Abstract
Background
As the second most frequent factor of brain metastasis worldwide, breast cancer and its pathogenesis have been researched intensively. Nevertheless, the molecular mechanisms of brain metastasis from breast cancer (BMBC) remain uncertain. The purpose of this study was to explore the key genes concerning the prognosis of BMBC and identify their predictive value.
Material/Methods
Obtained from the Gene Expression Omnibus (GEO) database, microarray datasets GSE125989, GSE52604, and GSE159956 were used to identify the differentially expressed genes (DEGs) and perform function enrichment analysis.
Results
Of a total of 240 DEGs, 113 genes were upregulated and 127 genes were downregulated. The protein–protein interaction (PPI) was performed through STRING, and 29 hub genes were screened through Cytoscape. After being examined through the cBioportal online platform and the Oncomine database, 8 key genes were finally obtained, including COL14A1, COL3A1, COL6A3, THY1, MMP14, GAP43, PTPRN, and SNAP25. In the validation dataset GSE46928, COL14A1 was shown to have predictive significance of brain metastasis in breast cancer.
Conclusions
The key genes explored in this article could assist in identifying the molecular mechanism of BMBC. Also, COL14A1, COL3A1, COL6A3, THY1, MMP14, GAP43, PTPRN, and SNAP25 might be candidate targets for diagnosis and treatment of BMBC, and COL3A1 might have predictive value.
Keywords: Brain Diseases, Breast Neoplasms, Medical Informatics
Background
In the last decades, bioinformatics analysis has been used to identify genetic alternations to help discover key genes and interrelated pivotal pathways in brain metastasis from breast cancer. The Gene Expression Omnibus (GEO) database, as a well-known public platform, can store chip, second-generation sequencing, and other high-throughput sequencing data. With this database, experimental sequencing data uploaded by other researchers can be retrieved.
As the most common malignant lesion in women worldwide, breast cancer is the second most common cause of brain metastasis [1]. Approximately 12% of patients have metastases, and the brain is the initial location [2]. As previously reported, for advanced breast cancer, the incidence rate of brain metastases rises from 10% to 30% [3]. Although it has been verified that local therapies are effective for brain metastases from breast cancer (BMBC), and systematic therapies mitigating extracranial symptoms are becoming increasingly used, the blood-brain barrier could prevent the penetration of anticancer drugs, which could reduce drug delivery to the site of metastasis and lessen drug effects [4]. Therefore, specific measures targeting BMBC remain unsolved, and the prognosis of BMBC is still disappointing. Therefore, it is important to identify the underlying biomarkers of BMBC.
Material and Methods
Microarray Data
Three datasets, GSE125989, GSE52604, and GSE159956, were downloaded from the National Center of Biotechnology Information (NCBI) GEO database (https://www.ncbi.nlm.nih.gov/geo/; GPL571, GPL6480, and GPL2567 platforms). An analysis from the database was designed with the approval of local Ethics Committee of the Fudan University Shanghai Cancer Center.
Identification of Differentially Expressed Genes
The differentially expressed genes (DEGs) between primary breast cancer and BMBC were inspected by the LIMMA package of R in datasets GSE125989 and GSE52604. Genes that lacked specific gene symbols were deleted, and genes with more than 1 probe were averaged. Adjusted P values were used to reduce the false-positive rate. The criteria of differential genes were set as genes with |log2fold change| >1 and an adjusted P value <0.05. For visualizing the DEGs, ggplot2 and Venn Diagram packages of R were used to build volcano plots and Venn diagrams.
Enrichment Analyses of DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov; version 6.8), as an online bioinformatic database, includes 5 functional annotation tools aiming to provide functional interpretation of series of genes with Gene Ontology (GO) at the cellular level and Kyoto Encyclopedia of Genes and Genomes (KEGG) at the pathway level. P<0.05 was considered to be statistically significant.
Protein–Protein Interaction Network Construction and Module Analysis
With a combined interaction score of >0.4 considered significant, the protein–protein interaction (PPI) network was built to search the pivotal interactions through the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org; version 10.0) online database, which provides insights into the mechanisms of brain metastasis. Also, the inside-software plugin cytoHubba of Cytoscape (version 3.4.0), an open source platform for complex network analysis, was used to identify hub genes among the genes with 12 different arithmetic methods. The top 10 nodes were ranked by every algorithm and were selected in gene collection.
Survival Analysis and Hub Genes Selection
The Kaplan-Meier curve on the cBioPortal (http://www.cbioportal.org) online platform and the box line diagram on the Oncomine database (http://www.oncomine.com) were used to explore the relationship between expression patterns and metastasis and to determine the final hub genes.
Validation Study
The dataset GSE52604 was used to reduce false positives through the LIMMA package of R. Finally, the validation dataset GSE159956 was used to conduct survival analysis to predict the value of key genes with the Survival and Survminer packages of R.
Results
Identification of DEGs
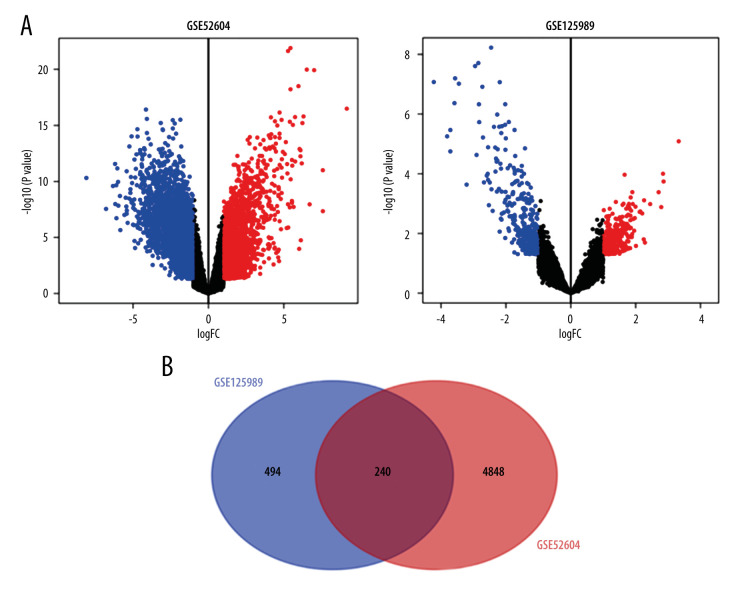
The GSE125989 dataset included 16 paired samples of primary breast cancer and brain metastases; the GSE52604 dataset contained 55 unpaired samples, including 10 primary breast cancer samples, 10 non-neoplastic brain samples, and 35 BMBC samples; and the GSE159956 dataset contained 285 primary breast cancer patients. Differential genes (734 in GSE125989 and 5088 in GSE52604) were identified and represented in the volcano plots, and Venn diagram displayed the overlap between the 2 datasets, containing 240 genes, of which 113 genes showed upregulation and 127 genes showed downregulation between primary breast cancer and BMBC (Figure 1).
Figure 1.
(A) Volcano plots and (B) Venn diagram of differentially expressed genes of 2 datasets (GSE52604 and GSE125989) by ggplot2 and Venn Diagram packages of R (version 4.1.1).
Enrichment Analyses of DEGs
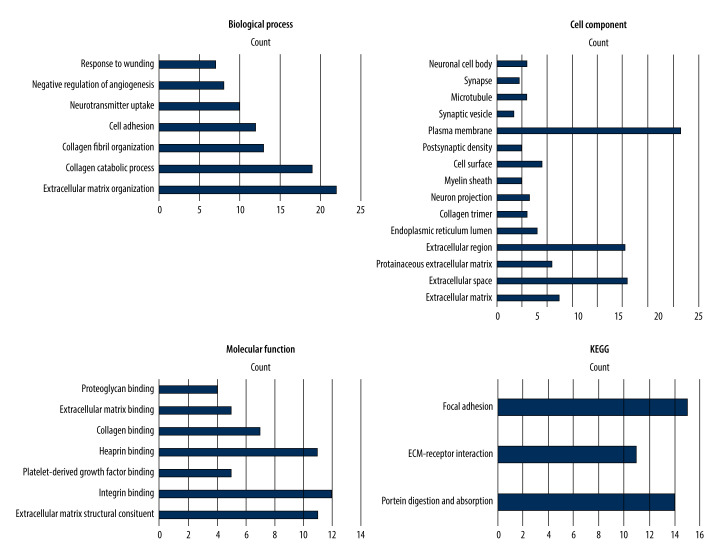
To further analyze the DEGs, GO and KEGG analysis were constructed through DAVID. According to the outcomes in the GO database, the changes in biological processes were significantly enriched in extracellular matrix organization, collagen fibril organization, cell adhesion, neurotransmitter uptake, negative regulation of angiogenesis, and reaction to wounding. Additionally, in the case of the molecular function analysis, the genes were mainly interrelated to extracellular matrix structural constituent, integrin binding, platelet-derived growth factor binding, and heparin binding. Moreover, for the cell component analysis, the differential genes were predominantly involved in the extracellular matrix, extracellular space, proteinaceous extracellular matrix, and plasma membrane. Furthermore, KEGG pathway analysis demonstrated that the DEGs were mostly enriched in protein digestion and absorption, extracellular matrix-receptor interaction, focal adhesion, platelet activation, PI3K-Akt signaling pathway, and phagosome. The above results are shown in Figure 2.
Figure 2.
Results of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the differentially expressed genes obtained from the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov) (version 6.8).
PPI Network Construction
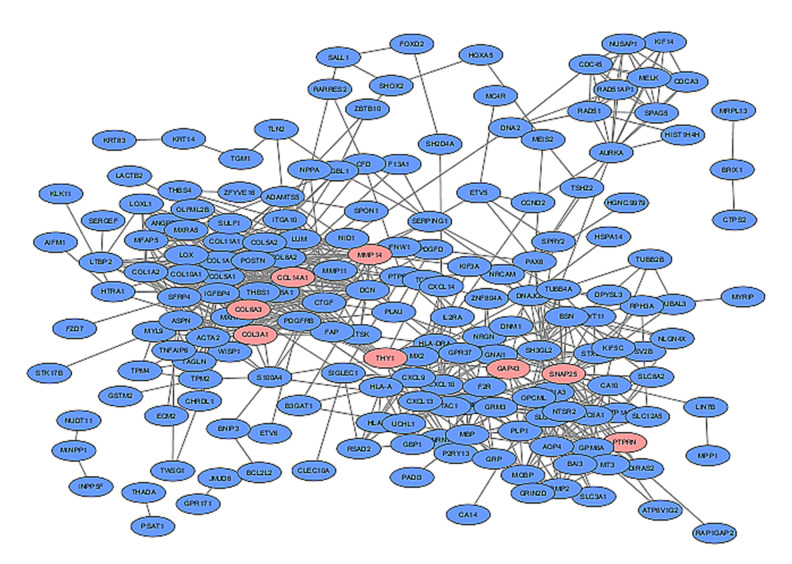
For the PPI network, 36 nodes and 722 edges were exhibited based on the STRING database, with a local clustering coefficient of 0.437 and P<1.0e-16. The 29 related genes were as follows: COL14A1, COL15A1,COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL6A3, DCN, GAP43, LOX, LOXL1, LUM, MMP14, NID1, PLAU, POSTN, PTPRN, SNAP25, SPON1, THY1, TPM4, TUBB4A, PAX6, COL8A2, COL10A1, THBS1, MXRA5, and ITGBL1 (Figure 3).
Figure 3.
In the protein–protein interaction network exhibited by the Cytoscape database (version 4.3.0), blue nodes represent associated products of differentially expressed genes, pink nodes represent 8 key genes identified below, and edges represent the protein–protein associations.
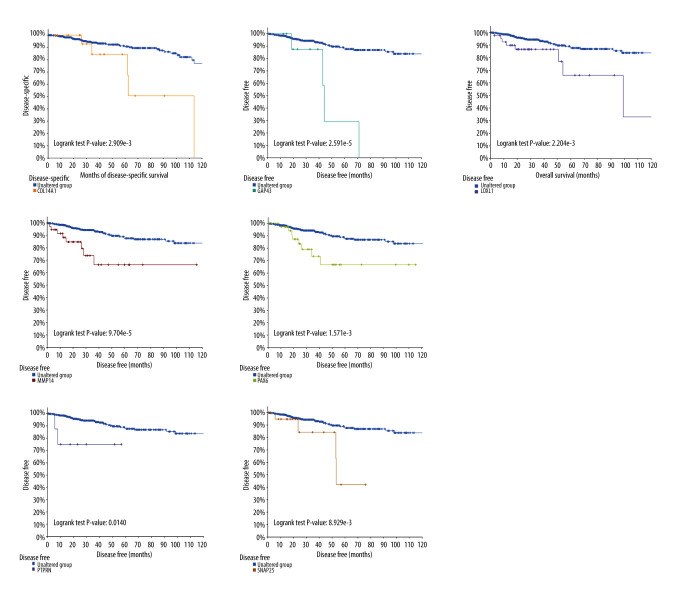
Survival Analysis and Hub Genes Selection
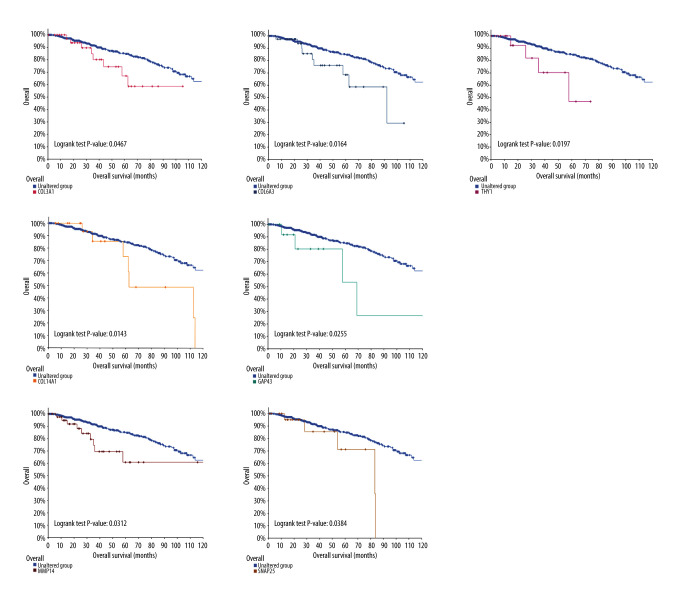
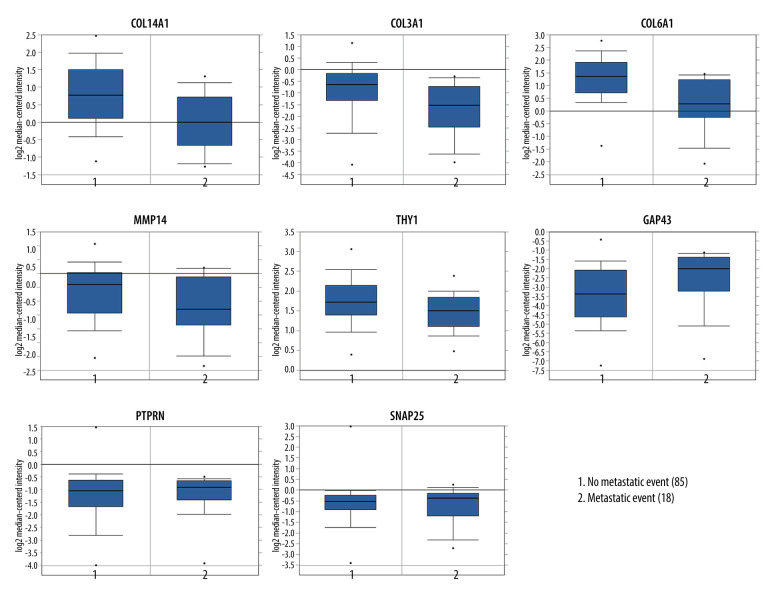
Subsequently, survival analysis was done to select the hub genes, as shown in Figures 4 and 5. Breast cancer patients with COL14A1, COL3A1, COL6A3, GAP43, MMP14, SNAP25, and THY1 alterations had worse overall survival. Meanwhile, breast cancer patients with GAP43, LOXL1, MMP14, PAX6, PTPRN, and SNAP25 alteration had worse disease-free survival. Overall, among these selected hub genes, 10 genes tested as significant, of which COL14A1, COL3A1, COL6A3, LOXL1, MMP14, and THY1 were downregulated, and GAP63, PAX6, PTPRN, and SNAP25 were upregulated. These 10 genes were further examined in an independent dataset (Symmans Breast 2 dataset) of the Oncomine database. In the Symmans Breast 2 dataset, lower mRNA levels of downregulated genes (COL14A1, COL3A1, COL6A3, THY1, and MMP14) were associated with a metastatic event, while higher mRNA levels of upregulated genes (GAP43, PTPRN, and SNAP25) were related to a metastatic event, which indicated that these 8 genes may play prominent parts in brain metastasis of breast cancer (Figure 6).
Figure 4.
Overall survival analysis on cBioPortal online platform.
Figure 5.
Disease-free survival analysis on cBioPortal online platform.
Figure 6.
Association between expression patterns of hub genes and metastatic event status by the Symmans Breast 2 dataset.
Validation Study
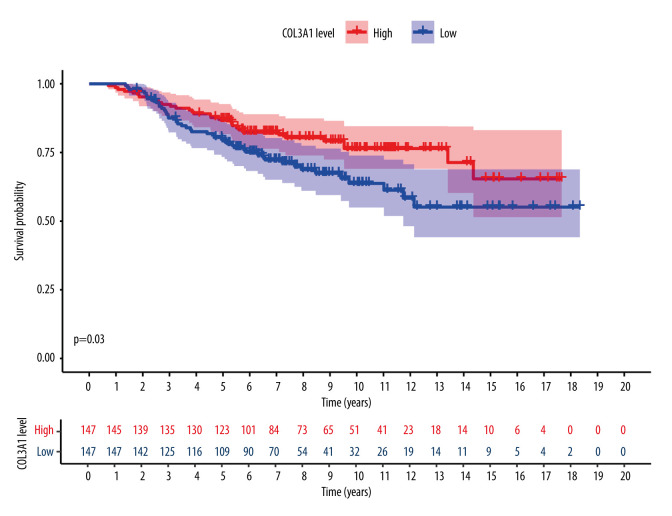
To reduce false positives, the comparison between normal brain tissues and tissues of brain metastases from breast cancer was performed with the GSE52604 dataset, and 4881 differentially expressed genes were identified, in which the final hub genes mentioned above were included. Finally, to judge the predictive value of the 8 identified hub genes, we performed survival analysis on the GSE159956 dataset and found that COL3A1 was statistically significant, as shown in Figure 7.
Figure 7.
COL3A1 was shown to have predictive value with statistical significance in the validation study. The survival curve was created with Survival and Survminer packages of R.
Discussion
As the most common carcinoma for women, breast cancer is also the second most common cause of brain metastases [5]. In recent years, BMBC showed a younger trend and more frequently occurs in patients with a larger tumor size or higher grade in some subtypes [6]. However, the mechanisms of BMBC remain poorly understood. Because it is extremely difficult for chemotherapy drugs to infiltrate the blood-brain barrier, novel biological therapy, instead of chemotherapy, is needed to prolong survival [7]. With the progress of bioinformatics analysis, DEGs associated with BMBC have become more accessible, providing an efficacious mode to identify potential targets in dealing with BMBC [5].
In the present study, to discover the DEGs between primary breast cancer and BMBC, 2 mRNA datasets were explored, resulting in 240 DEGs identified between the 2 datasets, of which 113 genes were upregulated and 127 genes were downregulated. According to functional analysis, these differential genes were chiefly related to biological processes, such as extracellular matrix organization, collagen fibril organization, cell adhesion, chemical synaptic transmission, and negative regulation of angiogenesis. In addition, the KEGG pathways of differential genes were mostly enriched in protein digestion and absorption, focal adhesion, platelet activation, PI3K-Akt signaling pathway, and phagosome. Moreover, the PPI network was created, with 29 hub genes being identified. After being examined through the cBioportal online platform and the Oncomine database, 8 key genes were finally obtained, including COL14A1, COL3A1, COL6A3, THY1, MMP14, GAP43, PTPRN, and SNAP25, of which COL3A1 and COL6A3 were also identified to play potentially important roles in BMBC in patients with breast cancer in a recent study [8].
COL14A1 is related to the regulation of fibrillogenesis and interacts with the fibril surface. Compared with in patients without metastases, COL14A1 is highly expressed in patients with metastasis, and further investigation is desired to verify its function in metastases diagnosis and prediction of prognosis [9]. Also, several studies have revealed that COL3A1 is vital for tumorigenesis and metastasis of invasive breast cancer [10–12]. In addition, m6A methylation-mediated COL3A1 upregulation reduces the expression level of METTL3, which assists metastasis of triple-negative breast cancer [13].
The encoded protein of THY1 is bound with cell adhesion and communication in numerous cells, but particularly in immunocytes and neurocytes [14,15]. One study demonstrated that immune-related THY1 is a potential therapeutic target and prognostic marker for breast cancer with brain metastasis [16]. Owing to its function on tumor progression and metastasis, MMP-14 is a vital target with clinical significance in metastatic cancers [17–20]. Since active MMP-14 locates on the cellular surface, the antibody-mediated blockade was shown to be a novel idea in oncotherapy [17]. It has been reported that MMP14 is probably interrelated with breast cancer, and MMP-14 acts as a biological target and can block immunodepression, tumor progression, and metastasis in triple-negative breast cancer in immunotherapies [21–23]. Type VI collagen alpha3 (COL6A3) is a critical component in breast cancer development, and its 6-EMT gene signature at diagnosis can help identify patients with metastasis with triple-negative breast cancer who could not achieve pathological complete response to neoadjuvant chemotherapy and underwent surgery combined with adjuvant therapy [24].
Several studies have indicated that the relationship between BMBC and hub genes, including SNAP25, GAP43, and PTPRN, has not been widely explored. SNAP25 has been reported to be relevant to metastasis in endometrial carcinoma [25]. Autophagy is thought to promote tumor progression by means of activating SNAP25 transcription, showing that SNAP25 has a significant role in the later period of carcinoma [26]. GAP43 is an absolute biomarker with predictive value of non-small cell lung cancer brain metastasis and it could assist metastasis through the Rac1/F-actin pathway [27]. However, GAP43 has rarely been researched in breast cancer. As signaling molecules that regulate multiple processes, including cell growth, differentiation, and oncogenic transformation, the protein encoded by PTPRN has a crucial role in glioblastoma and ovarian cancer, but its function in breast cancer needs to be further studied [27–29].
It has been shown that the prognosis of patients with breast cancer with brain metastases is very poor, and brain metastases shorten survival [6]. However, the progress of research on effective treatments is still slow, mainly because the standard treatment options for brain metastasis are limited [6]. Therefore, it is necessary to identify sensitive biomarkers as new targets to improve the prognosis of patients with metastatic breast cancer. The findings of the present study may provide new insights into the understanding of brain metastases in breast cancer. However, this study has some limitations. First, the clinical data in the GEO database is incomplete, which affected the comprehensive survival analysis. More data libraries should be exploited for clinical data, such as The Cancer Genome Atlas, and a recently published study on this topic is a good example [30]. Future studies can provide larger analyses of real-life data and the risk of a higher rate of brain metastasis at presentation. Second, this study included only 3 datasets, which may have biased the results of the study. Third, the results of this study need further experimental verification. For example, western blotting and enzyme real-time polymerase chain reaction can be used to test the expression of the selected key genes. It is also necessary to construct a nude mouse brain metastasis model of breast cancer and observe the effects of the selected key factors on the phenotypes of genes in BMBC.
Conclusions
In conclusion, 8 key genes were identified as potential biomarkers for BMBC. COL3A1 was significant for predicting brain metastasis from breast cancer. Further in vivo and in vitro studies could confirm our results, and the discovery of DEGs could provide new information regarding cell pathways and targets for new drugs.
Acknowledgements
The authors thank the researchers at the Department of Medical Oncology, Fudan University Shanghai Cancer Center.
Abbreviations
- BMBC
brain metastasis from breast cancer
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
protein–protein interaction
- DEGs
the differentially expressed genes
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
- STRING
Search Tool for the Retrieval of Interacting Genes
Footnotes
Conflict of interest: None declared
Availability of Data and Material
The datasets used and/or analyzed during the current study are available from GEO (http://www.ncbi.nlm.nih.gov/geo/).
Code Availability
R software, SPSS, Cytoscape, free websites online.
Declaration of Figures’ Authenticity
All figures submitted have been created by the authors, who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: None declared
References
- 1.Rostami R, Mittal S, Rostami P, et al. Brain metastasis in breast cancer: A comprehensive literature review. J Neurooncol. 2016;127:407–14. doi: 10.1007/s11060-016-2075-3. [DOI] [PubMed] [Google Scholar]
- 2.Rades D, Lohynska R, Veninga T, et al. Evaluation of 2 whole-brain radiotherapy schedules and prognostic factors for brain metastases in breast cancer patients. Cancer. 2007;110:2587–92. doi: 10.1002/cncr.23082. [DOI] [PubMed] [Google Scholar]
- 3.Kanchan RK, Siddiqui JA, Mahapatra S, et al. microRNAs orchestrate pathophysiology of breast cancer brain metastasis: Advances in therapy. Mol Cancer. 2020;19:29. doi: 10.1186/s12943-020-1140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N, Mohammad AS, Saralkar P, et al. Investigational chemotherapy and novel pharmacokinetic mechanisms for the treatment of breast cancer brain metastases. Pharmacol Res. 2018;132:47–68. doi: 10.1016/j.phrs.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan AJ, Giannoudis A, Palmieri C. The genomic landscape of breast cancer brain metastases: A systematic review. Lancet Oncol. 2021;22:e7–e17. doi: 10.1016/S1470-2045(20)30556-8. [DOI] [PubMed] [Google Scholar]
- 6.Kodack DP, Askoxylakis V, Ferraro GB, et al. Emerging strategies for treating brain metastases from breast cancer. Cancer Cell. 2015;27:163–75. doi: 10.1016/j.ccell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Custódio-Santos T, Videira M, Brito MA. Brain metastasization of breast cancer. Biochim Biophys Acta Rev Cancer. 2017;1868:132–47. doi: 10.1016/j.bbcan.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Wang L, Yang H, et al. Identification of potential genes related to breast cancer brain metastasis in breast cancer patients. Biosci Rep. 2021;41:BSR20211615. doi: 10.1042/BSR20211615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto R, Nakamura Y, Takami T, et al. Quantitative LC-MS/MS analysis of proteins involved in metastasis of breast cancer. PLoS One. 2015;10:e0130760. doi: 10.1371/journal.pone.0130760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong G, Deng L, Zhu J, et al. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer. 2014;14:1. doi: 10.1186/1471-2407-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Fu L, Li J, et al. Microarray analysis for differentially expressed genes between stromal and epithelial cells in development and metastasis of invasive breast cancer. J Comput Biol. 2020;27:1631–43. doi: 10.1089/cmb.2019.0154. [DOI] [PubMed] [Google Scholar]
- 12.Srour MK, Gao B, Dadmanesh F, et al. Gene expression comparison between primary triple-negative breast cancer and paired axillary and sentinel lymph node metastasis. Breast J. 2020;26:904–10. doi: 10.1111/tbj.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Zheng C, Jin Y, et al. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6A methylation-mediated COL3A1 up-regulation. Front Oncol. 2020;10:1126. doi: 10.3389/fonc.2020.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker TH, Hagood JS. Getting a grip on Thy-1 signaling. Biochim Biophys Acta. 2009;1793:921–23. doi: 10.1016/j.bbamcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetzel A, Chavakis T, Preissner KT, et al. Human Thy-1 (CD90) on activated endothelial cells is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18) J Immunol. 2004;172:3850–59. doi: 10.4049/jimmunol.172.6.3850. [DOI] [PubMed] [Google Scholar]
- 16.Lu WC, Xie H, Yuan C, et al. Genomic landscape of the immune microenvironments of brain metastases in breast cancer. J Transl Med. 2020;18:327. doi: 10.1186/s12967-020-02503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen AT, Chia J, Ros M, et al. Organelle specific O-glycosylation drives MMP14 activation, tumor growth, and metastasis. Cancer Cell. 2017;32:639–653e6. doi: 10.1016/j.ccell.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Wu X, Gardashova G, et al. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis. Sci Transl Med. 2020;12:eaaz2878. doi: 10.1126/scitranslmed.aaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claesson-Welsh L. How the matrix metalloproteinase MMP14 contributes to the progression of colorectal cancer. J Clin Invest. 2020;130:1093–95. doi: 10.1172/JCI135239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhtar N. Hijacking a morphogenesis proteinase for cancer cell invasion. Dev Cell. 2018;47:135–37. doi: 10.1016/j.devcel.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Ling B, Watt K, Banerjee S, et al. A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models. Oncotarget. 2017;8:58372–85. doi: 10.18632/oncotarget.17702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H, Hu L, Yu L, et al. KLF8 and FAK cooperatively enrich the active MMP14 on the cell surface required for the metastatic progression of breast cancer. Oncogene. 2014;33:2909–17. doi: 10.1038/onc.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGowan PM, Duffy MJ. Matrix metalloproteinase expression and outcome in patients with breast cancer: Analysis of a published database. Ann Oncol. 2008;19:1566–72. doi: 10.1093/annonc/mdn180. [DOI] [PubMed] [Google Scholar]
- 24.Wei LY, Zhang XJ, Wang L, et al. A six-epithelial-mesenchymal transition gene signature may predict metastasis of triple-negative breast cancer. Onco Targets Ther. 2020;13:6497–509. doi: 10.2147/OTT.S256818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu L, Shu Z, Sun X. Bioinformatic analysis of four miRNAs relevant to metastasis-regulated processes in endometrial carcinoma. Cancer Manag Res. 2018;10:2337–46. doi: 10.2147/CMAR.S168594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu Y, Yan X, Li D, et al. NUPR1 maintains autolysosomal efflux by activating SNAP25 transcription in cancer cells. Autophagy. 2018;14:654–70. doi: 10.1080/15548627.2017.1338556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Ying L, Jin J, et al. GAP43, a novel metastasis promoter in non-small cell lung cancer. J Transl Med. 2018;16:310. doi: 10.1186/s12967-018-1682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delsite R, Kachhap S, Anbazhagan R, et al. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloom AP, Jimenez-Andrade JM, Taylor RN, et al. Breast cancer-induced bone remodeling, skeletal pain, and sprouting of sensory nerve fibers. J Pain. 2011;12:698–711. doi: 10.1016/j.jpain.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Liu J, Qian X, He X. Identification of markers associated with brain metastasis from breast cancer through bioinformatics analysis and verification in clinical samples. Gland Surg. 2021;10:924–42. doi: 10.21037/gs-20-767. [DOI] [PMC free article] [PubMed] [Google Scholar]