ABSTRACT
The rapid antiphage mutation of pathogens is a big challenge often encountered in the application of phages in aquaculture, animal husbandry, and human disease prevention. A cocktail composed of phages with different infection strategies can better suppress the antiphage resistance of pathogens. However, randomly selecting phages with different infection strategies is time-consuming and labor intensive. Here, we verified that using a resistant pathogen quickly evolved under single phage infection, as the new host can easily obtain phages with different infection strategies. We randomly isolated two lytic phages (i.e., Va1 and Va2) that infect the opportunistic pathogen Vibrio alginolyticus. Whether they were used alone or in combination, the pathogen easily gained resistance. Using a mutated pathogen resistant to Va1 as a new host, a third lytic phage Va3 was isolated. These three phages have a similar infection cycle and lytic ability but quite different morphologies and genome information. Notably, phage Va3 is a jumbo phage containing a larger and more complex genome (240 kb) than Va1 and Va2. Furthermore, the 34 tRNAs and multiple genes encoding receptor binding proteins and NAD+ synthesis proteins in the Va3 genome implicated its quite different infection strategy from Va1 and Va2. Although the wild-type pathogen could still readily evolve resistance under single phage infection by Va3, when Va3 was used in combination with Va1 and Va2, pathogen resistance was strongly suppressed. This study provides a novel approach for rapid isolation of phages with different infection strategies, which will be highly beneficial when designing effective phage cocktails.
IMPORTANCE The rapid antiphage mutation of pathogens is a big challenge often encountered in phage therapy. Using a cocktail composed of phages with different infection strategies can better overcome this problem. However, randomly selecting phages with different infection strategies is time-consuming and labor intensive. To address this problem, we developed a method to efficiently obtain phages with disparate infection strategies. The trick is to use the characteristics of the pathogenic bacteria that are prone to develop resistance to single phage infection to rapidly obtain the antiphage variant of the pathogen. Using this antiphage variant as the host results in other phages with different infection strategies being efficiently isolated. We also verified the reliability of this method by demonstrating the ideal phage control effects on two pathogens and thus revealed its potential importance in the development of phage therapies.
KEYWORDS: pathogen control, phage cocktail, phage identification, antiphage mutation, genomic characterization
INTRODUCTION
Opportunistic pathogens such as Vibrio alginolyticus, Vibrio harveyi, and Aeromonas salmonicida often cause serious diseases in aquaculture, which result in tremendous economic loss (1–3). Typically, antibiotics are utilized to prevent these bacterial diseases. However, their long-term and excessive use results in the emergence and spread of multidrug-resistant bacteria, which are a huge threat to disease prevention in the aquaculture industry (4, 5). Therefore, alternative methods to antibiotics are urgently needed for the effective long-term prevention of pathogenic infections (6–8).
Phages are the most abundant organisms on the planet (9, 10). The high infection specificity (and rapid proliferation) of phages to their host bacteria make them a promising potential substitute to antibiotics in the control of these pathogens (11–13). However, pathogens frequently acquire resistance under the stress of phage infection, and this, in turn, seriously weakens the antibacterial effect of the phages. This is the main challenge that prevents phages from being widely used as antibiotic alternatives at present (14, 15).
The simultaneous use of several different phages (i.e., a phage cocktail) can, to a certain extent, circumvent the rapid antiphage evolution of a pathogen, although the actual bactericidal effects vary considerably (16–18). One reason why some phage cocktails are relatively inefficient at suppressing bacterial infections is due to the development of cross-resistance by the pathogenic host to different phages with similar infection mechanisms (19, 20). Only cocktails containing phages with different infection strategies (i.e., which target different surface receptors or have different infection mechanisms) are effective. For example, Chen et al. compared the bactericidal activity of 24 phage cocktails and developed an efficient cocktail containing just two phages that had vastly different genomes (19). In addition, Yang et al. constructed an effective cocktail by combining the ancestor phage PaoP5 and the trained phage PaoP5-m1, which have different receptor binding proteins (RBPs) (20). However, the procedures used to generate these cocktails were labor intensive and time-consuming, which increases the cost of developing and making new cocktails and thus hinders their commercial application.
We speculated that by using rapidly evolved phage-resistant bacteria under infection by a single phage as a host, it might be possible to isolate additional phages with different infection strategies. We thus explored the possibility of utilizing this type of shortcut for designing an effective phage cocktail. At the same time, the physiological and genomic characteristics of the phages isolated were analyzed to understand their potential infection characteristics. We suggest that phage cocktails designed through this strategy will help avoid bacterial cross-resistance to different phages.
RESULTS
Biological characterization of Va1, Va2, and Va3.
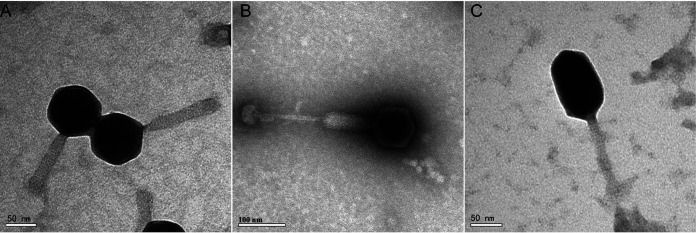
(i) Morphology of the phages. The lytic phages Va1 and Va2 were isolated from the coastal waters of the Yellow Sea, using wild-type V. alginolyticus 283T (ATCC 17749) as the host. The transmission electron microscopy (TEM) images show that both of these phages have icosahedral heads of 85 ± 5 nm in diameter and contractile tails of 117 ± 5 nm (Va1) and 250 ± 5 nm (Va2) in length. In addition, the tail of Va2 contains a knoblike appendage extending from the baseplate (Fig. 1A and B). Phage Va3 was also isolated from the coastal waters of the Yellow Sea, using the antiphage mutant V. alginolyticus 283R (which was resistant to infection by Va1) as the host (Fig. S4 in the supplemental material). Va3 was shown to have a prolate head of 107 ± 4 nm in length and 71 ± 3 nm in diameter, as well as a contractile tail of 95 ± 8 nm in length (Fig. 1C), and it formed tiny plaques on the double-layer plate (Fig. S1). The dimensions of the three phages are summarized in Table 1. Based on their morphology, the three phages were classified as being members of the Myoviridae.
FIG 1.
Transmission electron microscopy images of Va1 (A), Va2 (B), and Va3 (C).
TABLE 1.
Dimensions of the Va1, Va1, and Va2 phages obtained from the TEM images
| Phage | Head morphology | Head length (nm) | Head diam (nm) | Tail length (nm) |
|---|---|---|---|---|
| Va1 | Icosahedral | 85 ± 5 | 85 ± 5 | 117 ± 5 |
| Va2 | Icosahedral | 85 ± 5 | 85 ± 5 | 250 ± 5 |
| Va3 | Prolate | 107 ± 4 | 71 ± 3 | 95 ± 8 |
(ii) One-step growth curves.
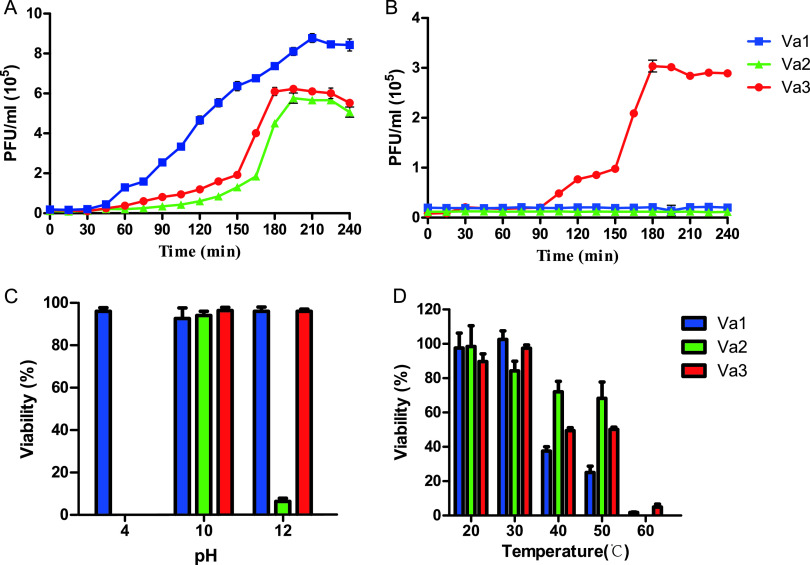
One-step growth curves were constructed to determine the life cycle characteristics of the three phages. During incubation with V. alginolyticus 283T, the lytic cycle lasted ∼210 min, ∼195 min, and ∼180 min for Va1, Va2, and Va3, respectively. In addition, Va1, Va2, and Va3 all had a latent period of ∼30 min and comparable burst sizes (i.e., 46, 50, and 40 PFU/cell, respectively) (Fig. 2A). In contrast, V. alginolyticus 283R was resistant to both Va1 and Va2, but Va3 exhibited lytic activity with a latent period of 90 min and a burst size of 35 PFU/cell (Fig. 2B). The simultaneous sensitivity of V. alginolyticus 283T and resistance of V. alginolyticus 283R to both Va1 and Va2 indicates that these phages might utilize a similar infection strategy, which is distinct from Va3 (18).
FIG 2.
One-step growth curves of Va1, Va2, and Va3 infecting wild-type (283T) (A) and mutant (283R) (B) V. alginolyticus and the sensitivity of these phages to pH (C) and temperature (D). The three phages lost activity at pH 2 and temperature 70°C and 80°C; thus, no data are shown.
(iii) The influence of external factors on phage particle stability.
The impact of pH, temperature, and chloroform on the infectivity of Va1, Va2, and Va3 were examined to assess the stability of these phages. Under a wide range of different pH (i.e., from 4 to 12), the three phages exhibited different characteristics (Fig. 2C). With regard to Va1, ∼100% remained viable at pH 4, 10, and 12, but none were viable at pH 2. Va2 appeared to be more sensitive than Va1 to acidic pH conditions, such that none remained viable at pH 2 or 4. In addition, although ∼100% Va2 was viable at pH 10, the viability decreased at pH 12. Like Va2, Va3 was also sensitive to pH 2 and 4, but there was ∼100% viability at pH 10 and 12. When these phages were exposed to different temperatures between 20°C and 80°C, there was ∼50% loss in phage viability at temperatures of 40°C and 50°C and ∼100% loss at 60°C and above (Fig. 2D). These findings indicate that the phages had a relatively strong tolerance to different environmental conditions, and so, they might maintain their infective activities when they are applied to control the numbers of pathogens in an aquaculture setting (16).
Moreover, all three of the phages were resistant to chloroform, which indicates that, in each case, the capsid was lipid free.
Genomic characterization of Va1, Va2, and Va3 and their phylogenetic analysis.
Our genomic characterization of the three phages indicated that Va1 and Va3 had circular double-stranded DNA genomes (of 194,900 and 247,567 bp, respectively), whereas Va2 had a linear double-stranded DNA genome (of 128,360 bp). These three phages were also shown to exhibit striking differences in their genome structure (Fig. 3A). The GC content of phage Va2 (40.7%) and Va3 (41%) was similar to that of their host V. alginolyticus 283T (44.6%), but the GC content of phage Va1 (35%) was lower than its host. In addition, although a total of 327,136, and 383 open reading frames (ORFs) were identified in Va1, Va2, and Va3, respectively, specific functions could only be assigned to 19.3%, 24.3%, and 24.3% of the ORFs, respectively. These functions included DNA replication (e.g., helicase), DNA metabolism (e.g., exonuclease), DNA packaging (e.g., terminase large/small subunit), and structure formation (e.g., capsid). In addition, auxiliary metabolic genes encoding proteins such as phosphate starvation protein, phosphoesterase, and dihydrofolate reductase were also present in the genomes of three phages. Moreover, the Va1 genome encoded 23 tRNAs (containing anticodon of 20 amino acids), whereas Va2 only encoded 3 tRNAs (containing anticodon of 2 amino acids). However, Va3 encoded the largest number of tRNAs (i.e., 34 in total) containing an anticodon of 25 different amino acids. These basic characteristics and the functional ORFs of the three phage genomes are summarized in Table 2 and Tables S1 to S3. As no virulence gene, antimicrobial resistance gene, or lysogenic genes (e.g., integrase, excisionase, and transposase gene) were detected in the genomes of Va1, Va2, or Va3, this indicates they should be considered to be obligate lytic phages, which meet the prerequisite for phage therapy candidates (21, 22).
FIG 3.
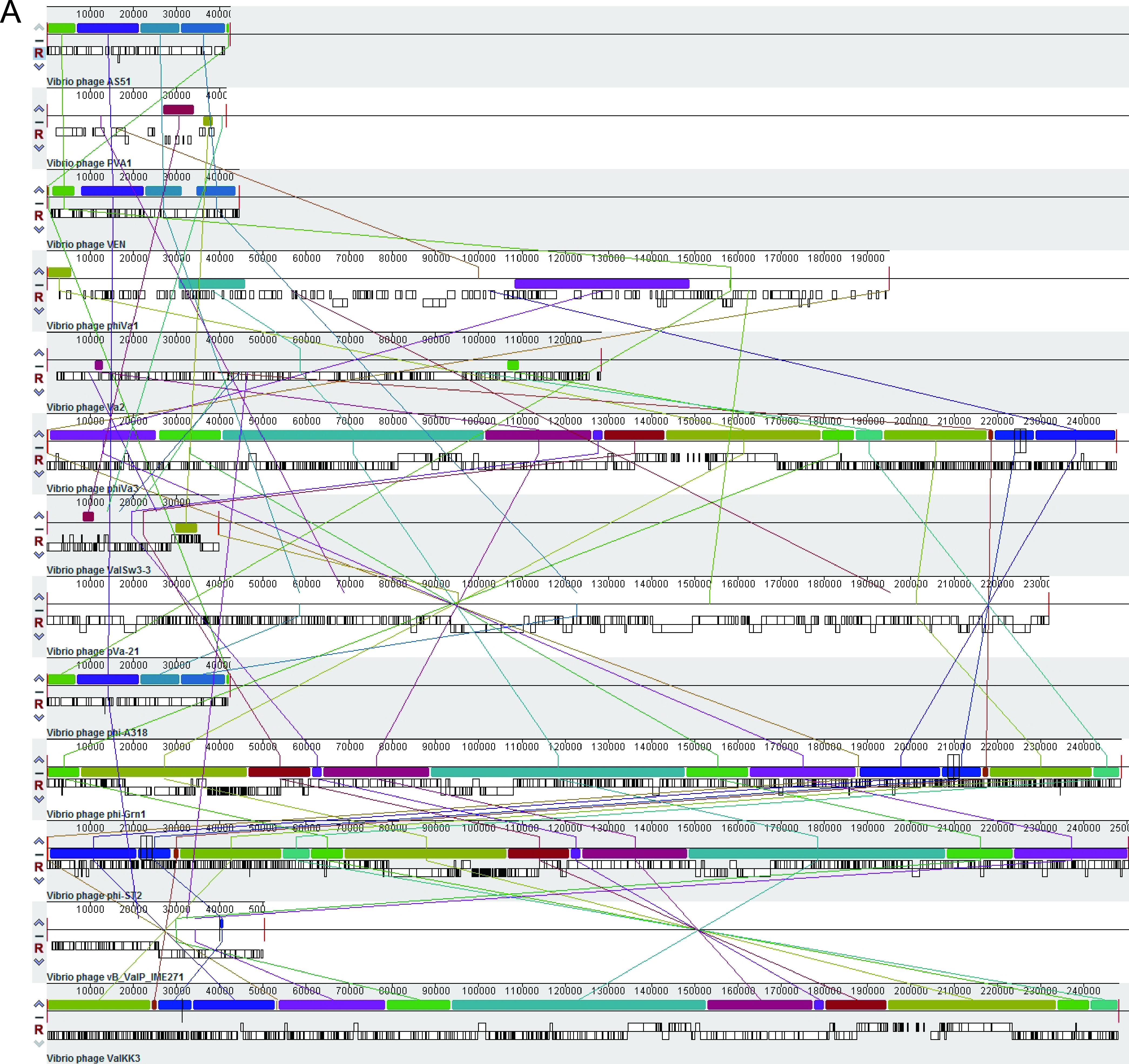
(A) Multiple-genome alignments of the whole genomes of the V. alginolyticus phages using the Mauve algorithm. (B) A phage proteomic tree to show Va1, Va2, Va3, and their closest relative phages, constructed using ViPTree.
TABLE 2.
Basic genome characteristics of Va1, Va2, and Va3
| Characteristic | Va1 | Va2 | Va3 |
|---|---|---|---|
| Genome length (bp) | 194,900 | 128,360 | 247,567 |
| No. of GC content (%) | 35 | 40.7 | 41 |
| tRNA | 23 | 3 | 34 |
| ORFs | 327 | 136 | 383 |
| Functionally annotated ORF | 63 | 33 | 93 |
| Tail fiber protein | 1 | 1 (tail sheath protein) | 5 |
| Baseplate protein | 1 | 0 | 7 |
It should also be noted that Va3 belongs to the giant phage group, whose members have genome sizes ranging from 200 to 500 kbp (23, 24). Giant phages are known to have a plethora of functional genes. They also frequently have a variety of strategies to counter host defenses and can withstand a range of physiological limitations caused by environmental conditions (23, 24). The large genome size of Va3 and the fact that it expressed more functional genes than Va1 and Va2 (Table S3) suggest that it might contain different infection strategies from these smaller phages.
Multiple-genome alignment (Fig. 3A) revealed that the three phages (especially Va1 and Va2) had little similarity with the genomes of other reported V. alginolyticus phages. In addition, BLAST analysis against the NCBI database revealed that the closest phage genome to Va1 was the Vibrio phage River4 (with 88.99% identity), but there was no phage genome match for Va2 and Va3, indicating the novelty of these three phages (24). Furthermore, phylogenetic analysis based on the phage proteome (Fig. 3B) revealed that Va1 and Va3 were most closely related to viruses affiliated with the Firehammervirus and Schizotequatrovirus, respectively, indicating that they are novel species of these genera (25). Moreover, Va2 formed a distinct branch from known Myoviridae members, which indicates that it is a novel genus in this family (25).
In vitro antibacterial characterization of different phages and their combinations.
The growth curves of V. alginolyticus 283T infected by phage Va1, Va2, and Va3 (Fig. 4A and Fig. S2) showed that, individually, the phages were all highly effective against V. alginolyticus 283T within the first 8 h. However, after 24 h of incubation, their antibacterial effects were significantly weakened, such that the optical density at 600 nm (OD600) value increased to 1.0 to 1.1, likely due to the emergence of antiphage bacterial mutants.
FIG 4.
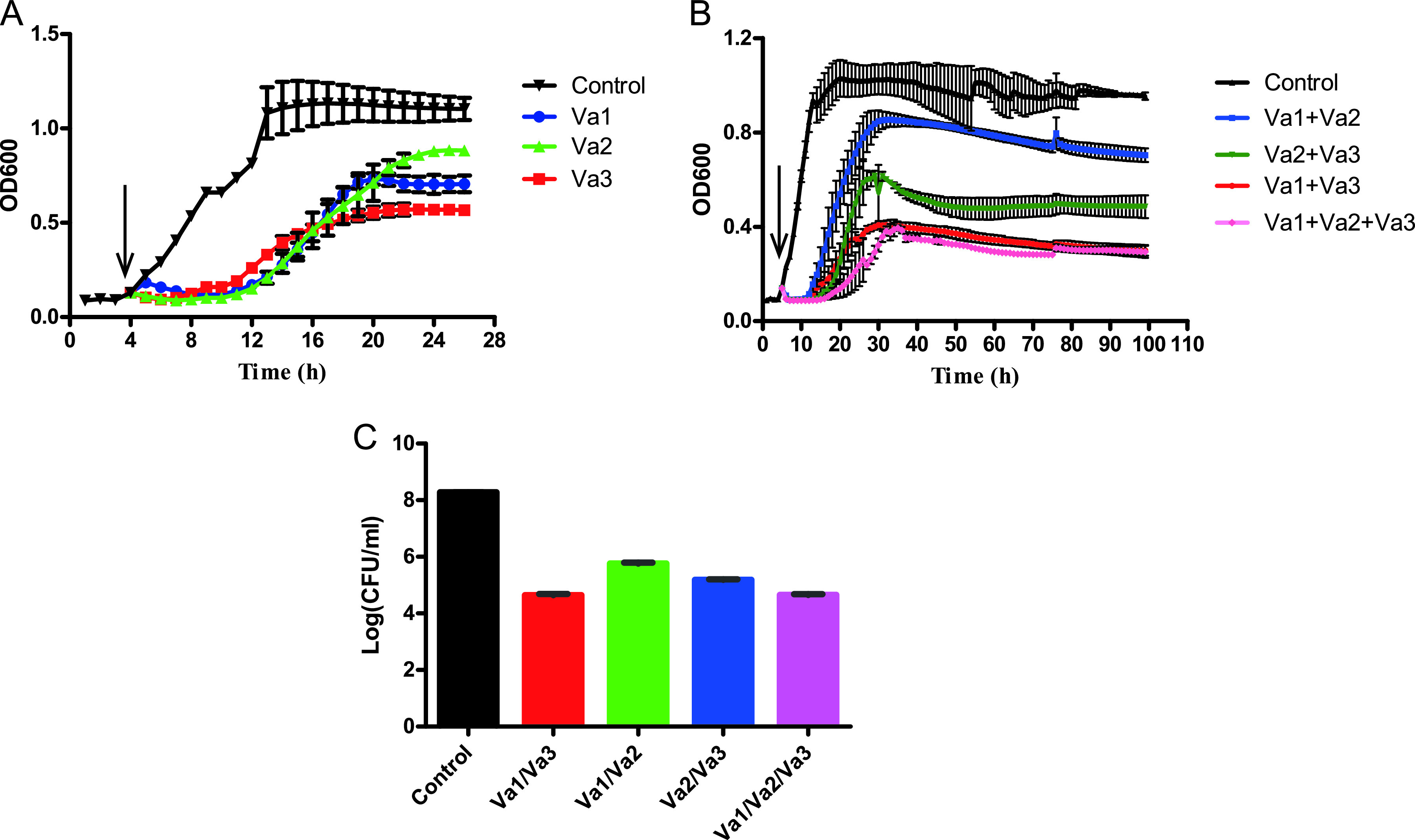
(A and B) Growth curves of V. alginolyticus 283T infected by Va1, Va2, and Va3 alone (A) and combinations of these three phages at an MOI of 1 (B). (C) The number of V. alginolyticus 283T phages in each group after treatment for 96 h. The arrows indicate the time point when the phages were added in the bacterial medium. The data are shown as mean ± SD, and the P values (calculated by one-way ANOVA and Student's t test) are shown in Tables S5 to S7 in the supplemental material.
Four different phage combinations were then designed using these three phages, and their ability to control the phage-resistant bacteria was investigated. At a multiplicity of infection (MOI) of 1 (Fig. 4B), the bacterial killing curve of Va1 plus Va2 failed to control the growth of V. alginolyticus 283T, as it regrew to an OD600 of 0.8 within 30 h after the initial 8-h inhibition period. In contrast, cocktails containing Va3 had a much better inhibitory effect on the growth of V. alginolyticus 283T, and the OD600 was always less than 0.6. Notably, cocktails consisting of Va1 plus Va3 or Va1 plus Va2 and Va3 successfully suppressed bacterial growth (with an OD600 of <0.4) throughout the 96-h duration of the experiment. Correspondingly, the maximum suppression effect of Va1 with Va3 or Va1 with Va2 and Va3 on V. alginolyticus 283T was 3.7- and 3.66-log CFU/mL, respectively. These values are both significantly higher than that obtained by the Va1 with Va2 (i.e., 2.8-log CFU/mL) (Fig. 4C).
We also showed that the antibacterial effects of phage infection with an MOI of 10 was comparable to that of MOI of 1 (Fig. S2). This suggests that Va3 likely played a dominant role in inhibiting bacterial growth in the cocktails. In addition, we suggest that a cocktail composed of Va1 plus Va3 is most promising for controlling V. alginolyticus 283T since the antibacterial activity of these two phages was similar to that of Va1 with Va2 and Va3. This suggests that the effectiveness of a phage cocktail does not depend on the number of phage participants, but it does depend on their efficiency.
Finally, we determined the host range of each phage cocktail (Table S4). We demonstrated that Va1 with Va2 could only infect three of the 15 tested Vibrio strains, whereas Va1 with Va3, Va2 with Va3, and Va1 with Va2 and Va3 could infect five of these strains. These findings indicate that Va3 also contributed to expanding the host range of the phage cocktails.
DISCUSSION
Phage cocktails have attracted much attention in recent years due to their potential in suppressing multidrug-resistant bacteria (26–28). However, methods for developing effective cocktails containing phages with distinct infection strategies are always time-consuming and labor intensive (19, 20). Here, to address this challenge, we explored the possibility of designing efficient phage cocktails using a shortcut method. Thus, we isolated novel phages using wild-type bacterial strain and its resistant variant under single phage infection as hosts.
In previous studies, phage cocktails have been constructed by isolating phages in a labor-intensive manner to prevent the rapid antiphage evolution of a pathogen (16, 19, 20, 26). Here, we used a mutant version (V. alginolyticus 283R) of the wild-type host bacterium V. alginolyticus 283T, which was resistant to the phages Va1 and Va2, to isolate another phage, Va3. Biological characterization of these phages showed that Va1 and Va2 (randomly isolated using the wild-type host strain) and Va3 had a short latent period and strong lysis activity, indicating that they were all potential candidates for phage therapy against V. alginolyticus (29, 30). In addition, the tolerance of these phages to relatively high temperatures and a broad pH range suggested that they can maintain their activity in different environmental conditions and thus have good application potential (16).
It has previously been reported that the phage that infects the wild-type strain of Salmonella and the phage that infects the mutant strain of this pathogen had different receptors (i.e., an outer core oligosaccharide versus an O-antigen) (18). In this study, we showed that Va1 and Va2 can only infect V. alginolyticus 283T, whereas Va3 can infect both V. alginolyticus 283T and V. alginolyticus 283R. These findings indicate that Va3 might utilize a different infection strategy from that of Va1 and Va2.
Our genomic characterization of the three phages indicated that Va1 and Va3 belong to the Firehammervirus and Schizotequatrovirus genera, respectively, whereas Va2 represents a novel genus of Myoviridae. Notably, Va3 was shown to contain considerably more genes encoding tail fiber proteins and baseplate proteins than the other two phages. We suggest that these might act as RBPs, which help Va3 bind to a greater variety of receptors on the cell surface of the host at the beginning of infection (31, 32). Moreover, Va3 was shown to possess 34 types of tRNA and 2 genes (i.e., gene_252 and gene_295) which might be involved in NAD+ synthesis. The diverse and abundant tRNAs in phage genome could substitute for the cleaved host tRNAs when bacteria limit phage replication by targeting their endogenous tRNA and allowing phage protein translation to continue (33, 34). In addition, the adequate tRNAs in a phage genome are considered to correspond to codons that are frequently used in phage-specific genes to increase their translation efficiency (34). This makes the postinfection progress of a phage exhibit a high level of independence on the molecular machinery of the host (35). Consequently, this independence appears to endow phages with extended host ranges, which is a well-known characteristic of an appropriate phage for cocktail construction (36). On the other hand, NAD+ is an indispensable and universal redox cofactor in many organisms (37). Therefore, phages that carry enzymes for NAD+ synthesis likely parallel similar biosynthetic pathways in the bacterial genomes, while essential NAD+ metabolites are suppressed by the host bacteria to limit viral replication (38). This provides functional backup to support phage development (39). In general, the greater variety of RBP and tRNA genes, as well as genes related to NAD+ synthesis in the Va3 genome, might equip this phage with different infection strategies from Va1 and Va2.
Bacteria have evolved a variety of immune mechanisms, including receptor mutations, masking phage receptors, CRISPR/Cas, and restriction-modification systems, which help protect them from attack by phages. In this way, the bacteria become highly diverse in both their morphological and physiological characteristics and thus become resistant to attack by phages (40). Phages that have diverse infection strategies can counter the different bacterial immune system mechanisms and perform in a complementary manner during infection. Thus, a cocktail of two or more of such phages has an enhanced effect on suppressing phage-resistant bacteria (20). In addition, from an evolutionary perspective, the combination of complementary phages will provide multiple selection pressures to the host bacteria. This will have a maximum cost to the fitness of the bacteria and thus suppress its evolutionary trajectory and promote the long-term efficacy of phage therapy (41, 42).
Here, we revealed that any phage cocktail containing Va3 was significantly better at impeding the emergence of phage-resistant bacteria than phage cocktail containing only Va1 and Va2. Thus, we propose that by utilizing a quickly evolved antiphage variant of bacteria as a new host (following infection by a single phage), this is more likely to isolate phages with different infection strategies. Subsequent cocktails prepared containing phages with different infection strategies will be more likely to prevent antiphage mutation of the pathogen. Indeed, we also used this method to construct phage cocktails against another pathogen (i.e., Vibrio parahaemolyticus MCS) and obtained similar antibacterial results (Fig. S3). In this case, any phage cocktail containing the phage VP4 more effectively inhibited V. parahaemolyticus than those consisting of phages isolated using the wild-type host. VP4 was isolated using a mutated version of the host, which was resistant to infection by VP1 (Fig. S4). These data further confirm the reliability of our method.
In this study, we revealed that if bacterial antiphage mutation was used properly, an efficient cocktail containing fewer phages with different infection strategies can quickly be obtained; thus, it has great potential for the development of phage therapies against bacterial pathogens.
MATERIALS AND METHODS
Phage isolation and purification.
Phages Va1 and Va2 were isolated using V. alginolyticus 283T (ATCC 17749; biosafety level 1 [BSL1]) (43) as the host. In brief, a 10-mL water sample was filtered through a 0.22-μm sterile microfilter and then incubated with 50 mL log-phase host culture in RO broth (comprising artificial seawater containing 1 g/L yeast extract, 1 g/L tryptone, and 1 g/L sodium acetate, pH 7.8 to 8.0) at 28°C with shaking. Phage samples were collected from the culture every 24 h for 7 days and were filter sterilized as described above. The presence of phages in the filtrate was determined by the double-layer agar method. This involved mixing and plating 4 mL molten RO soft agar (0.5%) and 500 μL of the filtrate that had been inoculated with 500 μL host culture for 30 min at 28°C on solidified 1.5% RO agar plates. A single plaque was selected with a sterile pipette tip and eluted in SM buffer (i.e., 50 mM Tris-HCl [pH 7.5] containing 100 mM NaCl and 10 mM MgSO4). This phage stock was then plated on semisolid agar medium again for plaque formation. This procedure was repeated five times to purify single phages (21).
Mutant bacteria that were resistant to the Va1 phage were selected. The early log-phase culture of V. alginolyticus strain 283T (108 CFU/mL) was infected with Va1 (108 PFU/mL) at a multiplicity of infection (MOI) of 0.1. Once the cloudy bacterial suspension became transparent due to the lytic activity of the phage, the surviving bacteria were selected by the streak-plating method. The resulting single colonies were collected and tested for their susceptibility to the phage using spot assays (44). Bacterial colonies that grew in the presence of Va1 were considered to exhibit resistance to the phage. These resistant bacterial colonies were verified by 16S rRNA gene sequencing and stored as phage-resistant mutant bacteria. One of them was named V. alginolyticus 283R, and it was used as the new host to isolate the novel phage Va3, following the procedure described above. All the bacterial strains and phages were maintained in a 0.85% NaCl solution containing 15% glycerol at −80°C for long-term preservation.
Preparation of high-titer phage suspensions.
To propagate the purified phages, a log-phase culture of host bacteria was infected with the phage at an MOI of 0.1 and incubated at 28°C for 24 h. The mixture was centrifuged (at 10,000 × g for 15 min), filtered through a 0.22-μm sterile microfilter, and treated with DNase I (2 ng/L) and RNase A (2 ng/L) at room temperature for 2 h. Phage particles in the treated filtrate were precipitated overnight by mixing with 10% (wt/vol) polyethylene glycol (PEG) 8000 in 1 M sodium chloride (final concentration). Finally, the phages were purified by CsCl density gradient ultracentrifugation (gradient density, 1.5 g · mL−1, 200,000 × g, 8 h, 4°C). The clear blue band consisting of phage particles was collected, dialyzed in SM buffer using 30-kDa superfilters (UFC5030), and stored at 4°C until further use (45).
Observing phage morphology using TEM.
CsCl-purified phages (in SM buffer) were examined by transmission electron microscopy (TEM). The phage suspension was deposited on a copper grid and negatively stained with 2% uranyl acetate for 3 min. Phages were subsequently observed using TEM (JEM-1010; Jeol, Tokyo, Japan) at an accelerating voltage of 80 kV (46).
One-step growth curves.
One-step growth curves were constructed to analyze the life cycle of the phages (29, 47). In brief, 1 mL of log-phase host bacteria was infected with the phage at an MOI of 0.01. After adsorption at 28°C for 20 min, the mixture was centrifuged at 6,000 × g for 5 min to remove the nonabsorbed phage particles. The pellet was then resuspended in 1 mL fresh RO medium, and 50 μL of the suspension was transferred into 50 mL RO medium and incubated at 28°C with continuous shaking. Samples were collected every 15 min, and the viral abundance was titrated using the double-agar plaque assay.
Analysis of phage stability.
The stability of the phages Va1, Va2, and Va3 under various pH and thermal conditions was assessed according to the methods described by Verma et al. (16) with some modifications. In brief, for the pH stability test, 100 μL phage suspension (108 PFU/mL) was mixed with 900 μL RO medium, and then the pH of the mixture was adjusted to 2, 4, 10, and 12, using NaOH or HCl. After incubation at 28°C for 1 h, the pH of each mixture was adjusted to neutral, and then phage titrations were determined through the double-layer agar method. In the thermostability test, the phage lysate (108 PFU/mL) was incubated at 20, 30, 40, 50, 60, 70, and 80°C for 1 h, and the phage titer was subsequently measured using the double-layer agar method. All the experiments were repeated three times.
To determine the sensitivity of the phages to chloroform (48), the phage suspension was mixed with chloroform such that the final concentration of the latter was 0%, 0.2%, 2%, and 20% (vol/vol). After shaking vigorously for 1 min, the mixture was incubated at room temperature for 30 min and centrifuged at 6,000 × g for 5 min. A drop of the supernatant was then placed on a V. alginolyticus 283T agar plate, and the plates were observed for the formation of plaques.
Phage DNA extraction, genome sequencing, and analysis.
For DNA extraction, 1 mL CsCl-purified phage suspension was incubated with DNase I (2 ng·L−1) and RNase A (2 ng·L−1) at room temperature for 2 h to digest any contaminating DNA and RNA from the host. The phage suspension was then treated with proteinase K (50 μg · mL−1), EDTA (20 mM), and SDS (0.5% [wt/vol]) at 55°C for 3 h with gentle mixing every 30 min. This was followed by DNA extraction with phenol/chloroform/isoamyl alcohol solution (at a ratio of 25:24:1 [vol/vol]) and chloroform/isoamyl alcohol (at a ratio of 24:1 [vol/vol]). The DNA was precipitated with 50% isopropanol (vol/vol) and centrifuged (12,000 × g, 15 min, 4°C). The precipitated pellet was air-dried and then resuspended in sterile TE buffer (10).
The extracted genome was treated, respectively, with DNase and RNase, after which gel electrophoresis was conducted to confirm whether it was DNA or RNA. In addition, S1 nuclease was used to identify if the DNA genome was single stranded or double stranded.
High-throughput sequencing (49) was conducted using the Illumina HiSeq 2500 platform by Oebiotech Co. (Qingdao, China) and Allwegene Co. (Beijing, China). After processing, the quality-filtered reads were assembled in a single contig using SPAdes genome assembler (v3.12.0). In addition, the GeneMarks online server and ORF Finder were used to predict ORFs in the phage genome (16), after which translated ORFs were annotated by the BLASTP algorithm against the nonredundant (nr) protein database of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) with an E value of <0.001. tRNAs in each genome were detected by tRNAscan-SE (50). Meanwhile, the web tools VirulenceFinder (51) and ResFinder server (52) were used to predict potential genes coding for virulence factors and antibiotic resistance. The three phage genomes (Va1, Va2, and Va3) were uploaded to the ViPTree (https://www.genome.jp/viptree) for phylogenetic analysis. The progressive Mauve algorithm was then used to compare these phages with other reported V. alginolyticus phages (53).
After genome sequencing, PCR amplifications were carried to check whether these genomes were circular or linear using the following primers: Va1-F (GTAACGATCCAACCTCTGCGGAAC), Va1-R (CGGTGATGGATTTAGTATAACATC), Va2-F (ACCAAAGTTGACTCGAAGTTCGC), Va2-R (TCTGGAGAAACGTGGATACAA), Va3-F (TGGTATGCATTCCCGAAAGG), and Va3-R (CAGTGTAGTAATTCAGCAT). In each case, the F and R primers extended out from the two termini of the “linear” phage genomes. If the phage genome was linear, then no PCR product would be detected, whereas if it was circular, then a PCR product would be generated, and its sequence would be consistent with the phage genome.
Bacterial growth reduction curve of individual phages and cocktails.
In a 48-well plate (Nest Biotechnology, China), phage suspensions were coincubated with the bacteria when they grew in the logarithmic phase (initial concentration of 108 CFU/mL) at different MOIs (i.e., 1 or 10). A control sample without any phage suspension was also prepared. OD600 measurements were then acquired at 1-h intervals using a growth curve instrument (Infinite M200 Pro; Tecan, Switzerland). The number of viable bacteria following treatment with a single phage or a phage cocktail were quantified at 24 h and 96 h, respectively, using the plate counting method. The ability of single phages (Va1, Va2, or Va3), or phage cocktails (Va1 with Va2, Va1 with Va3, Va2 with Va3, or Va1 with Va2 and Va3) to inactivate V. alginolyticus 283T was quantified by subtracting the number of bacteria in each phage infection treatment group by those in the bacteria control group. All assays were performed in triplicate.
Host range of different phage combinations.
Spot tests (44) were performed on several of the Vibrio sp. strains listed in Table S4 in the supplemental material to investigate the host ranges of Va1 plus Va2, Va1 plus Va3, Va2 plus Va3, and Va1 plus Va2 and Va3. In brief, 1 mL of a log-phase bacterial aliquot was mixed with 4 mL molten RO (containing 0.5% agar) and spread onto a solid RO agar plate (containing 1.5% agar). The phages were then mixed in equal proportions, and 5 μL of each phage lysate mixture (108 PFU/mL) was spotted on the surface of the agar and incubated at 28°C for 12 h. The bacterial sensitivity to the phage combination was then assessed by the presence of a lysis clear zone at the spot.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 5. Significant differences among the different treatment groups were assessed by one-way analysis of variance (ANOVA) or the Student's t test, and a P value of <0.05 was considered to be statistically significant.
Ethics statement.
This article does not contain any studies with human or animal subjects performed by any of the authors. All the bacteria used in this study do not cause disease in healthy adults and therefore present minimal potential hazard to the laboratory personnel and environment.
Data availability.
The complete genome sequences of Va1, Va2, and Va3 were deposited in the GenBank database under accession numbers MK387337, MW073017, and MK568540, respectively.
ACKNOWLEDGMENTS
This work was supported by the NSFC projects (41876174, 42106107, 42006093), Shandong provincial key research and development plan (2021ZDSYS29), and the open task of Qingdao National Laboratory for Marine Science and Technology (QNLM2016ORP0311).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Yongyu Zhang, Email: zhangyy@qibebt.ac.cn.
Nicole R. Buan, University of Nebraska-Lincoln
REFERENCES
- 1.Rorbo N, Ronneseth A, Kalatzis PG, Rasmussen BB, Engell-Sorensen K, Kleppen HP, Wergeland HI, Gram L, Middelboe M. 2018. Exploring the effect of phage therapy in preventing Vibrio anguillarum infections in cod and turbot larvae. Antibiotics-Basel 7:42. 10.3390/antibiotics7020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Cao Z, Zhen L, Wang L, Li H, Wu F, Jin L, Li X, Li S, Xu Y. 2015. Effect of bacteriophages on Vibrio alginolyticus infection in the sea cucumber, Apostichopus japonicus (Selenka). J World Aquacult Soc 46:149–158. 10.1111/jwas.12177. [DOI] [Google Scholar]
- 3.Wang Y, Barton M, Elliott L, Li X, Abraham S, O'Dea M, Munro J. 2017. Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473:251–258. 10.1016/j.aquaculture.2017.01.003. [DOI] [Google Scholar]
- 4.Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 28:591–595. 10.1016/j.tibtech.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Letchumanan V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Goh B, Ab Mutalib N, Lee L. 2016. Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 7:1114. 10.3389/fmicb.2016.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco L, Ambroa A, Lopez M, Fernandez-Garcia L, Bleriot I, Trastoy R, Ramos-Vivas J, Coenye T, Fernandez-Cuenca F, Vila J, Martinez-Martinez L, Rodriguez-Baño J, Pascual A, Cisneros JM, Pachon J, Bou G, Tomas M. 2019. Combined use of the Ab105-2 φΔCI lytic mutant phage and different antibiotics in clinical isolates of multi-resistant Acinetobacter baumannii. Microorganisms 7:556. 10.3390/microorganisms7110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Defoirdt T, Boon N, Sorgeloos P, Verstraete W, Bossier P. 2007. Alternatives to antibiotics to control bacterial infections: luminescent vibriosis in aquaculture as an example. Trends Biotechnol 25:472–479. 10.1016/j.tibtech.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Defoirdt T, Sorgeloos P, Bossier P. 2011. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol 14:251–258. 10.1016/j.mib.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y, Zhang Y, Zhou C, Chen Z, Yang S, Yan C, Jiao N. 2016. Complete genome sequence of the siphovirus Roseophage RDJL 2 infecting Roseobacter denitrificans OCh114. Mar Genomics 25:17–19. 10.1016/j.margen.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Garcia P, Martinez B, Obeso JM, Rodriguez A. 2008. Bacteriophages and their application in food safety. Lett Appl Microbiol 47:479–485. 10.1111/j.1472-765X.2008.02458.x. [DOI] [PubMed] [Google Scholar]
- 12.Kalatzis PG, Castillo D, Katharios P, Middelboe M. 2018. Bacteriophage interactions with marine pathogenic vibrios: implications for phage therapy. Antibiotics-Basel 7:15. 10.3390/antibiotics7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luong T, Salabarria AC, Edwards RA, Roach D. 2020. Standardized bacteriophage purification for personalized phage therapy. Nat Protoc 15:2867–2890. 10.1038/s41596-020-0346-0. [DOI] [PubMed] [Google Scholar]
- 14.Castillo D, Rorbo N, Jorgensen J, Lange J, Tan D, Kalatzis PG, Lo Svenningsen S, Middelboe M. 2019. Phage defense mechanisms and their genomic and phenotypic implications in the fish pathogen Vibrio anguillarum. FEMS Microbiol Ecol 95:fiz004. 10.1093/femsec/fiz004. [DOI] [PubMed] [Google Scholar]
- 15.Nakai T, Park SC. 2002. Bacteriophage therapy of infectious diseases in aquaculture. Res Microbiol 153:13–18. 10.1016/S0923-2508(01)01280-3. [DOI] [PubMed] [Google Scholar]
- 16.Stalin N, Srinivasan P. 2017. Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet Microbiol 207:83–96. 10.1016/j.vetmic.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Islam MS, Zhou Y, Liang L, Nime I, Liu K, Yan T, Wang X, Li J. 2019. Application of a phage cocktail for control of Salmonella in foods and reducing biofilms. Viruses-Basel 11:841. 10.3390/v11090841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Kim S, Park B, Ryu S. 2014. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl Environ Microbiol 80:1026–1034. 10.1128/AEM.03494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Yuan S, Liu Q, Mai G, Yang J, Deng D, Zhang B, Liu C, Ma Y. 2018. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front Microbiol 9:1476. 10.3389/fmicb.2018.01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Shen W, Zhong Q, Chen Q, He X, Baker J, Xiong K, Jin X, Wang J, Hu F, Le S. 2020. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front Microbiol 11:327. 10.3389/fmicb.2020.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Wang Z, Zhao J, Wang L, Xie G, Huang J, Zhang Y. 2021. A novel Vibriophage vB_VcaS_HC containing lysogeny-related gene has strong lytic ability against pathogenic bacteria. Virol Sin 36:281–290. 10.1007/s12250-020-00271-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlota B, Joan C, Spricigo DA, Otero J, Sanchez-Osuna M, Cortes P, Llagostera M. 2016. Genomics of three new bacteriophages useful in the biocontrol of Salmonella. Front Microbiol 7:545. 10.3389/fmicb.2016.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, Gao M. 2017. Jumbo bacteriophages: an overview. Front Microbiol 8:403. 10.3389/fmicb.2017.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malone LM, Warring S, Jackson SA, Warnecke C, Gardner PP, Gumy LF, Fineran PC. 2020. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol 5:48–55. 10.1038/s41564-019-0612-5. [DOI] [PubMed] [Google Scholar]
- 25.Adriaenssens EM, Brister JR. 2017. How to name and classify your phage: an informal guide. Viruses-Basel 9:70. 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateus L, Costa L, Silva YJ, Pereira C, Cunha A, Almeida A. 2014. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 424-425:167–173. 10.1016/j.aquaculture.2014.01.001. [DOI] [Google Scholar]
- 27.Naghizadeh M, Torshizi MAK, Rahimi S, Dalgaard TS. 2019. Synergistic effect of phage therapy using a cocktail rather than a single phage in the control of severe colibacillosis in quails. Poult Sci 98:653–663. 10.3382/ps/pey414. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y, Wang L, Li X, Tan D, Cong C, Xu Y. 2019. Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii. Virus Res 272:197734. 10.1016/j.virusres.2019.197734. [DOI] [PubMed] [Google Scholar]
- 29.Cai R, Wang Z, Wang G, Zhang H, Cheng M, Gu Z, Ji Y, Xi H, Wang X, Xue Y, Rahman S, Sun C, Feng X, Lei L, Tong Y, Han W, Gu J. 2019. Biological properties and genomics analysis of vB_KpnS_GH-K3, a Klebsiella phage with a putative depolymerase-like protein. Virus Genes 55:696–706. 10.1007/s11262-019-01681-z. [DOI] [PubMed] [Google Scholar]
- 30.Kreienbaum M, Dörrich AK, Brandt D, Schmid NE, Leonhard T, Hager F, Brenzinger S, Hahn J, Glatter T, Ruwe M, Briegel A, Kalinowski J, Thormann KM. 2020. Isolation and characterization of Shewanella phage Thanatos infecting and lysing Shewanella oneidensis and promoting nascent biofilm formation. Front Microbiol 11:573260. 10.3389/fmicb.2020.573260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MDT, de la Fuente-Nunez C, Lu TK. 2019. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179:459–469. 10.1016/j.cell.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai K, Iwazaki T, Yamashita E, Nakagawa A, Sakuraba F, Enomoto A, Inagaki M, Takeda S. 2019. Observation of unexpected molecular binding activity for Mu phage tail fibre chaperones. J Biochem 166:529–535. 10.1093/jb/mvz068. [DOI] [PubMed] [Google Scholar]
- 33.Leskinen K, Blasdel BG, Lavigne R, Skurnik M. 2016. RNA-sequencing reveals the progression of phage-host interactions between φR1–37 and Yersinia enterocolitica. Viruses-Basel 8:111. 10.3390/v8040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiljunen S, Hakala K, Pinta E, Huttunen S, Pluta P, Gador A, Lonnberg H, Skurnik M. 2005. Yersiniophage φR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology (Reading) 151:4093–4102. 10.1099/mic.0.28265-0. [DOI] [PubMed] [Google Scholar]
- 35.Ceyssens PJ, Minakhin L, Van den Bossche A, Yakunina M, Klimuk E, Blasdel B, De Smet J, Noben JP, Bläsi U, Severinov K, Lavigne R. 2014. Development of giant bacteriophage kz is independent of the host transcription apparatus. J Virol 88:10501–10510. 10.1128/JVI.01347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshikawa G, Askora A, Blanc-Mathieu R, Kawasaki T, Li Y, Nakano M, Ogata H, Yamada T. 2018. Xanthomonas citri jumbo phage XacN1 exhibits a wide host range and high complement of tRNA genes. Sci Rep 8:4486. 10.1038/s41598-018-22239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorci L, Martynowski D, Rodionov DA, Eyobo Y, Zogaj X, Klose KE, Nikolaev EV, Magni G, Zhang H, Osterman AL. 2009. Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis. Proc Natl Acad Sci USA 106:3083–3088. 10.1073/pnas.0811718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skjerning RB, Senissar M, Winther K, Gerdes K, Brodersen DE. 2019. The RES domain toxins of RES-Xre toxin-antitoxin modules induce cell stasis by degrading NAD+. Mol Microbiol 111:221–236. 10.1111/mmi.14150. [DOI] [PubMed] [Google Scholar]
- 39.Lakshminarayan MI, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. 2021. Jumbo phages: a comparative genomic overview of core functions and adaptions for biological conflicts. Viruses-Basel 13:63. 10.3390/v13010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai R, Wu M, Zhang H, Zhang Y, Cheng M, Guo Z, Ji Y, Xi H, Wang X, Xue Y, Sun C, Feng X, Lei L, Tong Y, Liu X, Han W. 2018. A smooth-type, phage-resistant Klebsiella pneumoniae mutant strain reveals that OmpC is indispensable for infection by phage GH-K3. Appl Environ Microbiol 84:e01585-18. 10.1128/AEM.01585-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright RCT, Friman VP, Smith MCM, Brockhurst MA. 2019. Resistance evolution against phage combinations depends on the timing and order of exposure. mBio 10:e01652-19. 10.1128/mBio.01652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaitekenas A, Tai A, Ramsay J, Stick S, Kicic A. 2021. Pseudomonas aeruginosa resistance to bacteriophages and its prevention by strategic therapeutic cocktail formulation. Antibiotics-Basel 10:145. 10.3390/antibiotics10020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KK, Yu SR, Yang TL, Liu PC, Chen FR. 1996. Isolation and characterization of Vibrio alginolyticus isolated from diseased kuruma prawn, Penaeus japonicus. Lett Appl Microbiol 22:111–114. 10.1111/j.1472-765x.1996.tb01121.x. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Zhang Y, Jiao N. 2010. Phage resistance of a marine bacterium, Roseobacter denitrificans OCh114, as revealed by comparative proteomics. Curr Microbiol 61:141–147. 10.1007/s00284-010-9588-3. [DOI] [PubMed] [Google Scholar]
- 45.Olszak T, Danis-Wlodarczyk K, Arabski M, Gula G, Maciejewska B, Wasik S, Lood C, Higgins G, Harvey B, Lavigne R, Drulis-Kawa Z. 2019. Pseudomonas aeruginosa PA5oct jumbo phage impacts planktonic and biofilm population and reduces its host virulence. Viruses-Basel 11:1089. 10.3390/v11121089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Liu Q, Fan J, Yan T, Zhang H, Yang J, Deng D, Liu C, Wei T, Ma Y. 2020. Characterization and genomic analysis of ValSw3-3, a new Siphoviridae bacteriophage infecting Vibrio alginolyticus. J Virol 94:e00066-20. 10.1128/JVI.00066-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathias M, Amy MC, Sif KB. 2010. Isolation and life cycle characterization of lytic viruses infecting heterotrophic and cyanobacteria, p 118–133. In Wilhelm SW, Weinbauter MG, Suttle CA (ed), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX. [Google Scholar]
- 48.Wang Z, Zhao J, Wang L, Li C, Liu J, Zhang L, Zhang Y. 2019. A novel benthic phage infecting Shewanella with strong replication ability. Viruses-Basel 11:1081. 10.3390/v11111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Yang L, Sun E, Song J, Wu B. 2019. Characterisation of a newly detected bacteriophage infecting Bordetella bronchiseptica in swine. Arch Virol 164:33–40. 10.1007/s00705-018-4034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Jiao N. 2009. Roseophage RDJL Phi1, infecting the aerobic anoxygenic phototrophic bacterium Roseobacter denitrificans OCh114. Appl Environ Microbiol 75:1745–1749. 10.1128/AEM.02131-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Tables S1 to S11. Download aem.02323-21-s0001.pdf, PDF file, 0.8 MB (787.3KB, pdf)
Data Availability Statement
The complete genome sequences of Va1, Va2, and Va3 were deposited in the GenBank database under accession numbers MK387337, MW073017, and MK568540, respectively.