SUMMARY
Inherited pathogenic succinate dehydrogenase (SDHx) gene mutations cause the hereditary pheochromocytoma and paraganglioma tumor syndrome. Syndromic tumors exhibit elevated succinate, an oncometabolite that is proposed to drive tumorigenesis via DNA and histone hypermethylation, mitochondrial expansion, and pseudohypoxia-related gene expression. To interrogate this prevailing model, we disrupt mouse adrenal medulla SDHB expression, which recapitulates several key molecular features of human SDHx tumors, including succinate accumulation but not 5hmC loss, HIF accumulation, or tumorigenesis. By contrast, concomitant SDHB and the neurofibromin 1 tumor suppressor disruption yields SDHx-like pheochromocytomas. Unexpectedly, in vivo depletion of the 2-oxoglutarate (2-OG) dioxygenase cofactor ascorbate reduces SDHB-deficient cell survival, indicating that SDHx loss may be better tolerated by tissues with high antioxidant capacity. Contrary to the prevailing oncometabolite model, succinate accumulation and 2-OG-dependent dioxygenase inhibition are insufficient for mouse pheochromocytoma tumorigenesis, which requires additional growth-regulatory pathway activation.
In brief
SDHB knockout in mouse chromaffin cells is not tumorigenic despite ample succinate accumulation and conveyance of cardinal features of SDHB-deficient pheochromocytomas. Armstrong et al. find that dual knockout of SDHB and NF1 yields SDHB-like tumors with accumulation of succinate and H3K9me3, in addition to swollen mitochondria and loss of 5hmC.
Graphical Abstract

INTRODUCTION
Succinate belongs to a family of so-called “oncometabolites,” a collection of TCA cycle-derived cellular metabolites that can pathologically accumulate to promote cellular metabolic reprogramming and drive neoplasia (Dang et al., 2009; Yang et al., 2013). Normally, succinate is oxidized to fumarate by the mitochondrial enzyme succinate dehydrogenase (SDH), complex II of the electron transport chain and a component of the TCA cycle. Germline loss-of-function mutations in succinate dehydrogenase (SDH) genes (SDHA, SDHB, SDHC, SDHD, or SDHAF2) cause the autosomal dominant hereditary pheochromocytoma and paraganglioma tumor syndrome (hPPGLs) (Baysal et al., 2000). The hPPGL syndrome exhibits incomplete penetrance, as stochastic loss of the wild-type allele, generally in the context of a partial chromosomal deletion, is required for tumor formation (Fishbein et al., 2017). Tumors with SDHx mutations exhibit impaired succinate-to-fumarate oxidase activity (Rapizzi et al., 2012; Favier et al., 2009) and accumulate succinate to levels as high as 100-fold normal (Lendvai et al., 2014). The current model of SDHx tumorigenesis proposes that high levels of succinate inhibit 2-oxoglutarate (2-OG)-dependent enzymes, leading to epigenetic changes and pseudohypoxia that promote tumorigenesis. Whether succinate accumulation is sufficient to promote tumor formation and how the loss of SDH enzymatic activity leads to tumorigenesis remains uncertain. It is well demonstrated that succinate levels in SDHx-PPGL tumor tissue are substantially elevated relative to non-SDHx tumors and normal organ levels (Lussey-Lepoutre et al., 2016) (Richter et al., 2014). In vitro data show that knockdown of SDHx subunits or chemical inhibition of SDH leads to succinate accumulation in diverse cell lines, although the extent of succinate elevation varies substantially (Xiao et al., 2012; Letouze et al., 2013). Consequently, the prevailing hypothesis, based on analysis of SDHx and other mitochondrial protein mutation-caused tumors (isocitrate dehydrogenase [IDH]1/2- and fumarate hydratase [FH]-related glioblastoma and renal cell carcinoma, respectively), is that succinate accumulation acts as an “oncometabolite” by competitively inhibiting 2-OG-dependent enzymes, which leads to DNA and histone hypermethylation, and hypoxia-inducible factor (HIF) stabilization. The importance of these pathways in PPGL tumorigenesis is also supported by the overlapping phenotype caused by mutation of other enzymes in these pathways: VHL, DNA methyltransferase-3α (DNMT3A), and HIF2α, which similarly augment HIF and DNA methylation (Remacha et al., 2018; Fishbein et al., 2017).
The 2-OG-dependent dioxygenase enzymes include the PHD, TET, and KDM/JMJ families (Losman et al., 2020). These enzymes, which modify the epigenetic landscape (TETs and KDM/JmJs) and regulate the adaptive cellular response to hypoxia via HIF1/2α stability (PHDs), utilize Fe2+, O2, and ascorbate as co-factors and hydroxylate or demethylate their substrates via concomitant oxidation of 2-OG to succinate. The TET enzyme family (TET1, TET2, TET3) are DNA demethylases that mediate the conversion of 5-methylcytosine (5mc) to 5-hydroxymethylcytosine (5hmC). TET inhibition by succinate manifests histologically in SDHx tumors as a loss of 5hmC immunostaining (Hoekstra et al., 2015a), while epigenetic DNA analysis reveals a substantial increase in CpG Island methylation (Letouze et al., 2013). The JmjC-domain enzyme family are histone demethylases that mediate the removal of histone lysine methyl groups (Laukka et al., 2018). These post-translational modifications influence chromatin structure, gene expression, and DNA replication/repair. Succinate inhibition of JmjC enzymes leads to an increase in multiple histone 3 lysine methyl marks, such as H3K9me3, H3K27me3, and H3K4me3 (Letouze et al., 2013; Hoekstra et al., 2015a).
While there are conflicting data with respect to HIF activation in PPGLs, it is clear that SDHx tumors exhibit a pseudohypoxic transcriptional signature, which is suggestive of HIF1/2 activation but does not exclude alternative mechanisms. Indeed, transcriptional analysis of PPGLs may be used to group tumors into two predominant clusters, reflecting the underlying driver mutation. Cluster 1 tumors, which includes several tricarboxylic acid cycle enzyme genes (SDHx, IDH1/2, FH, and MDH2), VHL, EPAS1, and EGLN1/2 mutations, all share a pseudohypoxic transcriptional profile and are hypermethylated. By contrast, Cluster 2 tumors, including NF1, RET, and HRAS mutations, are enriched in kinase-signaling transcripts and have a lower degree of DNA methylation (Fishbein et al., 2017; Dahia et al., 2005). Although the transcriptomics suggest succinate-mediated HIF stabilization, HIF1α immunostaining of human pheochromocytomas does not distinguish between SDHx-mutated and wild-type (wt)-SDHx tumors (Favier et al., 2009; Bernardo-Castineira et al., 2019) and HIF2α immunostaining is not universally positive in SDHx pheochromocytomas (Pollard et al., 2006). The role of succinate inhibition of PHDs and its relationship to the pseudohypoxic transcriptional profile remains incompletely understood.
Given the mechanistic uncertainty of SDHx tumorigenesis, numerous efforts to generate mouse models of Cluster 1 PPGLs have been made; however, these attempts have been unsuccessful, leading some to suggest its impossibility (reviewed in Lepoutre-Lussey et al., 2016; Lussey-Lepoutre et al., 2018). Approaches taken include global and conditional Sdhd disruption (Piruat et al., 2004; Diaz-Castro et al., 2012), conditional Sdhb deletion, and combined dual Sdhb/Pten loss with PSA-Cre (Lepoutre-Lussey et al., 2016) as well as Sdhc/p53 loss with Th-Cre (Al Khazal et al., 2020). All of these attempts failed to demonstrate elevated succinate in viable SDHx-deficient mouse chromaffin cells. Despite this history, we endeavored to address the question of whether the oncometabolite succinate is sufficient to initiate tumorigenesis via 2-OG enzyme inhibition by conditionally disrupting SDHB expression in the mouse chromaffin cells and studying the metabolic, ultrastructural, and transcriptomic consequences.
RESULTS
Loss of SDHB in mouse chromaffin cells is neither lethal nor proliferative
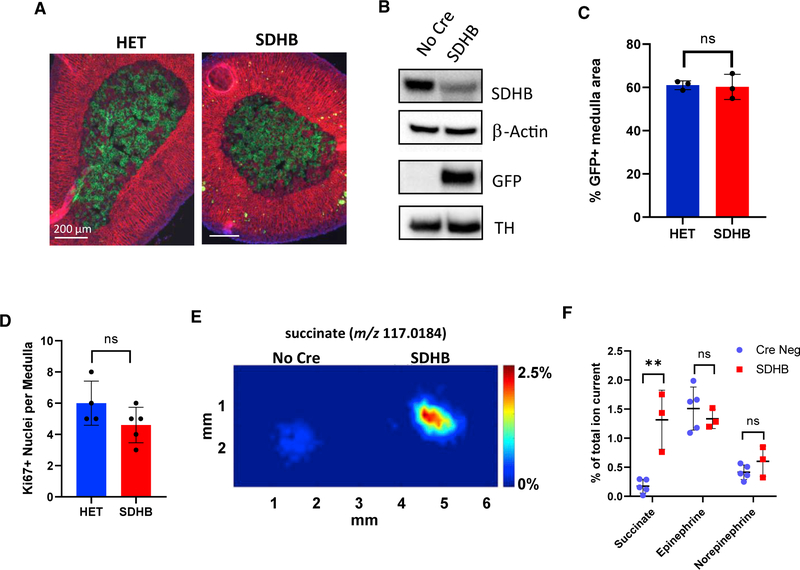
To conditionally disrupt Sdhb in the mouse adrenal medulla, we used mice with LoxP sites flanking exon 3 and bred them to animals expressing Cre recombinase driven by the tyrosine hydroxylase promoter (Th-Cre) (Savitt et al., 2005). We bred Th-Cre SDHBf/f animals to the mT/mG Cre-reporter mice (Muzumdar et al., 2007) to rigorously follow the recombined cell population. As previously reported (Savitt et al., 2005), incomplete Th-Cre-driven recombination occurred in adrenal gland chromaffin cells. To determine the impact of SDHB loss on chromaffin cell survival, we compared the percentage of GFP+ medulla areas between SDHBf/+Rosamtmg/+Th-Cre (HET) and SDHBf/fRosamtmg/+Th-Cre (SDHB) in adult (6-month-old) animals (Figure 1A). Males from both genotypes had 60% GFP+ medulla area (Figure 1C). Loss of SDHB expression was verified by performing western blots on surgically isolated adrenal medullas (Figure 1B). As anticipated from incomplete recombination, modest residual SDHB expression was present in SDHB medulla. Notably, Ki-67 immunostaining was not significantly different between HET and SDHB medullas (Figure 1D). Therefore, in contrast to expectations and prior mouse modeling attempts, SDHB loss neither provoked nor inhibited chromaffin cell proliferation.
Figure 1. Loss of SDHB in mouse adrenal medulla was neither lethal nor proliferative and led to succinate accumulation.
(A) Representative images of intrinsic mTomato/mGFP (mT/mG) fluorescence from 22-week-old HET and SDHB adrenal glands. Nuclei were stained with DAPI.
(B) Western blot of surgically isolated adrenal medullas.
(C) %GFP+ medulla area calculated from serially sectioned HET and SDHB adrenal glands using mT/mG fluorescence (n = 3 mice per genotype).
(D) Quantification of total Ki-67+ nuclei per medulla section (n = 4–5 mice per genotype).
(E) Representative adrenal gland DESI-MSI images of succinate from 28-week-old female animals. Images were normalized as a percentage of the total ion current (%TIC).
(F) Average abundance of succinate and catecholamines in the medulla as measured by DESI-MSI (n = 3–5 animals). Data are presented as mean ± SD and analyzed using an unpaired t test. **p < 0.01. SDHB mice in (F) were on a NF1f/+ background.
To determine if loss of SDHB resulted in succinate accumulation, we measured abundance of succinate in the adrenal medulla by desorption electrospray ionization-mass spectrometry imaging (DESI-MSI) (Figure 1E). DESI-MSI is a nondestructive, ambient ionization method that preserves tissue metabolite distribution, enables detection of minute metabolite alterations, and discriminates between the cortex and medulla of the small murine adrenal gland (Gouw et al., 2019; Wiseman et al., 2006; Vijayalakshmi et al., 2020). Because whole-gland metabolomics gives an averaged chemical footprint across the cortex and medulla, DESI-MSI was crucial to spatially resolve the distributions of succinate and other metabolite levels between these two adrenal compartments. Indeed, succinate was selectively increased (7.6-fold by averaged abundance) in the medulla of SDHB adrenals compared with controls (Figure 1F). Given that each DESI-MSI pixel (~120 μm2) included 60–100 cells, and approximately 60% of those cells were Cre-recombined, the actual succinate accumulation per SDHB−/− cell was likely higher. The DESI-MSI spectra also contained peaks for epinephrine and norepinephrine; however, these metabolites were un-changed (Figure 1F). These data provided definitive evidence that mouse SDHB-deficient chromaffin cells were viable and exhibited substantial succinate accumulation.
Having verified that SDHB loss in mouse chromaffin cells was viable and led to succinate accumulation, we analyzed the effects of SDHB loss at an ultrastructural and an immunohistochemical level. Transmission electron micrographs (TEM) of SDHB medulla revealed large clusters of swollen mitochondria in a subset of chromaffin cells (Figures 2A and S1A). Importantly, these clusters were never observed in HET adrenals. This mitochondrial phenotype occurred in SDHB-null cells but not adjacent SDHB-intact cells, reflecting incomplete Th-Cre-mediated recombination (Figure S1A). Notably, similar mitochondrial ultrastructural changes are characteristically observed in SDHx tumors (Douwes Dekker et al., 2003; Szarek et al., 2015). The TEM findings led us to predict that the mitochondrial clusters represented a general increase in mitochondrial protein that could be used as an immunohistochemical marker for SDHB deficiency. Direct staining for SDHB protein was inconsistent, given a low basal expression level in normal mouse chromaffin cells, where cortical SDHB expression is easily visualized (Figure S1B). Immunostaining of HET and SDHB adrenal glands for several mitochondrial proteins, including cytochrome c and TFAM, demonstrated robust induction in GFP+ cells of SDHB medulla, but not the HET medulla (Figures 2B and S1C). Hence, SDHB disruption caused mitochondrial swelling, clustering, and an apparently expanded mass that was associated with increased mitochondrial protein expression, including the master mitochondrial mass regulator TFAM.
Figure 2. Altered mitochondrial ultrastructure, cytochrome c expression, and histone methylation in SDHB medullas.
(A–C) (A) TEM images of HET and SDHB chromaffin cells (120,00× magnification). Co-staining with GFP and (B) cytochrome c (CytC) and (C) H3K9me3 in HET and SDHB GFP+ chromaffin cells and associated plots of ratio of fluorescence intensity in GFP+/GFP− cell populations. Data are presented as mean ± SD and analyzed using an unpaired t test. **p < 0.01; n = 3–5 mice per genotype.
To address the question of whether succinate accumulation in SDHB animals resulted in inhibition of TET, PHD, and KDM enzymes yielding the predicted changes in histone and DNA methylation as well as HIF protein stabilization, we performed a series of co-staining experiments. We co-stained GFP with H3K9me3, 5hmC, and HIF1α and compared the staining intensity of each marker between the GFP+ and GFP− populations within a section (Figures 2C, S1D, and S1E). We also looked for HIF2α but failed to detect nuclear HIF2α in any sample. As shown in Figure 2C, H3K9me3 immunostaining was specifically increased in GFP+ cells from SDHB animals but not HET animals, consistent with succinate-mediated inhibition of a KDM4 enzyme. 5hmC staining of HET controls revealed substantial variability in staining intensity throughout the medulla, with some cell clusters staining intensely and others weakly (Figure S1D). Calculation of the ratio of the 5hmC intensity in GFP+/GFP− medulla populations did not indicate a loss of 5hmC in GFP+ SDHB cells. This result was consistent with the observations that robust changes in 5hmC required cellular division (Lu et al., 2012), a phenotype not substantiated by Ki-67 staining (Figure 1D). Surprisingly, we found substantial HIF1α expression in a subset of normal chromaffin cells and a paradoxical loss of HIF1α expression in the GFP+ SDHB population (Figure S1E).
Concomitant loss of SDHB and NF1 induces chromaffin cell proliferation
Aged (>12 months) SDHB animals failed to develop tumors despite succinate accumulation. Therefore, we sought to promote SDHB cell proliferation by providing an additional driving force. While several Cluster 2 genes, such as neurofibromin 1 (NF1) and RET, have been modeled in mice, modeling of Cluster 1 gene (SDHx, VHL, EPAS1)-related PPGLs has been unsuccessful. Accordingly, we asked whether combining Cluster 1 and Cluster 2 gene disruption would enable development of SDHx-like pheochromocytomas. Toward this end, we evaluated publicly available TCGA transcriptomic data from five SDHx and six NF1 (germline mutation) human pheochromocytomas (Fishbein et al., 2017). Interestingly, two of the six germline NF1 pheochromocytomas were transcriptionally classified as Cluster 1 (pseudohypoxic), illustrating phenotypic overlap between some NF1 and SDHx pheochromocytomas. Additionally, loss of the NF1 locus (17q11) has been observed in some SDHB PPGLs (Fishbein et al., 2017). These data, combined with the knowledge that SDHB and NF1 tumors both exhibit loss of the 1p chromosome, suggested possible overlapping tumorigenic mechanisms. On the basis of apparently overlapping mechanisms of NF1 and SDHx pheochromocytoma formation, we tested whether combined NF1 and SDHB disruption could drive SDHx-like pheochromocytoma formation in mice.
NF1 is a GTPase-activating protein that negatively regulates the RAS/MAPK and PI3K/mTOR pathways (Kiuru and Busam, 2017). In prior work, approximately 18% of mice heterozygous for a NF1 loss-of-function mutation developed pheochromocytomas (Jacks et al., 1994). We reasoned that if the SDHB mice failed to develop pheochromocytomas as a consequence of a missing replication trigger, simultaneous loss of NF1 would be sufficient to induce tumor formation. Toward this end, we bred SDHBf/f and NF1f/f animals (Zhu et al., 2001) to generate the following experimental genotypes: SDHBf/+NF1f/+Rosamtmg/+Th-Cre (HET/HET), SDHBf/fNF1f/+Rosamtmg/+Th-Cre (SDHB/HET), SDHBf/+NF1f/fRosamtmg/+Th-Cre (HET/NF1), and SDHBf/fNF1f/fRosamtmg/+Th-Cre (SDHB/NF1).
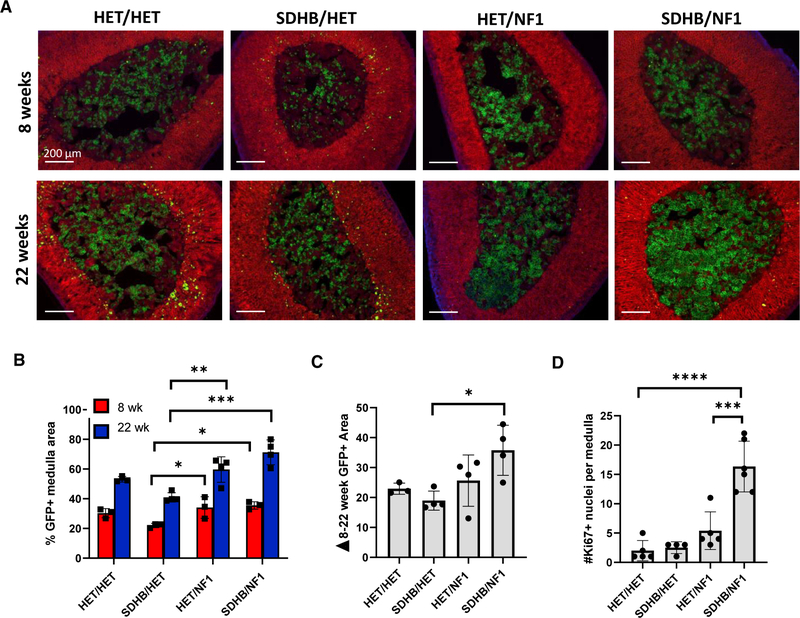
When adrenal medullas from all four genotypes were examined at 8 weeks of age, both SDHB/NF1 (35.5%) and HET/NF1 (34.1%) glands had significantly more GFP+ area compared with SDHB/HET glands (22%) (Figures 3A–3C). The difference between HET/HET and SDHB/HET %GFP+ area at 8 weeks was not statistically different. When animals were aged to 22 weeks, differences were more prominent. At 22 weeks, 71% of the SDHB/NF1 medulla area was GFP+ compared with 53% in HET/HET and 60% in HET/NF1 medullas. Notably, the patterns of SDHB/NF1 and HET/NF1 GFP+ cell expansion were distinct. In the SDHB/NF1 medullas, GFP+ cell clusters were uniformly distributed; however, GFP+ cell clusters in HET/NF1 medullas were mostly similar to HET/HET medullas with discrete areas of densely packed GFP+ cells, suggestive of nodular hyperplasia (Figure 3A). Importantly, SDHB/NF1 medulla exhibited the highest replication activity (Ki-67+ nuclei; Figure 3D), demonstrating a cooperative proliferative effect of concurrent SDHB and NF1 gene disruption. The increased SDHB/NF1 medulla replication (Ki-67) and expanded GFP+ area led to increased adrenal gland mass. Both SDHB/NF1 and HET/NF1 glands were approximately 24% heavier than control glands (Figure S2A) at 22 weeks. Paradoxically, SDHB/HET glands were 47% larger than those of controls; however, this was related to increased cortical area and fat accumulation (see below).
Figure 3. Simultaneous knockout of NF1 forced proliferation of SDHB chromaffin cells.
(A) Representative mT/mG fluorescence images of the indicated genotype for male animals at 8 and 22 weeks of age.
(B) Plot of percentage of the medulla area that is GFP+ for each genotype at 8 and 22 weeks (n = 3–4 mice per genotype).
(C) Plot of the increased %GFP+ area between 8 and 22 weeks.
(D) Quantification of Ki-67+ medulla nuclei per medulla area for the indicated genotypes. Data are presented as mean ± SD and analyzed using one-way ANOVA and Tukey’s multiple comparison test (n = 4–6 mice per genotype). ***adjusted p value < 0.001, **** < 0.0001.
SDHB/NF1 adrenal glands accumulated succinate and were hypermethylated
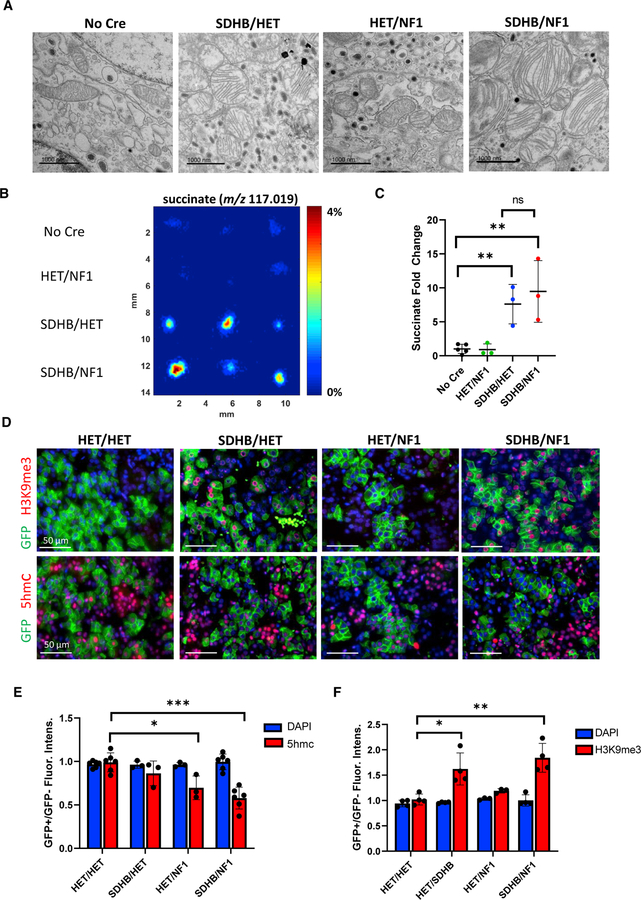
Next, we investigated whether SDHB/NF1 medullary cells, like their SDHB counterpart, exhibited succinate accumulation and the striking mitochondrial phenotype. Indeed, TEM ultrastructure analysis revealed that HET/NF1 mitochondria were indistinguishable from Cre negative controls, whereas SDHB/NF1 cells contained large clusters of swollen mitochondria with remarkably intact cristea, as observed in SDHB/HET medullas (Figure 4A). DESI-MSI analysis of HET/NF1 and SDHB/NF1 adrenal sections confirmed elevated succinate abundance in the SDHB/NF1 but not HET/NF1 medullas, which were indistinguishable from those of controls (Figures 4B and 4C). Catecholamine levels in these 5-month-old animals were not significantly changed from those of controls (Figure S2C). These data indicated a dominant effect of the SDHB-associated phenotype in the context of concurrent NF1 deficiency.
Figure 4. SDHB/NF1 chromaffin cells displayed swollen mitochondria, excess succinate, and hypermethylation.
(A) Representative TEM images of chromaffin cell mitochondria for each genotype imaged at 120,00× magnification. Scale bar, 1,000 nm.
(B) Representative DESI-MSI images of succinate for 3 samples from each genotype. Scale bar is percentage of total ion current.
(C–F) Fold change in average abundance of succinate in the medulla, based on data from (B). Animals used in this figure were 5–6 months old. (n = 3–5, **p < 0.01).
(D) Representative images of GFP staining with 5hmC and H3K9me3 in adrenal glands from 22-week-old animals. Quantification of (E) 5hmC and (F) H3K9me3 fluorescence expressed as the ratio of fluorescence intensity in the GFP+ cell population over the GFP− cell population within the same gland. DAPI intensity, which should not change, is used as an internal control. Data are presented as mean ± SD and analyzed using one-way ANOVA and Tukey’s multiple comparison test. ***adjusted p value < 0.001, **** < 0.0001; n = 4–6 mice per genotype.
Next, we explored whether the hypermethylation phenotype of replicating SDHB/NF1 cells was distinct from SDHB chromaffin cells. In contrast to SDHB/HET adrenal glands, HET/NF1 and SDHB/NF1 samples demonstrated significantly reduced 5hmC staining intensity in GFP+ nuclei relative to the GFP− nuclei (Figures 4D and 4E). However, at that age, loss of 5hmC in HET/NF1 (30%) and SDHB/NF1 (42%) was not statistically different. Notably, as observed in the SDHB medullas, H3K9me3 levels were elevated in the SDHB/NF1 GFP+ cell population but un-changed in HET/NF1 medullas (Figures 4D and 4F). Thus, loss of NF1 is sufficient to reduce 5hmC but does not promote histone hypermethylation, which selectively occurs in SDHB-deficient medullas.
Aged SDHB/NF1 animals developed medullary hyperplasia and pheochromocytomas
To ascertain whether the increased Ki-67, GFP+ -cell expansion and adrenal weights observed in the 22-week-old SDHB/NF1 animals were indicative of early stages of tumor formation, we aged (12–20 months) more than 100 genotypically diverse (No Cre, HET/HET, HET/NF1, SDHB/HET, and SDHB/NF1) animals. Aged (12+ months) HET/NF1 animals exhibited decreased body weight (averaging 18.3 g), whereas SDHB/HET animals developed obesity (averaging 57.5 g) (Figure S3A). Notably, obese SDHB/HET animals did not develop diabetes, apparently because of compensatory islet mass expansion and increased insulin secretion capacity (Figure S3B–S3F). This non-diabetic obese phenotype was similarly observed in Sdhc-deficient mice (Al Khazal et al., 2020) and was attributed to a loss of hypothalamic neurons.
The adrenal gland mass of SDHB/NF1 animals increased significantly in aged mice (Figure 5A). We defined a tumor as an adrenal gland over 6 mg. Using this cutoff, we found tumors in 76% of SDHB/NF1 and in 53% of HET/NF1 adrenal glands (Figures 5A and 5B). Among animals with tumors, 58% of SDHB/NF1 animals had bilateral tumors compared with only 25% of HET/NF1 animals, indicating higher pheochromocytoma penetrance in the double-knockout mice compared with HET/NF1 mice. There was no significant difference in the average weight of SDHB/NF1 and HET/NF1 tumors. Histologic analysis confirmed tumors to be pheochromocytoma (Figures 5C and S4A) with solid growth, usually as organoid clusters of epithelioid to polygonal cells with abundant eosinophilic to amphophilic granular cytoplasm and round nuclei with smooth contours. Occasional tumors demonstrated increased nuclear atypia and ganglion-like cell morphology, which are not uncommon in human pheochromocytoma. Notably, we found one tumor among SDHB/HET adrenal glands and two among HET/HET adrenal glands; potentially related to prior work reporting pheochromocytoma formation in heterozygous NF1 mice (Jacks et al., 1994). We did not observe metastases in the liver or lungs, although an exhaustive search for micro-metastases was not performed. We conclude that SDHB/NF1 disruption in mouse adrenal medulla was tumorigenic and that NF1-driven tumorigenesis was enhanced by loss of SDHB.
Figure 5. Pheochromocytoma development in 12+-month-old SDHB/NF1 mice.
(A) Log2 plot of adrenal weight by animal genotype for animals aged over 1 year. The dashed line marks the 6-mg cutoff for tumors.
(B) Table of tumor findings from the aged cohort.
(C) H&E staining of representative adrenals from each genotype. Normal adrenal glands (HET/HET) contain a small amount of central medullary adrenal tissue surrounded by the adrenal cortex, which is non-compressed. In SDHB/HET adrenal glands, there was slight enlargement of the gland but no tumor formation. In contrast, the adrenal glands from HET/NF1 and SDHB/NF1 mice have expansive growth of the adrenal medulla, consistent with pheochromocytoma, resulting in compression of the surrounding adrenal cortex.
(D) DESI-MSI images of succinate (m/z = 117.0189) and PI(38:4) (m/z = 885.5440) in human PPGLs and a normal adrenal.
(E) DESI-MSI images of succinate in adrenals from aged control, HET/NF1, and SDHB/NF1 animals. PI(38:4) was used as a positive control to show the tissue outline. All images were normalized as %TIC.
(F) Fold change in the average abundance of succinate in the medulla of 12-month-old animals. Data are presented as mean ± SD and analyzed using a one-way ANOVA. ***Adjusted p value < 0.001; n = 4–6 mice per genotype.
(G) Representative images of mouse and human tumors IHC with cytochrome c, 5hmC, and H3K9me3.
Next, we evaluated the molecular differences between HET/NF1 and SDHB/NF1 tumors. We compared succinate levels in the adrenal glands of controls, HET/NF1 tumors, and SDHB/NF1 tumors by DESI-MSI. Indeed, SDHB/NF1 tumors exhibited an average ~28-fold increase in medullary succinate abundance relative to aged control adrenal glands (Figures 5D and 5E). None of the HET/NF1 tumors had elevated succinate. For comparison, we also measured succinate abundance in three human SDHx PGLs, two apparently sporadic PPGLs, and one normal adrenal gland (Figure 5F and Table S1). In the three human SDHx PGLs, tissue-averaged succinate abundances were 33- to 137-fold higher relative to the normal adrenal. Interestingly, PGL-6 came from a young woman diagnosed with Carney Triad (CT) on the basis of multiple PGLs, wild-type GIST, and negative SDHx germline testing. This tumor exhibited equivocal SDHB staining, strong cytochrome c staining, and a 75-fold increase in succinate abundance. These findings highlighted the utility of DESI-MSI for identifying somatic SDHx-related tumors; indicating fallibility of SDHB immunohistochemistry, anticipated to be lost in CT as a result of epigenetic SDHC silencing, or the existence of an undescribed succinate-associated tumorigenic mechanism with retained SDHB expression.
Immunohistochemical analysis revealed that SDHB/NF1 tumors had virtually complete loss of 5hmC, which was retained by intervening sustentacular cells and occasional unrecombined (GFP−) chromaffin cells (Figure 5G). In contrast, HET/NF1 tumors exhibited weak, but persistent, 5hmC immunostaining. Although H3K9me3 staining was detected in both SDHB/NF1 and HET/NF1 tumors, it was more intense throughout double-knockout tumors (Figure 5G). Interestingly, 12-month-old SDHB animals, which did not have an expanded GFP+ cell population, had the same methylation pattern observed in 6-month-old SDHB animals (H3K9me3 induction without loss of 5hmC). These data again indicated that histone hypermethylation was insufficient for tumor initiation and that prolonged exposure to elevated succinate was not sufficient to cause loss of 5hmC. Perhaps the most striking difference between the SDHB/NF1 and HET/NF1 tumors was the induction of cytochrome c (Figure 5G). These data raised the possibility of leveraging cytochrome c staining as a surrogate marker for human SDHx tumors.
To ascertain how similar the characteristics of the SDHB/NF1 mouse tumors were to human SDHx tumors and if cytochrome c could reliably distinguish between SDHx and non-SDHx tumors, we applied the cytochrome c/H3K9me3/5hmC staining protocol to a panel of 30 genetically characterized human PPGLs (Table S2). As shown in Figure 5G, the human SDHB tumor cells stained robustly for cytochrome c and at the same time had almost complete loss of 5hmC immunostaining while inducing H3K9me3 in the cytochrome c-positive population. Intervening sustenacular cells were intensely 5hmC positive and only weakly stained for H3K9me3 and cytochrome c. The 15 non-Cluster 1 PPGLs had only background cytochrome c staining and weakly diffuse H3K9me3 and 5hmC. Indeed, Cluster 1 PPGLs (13 SDHx and 2 VHL) could be distinguished from non-Cluster 1 tumors on the basis of cytochrome c staining intensity (Figure S4B) indicating that cytochrome c staining intensity may be used to identify SDHx and, possibly, VHL PPGLs. Importantly, our mouse SDHB/NF1 tumors displayed a remarkably similar staining pattern to the human SDHx tumors, suggesting that it accurately models these histological aspects of the human disease.
SDHB/NF1 tumors were enriched in mitochondrial and ribosomal transcripts
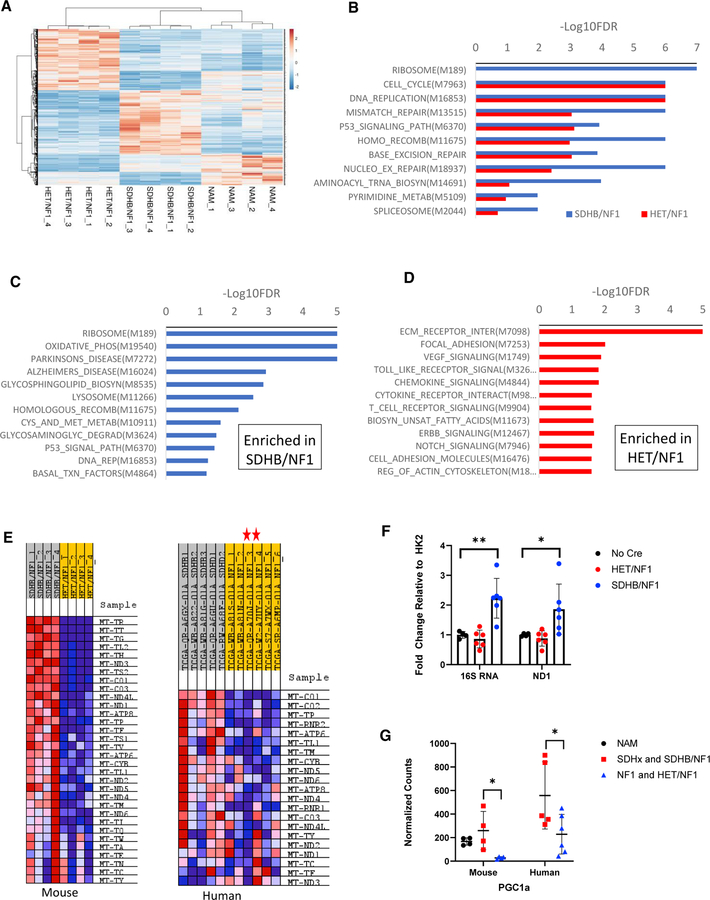
A hallmark and discriminating feature of human pheochromocytomas is their predominant clustering into two groups based on transcriptional signatures. Cluster 1 tumors, including SDHx, VHL, and EPAS1, are characterized by hypoxia-related transcript enrichment. Cluster 2 tumors, including NF1, RET, and HRAS, are defined by enrichment in kinase-signaling pathway transcripts. We sought to determine whether the SDHB/NF1 and HET/NF1 mouse tumors similarly formed transcriptional clusters, what pathways/genes defined these clusters, and how they compared with the human transcriptional signatures. We performed RNA-seq of SDHB/NF1 and HET/NF1 tumors, and age-matched normal adrenal medulla (NAM) controls. Principal component analysis revealed three distinct clusters, which separated according to genotype (Figures 6A and S4C): 3,679 genes were differentially expressed (p < 0.05) between SDHB/NF1 and HET/NF1 tumors. Importantly, the SDHB transcripts were significantly decreased by 57% and 89% in HET/NF1 and SDHB/NF1 tumors relative to NAM, respectively (Figure S4D), confirming the heterozygous and homozygous knockout of SDHB in these tumors.
Figure 6. Transcriptomics of mouse tumors revealed enrichment of mitochondrial and ribosomal gene transcripts.
(A) Heatmap and hierarchical clustering of top 1,200 most significantly up- and down-regulated transcripts between SDHB/NF1 and HET/NF1 tumor groups.
(B) KEGG gene set enrichment for SDHB/NF1 and HET/NF1 tumors relative to normal adrenal medulla. Plotted pathways had p < 0.01. Depicted pathways were the top 10 most enriched KEGG pathways in SDHB/NF1 tumors and their relative enrichment in HET/NF1 tumors. Numbers in parentheses are the systemic names for each KEGG pathway.
(C) KEGG gene set enrichment for SDHB/NF1 tumors relative to HET/NF1 tumors. Depicted pathways had p value < 0.03.
(D) KEGG gene set enrichment for HET/NF1 tumors relative to SDHB/NF1 tumors. Depicted pathways had p value < 0.01.
(E) Heatmap showing enrichment of mitochondrial DNA-encoded transcripts in mouse and human tumors. Red stars indicate the two NF1 pheochromocytomas, which were classified as pseudohypoxic.
(F) Quantification of mitochondrial DNA copy number by qPCR analysis (16S RNA and ND1/HK2) from mouse tumors and NAM (n = 4–6).
(G) Normalized counts of transcripts for PPARGC1a gene (PGC1a) from mouse and human tumors and mouse NAM. Data ae presented as mean ± SD and analyzed using an unpaired t test. **p < 0.01.
Next, we performed gene set enrichment analysis of SDHB/NF1 and HET/NF1 tumors relative to NAM to determine which KEGG pathways were enriched. As expected, both SDHB/NF1 and HET/NF1 tumors had significant enrichment within the CELL CYCLE and DNA REPLICATION pathways (Figure 6B). Unexpectedly, the RIBOSOME pathway was the most enriched pathway among SDHB/NF1 tumors but was not enriched in HET/NF1 tumors. Notably, SDHB/NF1 tumors also demonstrated significant alteration of multiple DNA repair pathways, which was substantially less in HET/NF1 tumors. These results were consistent with impaired DNA repair observed in human SDHx tumors (Sulkowski et al., 2020; Sulkowski et al., 2018). In human PPGLs, pathway analysis has generally compared differences between tumor types (Lopez-Jimenez et al., 2010). A similar analysis comparing SDHB/NF1 with HET/NF1 tumors showed that SDHB/NF1 tumors were most enriched in RIBOSOME and OXIDATIVE_PHOSPHORYLATION pathways (up-regulated expression), whereas HET/NF1 tumors were most significantly enriched for transcripts in the ECM_RECEPTOR_INTERACTION, vascular endothelial growth factor (VEGF), NOTCH, and Toll-like receptor (TLR) signaling pathways (Figures 6C and 6D). Remarkably, parallel analysis performed with the human SDHx and NF1 germline pheochromocytomas from the TCGA dataset (Fishbein et al., 2017) demonstrated overlapping enrichment in KEGG pathways, including ribosome and oxidative phosphorylation, in Cluster 1 tumors (Figure S4E). These data highlight the robust transcriptional similarity between human SDHx and mouse SDHB/NF1 pheochromocytomas.
Comparison of mouse SDHB/NF1 and human SDHx tumors with HET/NF1 (mouse) and NF1 (human) pheochromocytomas, respectively, demonstrated significant enrichment of oxidative phosphorylation and mitochondrial DNA-encoded transcripts (Figures 6C and 6E). These data, along with the large number of swollen mitochondria observed by EM, indicated a SDHB-associated expansion in mitochondria number. This conclusion was further substantiated by an ~2-fold increase in SDHB/NF1 tumor mitochondrial DNA copy number relative to HET/NF1 tumors and NAM (Figure 6F). Consistent with these findings were 9- and 2.4-fold increases in PPARGC1A gene expression in SDHB/NF1 and human SDHx pheochromocytomas, respectively (Figure 6G). Notably, the mitochondrial DNA-encoded signature was dissociated from the hypoxia signature in the human samples we analyzed; the two NF1 tumors that contained a pseudohypoxia signature did not contain a mitochondrial DNA gene expression signature (Figure 6E).
Our pathway analysis did not uncover significant enrichment of either hypoxia or HIF-regulated pathways when we compared SDHB/NF1 with HET/NF1 mouse tumors. By contrast, SDHx human tumors were significantly enriched in Hallmark_Hypoxia transcripts. It was unclear why SDHB/NF1 tumors lacked a pseudohypoxic signature; however, we suspect that pseudohypoxia may be acquired during later stages of tumor progression, since two of the four SDHB/NF1 samples exhibited increased expression of some HIF-regulated transcripts such as Cox4i2 and EPAS1.
Ascorbate depletion was lethal to SDHB-null chromaffin cells
The adrenal gland contains one of the highest deposits of ascorbate in the body, where it is found at millimolar levels in both mice and humans (Kim et al., 2012). Within the adrenal, DESI-MSI images of ascorbate generally showed higher abundance in the medulla and around the corticomedullary border (Figures 7 and S5). The effect of physiologic ascorbate levels on tumorigenesis are incompletely elucidated but believed to oppose cancer cell growth, in part, via activation of 2-OG-dependent enzymes to reduce HIF levels and reverse hypermethylation (Kuiper and Vissers, 2014). In addition to using 2-OG as a substrate, these enzymes require O2, Fe2+, and ascorbate for maximal activity; ascorbate is required for catalyst (iron) recycling (Kuiper and Vissers, 2014). Interestingly, DESI-MSI of 12+-month mouse tumors showed a trend toward reduced ascorbate abundance, whereas human tumors exhibited a pronounced ascorbate reduction (Figures 7A–7C). We hypothesized that ascorbate depletion in the context of SDHB disruption and elevated succinate would potentiate 2-OG enzyme inhibition and accelerate tumorigenesis in the absence of NF1 mutation. Toward this end, we bred SDHB animals to GULO−/− animals to deplete ascorbate (Maeda et al., 2000). We supplemented ascorbate into the diet of GULO−/− animals during breeding; however, supplementation was removed upon weaning (4 weeks), and animals were ascorbate depleted for 6 more weeks before being euthanized. Importantly, ascorbate has a Km of ~170–300 μM for PHDs and TETs which is high enough to render the enzymes sensitive to manipulation of ascorbate levels (Hirsila et al., 2003).
Figure 7. Ascorbate levels decreased in mouse and human tumors, but ascorbate depletion was paradoxically lethal to developing SDHB cells.
(A) Representative DESI-MSI images of ascorbate for control, HET/NF1, and SDHB/NF1 glands at ~12 months.
(B) DESI-MSI images of ascorbate from normal human adrenal and PPGLs. All images were normalized as %TIC.
(C) Average abundance of ascorbate in the medulla, extracted from DESI-MSI data (n = 4–6 for murine; n = 1–5 for human).
(D) Representative mT/mG images of male HET and SDHB adrenals in GULO+/+ and GULO−/− animals.
(E) Quantification of %GFP+ medulla area for male 10-week-old animals. Data are presented as mean ± SD and analyzed using a one-way ANOVA. **Adjusted-p value < 0.01.
(F) Proposed model for SDHB tumorigenesis. Inherited pathogenic mutations in SDHB result in a largely normal cellular state. Spontaneous loss of the 1p arm leads to copy number loss with loss of heterozygosity (CNL-LOH) for SDHB and induces heterozygosity for an unidentified tumor suppressor gene(s) (TSG) on the 1p arm. If this occurs in a cell type with high ascorbate levels (or other permissive redox environments), then the cell can survive loss of SDHB; otherwise, cell death may occur. Loss of SDHB initially leads to accumulation of succinate, swelling of mitochondria, and histone hypermethylation. Neoplasia is established through progressive rounds of cellular division, as the remaining allele(s) of the TSG is silenced via promoter methylation leading to an acceleration of replication and gradual loss of ascorbate and induction of hypoxia transcripts.
At 10 weeks, the percentage of GFP+ medulla area was determined in SDHB and HET ascorbate-depleted animals. DESI-MSI analysis confirmed that depleted GULO−/− adrenals had virtually undetectable ascorbate levels (Figure S5). In contrast to our hypothesis that ascorbate depletion would exacerbate 2-OG dioxygenase inhibition and potentiate tumorigenesis, the percent GFP+ cell area was substantially reduced in SDHB/GULO−/− medullas relative to control genotypes (Figures 7D and 7E). GULO+/+ male SDHB and HET animals both had an average GFP+ area of ~45% at 10 weeks. Ascorbate-depleted GULO−/− HET and SDHB mice had 30.5% and 12.9% GFP+ area, respectively. These data suggested that the high levels of ascorbate in the medulla were permissive for SDHB-deficient chromaffin cell survival, perhaps by maintaining a favorable redox environment. These results indicated that ascorbate depletion did not accelerate SDHB-deficient cell expansion but did not exclude the possibility of delayed tumor formation.
DISCUSSION
Attempts to model the SDHx PPGL syndrome in mice have been ongoing for over 15 years. Until now it was unclear whether the failure to develop tumors was due to lack of succinate accumulation and/or cellular lethality associated with SDH deficiency. Here, we combined a Cre-reporter system with DESI-MSI to rigorously phenotype and follow the SDHB−/− population in the mouse adrenal medulla. We show that SDHB-deficient chromaffin cells are viable, but not tumorigenic, despite substantially increased succinate levels and manifestation of cardinal epigenetic features of human SDHx tumors. Additionally, SDHB−/− chromaffin cells have expanded clusters of swollen mitochondria that are remarkably similar to those found previously in human SDHx tumors. We demonstrate the utility of DESI-MSI for targeted tracking of metabolic changes in the adrenal medulla by spatially resolving the distinct molecular profiles of the cortex and medulla. However, after 1 year of aging, SDHB-deficient chromaffin cells do not exhibit increased proliferation, loss of 5hmC, HIF1/2α accumulation, or significant changes in catecholamine abundances. These data definitively show that, in mice, loss of SDHB in chromaffin cells is insufficient for tumorigenesis despite high levels of succinate.
There are several unique aspects to SDHx tumor genetics that suggest that loss of an additional tumor suppressor, proposed to be located in SDHx-adjacent regions of the genome, is required for initiation of tumorigenesis. The strongest evidence for this comes from SDHD mutations, which primarily promote PPGL formation after paternal inheritance. Since the SDHD locus is not imprinted, the prevailing hypothesis is that loss of an adjacent imprinted gene is required for tumor initiation (Hoekstra et al., 2015b). Indeed, loss of heterozygosity (LOH) for SDHD tumors occurs with loss of the maternal copy of chromosome 11. With respect to SDHB, which is located on 1p36.13, it is important to note that LOH for virtually all SDHB tumors occurs via loss of the 1p chromosome arm (Fishbein et al., 2017), and this has been shown to be an early event in tumor formation (Edstrom et al., 2000; Dannenberg et al., 2000). Indeed, those attempting to model IDH1/2 mutations (Sasaki et al., 2012; Philip et al., 2018) and FH knockout (Adam et al., 2011) similarly concluded that loss of additional genes was necessary for tumor initiation. All together, our data, plus those from IDH and FH models, and the underlying genetic events in SDHx tumors, support the hypothesis that oncometabolite accumulation is insufficient for tumor initiation. Hence, we propose copy number loss with LOH (CNL-LOH) as the genetic mechanism of SDHB-dependent tumorigenesis.
In this study, we used loss of NF1 to initiate tumorigenesis in the SDHB mouse model. However, it is important to note that there is no evidence that biallelic NF1 and SDHB mutations co-occur in human PPGLs and that most tumors arising from individuals with germline NF1 or SDHB mutations have many different characteristics (Fishbein et al., 2017). Despite the differences in SDHB and NF1 tumor phenotypes, there is reason to believe there are overlaps in their tumorigenic mechanism. Indeed, the 1p arm is also lost in most NF1 pheochromocytomas. Additionally, the two major pathways activated by loss of NF1, the Ras-Raf-Erk and PI3-Akt-mTOR pathways, are also activated in SDHx cells and tumors. For example, phospho-ERK1/2 has also been shown to accumulate in SDHB-null cells (Chen et al., 2014), and many mTORC pathway components are upregulated in SDHx tumors (Oudijk et al., 2017). Moreover, it has been proposed that germline inheritance of many pheochromocytoma susceptibility genes, such as SDHx and NF1, share a common pathway for inhibition of apoptosis, which culminates in inhibition of PHD3 activity (Lee et al., 2005). Furthermore, a direct effect of NF1 on the SDH complex has been identified (Masgras et al., 2017). It is informative to note that the combined deletion of SDHB and NF1 was successful, whereas many other SDHx plus tumor suppressor gene combinations failed to initiate tumors in mice. For example, the PSA-Cre SDHB/PTEN deletion produced tumors without elevated succinate, leading to the conclusion that only PTEN-recombined cells survived recombination. Tumors also did not arise from the combined loss of SDHC with p53 or hypoxia (10% O2) (Al Khazal et al., 2020). Indeed, prior studies have not found loss of p53 in chromaffin cells to be tumorigenic (Tonks et al., 2010) unless combined with loss of PTEN or RB1, suggesting that specific pathways need to be simultaneously targeted to effect proliferation. The ability to convert a Cluster 2 cell type into a Cluster 1 cell type, shown here, suggests that there are overlapping tumorigenic pathways in these clusters and that high succinate levels and subsequent hypermethylation are a main feature driving the phenotypic distinctions between these tumor types.
Hypermethylation is commonly described in human malignancies, and several mechanisms exist by which 5hmC levels and TET activity can be modulated (Pfeifer et al., 2014). In the case of SDHx, IDH1/2, and FH tumors, the proposed mechanism is oncometabolite-mediated TET inhibition. However, other mechanisms, such as nuclear exclusion of TET1 enzyme (Muller et al., 2012), TET silencing via promoter hypermethylation (Li et al., 2016), and decreased expression of IDH1 (Chen et al., 2016) have also been described. In our system we find that NF1-null chromaffin cells exhibit a reduction of 5hmC in pre-tumor adrenals, whereas the SDHB-null cells retain 5hmC immunostaining. The NF1 result is consistent with reports showing that almost all PPGLs have loss of 5hmC relative to adjacent sustenacular cells, while SDHx tumors have a more extreme shift in this ratio (see Hoekstra et al., 2015a). So, why does elevated succinate appear insufficient for TET inhibition in our SDHB-null cells but sufficient for KDM4 inhibition? The succinate IC50 values for TETs and KDMs are fairly similar (Laukka et al., 2018), suggesting these enzymes should be equivalently inhibited by succinate. We propose that TET inhibition does occur in SDHB-null cells, but in order to observe a robust change in 5hmC marks, multiple rounds of cellular division are required. Evidence of this phenomenon was seen in experiments with IDH1-R132H cells, which gained tri-methyl histone H3 marks within a few passages, but loss of 5hmC required at least 12 passages (Lu et al., 2012). Indeed, in SDHB/NF1 tumors there was virtually no detectable 5hmC immunostaining, whereas NF1 tumors retained a low level of 5hmC, demonstrating that cellular succinate levels were sufficient for TET inhibition but loss of 5hmC was manifested only in cycling cells, not arrested cells. This also suggests that DNA hypermethylation per se is unlikely to be a driver of tumorigenesis but rather an effector that exacerbates progression over time. Alternatively, it is possible that the methylation patterns are sufficiently different between mouse and human such that oncometabolite-driven hypermethylation followed by tumor initiation is not possible in mice (in other words, the “right” genes do not get silenced in mice).
A hallmark feature of SDHx tumors is the upregulation of hypoxia-related transcripts. However, SDHB/NF1 tumors do not significantly upregulate Hif1/2α or hypoxia transcripts. HIF stabilization is also absent from other oncometabolite mouse models. For example, HIF1α accumulation was not observed in IDH1(R132H) knockin mice targeted to the subventricular zone, although hypermethylation was evident (Bardella et al., 2016). The kidney cysts that resulted from loss of FH did express nuclear HIF1α however dual knockout of FH and HIF1/2α in the kidney did not ameliorate the cystic phenotype; suggesting that HIF was not phenotypically relevant (Adam et al., 2011). Ultimately, these data do not resolve the role of HIF activation in SDHx tumors; however, our data, and those of others, indicate that the proposed mechanism of succinate-mediated PHD inhibition is incomplete. Additional events such as ROS accumulation (Goncalves et al., 2021) and/or CNL-LOH may be necessary for acquisition of the pseudohypoxic phenotype late in the tumorigenic process.
Finally, in considering why succinate is insufficient to drive tumorigenesis in SDHB mice, we explored the hypothesis that high ascorbate levels in the adrenal medulla were sufficient to maintain residual diooxygenase enzyme activity and suppress tumor formation. This phenomenon has been observed in IDH1R132H cell lines where vitamin C treatment increased 5hmC marks despite the presence of 2-HG (Mingay et al., 2018). High-dose vitamin C can also overcome the loss of Tet2 in a mouse model for leukemia (Cimmino et al., 2017). Conversely, leukemogenesis is accelerated in ascorbate-depleted Gulo/− mice transplanted with Flt3ITDTet2+/−HSCs (Agathocleous et al., 2017). Given these data, we expected that ascorbate depletion would further inhibit activity of 2-OG enzymes and initiate tumor formation. However, to our surprise, ascorbate depletion in SDHB animals led to a significant and preferential loss of SDHB-deficient cell viability, showing that ascorbate is required for cellular survival after loss of SDH activity and, potentially, indicating why SDHx mutations are associated with PPGLs, cell types with relatively high ascorbate levels. In support of this hypothesis, C. elegans, which also lack a gulo gene, exhibit premature aging and early death when their sdhb gene is knocked-out. Remarkably, this effect was largely reversed by the addition of ascorbate to the growth medium (Huang and Lemire, 2009). Similarly, TH-SDHD mice displayed a preferential loss of substantia nigra dopaminergic neurons, which were partially rescued by the addition of an antioxidant in the drinking water (Diaz-Castro et al., 2012). The requirement for a plentiful cellular redox environment has also been shown in other models of loss of TCA cycle genes. In FH mouse models, the NRF2 antioxidant pathway was the most upregulated transcriptional pathway in FH renal cysts (Adam et al., 2011). In contrast to the initial requirement for abundant ascorbate, we measured a significant loss of ascorbate in human PPGLs relative to normal adrenals. We propose that during tumor progression metabolic reprogramming relieves the ascorbate-dependence, which in turn potentiates 2-OG enzyme inhibition. Indeed, it has recently been shown that ultra-high-dose ascorbate preferentially kills both SDHB cell lines (Goncalves et al., 2021) and SDHB allografts (Liu et al., 2020). Notably, the role of ascorbate in maintaining the viability of SDH-deficient chromaffin cells does not explain the increased risk of SDHx-associated renal cell carcinoma and gastrointestinal stromal tumor, where ascorbate levels are low and alternative mechanisms of remediating oxidative stress, perhaps via glutathione regulation, would be required.
In summary, loss of SDHB in the mouse adrenal medulla is compatible with cell survival which is enabled by the high levels of ascorbate in the adrenal gland. Chromaffin cells respond to loss of SDHB by accumulating succinate, clustered swollen mitochondria, and histone hypermethylation, but not increased proliferation. Our data strongly substantiate a requirement for a second, unspecified, genetic event in SDHB tumors that is necessary for cell-cycle entry and tumorigenesis. We suspect that, in humans, as in the mouse, an additional gene(s), probably located on the 1p arm, must be concurrently lost to initiate tumor formation. We propose that upon CNL-LOH, succinate accumulates, metabolic adaptation takes place (mitochondrial swelling), and histone methylation is initiated (Figure 7F). This tumor initiation stage includes a heterozygous state for the 1p co-tumor suppressor(s) which are presumed to be haploinsufficient at this stage of tumor initiation. Subsequently, we hypothesize, the remaining wildtype allele(s) are epigenetically silenced through successive rounds of cellular division. The final neoplastic stage is achieved when the remaining 1p tumor suppressor gene(s) are sufficiently silenced. We further hypothesize that neoplasia is enforced by ascorbate loss, which triggers a pseudohypoxic transcriptional phenotype. Our unique success from combining SDHB and NF1, when several other combinations have failed, provides important new insights into the molecular mechanisms of SDHx tumorigenesis and a valuable new resource. Although a variety of cellular in vitro and transplant models exist, these suffer from several fundamental deviations including an uncharacteristically high replication rate. The SDHB/NF1 mouse model of hPPGL is a valuable new experimental resource that may be leveraged to better understand SDHx tumorigenesis, uncover new therapeutic avenues, and, perhaps most importantly, establishes the feasibility of modeling SDHB-deficient tumors in mice.
Limitations of the study
A limitation to this study is the use of NF1 disruption as a driving force for SDHB-deficient tumor formation. Homozygous loss of SDHB and NF1 has not been described in the human PPGLs. Another limitation is that complete ascorbate depletion is lethal to Gulo−/− animals by 10 weeks of age. Hence, the tumorigenic potential of surviving SDHB-deficient chromaffin cells, in the context of ascorbate depletion, could not be assessed. To study the long-term consequences of ascorbate deprivation on SDHB cellular survival and tumor formation, studies need to be conducted with an ultra-low ascorbate diet for several months. Finally, we have not evaluated the effect of ascorbate on dual NF1- and SDHB-deficient mouse tumors; hence the selective lethality of ascorbate depletion toward mouse SDHB-deficient chromaffin cells should not be extrapolated to survival of mouse or human SDHB-deficient tumors.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Justin Annes (jannes@stanford.edu).
Materials availability
SDHBF/F mice will be made available on request, but may require payment for shipping/handling costs and a material transfer agreement if there is a potential for commercial application.
Data and code availability
RNA sequencing data has been deposited in NCBI’s Sequence Read Archive (SRA) under accession code PRJNA790868 and are publicly available as of the date of publication. This study does not report the development of any new code. Any additional information required to reanalyze the data in this paper will be made available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
Th-Cre (#008601), NF1F/F (#17640) and mT/mG (#007576) mice were obtained from The Jackson Labs. The Gulo−/− mice were obtained from UC Davis Mouse Biology Program. These mice have all been described previously (Muzumdar et al., 2007), (Savitt et al., 2005), (Zhu et al., 2001), (Maeda et al., 2000). SDHBF/F mouse lines were generated from ES cell clone Sdhbtm1a(EUCOMM)Hmgu (MGI:5085385) obtained from EuMMCR and injected into C57BL6/J blastocysts. After breeding of chimeras, pups carrying germline SDHB−/+ were identified by coat color and tail genotyping PCR. SDHBtm1a/+ animals were bred to FLP1 mice (JAX #009086) to generate SDHBF/+. All strains were genotyped using standard PCR methods. Mice were maintained in a mixed 129/Sv-C57BL/6 background. Mice were raised in a barrier facility, housed in ventilated cages with access to water and normal chow ad libitum and maintained on a 12 h light/dark cycle under controlled temperature and humidity. Both male and female mice were used except when noted. GULO animals were kept in a regular animal facility and were provided ascorbic acid (3.3 g/L) in their drinking water until weaning. All animal experiments were performed in compliance with the Institutional Animal Care and Use Committee and the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). Animals were used at 3 timepoints: 10 weeks, 5–6 months and 1+ year.
Human samples for DESI-MSI
One healthy adrenal and 5 PPGLs were obtained as either freshly dissected samples or as fresh frozen tissue from the Stanford Tumor Bank (Table S1). Two samples (PGL-5 and −6) overlapped with samples used in Cytochrome C IHC analysis.
Human samples for cytochrome C analysis
Thirty paraffin embedded human PPGL tumor blocks (15 PCCs and 15 PGLs) with known genetic testing results and/or SDHB immunohistochemistry (IHC) were retrieved from the Stanford Tumor Bank. PCC samples included four cluster 1 and eleven non-cluster 1 tumors (Table S2). PGL samples included ten cluster 1 and five non-cluster 1 PGLs (Table S2). PPGLs were classified as cluster 1 (pseudohypoxic) and non-cluster 1 tumors, based on genetic testing results and/or SDHB immunohistochemistry. If SDH mutation was classified as “benign” or ‘VUS” while the tumor exhibited “preserved” SDH by IHC, the tumor was categorized as non-cluster 1. Tumors with preserved cytoplasmic SDHB IHC were classified as non-cluster 1 tumors, excluding those tumors with known cluster 1 gene mutation such as VHL or FH. Genetic testing performed in this study cohort ranged from limited analyses of 3 genes for SDHx mutations (SDHB, C, & D) to more comprehensive sequencing analyses of up to 104 genes.
METHOD DETAILS
mT/mG fluorescence imaging and GFP area quantification
Adrenals were fixed for 1–2 hours in 4% paraformaldehyde (PFA) in 0.01 M phosphate-buffered saline (PBS) and rinsed in PBS before equilibration in 30% sucrose in PBS overnight at 4°C. Cryoprotected samples were immersed in O.C.T.™ compound and frozen at −80C. OCT blocks were mounted on a cryostat and the cortex was trimmed away. Once the medulla was exposed, every 2nd or 3rd 10um sections were mounted on charged slides for a total of ~30 sections per gland. Slides were washed in PBS to remove OCT and mounted with Vectashield Antifade plus DAPI. 5–7 sections per gland were imaged on a Leica fluorescent microscope. Images were processed in Volocity 6.3 by hand drawing the medulla outline and then calculating the area within the medulla covered by GFP+ cells. The percentage of GFP+ medulla area is reported as the average of 5–7 images from each animal. A minimum of 3 animals per genotype were used. Male animals were used for this data set as the female X-zone made defining the medulla less accurate.
Histopathology and immunohistochemistry
Freshly removed adrenals were fixed in 4% paraformaldehyde overnight at 4°C, transferred to 70% ethanol and then processed for paraffin embedding at the Stanford Animal Histology Core. For immunohistochemistry, 5μm thick sections were deparaffinized, rehydrated and antigen retrieved in 10mM citric acid at pH 6.0 in a pressure cooker for 10min. Slides were blocked for 1 hour in 5% normal donkey serum plus 0.1% Triton X-100 in PBS. Primary antibodies were diluted in blocking buffer and slides were incubated overnight at 4°C followed by 3 PBS washes and one hour incubation in appropriate fluorophore-conjugated secondary antibodies (Jackson Immunoresearch). Stained sections were mounted in DAPI antifade and imaged using the Leica DM IL LED microscope equipped with a digital Leica DFC425 camera (Wetzlar, Germany). Signal intensities of target antigens were quantified using Volocity 6.3 (PerkinElmer, Waltham, MA, USA). Quantification of the target antigen GFP+/GFP− fluorescence intensity was calculated by creating masks for GFP+ medulla area and inverting that mask for GFP− medulla areas. The average pixel intensity of both blue (DAPI) and red (epitope of interest) channels within each mask was measured per image. Images from at least 3 animals per genotype were averaged for each measurement (both genders included).
Human tumor cytochrome C immunostaining and image analysis
Cytochrome C immunostaining was ion software from Leica. Images were screened using 2.5X and 10X magnification to identify representative and viable tumor tissue areas. Image capture was done using tilescan function at 20X and 40X magnification; exposure time was set at 200 milliseconds for all samples. Representative images which include tumoral (T) and non-tumoral (NT) areas were chosen for subsequent image analysis. Image analysis software (Volocity 6.3) was used for quantification of cytochrome c, comparing its immunofluorescence intensities in tumoral areas versus non-tumoral areas. Tumoral areas were identified by their unique structural arrangements such as Zellballen/nest, trabecular, sheet like or solid architecture as well as synaptophysin positivity. Tumor capsule, adrenal cortex (in PCC samples), blood vessel wall and/or fibrovascular tissue were used as internal negative controls and designated as non-tumoral areas. Cytochrome c expression were designated as “strong”, “moderate” or “weak” or “absent” if T to NT ratio is >2, between 1.8–2, between 1.5–1.8 and <1.5 respectively. This cut-off value was empirically determined based on our experience with biomarker image analysis. Degenerative or necrotic tumor areas (reflected by weak or negative immunoreactivity of synaptophysin) and areas with extensive hemorrhage (indicated by falsely elevated intensities and bright background signals) were excluded from analysis.
Western blotting
Mouse medulla were surgically removed by manual trimming away of the cortical layer. Both medulla from one mouse were pooled and homogenized in 100ul of RIPA plus protease inhibitors. 10ug of protein were loaded per lane on a 10% SDS-PAGE. Proteins were transferred to nitrocellulose for immunoblotting, blocked in 5% non-fat milk and incubated overnight in the corresponding antibody. After incubation with HRP-conjugated antibodies, bands were visualized on a Bio-Rad ChemiDoc Imaging System.
DESI-MSI
DESI-MS imaging was performed using a lab-built probe constructed of two concentric, fused silica capillaries: an outer sheath gas capillary, 350/250 μm (O.D./I.D.), and an inner solvent capillary of 150/50 μm for the SDHB/NF1 tissue sets and 150/20 μm for the GULO and human tissue sets. A histologically compatible solvent mixture of 1:1 (v/v) dimethyl-formamide:acetonitrile (Eberlin et al., 2011) was used at a rate of 1 μL/min, and nebulization was aided with 160 psi of nitrogen sheath gas for the SDHB/NF1 tissue sets and 140 psi for the GULO and human sets. A spray voltage of −5 kV was applied to the syringe needle. The probe tip was angled at 56° from the horizontal plane and positioned 6–7 mm from the inlet and 2–3 mm above the surface of the slide.
Images were acquired using a custom-built moving stage coupled to a high-resolution LTQ Orbitrap XL mass spectrometer (Thermo Fisher) with a pixel size of 115 × 130 μm, 118 × 130 μm, and 181 × 200 μm for the SDHB/NF1, GULO, and human tissues, respectively. The pixel size was determined by the cycle time per scan, the lateral speed of the moving stage, and the vertical step size. The orbitrap was used as the mass analyzer with resolving power set to 60,000. Automatic gain control was turned off, and the maximum ion injection time was set to 100 ms per microscan, corresponding to one scan per pixel, for the SDHB/NF1 tissue sets, and 500 ms per microscan for the GULO and human tissue sets. The capillary temperature was 275°C. Spectra were collected over the m/z range 50–1,000. The capillary voltage was −65 V, and the tube lens voltage was −120 V.
Tissues were mounted in optimal cutting temperature (O.C.T.) compound before freezing on dry ice and cryosectioned in 10 μm sections for DESI-MSI. Slides were kept in a −80°C freezer and then thawed in a vacuum desiccator prior to imaging. All tissue sets were imaged on the same day to minimize variability, except for the 12+ month glands, which were measured over two sequential days. For the SDHB/NF1 tissue sets, a fluorescent image of the section imaged by DESI-MS was subsequently taken.
Identification of ion species by MS/MS
Collision induced dissociation (CID) of a tissue extract was used to accurately identify and assign peaks of interest. The spectra can be shared upon request. For extraction, one whole adrenal gland, approximately 3 mg of tissue, from a No Cre mouse was macerated and then suspended in 200 μL of 1:1 (v/v) methanol:water for 15 minutes with periodic vortexing. The mixture was spun down at 3300 rpm, and the supernatant removed for standard electrospray ionization (ESI). Tandem-MS was performed on the LTQ Orbitrap XL with a lab-built ESI spray constructed of a 365/100 μm (O.D./I.D.) fused silica capillary. The spray tip was positioned approximately 8 mm from the inlet; the tissue extract pump rate was 5 μL/min; and the nitrogen sheath gas pressure was 120 psi. The orbitrap was used as the mass analyzer with a resolution of 60,000. The capillary temperature, spray voltage, capillary voltage, and tube lens voltage were the same as described for the DESI-MSI experiments above.
On-tissue fragmentation was performed using the DESI-MSI setup to verify the succinate and ascorbate peaks on a GULO tissue and a human SDHB paraganglioma, as well as norepinephrine on a SDHB/HET tissue. These CID spectra were acquired while the DESI probe scanned the tissue with the same speed and parameters as used for imaging.
Fragmentation patterns were compared to those in standard databases, including Metlin (Scripps Research, La Jolla, CA) (Smith et al., 2005),(Guijas et al., 2018), Lipid Maps (Fahy et al., 2007), and the Human Metabolome Database (HMDB), to make peak assignments. We also compared measured accurate masses with previously published literature values. (Wu et al., 2010), (Sun et al., 2018).
DESI-MSI data analysis
Spectra were acquired in Xcalibur 2.5.5 (Thermo Fisher Scientific), with each row of an image comprising a separate .raw file. The .raw files were converted to the .mzML format using MSConvert (ProteoWizard tool) (Kessner et al., 2008), (Chambers et al., 2012) and then .mzML rows were compiled into one .imzML image file per sample using imzMLConverter (version 1.3, provided as part of the MSiReader package) (Race et al., 2012). Image files were then visualized in MSiReader version 1.01 (Robichaud et al., 2013), (Bokhart et al., 2018) using the jet color scheme, where red is the most and blue the least intense. All image intensities were scaled as a percentage of total ion current unless otherwise specified. Smoothed images were linearly interpolated. The m/z tolerance was set to ±5 ppm, in accordance with the mass accuracy of the orbitrap mass analyzer.
DESI-MSI, as conducted in these experiments, is semi-quantitative. Relative changes in metabolite intensities, normalized as a percentage of total ion current, were used to assess changes over the control. The mean intensity for a metabolite was found by averaging the normalized abundance values for all pixels within a region of interest (ROI). Zeros, when present, were included in the mean. The ROI intensity averages for each metabolite were then exported to GraphPad for further statistical analysis. All comparisons and tests of statistical significance were only made within a tissue set, e.g., 12+ month adrenals, GULO adrenals, or human tissues, to avoid batch effects.
For the SDHB/NF1 tissue sets, the medulla was the ROI, because it was the targeted location of genetic recombination. To guide selection of pixels in the adrenal medulla, fluorescence microscopy images (mt/mg) of the tissue sections imaged by DESI were overlaid onto the MS image in MSiReader. Microscopy images were first scaled by the tissue dimensions, and then a series of metabolites and lipids were used to correlate the fluorescent and MS images, including PI(38:4), FA(20:4), succinate, and ascorbate. For medulla-specific analysis, pixels that were 50% or more within the adrenal medulla, as determined by comparison with the fluorescent overlay, were designated as part of the medulla ROI.
For the GULO and human tissues, the ROI was the entire tissue, where PI(38:4) was used as a guide for the outer tissue border. In a few cases, some water-soluble metabolites leaked slightly from the tissue and may not have been included in the ROI, although they are visible in the DESI-MS images. In these cases, succinate intensities per tissue, and therefore fold-change, may have been even higher than that reported.
Transmission electron microscopy
Samples were fixed in Karnovsky’s fixative: 2% Glutaraldehyde (EMS Cat# 16000) and 4% paraformaldehyde (EMS Cat# 15700) in 0.1M Sodium Cacodylate (EMS Cat# 12300) pH 7.4 for 1 hr. The fix was replaced with cold/aqueous 1% Osmium tetroxide (EMS Cat# 19100) and were then allowed to warm to Room Temperature (RT) for 2 hrs rotating in a hood, washed 3X with ultrafiltered water, then en bloc stained in 1% Uranyl Acetate at RT 2hrs while rotating. Samples were then dehydrated in a series of ethanol washes for 30 minutes each @ RT beginning at 50%, 70% EtOH then moved to 4°C overnight. They were place in cold 95% EtOH and allowed to warm to RT, changed to 100% 2X, then Propylene Oxide (PO) for 15 min. Samples are infiltrated with EMbed-812 resin (EMS Cat#14120) mixed 1:2, 1:1, and 2:1 with PO for 2 hrs each with leaving samples in 2:1 resin to PO overnight rotating at RT in the hood. The samples are then placed into EMbed-812 for 2 to 4 hours then placed into molds w/labels and fresh resin, orientated and placed into 65° C oven overnight.
Sections were taken around 80nm using an UC7 (Leica, Wetzlar, Germany) picked up on formvar/Carbon coated 100 mesh Cu grids, stained for 40s in 3.5% Uranyl Acetate in 50% Acetone followed by staining in Sato’s Lead Citrate for 2 minutes. Observed in the JEOL JEM-1400 120kV. Images were taken using a Gatan OneView 4k × 4k digital camera.
RNA-seq
Adrenals used for RNA-seq came from 16–18M old animals. For tumors samples (SDHB/NF1 and HET/NF1 animals), pheochromocytomas were removed, weighted and immersed in ice-cold PBS while cortex was manually trimmed away. For normal aged medulla controls, cortex was manually removed before processing. Normal medulla tissue from 2 animals (4 total medulla) were pooled in order to get enough tissue for experimentation. Tissue was transferred to RNAlater and stored at −20C until processing. Total RNA and DNA were purified from tissue using the Qiagen AllPrep kit according to the manufacturer’s instructions. RNA quality was assessed by bioanalyzer. Library preps were carried out using an Illumina kit with PolyA selection. Sequencing was done on Illumina HiSeq (2 × 150bp) to a minimum depth of 50M reads per sample. Clustering was performed in Clustvis (https://biit.cs.ut.ee/clustvis/) and gene set enrichment analysis was carried out with GSEA 4.1.0 (Broad Institute).
Mitochondrial DNA copy number assay
Real-time qPCR on DNA extracted from mouse SDHB/NF1 and HET/NF1 tumors and aged-matched NAM was used to measure mitochondrial DNA copy number. Hexokinase 2 primers were used to measure nuclear DNA and ND1 and 16S RNA primers were used to quantify mitochondrial DNA. 100ng of DNA was input into each qPCR assay with Sybr Green and 1μM primers.
Assessment of glucose homeostasis
Glucose physiology experiments were performed on female SDHB/HET and No Cre 12+ old animals. Blood glucose levels were obtained with glucose meter (Contour next EZ, Parsippany, NJ, USA). For glucose tolerance test, mice were fasted for 16 h and blood glucose levels were measured at indicated times following intraperitoneal injection of 2 g glucose (Sigma) per kg of body weight. For insulin tolerance test, mice were fasted for 6 h and blood glucose levels were measured at indicated times following intraperitoneal injection of 1U Insulin (Humalog) per kg of body weight. Homeostasis model assessment-insulin resistance (HOMA-IR) was calculated to assess changes in insulin resistance: insulin (pmol/L) × glucose (mmol/L)/22.5. HOMA β-cell function as a percentage (HOMA-B%) was calculated to assess steady state β-cell function: insulin (pmol/L) × 20/ glucose (mmol/L) – 3.5. Both were calculated using fasting values.
QUANTIFICATION AND STATISTICAL ANALYSIS
Volocity was used for quantification of signal intensity for all immunohistochemistry images and all mt/mg images. GraphPad Prism software was used for all statistical analysis. For statistical analysis of mouse and human data, the mean ± SEM of independent subjects was used. For analysis of statistical significance, either unpaired t-tests, one-way ANOVA or two-way ANOVA was used. Statistical details (p-values) of experiments can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-GFP | Abcam | Cat# ab13970 |
| Anti-Cytochrome C | Santa Cruz | Cat# sc-13156 |
| Anti-H3K9me3 | Active Motif | Cat# 39062 |
| Anti-TFAM | GeneTex | Cat# GTX103231 |
| Anti-Hif1a | Abcam | Cat# ab51608 |
| Anti-Hif2a | Novus | Cat# NB100–122 |
| Anti-5hmC | Active Motif | Cat# 39069 |
| Anti-Ki67 | Abcam | Cat# ab16667 |
| Anti-Tyrosine Hydroxylase | Abcam | Cat# ab112 |
| Anti-Synaptophysin | Abcam | Cat# ab32127 |
|
| ||
| Biological samples | ||
|
| ||
| Human PPGLs and normal Adrenal Gland | Stanford Tumor Bank | n/a |
| Mouse RNA-seq data (NAM, SDHB/NF1, HET/NF1) | Sequence Read Archive | PRJNA790868 |
|
| ||
| Experimental models: Organisms/Strains | ||
|
| ||
| B6.129(Cg)-Gt(ROSA) 26Sortm4(ACTB-tdTomato,-EGFP)Luo/J |
The Jackson Lab | 007676 |
| B6.129(Cg)-Nf1tm1Par/J | The Jackson Lab | 017640 |
| B6.Cg-7630403G23RikTg(Th-cre)1Tmd/J | The Jackson Lab | 008601 |
| B6.129P2-Gulotm1Mae/Mmucd | MMRRC | 000015-UCD |
| B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ | The Jackson Lab | 009086 |
| SDHBF/F | This paper | n/a |
|
| ||
| Oligonucleotides | ||
|
| ||
| HK2 FWD: GCCAGCCTCTCCTGATTTTAGTGT | This paper | n/a |
| HK2 REV: GGGAACACAAAAGACCTCTTCTGG | This paper | n/a |
| ND1 FWD: CTAGCAGAAACAAACCGGGC | This paper | n/a |
| ND1 REV: CCGGCTGCGTATTCTACGTT | This paper | n/a |
| 16S RNA FWD: CCGCAAGGGAAAGATGAAAGAC | This paper | n/a |
| 16S RNA REV: TCGTTTGGTTTCGGGGTTTC | This paper | n/a |
|
| ||
| Software and algorithms | ||
|
| ||
| Volocity 6.3 | Perkin Elmer | n/a |
| GSEA 4.1.0 | Broad Institute/UC San Diego | n/a |
| GraphPad Prism v.8.0 | GraphPad Software | n/a |
| ClustVis | https://biit.cs.ut.ee/clustvis/ | n/a |
| MSiReader v1.02 | https://msireader.wordpress.ncsu.edu | n/a |
Highlights.
Generation and phenotypic characterization of chromaffin cell SDHB knockout mice
SDHB loss and succinate accumulation are insufficient for pheochromocytoma initiation
Dual SDHB/NF1 chromaffin cell knockout mice develop SDHx-like pheochromocytomas
Robust survival of SDHB-deficient chromaffin cells requires an ample redox environment
ACKNOWLEDGMENTS
This work was conducted with support from the Stanford Cancer Institute (an NCI-designated Comprehensive Cancer Center), George Fisher (Stanford Cancer Institute), NIH awards DK101530 and DK119955 (J.P.A.), and the Child Health Research Institute, Lucile Packard Foundation for Children’s Health, as well as the Stanford CTSA (UL1 TR001085).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110453.
REFERENCES
- Adam J, Hatipoglu E, O’flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. (2011). Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, Cowin BL, Bruner E, Murphy MM, Chen W, et al. (2017). Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Khazal F, Kang S, Nelson Holte M, Choi DS, Singh R, Ortega-Saenz P, Lopez-Barneo J, and Maher LJ 3rd. (2020). Unexpected obesity, rather than tumorigenesis, in a conditional mouse model of mitochondrial complex II deficiency. FASEB J. 35, e21227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardella C, Al-Dalahmah O, Krell D, Brazauskas P, Al-Qahtani K, Tomkova M, Adam J, Serres S, Lockstone H, Freeman-Mills L, et al. (2016). Expression of Idh1(R132H) in the murine subventricular zone stem cell niche recapitulates features of early gliomagenesis. Cancer Cell 30, 578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, Van Der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. (2000). Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851. [DOI] [PubMed] [Google Scholar]
- Bernardo-Castineira C, Saenz-De-Santa-Maria I, Valdes N, Astudillo A, Balbin M, Pitiot AS, Jimenez-Fonseca P, Scola B, Tena I, Molina-Garrido MJ, et al. (2019). Clinical significance and peculiarities of succinate dehydrogenase B and hypoxia inducible factor 1alpha expression in parasympathetic versus sympathetic paragangliomas. Head Neck 41, 79–91. [DOI] [PubMed] [Google Scholar]
- Bokhart MT, Nazari M, Garrard KP, and Muddiman DC (2018). MSiReader v1.0: evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J. Am. Soc. Mass Spectrom. 29, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X, et al. (2016). Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res 26, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu T, Zhang S, Zhou J, Wang Y, and Di W (2014). Succinate dehydrogenase subunit B inhibits the AMPK-HIF-1alpha pathway in human ovarian cancer in vitro. J. Ovarian Res. 7, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et al. (2017). Restoration of TET2 function blocks aberrant self-Renewal and leukemia progression. Cell 170, 1079–1095.e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, Kung AL, Sanso G, Powers JF, Tischler AS, et al. (2005). A HI-F1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 1, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg H, Speel EJ, Zhao J, Saremaslani P, Van Der Harst E, Roth J, Heitz PU, Bonjer HJ, Dinjens WN, Mooi WJ, et al. (2000). Losses of chromosomes 1p and 3q are early genetic events in the development of sporadic pheochromocytomas. Am. J. Pathol. 157, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Castro B, Pintado CO, Garcia-Flores P, Lopez-Barneo J, and Piruat JI (2012). Differential impairment of catecholaminergic cell maturation and survival by genetic mitochondrial complex II dysfunction. Mol. Cell Biol 32, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douwes Dekker PB, Hogendoorn PC, Kuipers-Dijkshoorn N, Prins FA, Van Duinen SG, Taschner PE, Van Der Mey AG, and Cornelisse CJ (2003). SDHD mutations in head and neck paragangliomas result in destabilization of complex II in the mitochondrial respiratory chain with loss of enzymatic activity and abnormal mitochondrial morphology. J. Pathol. 201, 480–486. [DOI] [PubMed] [Google Scholar]
- Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cheng L, and Cooks RG (2011). Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry. Chembiochem 12, 2129–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstrom E, Mahlamaki E, Nord B, Kjellman M, Karhu R, Hoog A, Goncharov N, Teh BT, Backdahl M, and Larsson C (2000). Comparative genomic hybridization reveals frequent losses of chromosomes 1p and 3q in pheochromocytomas and abdominal paragangliomas, suggesting a common genetic etiology. Am. J. Pathol. 156, 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, and Subramaniam S (2007). LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, W606–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier J, Briere JJ, Burnichon N, Riviere J, Vescovo L, Benit P, Giscos-Douriez I, De Reynies A, Bertherat J, Badoual C, et al. (2009). The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One 4, e7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T, et al. (2017). Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell 31, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J, Moog S, Morin A, Gentric G, Muller S, Morrell AP, Kluckova K, Stewart TJ, Andoniadou CL, Lussey-Lepoutre C, et al. (2021). Loss of SDHB promotes dysregulated iron homeostasis, oxidative stress, and sensitivity to ascorbate. Cancer Res. 81, 3480–3494. [DOI] [PubMed] [Google Scholar]
- Gouw AM, Margulis K, Liu NS, Raman SJ, Mancuso A, Toal GG, Tong L, Mosley A, Hsieh AL, Sullivan DK, et al. (2019). The MYC oncogene cooperates with sterol-regulated element-binding protein to regulate lipogenesis essential for neoplastic growth. Cell Metab 30, 556–572.e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. (2018). METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 90, 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, and Myllyharju J (2003). Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780. [DOI] [PubMed] [Google Scholar]
- Hoekstra AS, De Graaff MA, Briaire-De Bruijn IH, Ras C, Seifar RM, Van Minderhout I, Cornelisse CJ, Hogendoorn PC, Breuning MH, Suijker J, et al. (2015a). Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget 6, 38777–38788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra AS, Devilee P, and Bayley JP (2015b). Models of parent-of-origin tumorigenesis in hereditary paraganglioma. Semin. Cell Dev Biol 43, 117–124. [DOI] [PubMed] [Google Scholar]
- Huang J, and Lemire BD (2009). Mutations in the C. elegans succinate dehydrogenase iron-sulfur subunit promote superoxide generation and premature aging. J. Mol. Biol. 387, 559–569. [DOI] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, and Weinberg RA (1994). Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 7, 353–361. [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, and Mallick P (2008). Proteo-Wizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Bae S, Yu Y, Kim Y, Kim HR, Hwang YI, Kang JS, and Lee WJ (2012). The analysis of vitamin C concentration in organs of gulo(−/−) mice upon vitamin C withdrawal. Immune Netw. 12, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, and Busam KJ (2017). The NF1 gene in tumor syndromes and melanoma. Lab Invest 97, 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper C, and Vissers MC (2014). Ascorbate as a co-factor for fe- and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression. Front Oncol. 4, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukka T, Myllykoski M, Looper RE, and Koivunen P (2018). Cancer-associated 2-oxoglutarate analogues modify histone methylation by inhibiting histone lysine demethylases. J. Mol. Biol. 430, 3081–3092. [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG Jr., and Schlisio S (2005). Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell 8, 155–167. [DOI] [PubMed] [Google Scholar]
- Lendvai N, Pawlosky R, Bullova P, Eisenhofer G, Patocs A, Veech RL, and Pacak K (2014). Succinate-to-fumarate ratio as a new metabolic marker to detect the presence of SDHB/D-related paraganglioma: initial experimental and ex vivo findings. Endocrinology 155, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepoutre-Lussey C, Thibault C, Buffet A, Morin A, Badoual C, Benit P, Rustin P, Ottolenghi C, Janin M, Castro-Vega LJ, et al. (2016). From Nf1 to Sdhb knockout: successes and failures in the quest for animal models of pheochromocytoma. Mol. Cell Endocrinol 421, 40–48. [DOI] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. (2013). SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 23, 739–752. [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Mao H, Du Z, Chan WY, Murray P, Luo B, Chan AT, Mok TS, Chan FK, et al. (2016). Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci. Rep. 6, 26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pang Y, Zhu B, Uher O, Caisova V, Huynh TT, Taieb D, Hadrava Vanova K, Ghayee HK, Neuzil J, et al. (2020). Therapeutic targeting of SDHB-mutated pheochromocytoma/paraganglioma with pharmacologic ascorbic acid. Clin. Cancer Res. 26, 3868–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Jimenez E, Gomez-Lopez G, Leandro-Garcia LJ, Munoz I, Schiavi F, Montero-Conde C, De Cubas AA, Ramires R, Landa I, Leskela S, et al. (2010). Research resource: transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol. Endocrinol. 24, 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Koivunen P, and Kaelin WG Jr. (2020). 2-Oxoglutarate-dependent dioxygenases in cancer. Nat. Rev. Cancer 20, 710–726. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. (2012). IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussey-Lepoutre C, Bellucci A, Morin A, Buffet A, Amar L, Janin M, Ottolenghi C, Zinzindohoue F, Autret G, Burnichon N, et al. (2016). In vivo detection of succinate by magnetic Resonance spectroscopy as a hallmark of SDHx mutations in paraganglioma. Clin. Cancer Res. 22, 1120–1129. [DOI] [PubMed] [Google Scholar]
- Lussey-Lepoutre C, Buffet A, Morin A, Goncalves J, and Favier J (2018). Rodent models of pheochromocytoma, parallels in rodent and human tumorigenesis. Cell Tissue Res 372, 379–392. [DOI] [PubMed] [Google Scholar]
- Maeda N, Hagihara H, Nakata Y, Hiller S, Wilder J, and Reddick R (2000). Aortic wall damage in mice unable to synthesize ascorbic acid. Proc. Natl. Acad. Sci. U S A. 97, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masgras I, Ciscato F, Brunati AM, Tibaldi E, Indraccolo S, Curtarello M, Chiara F, Cannino G, Papaleo E, Lambrughi M, et al. (2017). Absence of neurofibromin induces an oncogenic metabolic switch via mitochondrial ERK-mediated phosphorylation of the chaperone TRAP1. Cell Rep 18, 659–672. [DOI] [PubMed] [Google Scholar]
- Mingay M, Chaturvedi A, Bilenky M, Cao Q, Jackson L, Hui T, Moksa M, Heravi-Moussavi A, Humphries RK, Heuser M, and Hirst M (2018). Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia 32, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Gessi M, Waha A, Isselstein LJ, Luxen D, Freihoff D, Freihoff J, Becker A, Simon M, Hammes J, et al. (2012). Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am. J. Pathol. 181, 675–683. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Oudijk L, Papathomas T, De Krijger R, Korpershoek E, Gimenez-Roqueplo AP, Favier J, Canu L, Mannelli M, Rapa I, Curras-Freixes M, et al. (2017). The mTORC1 complex is significantly overactivated in SDHX-mutated paragangliomas. Neuroendocrinology 105, 384–393. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Xiong W, Hahn MA, and Jin SG (2014). The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res 356, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, Welker AE, Angel SN, Tripp SR, Sonnen JA, et al. (2018). Mutant IDH1 promotes glioma formation in vivo. Cell Rep 23, 1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Pintado CO, Ortega-Saenz P, Roche M, and Lopez-Barneo J (2004). The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol. Cell Biol 24, 10933–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, El-Bahrawy M, Poulsom R, Elia G, Killick P, Kelly G, Hunt T, Jeffery R, Seedhar P, Barwell J, et al. (2006). Expression of HIF-1alpha, HIF-2alpha (EPAS1), and their target genes in paraganglioma and pheochromocytoma with VHL and SDH mutations. J. Clin. Endocrinol. Metab. 91, 4593–4598. [DOI] [PubMed] [Google Scholar]
- Race AM, Styles IB, and Bunch J (2012). Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 75, 5111–5112. [DOI] [PubMed] [Google Scholar]
- Rapizzi E, Ercolino T, Canu L, Giache V, Francalanci M, Pratesi C, Valeri A, and Mannelli M (2012). Mitochondrial function and content in pheochromocytoma/paraganglioma of succinate dehydrogenase mutation carriers. Endocr. Relat. Cancer 19, 261–269. [DOI] [PubMed] [Google Scholar]
- Remacha L, Curras-Freixes M, Torres-Ruiz R, Schiavi F, Torres-Perez R, Calsina B, Leton R, Comino-Mendez I, Roldan-Romero JM, Montero-Conde C, et al. (2018). Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet. Med. 20, 1644–1651. [DOI] [PubMed] [Google Scholar]
- Richter S, Peitzsch M, Rapizzi E, Lenders JW, Qin N, De Cubas AA, Schiavi F, Rao JU, Beuschlein F, Quinkler M, et al. (2014). Krebs cycle metabolite profiling for identification and stratification of pheochromocytomas/paragangliomas due to succinate dehydrogenase deficiency. J. Clin. Endocrinol. Metab. 99, 3903–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G, Garrard KP, Barry JA, and Muddiman DC (2013). MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J. Am. Soc. Mass Spectrom. 24, 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio Ii, Cairns RA, Mccracken S, Wakeham A, Haight J, et al. (2012). D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 26, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitt JM, Jang SS, Mu W, Dawson VL, and Dawson TM (2005). Bcl-x is required for proper development of the mouse substantia nigra. J. Neurosci. 25, 6721–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, and Siuzdak G (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. [DOI] [PubMed] [Google Scholar]
- Sulkowski PL, Oeck S, Dow J, Economos NG, Mirfakhraie L, Liu Y, Noronha K, Bao X, Li J, Shuch BM, et al. (2020). Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 582, 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski PL, Sundaram RK, Oeck S, Corso CD, Liu Y, Noorbakhsh S, Niger M, Boeke M, Ueno D, Kalathil AN, et al. (2018). Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 50, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wu Y, Nanba K, Sbiera S, Kircher S, Kunzke T, Aichler M, Berezowska S, Reibetanz J, Rainey WE, et al. (2018). High resolution tissue mass spectrometry imaging reveals a refined functional anatomy of the human adult adrenal gland. Endocrinology 159, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarek E, Ball ER, Imperiale A, Tsokos M, Faucz FR, Giubellino A, Moussallieh FM, Namer IJ, Abu-Asab MS, Pacak K, et al. (2015). Carney triad, SDH-deficient tumors, and Sdhb+/− mice share abnormal mitochondria. Endocr. Relat. Cancer 22, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks ID, Mould AW, Schroder WA, Cotterill A, Hayward NK, Walker GJ, and Kay GF (2010). Dual loss of rb1 and Trp53 in the adrenal medulla leads to spontaneous pheochromocytoma. Neoplasia 12, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalakshmi K, Shankar V, Bain RM, Nolley R, Sonn GA, Kao CS, Zhao H, Tibshirani R, Zare RN, and Brooks JD (2020). Identification of diagnostic metabolic signatures in clear cell renal cell carcinoma using mass spectrometry imaging. Int. J. Cancer 147, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Song Q, and Cooks RG (2006). Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl. 45, 7188–7192. [DOI] [PubMed] [Google Scholar]
- Wu C, Ifa DR, Manicke NE, and Cooks RG (2010). Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst 135, 28–32. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. (2012). Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Soga T, and Pollard PJ (2013). Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest 123, 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, and Parada LF (2001). Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15, 859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data has been deposited in NCBI’s Sequence Read Archive (SRA) under accession code PRJNA790868 and are publicly available as of the date of publication. This study does not report the development of any new code. Any additional information required to reanalyze the data in this paper will be made available from the lead contact upon request.