Abstract
Methicillin-resistant Staphylococcus aureus strains with decreased vancomycin susceptibility have been isolated from patients in the United States and Japan. The impact of decreased vancomycin susceptibility on the drug’s pharmacodynamic parameters has not been addressed. We studied the activity of vancomycin against three clinical strains of vancomycin intermediate-susceptible Staphylococcus aureus (VISA) under high- and low-inoculum conditions, with stationary- and logarithmic-growth-phase kill curves, and in postantibiotic effect (PAE) experiments. We also investigated the stability of the decreased vancomycin susceptibility by using population susceptibility profiles. The respective vancomycin microdilution MICs and MBCs for VISA strains HIP5836, 14379, and Mu50 were 8 and 8, 8 and 8, and 8 and 16 μg/ml. HIP5836 had the most homogeneous elevation of vancomycin MICs, because the MIC for nearly all bacteria in the inoculum was 8 μg/ml. The population MICs (defined as the lowest vancomycin concentration inhibiting 99.9% of growth) for the first serial passages of HIP5836, Mu50, and 14379 were 8, 4, and 2 μg/ml, respectively. After 10 passages, they decreased to 4, 2, and 1 μg/ml, respectively. The Mu50 population MIC increased to 12 μg/ml after five serial passages on vancomycin agar. In the low- and high-inoculum kill curves, time to 99.9% killing was significantly (P < 0.05) longer for both Mu50 and HIP5836 than that for 14379 and a control strain. However, colony counts at 24 h were similar to those of the vancomycin-sensitive strain for all VISA strains. The PAE (at 4× MIC) ranged from 1.3 h for 14379 to 2.0 h for HIP5836 and was similar to or greater than the PAE against the vancomycin-sensitive strain. In conclusion, we found that the decreased vancomycin susceptibility increased during persistent exposures to the drug and decreased upon removal of the selective pressure. The decreased vancomycin susceptibility decreased the rate of vancomycin killing, but did not affect the extent of killing or the PAE.
Staphylococcus aureus continues to be a significant pathogen and has demonstrated an impressive ability to become resistant to nearly all antistaphylococcal antimicrobial agents used in clinical practice. Resistance to penicillin (due to the production of β-lactamase) was reported in strains of S. aureus soon after its introduction and became commonplace by the 1950s (24). S. aureus isolates resistant to β-lactamase-stable agents (methicillin-resistant S. aureus [MRSA]) were first reported in 1961 (1) and had become a widespread problem in most hospitals in the United States by the 1980s. Vancomycin, a glycopeptide antibiotic, was introduced into clinical practice in 1956 but was not used widely due to its poorer perceived tolerability (14). With the increase in MRSA, its use rapidly escalated because it commonly was the only antibiotic treatment to which MRSA was susceptible (23). Although an alarming increase in the prevalence of vancomycin-resistant enterococci has occurred in the 1990s, resistance in clinical strains of staphylococci has remained rare. The isolation of vancomycin-intermediate S. aureus (VISA), for which MICs were 8 μg/ml, was reported from Japan in 1997 (5) and soon thereafter from two sites in the United States (3, 4). Each VISA strain was recovered from patients who had received extended treatment courses of vancomycin (3, 11).
The mechanism of decreased susceptibility to vancomycin in these S. aureus strains is not yet fully understood. Expression of reduced vancomycin susceptibility appears to be heterogeneous, and gene probe hybridization studies indicate no presence of vanA or vanB genes (2, 10–12, 16). These clinical VISA strains appear to share common characteristics with laboratory-derived staphylococcal mutants with decreased vancomycin susceptibility, including thickened cell walls, increased production of cell wall precursors, decreased autolysis, increased penicillin-binding proteins (PBPs), and slower growth than vancomycin-sensitive MRSA (7, 11, 16–18, 21, 22).
The recent isolation of these clinical strains of VISA highlights the need for other effective antimicrobials for MRSA infections. However, the impact of decreased vancomycin susceptibility on its pharmacodynamic parameters and its clinical efficacy have not been fully addressed. We studied the effect of reduced vancomycin susceptibility on its killing activity against high inocula, low inocula, and stationary- and logarithmic-growth-phase VISA, as well as its impact on postantibiotic effect (PAE). Additionally, we studied the stability of decreased vancomycin susceptibility and the effects of persistent suboptimal vancomycin exposure on the population susceptibility profiles of VISA.
MATERIALS AND METHODS
Bacterial strains.
The VISA strains tested in this investigation were 14379 (William Beaumont Hospital, Royal Oak, Mich. [16]), Mu50 (Juntendo Hospital, Tokyo, Japan [11]), and HIP5836 (Centers for Disease Control and Prevention, Atlanta, Ga. [3]). A clinical strain of vancomycin-susceptible MRSA, 494, was used for comparisons of vancomycin activity.
Antimicrobial agents and media.
Vancomycin (lot no. 35H040425) was commercially purchased from Sigma Chemical Company (St. Louis, Mo.). Mueller-Hinton broth (Difco, Detroit, Mich.) supplemented with calcium (25 mg/liter) and magnesium (12.5 mg/liter) (SMHB) was used for all microdilution susceptibility testing, logarithmic-phase time-kill curves, and PAE experiments. Phosphate-buffered saline (PBS) supplemented with 1.2 g of MHB (PBS-MHB [supplying 1 g of Casamino Acids per liter]) was used for the stationary-growth-phase analyses. All experimental samples were plated on tryptic soy agar (TSA [Difco]) to determine colony counts. Mueller-Hinton medium (Difco) was used to prepare vancomycin-containing agar plates for agar dilution MICs and for analysis of population susceptibility profiles.
Susceptibility testing.
Microdilution MICs and MBCs (in SMHB and PBS-MHB) and agar dilution MICs for vancomycin were determined for each drug by using a standard inoculum of 5 × 105 CFU/ml according to the guidelines of the National Committee for Clinical Laboratory Standards (18).
Population susceptibility profiles.
Vancomycin population susceptibility profiles were determined in duplicate by modifications of previously described methods (8, 22). Fifty microliters of an ∼1 × 109 to 1 × 1010 CFU/ml suspension of bacteria was placed on Mueller-Hinton medium containing doubling dilutions of vancomycin (range, 0.25 to 64 μg/ml) by using an autoplate spiral dispenser (model 3000; Spiral Bioscience, Frederick, Md.). After incubation at 35°C for 24 to 48 hours, colony counts were determined with a laser bacterial colony counter (model 500A; Spiral System Instruments, Inc.). The lowest vancomycin concentration inhibiting 99.9% of growth was defined as the population MIC (22). Cultures of all three VISA strains were repeatedly grown on TSA, and population susceptibility profiles were reassessed after 2, 5, and 10 serial passages to evaluate the stability of decreased vancomycin susceptibility. Additionally, one VISA strain (Mu50) was grown on agar containing vancomycin at 1/2× and 1× MIC, with reevaluation of the population susceptibility profile and MICs after five serial passes.
Time-kill curves.
Time-kill curves were determined in duplicate with starting inocula of 105 CFU/ml (low inoculum), 106 CFU/ml (stationary phase), or 107 CFU/ml (high inoculum). Fresh overnight growths of bacteria in SMHB were diluted as necessary to produce the desired starting inocula in 10 ml of medium (SMHB for exponentially growing high- and low-inoculum experiments and PBS-MHB for stationary-phase experiments). Vancomycin was tested at a concentration of 15 μg/ml to approximate typical mid-dose concentrations in serum. Samples (0.1 ml) were removed from each well at 0, 4, 8, and 24 h and diluted appropriately in cold 0.9% sodium chloride. Colony counts were determined by spotting 20-μl samples in triplicate onto TSA and incubation at 35°C for 24 h. We determined these methods to have a lower limit of reliable detection of 2 log10 CFU/ml. For the stationary-phase experiments with HIP5836, Mu50, and 14379, inclusion of all data points resulted in linear kill curves with r values of ≥0.95. Linear regression of the entire kill curves for these experiments was used to compute the time to 99.9% kill. Linear regression of kill-curve data from the 0-, 4-, and 8-h time points only was used to determine the time to 99.9% kill for the VISA 14379 and MRSA 494 high-inoculum experiments, the MRSA 494 stationary-phase experiments, and all low-inoculum experiments, since inclusion of the 24-h data points resulted in nonlinear kill curves (see Fig. 2). Growth controls also were sampled every hour until the media became cloudy, and doubling times were calculated by using linear regression of the growth control curve.
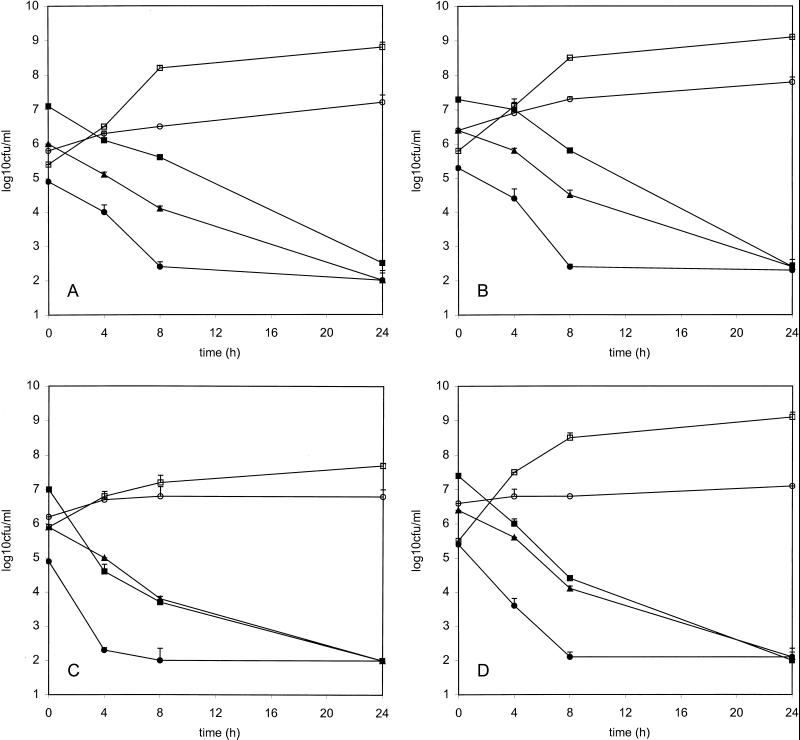
FIG. 2.
Time-kill curves for vancomycin (15 μg/ml) versus HIP5836 (A), Mu50 (B), 14379 (C), and MRSA 494 (D). ●, low inoculum; ■, high inoculum; ▴, stationary phase; □, growth control in SMHB; ○, growth control in PBS-MHB.
PAE.
The PAE was determined by methods described by Craig and Gudmundsson (6). Organisms were grown overnight in SMHB, diluted with prewarmed SMHB, and incubated for 2 to 4 h to allow for ∼7 log10CFU of exponentially growing bacteria per ml. One milliliter of this suspension was added to 9 ml of prewarmed SMHB containing vancomycin at concentrations of 1× and 4× MIC (8 and 32 μg/ml, respectively) for the VISA strains and 1×, 4×, and 8× MIC (1, 4, and 8 μg/ml, respectively) for MRSA 494. The 8× MIC (64 μg/ml) concentration was not tested against the VISA strains, because this concentration is not commonly achieved during usual dosing. Test tubes were sampled prior to and 1 h after antibiotic addition. Samples were diluted 1:1,000 into prewarmed SMHB after 1 h of exposure to vancomycin, and aliquots (0.1 ml) were removed immediately after this dilution and every hour thereafter for up to 7 h. Colony counts were determined as described in the time-kill curve section. The PAE was calculated by the equation PAE = T − C, where T was the time to achieve 1-log10CFU/ml growth for the antibiotic-exposed sample and C was the time to achieve 1-log10CFU/ml growth for the untreated control sample. For all PAE experiments, T and C were determined either by linear regression (if r ≥ 0.95) or by visual inspection of the regrowth curve. Each PAE experiment was performed in duplicate.
Statistical analyses.
The colony counts at 24 h and times to 99.9% killing were compared between groups by using analysis of variance with Tukey’s test for multiple comparisons, with a P value of ≤ 0.05 indicating statistical significance. All statistical analyses were performed with SPSS statistical software (release 6.1.3; SPSS, Inc., Chicago, Ill.).
RESULTS
Susceptibilities.
The respective vancomycin microdilution MICs and MBCs for VISA strains HIP5836, 14379, and Mu50 were 8 and 8, 8 and 8, and 8 and 16 μg/ml. The respective MIC and MBC for the vancomycin-sensitive clinical strain were 0.5 and 1 μg/ml. MICs and MBCs in PBS-MHB were 2 to 4 times lower than SMHB values when read at 18 to 24 h, but were similar to SMHB MICs and MBCs at 48 h. Vancomycin agar dilution MICs were similar to microtiter MICs for all strains.
Population susceptibility profiles.
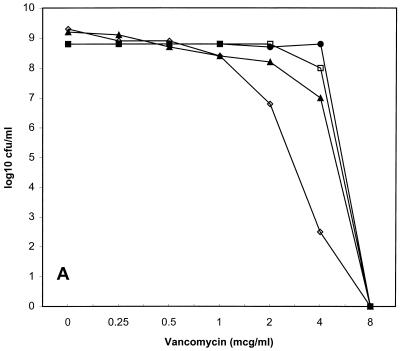
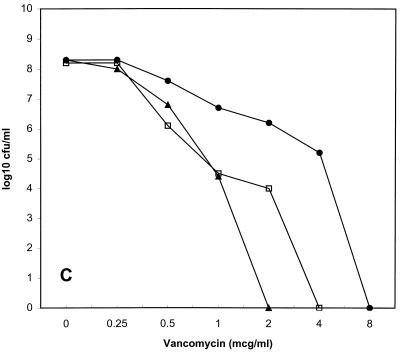
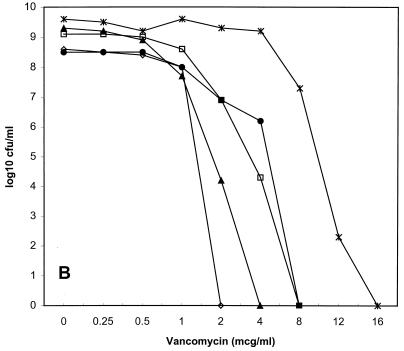
Population susceptibility profiles for the three strains of VISA are shown in Fig. 1. The population MICs for the first serial passes of HIP5836, Mu50, and 14379 were 8, 4, and 2 μg/ml; after passes 2, 5, and 10, they were 8, 8, and 4; 4, 2, and 2; and 1, 1, and 1, respectively. HIP5836 had the most homogeneous baseline elevation of vancomycin MICs, because the MIC for nearly all bacteria in the inoculum was 8 μg/ml.
FIG. 1.
Vancomycin population susceptibility profiles for HIP5836 (A), Mu50 (B), and 14379 (C). ●, first pass; □, second pass; ▴, fifth pass; ◊, tenth pass; ×, fifth pass on vancomycin agar for Mu50. mcg, micrograms.
The subpopulations with decreased vancomycin susceptibility were unstable for all three VISA strains tested. For HIP5836, there was a 10-fold decrease in subpopulations for which vancomycin MICs were 8 μg/ml after the second pass and a 100-fold decrease after the fifth pass. For Mu50, there was a 100-fold decrease in subpopulations for which vancomycin MICs were 8 μg/ml after the second pass, and no colonies grew on the 2-μg/ml plates after 10 passes. The MIC for the Mu50 population increased to 12 μg/ml after five passes on vancomycin agar. VISA 14379 was the least stable strain, because the MICs for less than 0.01% of the total inoculum were ≥2 μg/ml by the fifth pass. The population susceptibility profile for the tenth pass of VISA strain 14379 was not determined due to the absence of any subpopulations with decreased vancomycin susceptibility.
Growth characteristics and time-kill curves.
The doubling times for Mu50, HIP5836, 14379, and 494 were 34, 39, 75, and 29 min, respectively. VISA 14379 was the only strain that grew substantially slower than 494. Results from the time-kill curves are shown in Fig. 2 and Table 1. Colony counts at 24 h were similar for all of the strains tested. In the low- and high-inoculum kill curves, time to 99.9% killing was significantly longer (P < 0.05) for both Mu50 and HIP5836 than those for 14379 and 494. The times to 99.9% killing were similar for all strains in the stationary-phase kill curves.
TABLE 1.
Mean time to 99.9% killing for HIP5836, Mu50, 14379, and MRSA 494 in the time-kill curves
| Organism | Time to 99.9% killing (h)a
|
||
|---|---|---|---|
| Low inoculum | High inoculum | Stationary phase | |
| HIP5836 | 10.0 ± 0.4b | 15.7 ± 1.0c | 13.1 ± 1.1c |
| Mu50 | 8.7 ± 0.5b | 15.3 ± 0.5c | 13.5 ± 1.4c |
| 14379 | 5.9 ± 2.6b | 8.1 ± 2.0b | 11.6 ± 0.1c |
| MRSA 494 | 7.4 ± 0.9b | 8.1 ± 1.2b | 10.8 ± 0.4b |
Data represent the mean ± standard deviation from duplicate experiments.
Time to 99.9% kill determined by linear regression of the line formed from the 0-, 4-, and 8-h time points.
Time to 99.9% kill determined by linear regression of the line formed from all time points.
PAE.
Vancomycin PAEs are summarized in Table 2. Mean PAEs at 1× MIC ranged from 0.5 h for 14379 to 1.2 for HIP5836. PAEs at 4× MIC approximately doubled for each VISA isolate and ranged from 1.3 h for 14379 to 2.0 h for HIP5836. PAEs were not appreciably different against the VISA strains versus those for the vancomycin-sensitive strain.
TABLE 2.
PAEs for vancomycin against VISA strains
| Strain | PAE (h)a
|
||
|---|---|---|---|
| 1× MIC | 4× MIC | 8× MIC | |
| VISA NJ | 1.2 ± 0.1 | 2.0 ± 0.4 | NDb |
| VISA Mu50 | 0.8 ± 0.1 | 1.5 ± 0.1 | ND |
| 14379 | 0.5 ± 0.1 | 1.3 ± 0.1 | ND |
| MRSA 494 (control) | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 |
Values are means ± standard deviations.
ND, not done.
DISCUSSION
Therapeutic failures or slow responses with vancomycin therapy versus staphylococci have been previously attributed to such factors as a slower killing rate versus β-lactam antibiotics, decreased penetration to infected tissue sites, the presence of foreign bodies, or the underlying compromised health status of the patient (13, 14, 22). However, the recent reports of MRSA with decreased vancomycin susceptibility indicate that therapeutic failures against MRSA soon will be related to vancomycin resistance.
We evaluated this decreased vancomycin susceptibility and its potential to impact the pharmacodynamic parameters of vancomycin. We found that all three VISA strains expressed heterogeneous resistance to vancomycin (“hetero-VISA” or “hetero-VRSA” [vancomycin-resistant S. aureus]) (12). Each strain had different percentages of subpopulations expressing decreased vancomycin susceptibility, with HIP5836 being the strain closest to achieving homogeneous expression of resistance, followed by Mu50 and 14379. It is notable that our Mu50 population susceptibility profile differed appreciably from that previously described (12). In that investigation, nearly 100% of Mu50 cells grew on 4 μg of vancomycin agar per ml, and 0.001% of the total cells grew on 10 μg/ml. In contrast, we found that only 1% of the total Mu50 cell population grew on 4 μg of vancomycin agar per ml, while no colonies grew on concentrations ≥8 μg/ml. This discrepancy probably is explained best by our finding of diminished subpopulations with decreased vancomycin susceptibility for Mu50 (as well as for the other two VISA strains) with serial passages. We do not know the exact number of serial passages of Mu50 on antibiotic-free media prior to our acquisition of this isolate, but at least two passages occurred prior to the generation of the frozen bacterial stock used for all subsequent experiments. These observations are important and need to be considered during future study of VISA strains.
The potential for the development of glycopeptide resistance in staphylococci has been studied for well over a decade. During this time, it has been reported that sequential in vitro exposures select for teicoplanin resistance more readily than for vancomycin resistance (20, 25). As well, decreased vancomycin susceptibility in staphylococci has been reported to be both stable (7, 10, 12, 22) and unstable (9, 25). These discrepancies in resistance stability could be related to different selection methods used (broth versus agar, continuous passes in the presence of drug versus alternating passes in drug-containing and drug-free media, etc.), different resistance assessment methods (broth dilution MICs versus population susceptibility profiles), or differences in resistance mechanisms between strains. Our findings of increased vancomycin susceptibility after repeated serial passages under antibiotic-free conditions and the emergence of further vancomycin-resistant subpopulations after repeated passages on vancomycin agar both suggest that decreased susceptibility may result from the selection of cells which have adapted to survive in the presence of the drug.
The exact mechanisms involved in the production and regulation of decreased vancomycin susceptibility in staphylococci are not completely determined. Studies of vancomycin resistance in enterococci have shown that a family of van genes allow the bacteria to replace the peptidoglycan d-alanyl-d-alanine terminus (the active binding site for vancomycin) with a d-alanyl-d-lactate terminus which has a 1,000-fold-decreased affinity for vancomycin (15). The presence of van genes has not been documented in VISA (11, 16), but increased production of cell wall precursors, thickened and irregular cell walls, decreased autolysis, increased quantities of PBPs, and slower growth have been described in both in vivo (10–12) and in vitro (7, 16, 17, 19, 21) resistant staphylococcal mutants. Although all of our VISA isolates grew more slowly than the vancomycin-sensitive strain, only one (14379) had substantially reduced growth. This VISA strain (which grows as small nonpigmented colonies) appears to be one of the small-colony variants, which usually are less susceptible to cell wall-active agents. Because vancomycin activity was appreciably decreased against all VISA strains and against all slow-growing bacteria (regardless of the vancomycin susceptibility), it appears that the decreased growth rates were not a primary factor decreasing vancomycin susceptibility.
Decreased vancomycin susceptibility significantly reduced the rate but not the extent of vancomycin killing for both HIP5836 and Mu50 compared to the vancomycin-sensitive MRSA. These decreases were noted without regard to inoculum. In contrast, 14379 was killed at a rate similar to that of the vancomycin-sensitive strain. These findings likely were explained by the population susceptibility profiles for each VISA strain. Because 14379 possessed much fewer cells with decreased susceptibility compared to HIP5836 and Mu50, the vast majority of cells in the test inoculum probably were killed similar to a vancomycin-susceptible strain. For all of these VISA strains, it appears that maintaining vancomycin concentrations at least 2× greater than the MIC could result in adequate killing activity.
Vancomycin PAEs against VISA were similar to or greater than the PAEs against the vancomycin-sensitive strain and also were within ranges previously reported for vancomycin-sensitive staphylococci (6). Interestingly, the PAEs for all three strains doubled upon increasing the concentration from 1 to 4× the MIC. This finding could be related to the “trapping” of vancomycin by the excess cell wall material which previously has been reported for vancomycin- and teicoplanin-resistant strains of S. aureus (21) and coagulase-negative staphylococci (22). These bacteria had the ability to quantitatively remove glycopeptide from the test media, which then was recovered from purified cell wall fractions in a biologically active form (21). Thus, the higher vancomycin concentrations could increase the PAEs against VISA in our experiments through better delivery of vancomycin to the site of active cell wall synthesis.
In conclusion, we found that decreased susceptibility to vancomycin appears to be a selective or inducible process that increased during persistent suboptimal exposure to the drug and decreased upon removal of the selective pressure. The decreased susceptibility to vancomycin decreased the rate but not the extent of vancomycin killing during exposure to concentrations approximately 2× greater than the MIC. The vancomycin PAE was more dependent on the concentration relative to the MIC against these strains. Because of the time-dependent killing activity for vancomycin, more frequent doses or continuous infusions of the drug might improve its activity against VISA until newer antibiotics become available.
REFERENCES
- 1.Barber M. Methicillin-resistant staphylococci. J Clin Pathol. 1961;14:385. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J M, Medeiros A A, Hiramatsu K. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) from the United States with subpopulations of cells with reduced susceptibility to vancomycin, abstr. LB-15; p. 11. [Google Scholar]
- 3.Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbid Mortal Weekly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Reduced susceptibility of Staphylococcus aureus to vancomycin—Japan, 1996. Morbid Mortal Weekly Rep. 1997;46:624–635. [PubMed] [Google Scholar]
- 6.Craig W A, Gudmundsson S. Postantibiotic effect. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: The Williams and Wilkins Co.; 1996. pp. 296–329. [Google Scholar]
- 7.Daum R S, Gupta S, Sabbagh R, Milewski W M. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J Infect Dis. 1992;166:1066–1072. doi: 10.1093/infdis/166.5.1066. [DOI] [PubMed] [Google Scholar]
- 8.DeLencastre H, Figueiredo A M S, Urban C, Rahal J, Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenwood D, Bidgood K, Turner M. A comparison of the responses of staphylococci and streptococci to teicoplanin and vancomycin. J Antimicrob Chemother. 1987;20:155–164. doi: 10.1093/jac/20.2.155. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Hanaki H, Boyle-Vavra S, Daum R S, Labischinski H, Tenover F C. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Isolation and characterization of MRSA clinical strains with reduced susceptibility to vancomycin, abstr. C-166; p. 74. [Google Scholar]
- 11.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogenously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 13.Houlihan H H, Mercier R-C, Rybak M J. Pharmacodynamics of vancomycin alone of in combination with gentamicin at various dosing intervals against methicillin-resistant Staphylococcus aureus-infected fibrin-platelet clots in an in vitro infection model. Antimicrob Agents Chemother. 1997;41:2497–2501. doi: 10.1128/aac.41.11.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karchmer A W. Staphylococcus aureus and vancomycin: the sequel. Ann Intern Med. 1991;115:739–741. doi: 10.7326/0003-4819-115-9-739. [DOI] [PubMed] [Google Scholar]
- 15.Leclercq R, Courvalin P. Resistance to glycopeptides in enterococci. Clin Infect Dis. 1997;24:545–556. doi: 10.1093/clind/24.4.545. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell J, Ionescu M, Farnaz D, Donabedian S, Perri M B, Thal L A, Sunstrum J, Chow J W, Smith T, Zervos M J. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Characterization of a unique isolate of vancomycin-intermediate Staphylococcus aureus (VISA), abstr. LB-14; p. 11. [Google Scholar]
- 17.Moreira B, Boyle-Vavra S, deJonge B L M, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard. NCCLS document M7-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 19.Pfeltz R F, Batten M A, Baranyk C, Jayaswal R K, Wilkinson B J. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Strain specificity and vancomycin-resistance in methicillin-resistant Staphylococcus aureus, abstr. A19; p. 41. [Google Scholar]
- 20.Schlaes D M, Schlaes J H. Teicoplanin selects for Staphylococcus aureus that is resistant to vancomycin. Clin Infect Dis. 1995;20:1071–1073. doi: 10.1093/clinids/20.4.1071. [DOI] [PubMed] [Google Scholar]
- 21.Sieradzki K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieradzki K, Villari P, Tomasz A. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical strains of staphylococci. Antimicrob Agents Chemother. 1998;42:100–107. doi: 10.1128/aac.42.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small P M, Chambers H F. Vancomycin for Staphylococcus aureus endocarditis in intravenous drug users. Antimicrob Agents Chemother. 1990;34:1227–1231. doi: 10.1128/aac.34.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spink W W, Ferris V. Quantitative action of penicillin inhibitor from penicillin-resistant strains of staphylococci. Science. 1945;102:221. doi: 10.1126/science.102.2644.221. [DOI] [PubMed] [Google Scholar]
- 25.Watanakunakorn C. In-vitro selection of resistance of Staphylococcus aureus to teicoplanin and vancomycin. J Antimicrob Chemother. 1990;25:69–72. doi: 10.1093/jac/25.1.69. [DOI] [PubMed] [Google Scholar]