represented countries accounted for 64.6% of all data collected (United States, 43.3%; China, 12.2%; Germany, 9.1%) (Fig. 5A, Fig. S5). To determine if the geographic location of laboratories affects N2 survival, we analyzed data from the 10 most represented countries in the dataset. Combined, these 10 countries account for 86.2% (878 lifespans) of our dataset. We did not analyze the remaining 13.8% of the experiments, as they were distributed amongst the remaining 26 countries and resulted in few experiments per country. We noticed the percentage of animals alive on days 15, 20, and 25 of adulthood at 20 °C was variable amongst these 10 countries (Fig. S6). We also observed notable deviation in N2 lifespan amongst the 5 most represented countries (Fig. 5B–F). Interestingly, the average reported mean lifespan of N2 worms was increased in experiments from Japan and Germany compared to other countries (p < 0.05) (Fig. 5G, Fig. S2F). We further performed a similar analysis considering individual states within the United States. In the entire data set, N2 lifespan information from experiments originating in the United States most often came from California (25.2%), Massachusetts (11.0%), Michigan (8.7%), and New York (7.5%) (Fig. S7). Our analysis showed that N2 lifespans were shorter lived when reported from New York-based laboratories as compared to other US locations (p < 0.05) (Fig. 5H, Fig. S2G). These findings indicate the geographic location of the laboratory may contribute to the variation observed in N2 lifespan experiments.
Fig. 5.
Distinct geographic locations show differences in N2 lifespan.
A. Analysis of countries experiments were performed in the entire dataset. B–F. Average survival curves for the United States (B), China (C), Germany (D), Republic of Korea (E), and Japan (F) at 20 °C on solid NGM. Yellow indicates 1 standard deviation from the mean and blue indicates 2 standard deviations from the mean. Survival above 100% and below 0% is not physiologically possible. G-H Tukey-method boxplots of the reported mean N2 lifespans of different countries (G) and states of the United States (H). CA: California; MA: Massachusetts; NY: New York; WA: Washington; TX: Texas; MI: Michigan. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
The nematode C. elegans is a well-established model system popular in the aging research community. The N2 strain is generally recognized as the wild-type reference control in C. elegans experiments. However, as we demonstrate here, survival and mean lifespan of published, peer-reviewed N2 lifespan experiments range from approximately 7 to over 35 days at comparable experimental conditions (Fig. 2A, Fig. S9). This lab-to-lab variance in N2 survival limits the inter-lab comparability of N2 lifespan results and likely affects experimental reproducibility. Considering N2 survival is commonly used as a benchmark to decide whether mutations or treatments alter C. elegans lifespans, the observed inconsistencies are concerning. This variance may lead to contradicting results from different labs regarding the role of particular genes in C. elegans lifespan regulation.
Our analysis revealed that, in addition to assay temperature, the usage of FUDR may influence N2 survival. The practice of using FUDR in lifespan experiments has been questioned previously due to a lack of complete comprehension for its mechanism of action and potential effects on worm development. Indeed, FUDR has been shown to alter worm proteostasis, a process critically involved in lifespan regulation (Feldman et al., 2014). The temporal administration of FUDR has been shown to impact N2 phenotypes and possibly senescence (Wang et al., 2019), and an extension of lifespan after FUDR administration to mutant C. elegans has been previously established (Aitlhadj and Stürzenbaum, 2010; Van Raamsdonk and Hekimi, 2011; Kato et al., 2016). Our results suggest the possibility that the use of FUDR may contribute to altering N2 lifespan as well. However, due to the large variability in our ± FUDR dataset, we do not find strong statistical support to define FUDR usage as a significant factor in N2 lifespan experiments.
Our analysis further indicates that the geographic location of the laboratory in which the experiment was performed contributes to the observed variance in N2 lifespan. Notably, wild C. elegans isolates from different geographic locations were shown to acquire specific mutations in response to differences in elevation, humidity, and temperature prevalent at sites of isolation (Evans et al., 2017). Unlike temperature, however, humidity, air quality, and air pressure are not explicitly monitored and/or regulated during C. elegans lifespan experiments. Experimental support for this hypothesis thus remains to be provided. Another possible explanation for inter-lab differences in N2 survival may lay in local sharing of N2 reference worms. Inheriting C. elegans strains from neighboring/nearby laboratories, for example when junior investigators become independent, facilitates the installment of high-passage number N2 strains as lab-specific references. However, continuous passaging of N2 worms is likely leading to the acquisition of spontaneous mutations as they are passaged. We thus suggest to regularly replenish N2 reference strains with fresh batches obtained from the Caenorhabditis Genetics Center (CGC).
Perhaps not surprising, our study highlights the variable and often insufficient disclosure of experimental details critical for inter-lab data comparisons and data reproduction. Our data collection strategy was focusing on experimental features we expected to be consistently reported in all publications. This approach had many benefits, as it was 1) technically simple; 2) unbiased towards specific journals and/or laboratories; and 3) high throughput. However, key experimental details were frequently missing in the published literature, as highlighted in Fig. 1. For example, much of our analysis relied on reported mean lifespan, which was only disclosed in ~57% of publications. These gaps in disclosing experimental details reduce the cross-comparability of results amongst laboratories. We also find that while most ANOM tests yielded in p-values of less than 0.05, group-specific confidence intervals show little separation or even overlap. This implies that despite a “significant” p-value, different groups may not be reproducibly distinct (Figs. S2, S3). Reducing the variability in our dataset would lead to a clearer distinction between group-specific confidence intervals, which is critical to draw well-supported conclusions regarding the significance of how individual factors affect N2 lifespan. A possible solution to this problem would be the establishment of a reporting standard for the performance and disclosure of C. elegans lifespan experiments. We thus propose a community-wide effort to define and implement a standardized set of protocols for lifespan experiments. Similar recommendations have been proposed by earlier multi-lab consortia, perhaps most prominently by the Caenorhabditis Intervention Testing Program (CITP) (Lucanic et al., 2017). Notably, the CITP demonstrated that the adherence to standardized experimental conditions is sufficient to reduce inter-lab N2 lifespan variance to approximately 10% (Lucanic et al., 2017). The remaining variability may arise from environmental stimuli (humidity, salinity, altitude, air pressure, etc.), acquired strain-specific background mutations, epigenetic traits, and/or experimenter-related bias. The possibility of variance in N2 lifespan arising due to experimental-related scoring bias may be diminished in the near future by the use of auto- mated lifespan scoring techniques (Felker et al., 2020; Le et al., 2020; Puchalt et al., 2020). Finally, we also have to consider that naturally occurring developmental variation may contribute to our observations. The production of different phenotypes in isogenic populations cultured in the same environment is well-documented (Molenaar et al., 1993; Vogt et al., 2008; Gärtner, 2012). To what extend this applies to C. elegans remains to be studied.
In conclusion, we believe the implementation of standardized and/or automated protocols paired with more succinct methodological reporting would lead to more reliable and reproducible data, benefiting biomedical research as a whole.
4. Materials and methods
4.1. Data collection
Data was collected during the period of May 2020 – April 2021 from published articles in peer-reviewed journals. Publications were found via Google or PubMed searches similar to “C. elegans lifespan experiments” and were chosen at random, typically from the top of the search results page downwards. As data collection continued and new experiments became increasingly more difficult to find, more specific search criteria (i.e. a specific disease/field, such as Alzheimer’s disease or neurodegeneration, or other key words such as a certain country) were added to the search criteria to widen the search. In order to be included for analysis, experiments must have 1) used wild-type N2 worms as a control, 2) had N2 worms grown on OP50 E. coli (hence N2 worms grown on e.g. E.coli HT115 were excluded), and 3) had N2 worm plates remained untreated by drugs or other experimental compounds. Lifespan curves were carefully measured using straight-edged rulers and reported accordingly for the percentage of animals alive on days 3, 5, 10, 15, 20, 25, 30, 40, and 50 of adulthood. Papers were then meticulously examined for details regarding growth media, experimental conditions, use of FUDR, geographic location, etc. and data was tabulated accordingly (Fig. 1). If data could not be found in the published article, the cell was left blank and disregarded from the analysis. Multiple experiments from the same publication were considered as separate data points, as long as the experiments were done independently. The ratio of labs to number of measurements is roughly 0.60 for countries and 0.45 for states. The number of values per lab ranges from 1 to 13 with a median of 1, indicating large diversity in the database. The database is accessible following this link: https://github.com/jpcav97/N2-Lifespans.
4.2. Data curation, analysis, and figure generation
Meta-analysis was done via custom-made Python script using Spyder (Version 4.51). The script can be found on GitHub.com, following this link: https://github.com/jpcav97/N2-Lifespans. Data was saved as a. csv file, then imported into GraphPad Prism (Version 9.1.0) for figure generation and data analysis. For analysis, we excluded information from lifespan assays that did not disclose temperature (n = 17), growth media (n = 59), both (n = 10), or included a temperature shift between larval development and adult lifespan assay conditions (n = 79). We further excluded experiments with worms constantly at 22 °C (n 10) and 21 (n = 3) due to lack of statistical power. Experiments using conditions which appeared less than 3 times were disregarded (n = 42). Data was placed into groups based on three experimental conditions: temperature maintained (hatching through larval stage 4), temperature assayed (adulthood), and growth media (solid nematode growth media or liquid S-medium). As such, each group contained data entries with identical temperatures and growth media. Conditions were added for different types of analysis, such as use of FUDR or worm manipulation. For most of the analysis, experiments maintained and assayed at 20 °C on solid nematode growth media (Assay Condition 1, Table 1) were used, unless otherwise stated. To generate a lifespan curve, the percent of worms alive was averaged over all experiments under the same experimental conditions for each day extracted. This technique allowed for analysis of each lifespan experiment in the desired group being analyzed. For grouped summary data, the data was extracted the same as for lifespan curves and then further analyzed based on the percentage of worms alive on a given day for a given bin size (Figs. S5 and S6). One and two standard deviations (SD) from the mean were calculated and plotted onto mean lifespan curves. For some lifespan curves, the theoretical mean ± 1SD and/or ± 2SD space resulted in some area coverage below 0% and/or above 100% survival. These values, while physiologically unachievable, serve to define the theoretical mean ± 2SD space and were not observed but mathematically calculated. Boxplots were generated and visually reported using the Tukey boxplot method by collating all reported mean lifespans for the target experimental group.
4.3. Statistical analysis
We performed a Shapiro-Wilks test to confirm that data is normally distributed. We further analyzed the data by performing a one-factor unbalanced Analysis of Mean (ANOM) test, which accounts for differences in sample size. All ANOM tests were performed using a 95% confidence level (α = 0.05). These analyzes were done using custom-made Python script using Spyder (Version 4.51). The results of all ANOM tests are graphically represented in Supplemental Fig. S2. For each Figure, the center horizontal line is the grand mean (the mean of all reported mean lifespans including all groups compared in that graph). The decision lines are specific to each group and are the dashed lines above and below the grand mean line. The mean of each group is represented by the black dot. If a group mean (black dot) is above or below the group’s decision line, that group mean is statistically higher or lower, respectively, from the grand mean (α = 0.05). A visual representation and matrix of mean lifespan data and supporting confidence intervals is provided in Supplemental Fig. S3 and Table S1, respectively.
Supplementary Material
Acknowledgements
The authors would like to thank Shannon M. Lacy for her feedback on the manuscript.
Funding Source(s)
This work was supported by the Systems and Integrative Biology Training Grant T32 GM832230 (NDU), AARG, MICHR UL1TR002240, and R35GM142561 (MCT), and the Undergraduate Research Opportunity Program of the University of Michigan.
Footnotes
CRediT authorship contribution statement
Nicholas D. Urban: Conceptualization, Investigation, Data curation, Formal analysis, Writing – original draft, Visualization, Funding acquisition, Project administration. Joseph P. Cavataio: Software, Data curation, Formal analysis. Yasmeen Berry: Investigation. Brandon Vang: Investigation. Anirudh Maddali: Investigation. Richard J. Sukpraphrute: Investigation. Santiago Schnell: Writing – review & editing. Matthias C. Truttmann: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.exger.2021.111622.
References
- Aitlhadj L, Stürzenbaum SR, 2010. The use of FUdR can cause prolonged longevity in mutant nematodes. Mech. Ageing Dev. 131 (5), 364–365. 10.1016/j.mad.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Altun Z, Hall D, 2009. Introduction. Worm Atlas. 10.3908/wormatlas.1.1. [DOI] [Google Scholar]
- Ankeny RA, 2001. The natural history of Caenorhabditis elegans research. Nat Rev Genet. 2 (6), 474–479. 10.1038/35076538. [DOI] [PubMed] [Google Scholar]
- Barbour V, 2002. Celebrating death—the 2002 Nobel prize in physiology or medicine. Lancet 360 (9340), 1117. 10.1016/S0140-6736(02)11240-2. [DOI] [PubMed] [Google Scholar]
- Brenner S, 1974. The genetics of Caenorhabditis elegans. Genetics 77 (1), 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Goldstein B, 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202 (3), 885–901. 10.1534/genetics.115.182162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KS, Zhao Y, Brady SC, Long L, McGrath PT, Andersen EC, 2017. Correlations of genotype with climate parameters suggest Caenorhabditis elegans niche adaptations. G3 GenesGenomesGenetics 7 (1), 289–298. 10.1534/g3.116.035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Kosolapov L, Ben-Zvi A, 2014. Fluorodeoxyuridine improves Caenorhabditis elegans proteostasis independent of reproduction onset. Sherman M, ed. PLoS ONE 9 (1), e85964. 10.1371/journal.pone.0085964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felker DP, Robbins CE, McCormick MA, 2020. Automation of C. elegans lifespan measurement. Transl. Med.Aging 4, 1–10. [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC, 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 (6669), 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gärtner K, 2012. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Int. J. Epidemiol. 41 (2), 335–341. 10.1093/ije/dyr219. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL, 2000. Defining wild-type life span in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 55 (5), B215–B219. 10.1093/gerona/55.5.B215. [DOI] [PubMed] [Google Scholar]
- Haghani A, Dalton HM, Safi N, et al. , 2019. Air pollution alters Caenorhabditis elegans development and lifespan: responses to traffic-related nanoparticulate matter. J. Gerontol. Ser. A 74 (8), 1189–1197. 10.1093/gerona/glz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miyaji M, Zhang-Akiyama Q-M, 2016. FUdR extends the lifespan of the short-lived AP endonuclease mutant in Caenorhabditis elegans in a fertility-dependent manner. Genes Genet. Syst. 91 (4), 201–207. 10.1266/ggs.15-00064. [DOI] [PubMed] [Google Scholar]
- Lai C-H, 2000. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10 (5), 703–713. 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le KN, Zhan M, Cho Y, Wan J, Patel DS, Lu H, 2020. An automated platform to monitor long-term behavior and healthspan in Caenorhabditis elegans under precise environmental control. Commun. Biol. 3 (1), 297. 10.1038/s42003-020-1013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Plummer WT, Chen E, et al. , 2017. Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects. Nat. Commun. 8 (1), 14256. 10.1038/ncomms14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DH, Stiles JW, Santelli J, Sanadi DR, 1979. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J. Gerontol. 34 (1), 28–36. 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- Molenaar PC, Boomsma DI, Dolan CV, 1993. A third source of developmental differences. Behav. Genet. 23 (6), 519–524. 10.1007/BF01068142. [DOI] [PubMed] [Google Scholar]
- Porta-de-la-Riva M, Fontrodona L, Villanueva A, Cerón J, 2012. Basic Caenorhabditis elegans methods: synchronization and observation. J. Vis. Exp. 64, 4019. 10.3791/4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalt JC, Sánchez-Salmerón A-J, Ivorra E, Genovés Martínez S, Martínez R, Martorell Guerola P, 2020. Improving lifespan automation for Caenorhabditis elegans by using image processing and a post-processing adaptive data filter. Sci. Rep. 10 (1), 8729. 10.1038/s41598-020-65619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D, Blumenthal T, Meyer B, Priess J, 1997. C. elegans II, 2nd edition. Cold Spring Harbor Laboratory Press. https://www.ncbi.nlm.nih.gov/books/NBK20127/. (Accessed 8 June 2021). [PubMed] [Google Scholar]
- Sterken MG, Snoek LB, Kammenga JE, Andersen EC, 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31 (5), 224–231. 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhr NL, Curran SP, 2020. Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Commun. Biol. 3 (1), 653. 10.1038/s42003-020-01379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Using C, 2015. Elegans for aging research. Invertebr. Reprod. Dev. 59 (sup1), 59–63. 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S, 2011. FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech. Ageing Dev. 132 (10), 519–521. 10.1016/j.mad.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren JR, Braeckman BP, 1999. Mechanisms of life span determination in Caenorhabditis elegans☆. Neurobiol. Aging 20 (5), 487–502. 10.1016/S0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJ, Schubart CD, 2008. Production of different phenotypes from the same genotype in the same environment by developmental variation. J. Exp. Biol. 211 (Pt 4), 510–523. 10.1242/jeb.008755. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao Y, Zhang Z, 2019. Age-dependent effects of floxuridine (FUdR) on senescent pathology and mortality in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 509 (3), 694–699. 10.1016/j.bbrc.2018.12.161. [DOI] [PubMed] [Google Scholar]
- Win MTT, Yamamoto Y, Munesue S, Han D, Harada S-I, Yamamoto H, 2013. Validated liquid culture monitoring system for lifespan extension of Caenorhabditis elegans through genetic and dietary manipulations. Aging Dis. 4 (4), 178–185. [PMC free article] [PubMed] [Google Scholar]
- WormBook, 2017. In: History of research on C. elegans and other free-living nematodes as model organisms, pp. 1–84. 10.1895/wormbook.1.181.1. Published online September 7. [DOI] [PMC free article] [PubMed]
- Zhang S, Li F, Zhou T, Wang G, Li Z, 2020. Caenorhabditis elegans as a useful model for studying aging mutations. Front. Endocrinol. 11, 554994 10.3389/fendo.2020.554994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.