Abstract
Polyethylene terephthalate (PET) is the most widespread synthetic polyester, having been utilized in textile fibers and packaging materials for beverages and food, contributing considerably to the global solid waste stream and environmental plastic pollution. While enzymatic PET recycling and upcycling have recently emerged as viable disposal methods for a circular plastic economy, only a handful of benchmark enzymes have been thoroughly described and subjected to protein engineering for improved properties over the last 16 years. By analyzing the specific material properties of PET and the reaction mechanisms in the context of interfacial biocatalysis, this Perspective identifies several limitations in current enzymatic PET degradation approaches. Unbalanced enzyme–substrate interactions, limited thermostability, and low catalytic efficiency at elevated reaction temperatures, and inhibition caused by oligomeric degradation intermediates still hamper industrial applications that require high catalytic efficiency. To overcome these limitations, successful protein engineering studies using innovative experimental and computational approaches have been published extensively in recent years in this thriving research field and are summarized and discussed in detail here. The acquired knowledge and experience will be applied in the near future to address plastic waste contributed by other mass-produced polymer types (e.g., polyamides and polyurethanes) that should also be properly disposed by biotechnological approaches.
Keywords: Hydrolase, enzymatic degradation, interfacial biocatalysis, plastic recycling, protein engineering, polyethylene terephthalate (PET), product inhibition, thermostability
1. Introduction
Scientists began to study biological degradation of plastic waste in the early 1970s.1 Around the same time, the distribution of plastic waste in the Pacific Ocean was first quantified.2 After nearly 50 years, climate change and environmental pollution caused by mass production, mass consumption, and improper end-of-life management of petroleum-based plastic products have become unprecedented challenges for human society.3−5 Consciousness of these issues spreading among governments and policymakers, industries producing or relying on plastics, and end-users of plastic products, has recently boosted research and development of novel plastic replacement materials and waste plastic valorization strategies to enable a transition from a linear to a circular plastic economy.6−9
To this end, biotechnological plastic recycling has become a thriving research area in recent years.6,9,10 The biodegradation of hydrophobic vinyl polymers, such as polyolefins and polystyrene, which represent over 80% of all conventional plastics produced,11 is still an intensively debated topic.9,12,13 In contrast, the biocatalytic degradation of polyester-type plastics, such as polyethylene terephthalate (PET), has evolved in the last two decades from verifying trace amounts of released monomers after weeks of incubation to highly efficient depolymerization within several hours.14−17 PET is a heteroatomic polymer consisting of terephthalic acid (TPA) and ethylene glycol (EG) connected by ester bonds. It is a semicrystalline thermoplastic composed of crystalline regions with uniformly packed molecules and amorphous regions with randomly arranged microstructures.18 With a first patent filed for its synthesis in 1941 and commercialization starting in the early 1950s,19 particularly as synthetic polyester fabric for the textile industry and as packaging materials for food and beverages,20,21 PET has become one of the most important mass-produced petrochemical plastics.22 The majority of PET products have a high crystalline fraction (usually 30–40% crystallinity)23,24 benefiting their durability against mechanical and chemical stress.25 Post-consumer PET accounts for a considerable fraction of the global solid waste stream, and its breakdown is a source of microplastics and microfibers that further pollute the environment.11,26−28
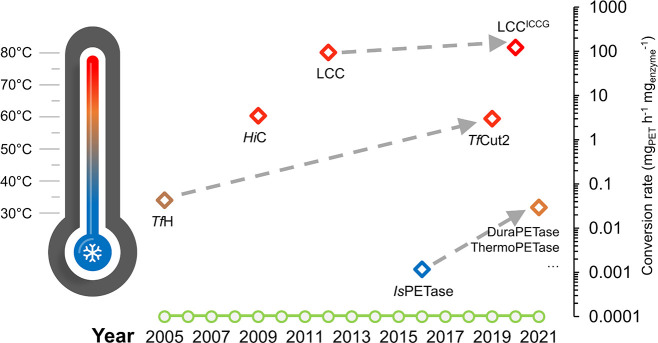
Tokiwa et al. suggested in the 1970s that aromatic polymers are resistant to enzymatic and microbial attack.29 Later research found, however, that crude enzyme preparations from specific fungal species can induce surface modifications on high-crystallinity PET materials.30 The first breakthrough was achieved with the cutinase TfH (from the filamentous actinomycete Thermobifida fusca) which caused >50% weight loss of melt-quenched post-consumer PET bottles (10% crystallinity) following 3 weeks of incubation.31 During 16 years of extensive research, the initial low PET degradation activity of T. fusca PET hydrolase has been considerably enhanced by more than an order of magnitude (Figure 1) as a result of both protein and process engineering.32 The development of metagenomic approaches has allowed for the identification of a more active leaf-branch-compost cutinase LCC,33 which has emerged as the most promising benchmark thermophilic PET hydrolase. In 2020, Nature published a breakthrough article on engineered LCC variants which can rapidly depolymerize amorphized (i.e., by lowering the crystallinity through a thermomechanical process to allow for enzymatic degradation) PET waste at an industrially relevant scale within 10 h, and the recovered monomers were readily used to synthesize virgin polymers, thereby closing the recycling loop.16IsPETase and the related monoester-hydrolyzing IsMHETase, both derived from the bacterium Ideonella sakaiensis, which was isolated from plastic-contaminated sediment samples, have also been intensively studied.34,35 The uniqueness of this bacterium in degrading and metabolizing highly amorphous PET led not only to the discovery of a tandem polymer degradation pattern by two distinct enzyme classes,36 but also to its utility in the efficient bio-upcycling of PET waste to other useful biopolymers when it was applied as a whole cell-based catalyst.37
Figure 1.
Selected milestones of a 16-year-long history of identifying and engineering PET hydrolases. Both optimal reaction temperatures (in varying colors) for PET degradation and normalized maximal conversion rates (diamonds corresponding to logarithmic values on the vertical axis) calculated based on various publications are denoted, regardless of the material properties of applied PET substrates. The successes in raising the degradation performance using certain benchmark enzymes by both protein and process engineering are indicated by arrows with broken lines. TfH: hydrolase from T. fusca DSM43793;31HiC: Humicola insolens cutinase;42 LCC: leaf-branch compost cutinase;33,52IsPETase: I. sakaiensis PET hydrolase;16,34TfCut2: T. fusca KW3 cutinase;53 LCCICCG: LCC variant with indicated quadruple substitutions;16 DuraPETase, ThermoPETase, etc.: various thermostabilized IsPETase variants.54−56
Based on >65 crystal structures so far elucidated,38 these bacterial PET hydrolases exhibit conserved structural properties and can be classified into a single subclass of the α/β-hydrolase fold enzyme superfamily as represented by the first solved Streptomyces exfoliates lipase structure (PDB ID: 1JFR).39,40 This subclass is distinct from the polyester-hydrolyzing fungal cutinases (EC 3.1.1.74) with shorter polypeptides and more compact structures.41Thermomyces insolens (formerly named Humicola insolens) cutinase HiC is a prominent member of the fungal PET hydrolase family. It has been successfully commercialized and also received broad attention in recent years by researchers working on PET waste degradation. However, here the emphasis was more on process engineering and improvement rather than protein engineering.42−45
This Perspective intends to provide a deeper understanding of important obstacles in the engineering of effective industrially applicable PET hydrolases, as well as to provide guidance for further efforts in this intriguing research field. Previous research has indicated that enzymatic polyester hydrolysis occurs preferentially in its amorphous parts rather than the well-ordered crystalline regions with poor chain mobility.46 Understanding the accessibility of amorphous polymer chains around the polymer glass transition temperature, in conjunction with tailoring the thermostability of biocatalysts, has thus become a focus of contemporary research.38,47 As a typical surface erosion process, it is not yet fully understood how the adsorption and desorption of biocatalysts onto the polymer surface influence the overall degradation kinetics and performance. This problem is reminiscent of the decades spent investigating the relationship between the binding of cellulases onto crystalline cellulose and catalytic activity.48 Altering their affinity to polymer substrates by protein engineering, on the other hand, has proven to be a viable approach to maximize the catalytic capacity of both cellulose and PET depolymerases. Similar to the aforementioned natural polymer-degrading enzymes such as cellulases, mass transfer of certain degradation intermediates that can function as inhibitors of the biocatalysts, has been identified as the third main bottleneck that must be addressed with a low-cost solution for adequate reaction efficiency.49−51 This Perspective summarizes and discusses special efforts devoted to the engineering of thermostability (Section 2), to study the de/adsorption properties, and to address the product inhibition (Sections 3 and 4).
2. Design of Thermostable PET Hydrolases Based on Material Properties
Earlier research on enzymatic degradation of aromatic and aliphatic copolyesters suggested that increasing the proportion of aromatic moieties raises the melting point, thus lowering the polymer chain mobility and biodegradability at the optimal reaction temperatures of the applied hydrolases.57,58 For a long time, enzymatic depolymerization of PET with a melting point over 260 °C was thought to be unachievable according to this idea, until, in 2005, Müller et al. showed significant weight loss of amorphized PET waste using TfH.31 This discovery drew attention to the preferential (or perhaps exclusive) degradation of amorphous PET, as well as the importance of its glass transition temperature (Tg) in understanding enzymatic degradation.42,46Tg is the temperature range in which the glass transition process occurs, i.e., the amorphous polymer structure transitions from a “glassy” (hard and brittle) state into a sticky-liquid or rubbery one as a function of increasing temperature.18,59 The Tg of bulk amorphous PET is in the range of 65–71 °C, estimated based on data collected with differential scanning calorimetry (DSC) at zero heating rate.60−62 This has so far been accepted as a standard value by research communities working on enzymatic PET hydrolysis.31,42,53,63−65 In addition to its role as both a solvent and a reactant in enzymatic PET degradation, water has a plasticization effect on the PET polymer, as reported since the early 1980s,66,67 by lowering the Tg of bulk polymer by up to 16 °C.60,68,69 This influence is more pronounced on the surface polymer layer of about 13 nm which has an intrinsically lower Tg (∼48.1 °C) than that of the bulk polymer even when water is not present.70 With both effects taken into account, the superficial PET layer exposed to enzymatic degradation in an aqueous milieu can indeed have a biocatalysis-relevant Tg ≲ 40 °C, explaining the considerably fast degradation of amorphous PET by, for example, I. sakaiensis and its IsPETase at ambient temperature.34 The enhanced segmental mobility of the surface polymer, on the other hand, may decrease to a level similar to that of the bulk polymer as a result of superficial crystallization.71 For example, as a result of UV-treatment-induced crystallization, the overall degradation performance could be significantly impaired.72
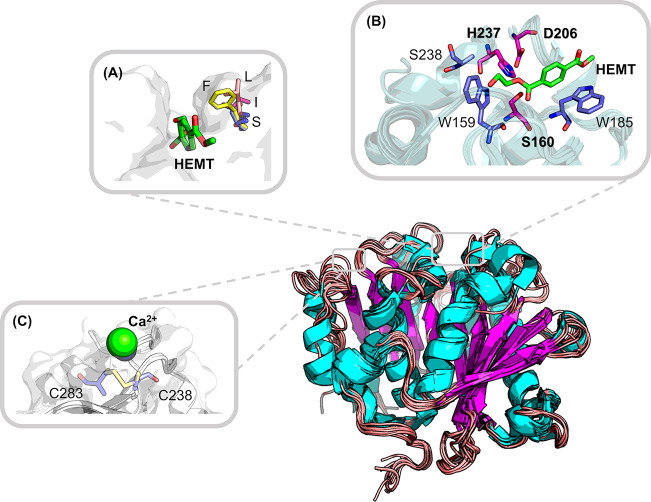
Based on these PET material characteristics, the advantage of using thermophilic and thermostable enzymes in depolymerization, over their mesophilic counterparts, is obvious and evident from preliminary studies.42,56,73,74 In this regard, searching for new thermophilic enzymes, such as those found in metagenomic libraries,33,75 as well as known thermostabilizing ones, provides viable options for improving degradation performance.74 In the presence of selected bivalent ions such as Ca2+ or Mg2+, the overall thermostability of many bacterial PET hydrolases has been improved, as indicated by increased midpoints of thermal denaturation (Tm) by 10–16 °C and enhanced optimal temperatures (Topt) for PET degradation by at least 10 °C.52,76−78 The probable Ca2+ binding site was found to be close to the catalytic triad based on cocrystallized structures (Figure 2) and molecular dynamics simulations.76−81 The interaction with Ca2+ controls the process of opening and closing the active site during substrate binding and unbinding by Cut190 from Saccharomonospora viridis.78,80 Because Ca2+ will precipitate terephthalate to generate insoluble byproducts from the reaction supernatants,82 relying on Ca2+ salts for high degradation efficiency is undesirable and can be minimized by tailoring the Ca2+-binding sites of the biocatalysts. Earlier research was carried out by substituting one major Ca2+-binding site with a salt bridge or a putative disulfide bond, resulting in an increased Tm of the T. fusca cutinase TfCut2 by up to 25 °C.77,79 The latter strategy was then adopted in engineering other homologous PET hydrolases, resulting in increased Tm by up to 26 °C,16,56,81,83 although the formation of this disulfide bond was indeed validated by protein crystallography for only two enzymes.16,81 More recently, Nakamura et al. have also introduced cysteine pairs to form putative disulfide bonds at alternative positions in the homologous hydrolase PET2, but this led only to a marginal increase of Tm by less than 3.1 °C.84
Figure 2.
Frequently reported mutation hot spots illustrated on the superposed crystal structures of known bacterial PET hydrolases. Backbones shown in the cartoon are derived from the structural superposition with selected homologous enzymes. The catalytic triad (S160, D206, and H237) as well as two aromatic residues (W159 and W185) are involved in the interaction with the monomer analogue 1-(2-hydroxyethyl)-4-methyl terephthalate (HEMT) (A, B) based on the IsPETase structure (PDB ID: 5XH3; the numbering of residues is modified consistently with other structures solved later for easy comprehension). (A) One frequently reported mutation hotspot equivalent to S209 (B) in IsPETase can adopt various residues which might influence the widths of the binding pocket:65,90 F found in 4CG1, 4EB0, and 7OSB and also conserved in many other PET hydrolases; S found in the IsPETase structure 5XH3; I found in an LCC mutant 6THT; L found in another PET hydrolase 7CUV. (C) One of the putative Ca2+ binding sites revealed by cocrystallized structures such as 4WFJ, 5LUL, and 5ZNO can be replaced by a disulfide bridge (6THT, 7CTS, and 7CTR) to thermostabilize several PET hydrolases.
Enhanced thermostability of PET hydrolases has also been reported when they were recombinantly expressed in alternative hosts other than Escherichia coli, for example, Bacillus subtilis(53,85) or Pichia pastoris.86 Compared to the wild-type LCC obtained from E. coli, an increased Tm of >4 °C or >12 °C has been described when it was expressed in B. subtilis or P. pastoris, respectively.85,86 The remarkable improvement with the latter host was attributed to glycosylation which mitigated the enzyme thermal aggregation at high reaction temperatures.86
Despite the fact that IsPETase is distinguished by its high PET hydrolyzing activity at ambient temperature, an effect which is presumably due to its more flexible and open substrate binding cleft,87 efforts to thermostabilize this enzyme have been widely reported recently, with the goal of outperforming other cutinase-like PET hydrolases also at higher temperature ranges (Table 1). A ThermoPETase mutant with three residue substitutions, generated based on a structure-based engineering strategy, exhibited an increased Tm by 8.8 °C and up to 14-fold improved PET hydrolyzing activity compared to the wild-type IsPETase at 40 °C.54 DuraPETase is an IsPETase variant with ten mutated residues, discovered by a novel GRAPE (greedy accumulated strategy for protein engineering) computational method.55 Compared to wild-type IsPETase, DuraPETase showed a Tm increase of 31 °C and over 300-fold higher hydrolytic activity against high-crystallinity PET powder. By applying a convolutional neural network, MutCompute, trained for stability optimization, Lu et al. reported in a recent preprint the most thermostable IsPETase variant based on a single N233K substitution in addition to the DuraPETase, resulting in a Tm of 83.5 °C.88 FAST-PETase, the most promising variant based on additional mutations of ThermoPETase, demonstrated significantly improved PET degradation activity against low-crystallinity PET waste at 50 °C compared to the wild-type and other known mutants of IsPETase. By screening a randomized IsPETase library based on error-prone PCR, at least one of their computationally targeted stability-related hot spots was also discovered.56 Further substitutions related to thermostabilizing IsPETase found by other protein engineering strategies have been reported to be distributed across the entire sequence (Table 1). In another recent preprint, >13,000 IsPETase variants were evaluated by applying catalytic activity at elevated temperatures as a primary selection pressure. This directed evolution procedure afforded a HotPETase variant with 21 mutations compared to wild-type IsPETase and a Tm of 82.5 °C.89 Therefore, the thermostability of IsPETase and other homologous enzymes depends on the interplay of many effects, necessitating further comprehensive research.
Table 1. Selected IsPETase Engineering Studies Designed to Improve Thermal Stability.
| nomenclature and introduced mutations | improved stabilitya | design approach and interpretation | refs |
|---|---|---|---|
| S238F/W159H | Tm = 56.5 °C, ΔTm = +9.7 °C | Structural and sequence comparison with homologous PET hydrolases | (65,90) |
| ThermoPETase: S121E/D186H/R280A | Tm = 57.6 °C, ΔTm = +8.8 °C | Structure-based design; Water-mediated hydrogen bond between E121 and H186 | (54) |
| DuraPETase: A214H/I168R/W159H/S188Q/R280A/A180I/G165A/Q119Y/L17F/T140D | Tm = 77.0 °C, ΔTm = +31.0 °C | Structure-based design and machine learning approach; Multiple stabilizing interactions | (55) |
| W159H/F229Y | ΔTm = +10.4 °C | Sequence comparison with other homologous PET hydrolases | (91) |
| DuraPETase+N233K | Tm = 83.5 °C, ΔTm = +38.4 °C | Machine learning; Introduction of salt bridge between K233 and E204 | (88) |
| FAST-PETase: ThermoPETase+R224Q/N233K | Tm= 67.4 °C, ΔTm = +22.3 °C | Machine learning; Introduction of a hydrogen bond between Q224 and S192 | (88) |
| TS-PETase: ThermoPETase+N233C/S282C | Tm = 69.4 °C, ΔTm = +22.3 °C | Structural comparison with other homologous PET hydrolases | (83) |
| TM3: ThermoPETase+K95N/F201I/N233C/S282C | Tm = 70.8 °C, ΔTm = +25.8 °C | Random mutagenesis based on error prone PCR and structural comparison with LCCICCG mutant | (56) |
| D1: DuraPETase+N233C/S282C | Tm = 81.1 °C, ΔTm = +36.1 °C | Structural comparison with LCCICCG mutant | (56) |
| HotPETase: TS-PETase+P181V/S207R/S214Y/Q119K/S213E/R90T/Q182M/N212K/R224L/S58A/S61V/K95N/M154G/N241C/K252M/T270Q | Tm= 82.5 °C, ΔTm = +37.5 °C | Directed evolution | (89) |
ΔTm was estimated compared to Tm of the wild-type IsPETase either determined in the same publication or that of 45.1 °C determined by Brott et al.56Tm values were determined by circular dichroism, differential scanning calorimetry, or differential scanning fluorimetry.
By raising the reaction temperature from 65 to 75 °C, the transition of the polymer microstructure from a less-ordered amorphous state to a nondegradable crystalline state can occur significantly earlier and faster.16 This “physical aging” reaction competes with the enzymatic depolymerization and thus becomes the main determining factor of the total achievable degradation level.53 As a result, using biocatalysts with only very high thermostability, such as the BhrPETase with a Tm of 101 °C,85 is not always a good way to improve the degradation performance. As thermostabilizing an enzyme usually comes at the expense of lowering the flexibility of certain catalysis-relevant structural segments, a more balanced protein engineering strategy focusing on a thermo-active biocatalyst with Topt at 75 °C and sufficient long-term robustness will hold promise for industrial applications. A recent techno-economic analysis of a simulated enzymatic PET recycling process revealed that the reaction duration and enzyme price have a greater influence on process costs than the energy cost to maintain an operating temperature of up to 80 °C.92 Therefore, a commercially viable biocatalytic PET degradation process will rely on highly efficient enzymes rather than more heat-stable ones, which have, for example, Topt in a higher temperature range but are less active at 75 °C.
3. Understanding Interfacial Enzymatic PET Hydrolysis from a Structure–Function Perspective
Enzymatic PET hydrolysis is a surface erosion process characterized by the primary degradation of the exterior polymer to expose the inside of the material.42,46,93 Because neither water (as both a solvent and a reactant) nor the biocatalysts can permeate the inner core of the polymer, only a limited number of superficial ester bonds can be accessed, implying that the reaction occurs primarily under conditions using an excess of enzyme.94 As a consequence of this, research on PET hydrolysis kinetics has commonly employed an inverse Michaelis–Menten equation90,95,96 or its mathematically equivalent expression based on the derivation of the Langmuir adsorption isotherm.42,63,97−99 By simultaneous analysis using the conventional Michaelis–Menten equation, which is typically used to analyze bulk reactions on soluble substrates in great excess to enzymes, the accessible enzyme attack sites on the PET surface can be estimated using the ratio of parameters derived from these swapped kinetic models.90,96
The degradation-relevant binding of these PET hydrolases and variants was investigated by various biophysical approaches, such as quartz crystal microbalance,100−102 chemiluminescence,100 and fluorescence.103 Binding isotherms have been directly estimated based on the concentrations of free enzymes determined in the supernatant after incubation with PET.104−106 Selected PET hydrolases were shown to have a high affinity for the PET surface, as evidenced by the rapid formation of a monolayer, although this was thought to be mainly contributed by nonspecific adsorption.106 A related kinetic study indicated that PET polymers were hydrolyzed at a remarkably lower rate than PET oligomers, regardless of their water solubility.96 The conversion rates of these oligomers were found to be comparable to those of small-molecule para-nitrophenyl esters. This suggests that the complexation or dissociation with the degradable polyester strands requires large activation barriers, determining the overall conversion rate rather than the chemical catalysis itself, which showed, in addition, a similar hydrolysis enthalpy to those reported in the literature for other readily accessible esters.95 Recently, the hydrolysis reaction mechanism of several PET hydrolases against PET-related oligomers was investigated using QM/MM molecular dynamics (MD) simulations or adiabatic mapping to resemble polyester degradation.107−110 Because there is only one cocrystallized IsPETase structure in a complex with a PET monomer analogue available so far, molecular docking of oligomeric aromatic esters has been widely used to study the substrate interaction in the binding groove as well as with the catalytic triad of various PET hydrolases.16,65,87,111−113 The protein surface landscapes of bacterial PET hydrolases are comparable in close vicinity to the catalytic triad, requiring a high conformational selectivity of accommodated PET repeating units directly around the target ester bond at favored twisting angles.112−114 In contrast, it is unclear and unlikely if distal PET repeating units are involved in a catalysis-related interaction with additional surface residues which requires a long PET strand to adopt a specific conformation.113,115 Nonetheless, the defined polymer segment conformation directly next to the target ester bond will require a steric reconfiguration of the polymer chains with adequate mobility in order to form the productive tetrahedral intermediate via the initial nucleophilic attack by the catalytic serine.115 This is evidenced by the highest activation free-energy barrier determined by QM/MM adiabatic mapping, suggesting the nucleophilic attack as a probable rate-limiting step (Figure 3).107,108 Further reaction steps of the PET hydrolysis mechanism are canonically the same as for other conventional ester hydrolases, in agreement with the recent findings in kinetic studies.96
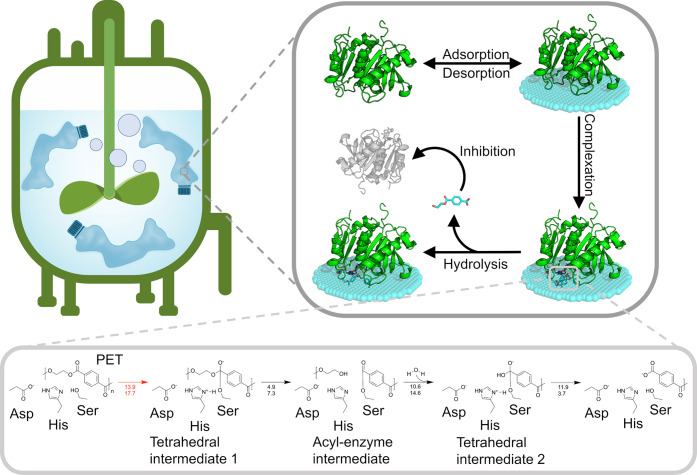
Figure 3.
Interfacial biocatalytic hydrolysis of PET and its reaction mechanism. The states of a PET hydrolase are schematically illustrated in the upper right panel. In the lower panel, individual steps of the hydrolysis reaction are schematically shown in line with their activation free-energy barriers in kcal·mol–1 summarized based on different studies.107,108 The reaction is initiated by a nucleophilic attack by a catalytic serine resulting in a tetrahedral intermediate stabilized by a catalytic histidine, an aspartic acid, and the oxyanion hole, followed by breakdown of the tetrahedral intermediate 1 into an acyl–enzyme intermediate and release of an alcohol. The aspartate–histidine pair activates the water for attack on the acyl–enzyme intermediate carbonyl, resulting in the formation of the second tetrahedral intermediate. The deacylation of this tetrahedral intermediate releases the carboxylic acid product. The rate-limiting step is regarded as the initial nucleophilic attack and highlighted in red with two free-energy activation barriers denoted. The top number is the Boltzmann-weighted average from 20 QM/MM MD simulations,108 and the bottom number comes from adiabatic mapping studies.107
PET hydrolases are thought to possess dynamic catalytic sites with aromatic subsites which are locally stabilized upon substrate binding.65,112,116 For example, the conserved W185 in IsPETase was also found in many other known PET hydrolases (Figure 2B).116,117 A substitution of this residue usually resulted in a drastically reduced hydrolytic activity on PET.112,118 Recently, Chen et al. identified that S214 and I218 are uniquely present in IsPETase to reduce the steric hindrance of W185, therefore enabling a higher flexibility of the indole side chain, allowing a higher polymer hydrolysis rate.119 Introduction of the corresponding double-residue substitutions into homologous polyester hydrolases indeed boosted PET hydrolyzing activity, albeit at the expense of stability at temperatures >60 °C.
Large polymer segments may be unable to fit into a binding cleft that is too narrow. The overall catalytic properties of several PET hydrolases can be changed by widening or narrowing the binding clefts.97,120 Interestingly, the free energy barrier and hydrolytic reaction thermodynamics do not differ significantly between the thermophilic LCCIICCG mutant and the mesophilic IsPETase.107 This suggests that the high activity of thermostable PET hydrolases is attributable mostly to the increased accessibility of PET polymer to nucleophilic attack at elevated temperatures, rather than to the fundamental interaction affinity. Austin et al. measured the width of the substrate binding cleft of IsPETase via the residue pair T88/S238.65 By substituting S238 to F, which is highly conserved in homologous cutinase-like PET hydrolases, increased hydrolytic activity on PET was determined when accompanied by the mutation W159H, although the binding cleft width appeared to be narrowed. This phenomenon suggests that the polymer binding in the surface groove is more likely a dynamic rather than static process, as supported by an NMR analysis of PET chain mobility in the context of enzymatic hydrolysis.115,121 The residue equivalent of the IsPETase S238 is F in TfCut2 and LCC.52,111 By substitution to A, I, and W, significantly increased PET hydrolysis activity was reported.16,122 These findings suggest that further systematic iterative engineering of all variable residues in the PET hydrolase binding pocket may be beneficial for gaining a thorough understanding of the rate-limiting interactions with aromatic polyesters and, as a result, improving the overall degradation performance.
Mono(2-hydroxyethyl) terephthalate (MHET) and bis(2-hydroxyethyl) terephthalate (BHET) are degradation intermediates well-known to inhibit the efficiency of depolymerization catalyzed by various PET hydrolases, except for HiC and Cut190.50,78,123,124 MHET has a stronger inhibitory effect and can hardly be hydrolyzed by the wild-type IsPETase.96 Thus, I. sakaiensis produces a distinct MHETase to yield TPA and EG for its growth.34 This serves as a template for developing a dual enzyme system for effective PET degradation containing an oligo-ester-specific helper enzyme.123 Recently, a chimeric dual-enzyme system with MHETase linked to IsPETase resembling their natural microbial host was established, resulting in PET degradation efficiency improved by at least 2.8-fold.125 Additional MHETase-like enzymes from Comamonas thiooxydans and Hydrogenophaga sp. PML113125 and an enzyme from a marine consortium126 with similar structures but considerably lower substrate affinity for MHET than the IsMHETase127 were also identified. This distinction in catalytic characteristics may provide a wide range of choices for combining with other depolymerizing enzymes, particularly for the use in whole cell-based multiple enzyme systems.128,129 While IsPETase has been successfully stabilized, additionally thermostabilizing MHETase-like enzymes would provide a robust two-enzyme system with industrial promise. Other technical solutions have been proposed in this regard, such as employing enzyme mutants that are less susceptible to product inhibition99 or a membrane reactor for continuous removal of the inhibitors.130
4. Engineering Enzymatic De/Adsorption onto PET Surfaces by Including Binding Modules
To resemble other natural polymer degrading enzymes, specific polymer binding modules were either directly added in the reaction mixture or covalently fused to appropriate PET hydrolases. Among these, the cellulose-binding domains (CBM) CenA from Cellulomonas fimi(104) and that of cellobiohydrolase I from Trichoderma reesei,100,131 the polyhydroxyalkanoate (PHA)-binding module of PHA depolymerase from Alcaligenes faecalis,100 and the chitin-binding module from the chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBCH1103 increased the degradation performance when they were fused to the C-termini of PET hydrolases (Table 2). Selected tryptophan residues in CBMs were identified as mutation hot spots for enhanced hydrogen bond formation with aromatic moieties at the PET surface.104,132
Table 2. Selected PET Hydrolase Engineering Studies in the Presence of Binding Modules.
| binding module | enzyme | fold improvement | reaction condition | ref |
|---|---|---|---|---|
| PcAA14A | ThermoPETase | 1.3 | PET granules (3 g/L) incubated with 8.3 mgEnzyme/gPET in glycine-NaOH buffer (pH 9) for 5 days | (137) |
| RolA | IsPETase | 1.2 (by weight loss) | Preincubation with RolA for 3 h, then degradation of PET fiber waste and high-crystallinity PET powder (15 g/L) at pH 8, 30 °C with 1.3 gEnzyme/gPET for 5 days | (136) |
| 1.5 (by HPLC) | ||||
| HGFI | IsPETase | 1.3 (by weight loss) | ||
| 1.6 (by HPLC) | ||||
| Zwitterionic polypeptide (EK) | IsPETase | 11 | Degradation of amorphous Goodfellow PET film and high-crystallinity (45.2%) beverage bottle with 200 nM enzyme in glycine-NaOH-buffer at pH 9 and 30–40 °C for up to 6 days | (138) |
| CBM | A cutinase from T. fusca | 1.4–1.5 (only affinity) | Incubated with PET fiber and 50 μM enzyme in 50 mM Tris-buffer (pH 8) at 50 °C for 24 h | (104) |
| ChBD | LCCICCG | 1.2 | With 0.6 g/L amorphous Goodfellow PET film, post-consumer waste PET (16% crystallinity), or high crystallinity (40%) PET at 65 °C for 12 h | (103) |
| CBM | LCCICCG | 1.3 | ||
| CBM | ThC_Cut1 | 1.4 | With amorphous Goodfellow PET film and 25 mM enzyme in 100 mM potassium phosphate buffer (pH 7) at 50 °C | (100) |
| PBM | ThC_Cut1 | 3.8 |
Hydrophobins are small amphiphilic proteins (about 20 kDa) produced by filamentous fungi with surfactant-like activity.133 Although the highest boosting effect with hydrophobins was discovered when fused to a PET hydrolase, they can already improve the enzymatic binding affinity to the PET surface when introduced to the degradation mixtures as free molecules or applied to material as a pretreatment.134−136
In this regard, the lytic polysaccharide monooxygenase (LPMO) from the white-rot fungus Pycnoporus coccineus (PcAA14A)137 and a zwitterionic polymer based on positively charged lysine and negatively charged glutamate138 were both recently shown to improve PET degradation performance when added to IsPETase. LPMO and the so-called EKylation, on the other hand, appear to promote the enzymatic degradation via different mechanisms than hydrophobins or other binding modules.
While the necessity of binding modules for effective cellulase-catalyzed cellulose degradation has been questioned,139 their presence was recently found to alter the binding and hydrolysis kinetics at various solid loading levels.140 The CBM was shown to favor low solid loading level in an adsorption-controlled scenario where higher catalytic efficiency is associated with tighter substrate binding but disfavored at high substrate concentrations, as shown by a so-called “volcano plot” of specific reaction rates against the binding strengths.140 Accordingly, optimal PET hydrolases are suspected to also follow this Sabatier principle and will function at an intermediate substrate binding strength. Future research should be conducted to confirm this hypothesis based on comprehensive binding and hydrolysis kinetic studies with various PET hydrolases, as well as those containing artificially introduced polymer binding modules, to clarify their roles for the interfacial biocatalysis, which is obviously missing so far. This is of great interest for potential industrial applications, where a very high solid loading level is generally preferred, in terms of an economical use of biocatalysts.
5. Concluding Remarks and Future Perspectives
More than 16 years have elapsed since the first microbial PET hydrolase was reported. Many research efforts have been expended in the quest for new PET hydrolases as well as in the optimization of known enzymes. A large number of high-quality studies have been conducted using a few benchmark enzymes, such as cutinase(-like) enzymes from Thermobifida species,141 the plant compost-derived LCC cutinase, and IsPETase. Different research groups employed varying experimental conditions, such as pH, buffer compositions (different salts) and concentrations (different ionic strengths), and agitation techniques and speeds, making a balanced comparison of the degradation data very challenging, even when the same benchmark enzyme was used. Selected experimental parameters can also have a significant impact on the degradation performance as well as the related data interpretation in terms of interfacial biocatalysis mechanisms. For example, buffer salts can influence the enzyme performance142 and the solubility of degradation products or inhibitors,143 and they can also fundamentally modify the water interaction with polymeric substrates.144 When simultaneous mechanical treatment is occurring, the ideal water volume for enzymatic PET degradation can vary dramatically.44 More crucially, studies employed PET materials from various suppliers, with varying crystallinity (even that of the widely used amorphous Goodfellow PET film may differ from batch to batch) and in various forms (powder, foil, sheet, etc.). As a result, standardization of PET hydrolysis tests and conditions using polymer samples with defined material sources and properties should be applied in future investigations to allow for a straightforward data comparison. Several recent studies used manually picked post-consumer low-crystallinity PET packaging in order to achieve striking enzymatic degradation performance at a small laboratory scale.88,145 However, in a real-world scenario, post-consumer PET packaging with all crystallinity patterns ends up in mixed household plastic waste which is collected and then separated in an industrial plastic sorting system according to polymer type and color, but not crystallinity.146,147 To remove the contaminants which may hamper the subsequent recycling process, a washing step (e.g., with hot soda water) is usually applied prior to thermomechanical reprocessing (e.g., shredding and drying).148 These processes will age the sorted waste polymer considerably and consequently eliminate the low-crystallinity fraction of PET. Following additional amorphization and micronization steps, these waste plastic flakes can be readily used for biotechnological recycling.16,92 Therefore, for lab-scale research in the field of PET hydrolases, using equally pretreated real-world PET waste (e.g., with the same amorphization and micronization equipment and protocols) instead of biased selection of a low-crystallinity fraction will deliver more reliable standardized data on the enzymatic degradation performance of industrial relevance.
As an alternative to the energy-consuming amorphization step,92 the challenge in lowering the polymer crystallinity is envisaged to be possibly solved with a specific crystalline PET depolymerase, like those found in natural cellulase systems to decrystallize cellulose.149 To this end, continuously growing PET hydrolase sequence data from large-scale sequencing projects can be used for exploring this missing enzyme activity and other novel PET hydrolases in genomic and metagenomic databases. EnzymeMiner and other available in silico tools provide useful computational platforms for automated identification of promising enzyme candidates for experimental validation.150 Pan-genome analysis focusing on microbes which produce known PET hydrolases, e.g., in the Thermobifida and Ideonella genera, could facilitate finding yet-unknown MHETase-like enzymes to address the product-inhibition problem. Nevertheless, as the substrates of identified PET hydrolases and carbon sources for the host strains are unlikely to be synthetic polyester in their natural habitats, the probability of success of such a research activity is hard to predict. The explored sequence space can also be used to predict enzymes with improved properties by machine learning and ancestral sequence reconstruction.151−153 Robust proteins can be designed by sequence-based approaches and fully automated computational workflows, like FireProtASR, making complex protocols of ancestral inference accessible to nonexperts.154 Recent developments of highly accurate protein structure prediction methods, like AlphaFold2 and RoseTTAFold, can assist a structure-based design of novel PET hydrolases identified by database searches.155−157 While hydrolytic biocatalysis might fundamentally be unable to crack the well-ordered crystalline PET, oxidative enzymes, which may have a mechanistically comparable function to LPMOs known from lignin and cellulose degrading systems,158 can offer further options for mining for polyester decrystallizing activities.
High-quality structures incorporating PET relevant ligands will be required for further understanding of the structure–function relationship and related engineering of PET hydrolases. Application of novel cryoEM techniques to examine the binding of bigger polymer substrates may be required for a thorough understanding of the substrate–enzyme interaction at the solid–liquid interface. To date, these mechanistic insights have primarily been derived from docking and simulation experiments using flexible PET oligomers, which overlook the inherent mobility and steric constraints given by neighboring polymers under realistic reaction conditions. The utilization of particular force fields designed for interface reactions, as well as taking into account real-world material properties, will help in silico protein engineering even more.159 The calculated energy barriers for PET hydrolysis are balanced for individual reaction stages (Figure 3), implying that more than one catalytic step would need to be enhanced at the same time. This is typically difficult to address rationally, providing room for directed evolution and machine learning. Hence, the future development of suitable screening assays applicable to large enzyme libraries is of particular relevance in this regard. Due to technical limitations of individual approaches, achieving ultra-high-throughput screening specifically for PET degradation activity still remains a challenge (Figure 4).
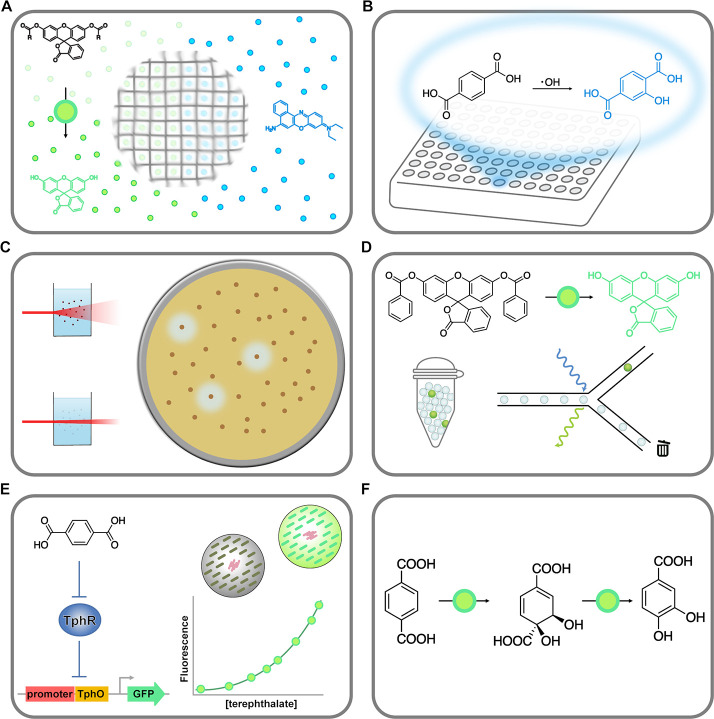
Figure 4.
Potential PET hydrolase screening methods arranged in order of increasing potential throughput. (A) Fluorogenic substrates like fluorescein dilaurate can be trapped in polyester films or particles175,176 and can be released and hydrolyzed upon polymer hydrolysis, generating a fluorescence signal. (B) Fluorimetric method based on the reaction of terephthalic acid with hydroxyl radicals to form the fluorophore 2-hydroxyterephthalic acid.177−179 Tens of thousands of clones can be screened using microtiter plate-based assays (A, B). (C) Agar plate assay based on the hydrolysis of polyester (PET) nanoparticles.75,180,181 Clear zones (halos) form around clones expressing active polyester-hydrolyzing enzymes, allowing simple visual identification. Millions of clones can easily be screened using this method. (D) Recently reported ultra-high-throughput droplet-based assay for PETase activity.182 The use of the fluorogenic surrogate substrate fluorescein dibenzoate indicates a low selectivity, since many other esterases would also be identified using this assay. Tens of millions of clones could be analyzed using this method. Combinations of the turbidimetric assay (C) and droplet-based methods (D) seem promising. (E) Ultra-high-throughput assay based on a terephthalic acid biosensor.183 Cells could be entrapped in hydrogel beads184 containing reporter cells that express GFP in response to terephthalic acid formed by clones expressing active PET-hydrolyzing enzymes. Because fluorescence-activated cell sorting (FACS) can be used to sort the beads, the throughput of this method is potentially in the hundreds of millions. (F) Envisaged growth selection approach based on the conversion of terephthalic acid to protocatechuic acid, which could be catabolized by engineered strains of E. coli or other model organisms.183,185 The throughput of this method would be limited only by library size and transformation efficiencies, making it one of the most attractive methods.
Furthermore, researchers may investigate the power and specific advantages of using biocatalysts over chemical catalysts, such as when combined with living cells as whole-cell catalysts, which can potentially allow for a one-pot process that simultaneously includes an enzyme cascade not only to degrade the plastics but also to convert the degradation intermediates into a variety of products with added value.32 The utility of mesophilic enzymes can potentially be reemphasized using such a procedure, because numerous metabolic pathways converting monomers to other value-added compounds are currently only available/engineerable in mesophilic microbial frameworks. Aside from the well-engineered frameworks such as Pseudomonas putida, E. coli, and P. pastoris,160−164I. sakaiensis has recently emerged as a new promising strain for genomic engineering with considerably high conversion efficiency from PET substrates to PHA biopolymers.37,129 On the other hand, thermophilic whole-cell biocatalysts such as Clostridium thermocellum can be easily employed for faster PET depolymerization, for example, targeting textile waste also containing cellulose, which may be degraded and valorized simultaneously.21,165 For more details regarding the biotechnological potential of enzymatic textile recycling, readers are referred to a recent comprehensive review by Jönsson et al.21 In this regard, designing the PET degrading multienzyme complex by combining the advantages of various enzyme classes to resemble an architecture similar to the natural cellulosome might be a viable option.128
A photosynthetic microalga, Phaeodactylum tricornutum, has been engineered to functionally express IsPETase and is able to degrade selected PET-related materials in a saltwater environment at 21–30 °C.166 This has been suggested to be a potential bioremediation approach for seawater polluted by PET microplastics, although the conversion rate was very low and the application of genetically engineered microorganisms in an open environment is currently strictly forbidden worldwide.9 Although a recent study has reported wild-type marine microbes with PET-metabolizing activity,167 microplastics present in aquatic ecosystems can hardly support microbial growth and will therefore remain as persistent xenobiotics that cannot be remediated easily.168
Scientists have thus far succeeded in identifying and creating biocatalysts that appear to fit the need for PET waste depolymerization on an industrial and commercial scale. The fundamental understanding of interfacial biocatalysis on PET should be extended and transferred to address the challenges associated with the biotechnological degradation of other more abundant plastics such as polyolefins or more similar plastics such as polyamides (PA) and polyurethanes (PUR) with hydrolyzable backbones.21,32,169 While the breakdown of carbon–carbon backbones in polyolefins can be energetically very challenging,170,171 chemical and thermal pretreatments have been shown to enable subsequent biochemical transformation172,173 and thus should be extensively studied in future research. On the other hand, the identification of putative PUR or PA hydrolases is envisioned as a result of collaborative large-scale research activities (e.g., the MIX-UP project174 and the upPE-T project) on plastic recycling. A combination of these novel biocatalysts will pave a new path for the valorization of unsorted mixed plastic waste that cannot be efficiently recycled via other disposal approaches at an industrial scale.
Acknowledgments
The authors gratefully acknowledge the financial support received from the European Union’s Horizon 2020 research and innovation program (MIX-UP, grant number 870294; upPE-T, grant number 953214; CETOCOEN Excellence, grant number 857560), from the Czech Ministry of Education (CZ.02.1.01/0.0/0.0/16_026/0008451) and the Alexander von Humboldt foundation.
The authors declare no competing financial interest.
References
- Brown B. S.; Mills J.; Hulse J. M. Chemical and Biological Degradation of Waste Plastics. Nature 1974, 250, 161–163. 10.1038/250161a0. [DOI] [PubMed] [Google Scholar]
- Wong C. S.; Green D. R.; Cretney W. J. Quantitative Tar and Plastic Waste Distributions in the Pacific Ocean. Nature 1974, 247, 30–32. 10.1038/247030a0. [DOI] [Google Scholar]
- Borrelle S. B.; Ringma J.; Law K. L.; Monnahan C. C.; Lebreton L.; McGivern A.; Murphy E.; Jambeck J.; Leonard G. H.; Hilleary M. A.; Eriksen M.; Possingham H. P.; Frond H. D.; Gerber L. R.; Polidoro B.; Tahir A.; Bernard M.; Mallos N.; Barnes M.; Rochman C. M. Predicted Growth in Plastic Waste Exceeds Efforts to Mitigate Plastic Pollution. Science 2020, 369, 1515–1518. 10.1126/science.aba3656. [DOI] [PubMed] [Google Scholar]
- MacLeod M.; Arp H. P. H.; Tekman M. B.; Jahnke A. The Global Threat from Plastic Pollution. Science 2021, 373, 61–65. 10.1126/science.abg5433. [DOI] [PubMed] [Google Scholar]
- Meys R.; Kätelhön A.; Bachmann M.; Winter B.; Zibunas C.; Suh S.; Bardow A. Achieving Net-Zero Greenhouse Gas Emission Plastics by a Circular Carbon Economy. Science 2021, 374, 71–76. 10.1126/science.abg9853. [DOI] [PubMed] [Google Scholar]
- Ellis L. D.; Rorrer N. A.; Sullivan K. P.; Otto M.; McGeehan J. E.; Román-Leshkov Y.; Wierckx N.; Beckham G. T. Chemical and Biological Catalysis for Plastics Recycling and Upcycling. Nat. Catal. 2021, 4, 539–556. 10.1038/s41929-021-00648-4. [DOI] [Google Scholar]
- Kakadellis S.; Rosetto G. Achieving a Circular Bioeconomy for Plastics. Science 2021, 373, 49–50. 10.1126/science.abj3476. [DOI] [PubMed] [Google Scholar]
- Coates G. W.; Getzler Y. D. Y. L. Chemical Recycling to Monomer for an Ideal, Circular Polymer Economy. Nat. Rev. Mater. 2020, 5, 501–516. 10.1038/s41578-020-0190-4. [DOI] [Google Scholar]
- Wei R.; Tiso T.; Bertling J.; O’Connor K.; Blank L. M.; Bornscheuer U. T. Possibilities and Limitations of Biotechnological Plastic Degradation and Recycling. Nat. Catal. 2020, 3, 867–871. 10.1038/s41929-020-00521-w. [DOI] [Google Scholar]
- Cornwall W. The Plastic Eaters. Science 2021, 373, 36–39. 10.1126/science.373.6550.36. [DOI] [PubMed] [Google Scholar]
- Geyer R.; Jambeck J. R.; Law K. L. Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 2017, 3, e1700782 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderthal H.; Tai S. L.; Harrison S. T. L. Non-Hydrolyzable Plastics - an Interdisciplinary Look at Plastic Bio-Oxidation. Trends Biotechnol. 2021, 39, 12–23. 10.1016/j.tibtech.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Li Z.; Wei R.; Gao M.; Ren Y.; Yu B.; Nie K.; Xu H.; Liu L. Biodegradation of Low-Density Polyethylene by Microbulbifer hydrolyticus IRE-31. J. Environ. Manage. 2020, 263, 110402. 10.1016/j.jenvman.2020.110402. [DOI] [PubMed] [Google Scholar]
- Yoon M.-Y.; Kellis J.; Poulose A. J. Enzymatic Modification of Polyester. AATCC Review 2002, 2, 33–36. [Google Scholar]
- Wei R.; Zimmermann W. Biocatalysis as a Green Route for Recycling the Recalcitrant Plastic Polyethylene Terephthalate. Microb. Biotechnol. 2017, 10, 1302–1307. 10.1111/1751-7915.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier V.; Topham C. M.; Gilles A.; David B.; Folgoas C.; Moya-Leclair E.; Kamionka E.; Desrousseaux M. L.; Texier H.; Gavalda S.; Cot M.; Guémard E.; Dalibey M.; Nomme J.; Cioci G.; Barbe S.; Chateau M.; André I.; Duquesne S.; Marty A. An Engineered PET Depolymerase to Break Down and Recycle Plastic Bottles. Nature 2020, 580, 216–219. 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Kawai F. Emerging Strategies in Polyethylene Terephthalate Hydrolase Research for Biorecycling. ChemSusChem 2021, 14, 4115–4122. 10.1002/cssc.202100740. [DOI] [PubMed] [Google Scholar]
- Jog J. P. Crystallization of Polyethyleneterephthalate. J. Macromol. Sci. Part C 1995, 35, 531–553. 10.1080/15321799508014598. [DOI] [Google Scholar]
- Xin J.; Zhang Q.; Huang J.; Huang R.; Jaffery Q. Z.; Yan D.; Zhou Q.; Xu J.; Lu X. Progress in the Catalytic Glycolysis of Polyethylene Terephthalate. J. Environ. Manage. 2021, 296, 113267. 10.1016/j.jenvman.2021.113267. [DOI] [PubMed] [Google Scholar]
- Welle F. Twenty Years of PET Bottle to Bottle Recycling—an Overview. Resour. Conserv. Recycl. 2011, 55, 865–875. 10.1016/j.resconrec.2011.04.009. [DOI] [Google Scholar]
- Jönsson C.; Wei R.; Biundo A.; Landberg J.; Schwarz Bour L.; Pezzotti F.; Toca A.; M. Jacques L.; Bornscheuer U. T.; Syrén P.-O. Biocatalysis in the Recycling Landscape for Synthetic Polymers and Plastics Towards Circular Textiles. ChemSusChem 2021, 14, 4028–4040. 10.1002/cssc.202002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S. R.; Rorrer N. A.; Carpenter A. C.; Beckham G. T. Manufacturing Energy and Greenhouse Gas Emissions Associated with Plastics Consumption. Joule 2021, 5, 673–686. 10.1016/j.joule.2020.12.027. [DOI] [Google Scholar]
- Karacan I. An in Depth Study of Crystallinity, Crystallite Size and Orientation Measurements of a Selection of Poly(Ethylene Terephthalate)Fibers. Fibers Polym. 2005, 6, 186–199. 10.1007/BF02875642. [DOI] [Google Scholar]
- Bashir Z.; Al-Aloush I.; Al-Raqibah I.; Ibrahim M. Evaluation of Three Methods for the Measurement of Crystallinity of PET Resins, Preforms, and Bottles. Polym. Eng. Sci. 2000, 40, 2442–2455. 10.1002/pen.11376. [DOI] [Google Scholar]
- Webb H. K.; Arnott J.; Crawford R. J.; Ivanova E. P. Plastic Degradation and Its Environmental Implications with Special Reference to Poly(Ethylene Terephthalate). Polymers 2013, 5, 1–18. 10.3390/polym5010001. [DOI] [Google Scholar]
- Suaria G.; Achtypi A.; Perold V.; Lee J. R.; Pierucci A.; Bornman T. G.; Aliani S.; Ryan P. G. Microfibers in Oceanic Surface Waters: A Global Characterization. Sci. Adv. 2020, 6, eaay8493 10.1126/sciadv.aay8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Wei R.; Luo W.; Hu L.; Li B.; Di Y.; Shi H. Microplastic Pollution in Water and Sediment in a Textile Industrial Area. Environ. Pollut. 2020, 258, 113658. 10.1016/j.envpol.2019.113658. [DOI] [PubMed] [Google Scholar]
- Worm B.; Lotze H. K.; Jubinville I.; Wilcox C.; Jambeck J. Plastic as a Persistent Marine Pollutant. Annu. Rev. Environ. Resour. 2017, 42, 1–26. 10.1146/annurev-environ-102016-060700. [DOI] [Google Scholar]
- Tokiwa Y.; Suzuki T. Hydrolysis of Polyesters by Lipases. Nature 1977, 270, 76–78. 10.1038/270076a0. [DOI] [PubMed] [Google Scholar]
- Sato M. Deterioration of Filaments and Films of Polyethyleneterephthalate with Enzyme of Cladosporium Cladosporioides FERM J-8. Sen-i Gakkaishi 1983, 39, 67–77. 10.2115/fiber.39.5_T209. [DOI] [Google Scholar]
- Mueller R.-J.; Schrader H.; Profe J.; Dresler K.; Deckwer W.-D. Enzymatic Degradation of Poly(Ethylene Terephthalate): Rapid Hydrolyse Using a Hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. 10.1002/marc.200500410. [DOI] [Google Scholar]
- Tiso T.; Winter B.; Wei R.; Hee J.; de Witt J.; Wierckx N.; Quicker P.; Bornscheuer U. T.; Bardow A.; Nogales J.; Blank L. M. The Metabolic Potential of Plastics as Biotechnological Carbon Sources – Review and Targets for the Future. Metab. Eng. 2021, 1. 10.1016/j.ymben.2021.12.006. [DOI] [PubMed] [Google Scholar]
- Sulaiman S.; Yamato S.; Kanaya E.; Kim J. J.; Koga Y.; Takano K.; Kanaya S. Isolation of a Novel Cutinase Homolog with Polyethylene Terephthalate-Degrading Activity from Leaf-Branch Compost by Using a Metagenomic Approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. 10.1128/AEM.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S.; Hiraga K.; Takehana T.; Taniguchi I.; Yamaji H.; Maeda Y.; Toyohara K.; Miyamoto K.; Kimura Y.; Oda K. A Bacterium That Degrades and Assimilates Poly(Ethylene Terephthalate). Science 2016, 351, 1196–1199. 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- Kan Y.; He L.; Luo Y.; Bao R. IsPETase Is a Novel Biocatalyst for Poly(Ethylene Terephthalate) (PET) Hydrolysis. ChemBioChem. 2021, 22, 1706–1716. 10.1002/cbic.202000767. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U. T. Feeding on Plastic. Science 2016, 351, 1154–1155. 10.1126/science.aaf2853. [DOI] [PubMed] [Google Scholar]
- Fujiwara R.; Sanuki R.; Ajiro H.; Fukui T.; Yoshida S. Direct Fermentative Conversion of Poly(Ethylene Terephthalate) into Poly(Hydroxyalkanoate) by Ideonella sakaiensis. Sci. Rep. 2021, 11, 19991. 10.1038/s41598-021-99528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F.; Kawabata T.; Oda M. Current State and Perspectives Related to the Polyethylene Terephthalate Hydrolases Available for Biorecycling. ACS Sustain. Chem. Eng. 2020, 8, 8894–8908. 10.1021/acssuschemeng.0c01638. [DOI] [Google Scholar]
- Wei Y.; Swenson L.; Castro C.; Derewenda U.; Minor W.; Arai H.; Aoki J.; Inoue K.; Servin-Gonzalez L.; Derewenda Z. S. Structure of a Microbial Homologue of Mammalian Platelet-Activating Factor Acetylhydrolases: Streptomyces exfoliatus Lipase at 1.9 Å Resolution. Structure 1998, 6, 511–519. 10.1016/S0969-2126(98)00052-5. [DOI] [PubMed] [Google Scholar]
- Carr P. D.; Ollis D. L. Alpha/Beta Hydrolase Fold: An Update. Protein Pept. Lett. 2009, 16, 1137–1148. 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- Chen S.; Su L.; Chen J.; Wu J. Cutinase: Characteristics, Preparation, and Application. Biotechnol. Adv. 2013, 31, 1754–1767. 10.1016/j.biotechadv.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Ronkvist Ã. S. M.; Xie W.; Lu W.; Gross R. A. Cutinase-Catalyzed Hydrolysis of Poly(Ethylene Terephthalate). Macromolecules 2009, 42, 5128–5138. 10.1021/ma9005318. [DOI] [Google Scholar]
- de Castro A. M.; Carniel A.; Nicomedes Junior J.; da Conceição Gomes A.; Valoni É. Screening of Commercial Enzymes for Poly(Ethylene Terephthalate) (PET) Hydrolysis and Synergy Studies on Different Substrate Sources. J. Ind. Microbiol. Biotechnol. 2017, 44, 835–844. 10.1007/s10295-017-1942-z. [DOI] [PubMed] [Google Scholar]
- Kaabel S.; Therien J. P. D.; Deschênes C. E.; Duncan D.; Friščić T.; Auclair K. Enzymatic Depolymerization of Highly Crystalline Polyethylene Terephthalate Enabled in Moist-Solid Reaction Mixtures. Proc. Nat. Acad. Sci. U.S.A. 2021, 118, e2026452118 10.1073/pnas.2026452118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel A.; Gomes A. d. C.; Coelho M. A. Z.; de Castro A. M. Process Strategies to Improve Biocatalytic Depolymerization of Post-Consumer PET Packages in Bioreactors, and Investigation on Consumables Cost Reduction. Bioprocess Biosyst. Eng. 2021, 44, 507–516. 10.1007/s00449-020-02461-y. [DOI] [PubMed] [Google Scholar]
- Mueller R.-J. Biological Degradation of Synthetic Polyesters--Enzymes as Potential Catalysts for Polyester Recycling. Process Biochem. 2006, 41, 2124–2128. 10.1016/j.procbio.2006.05.018. [DOI] [Google Scholar]
- Maurya A.; Bhattacharya A.; Khare S. K. Enzymatic Remediation of Polyethylene Terephthalate (PET)–Based Polymers for Effective Management of Plastic Wastes: An Overview. Front. Bioeng. Biotechnol. 2020, 8, 602325. 10.3389/fbioe.2020.602325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S.; Ohno M.; Hidaka M.; Nakamura A.; Masaki H.; Uozumi T. Correlation between Cellulose Binding and Activity of Cellulose-Binding Domain Mutants of Humicola grisea Cellobiohydrolase 1. FEBS Lett. 2007, 581, 5891–5896. 10.1016/j.febslet.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Hsieh C.-W. C.; Cannella D.; Jørgensen H.; Felby C.; Thygesen L. G. Cellulase Inhibition by High Concentrations of Monosaccharides. J. Agric. Food Chem. 2014, 62, 3800–3805. 10.1021/jf5012962. [DOI] [PubMed] [Google Scholar]
- Barth M.; Oeser T.; Wei R.; Then J.; Schmidt J.; Zimmermann W. Effect of Hydrolysis Products on the Enzymatic Degradation of Polyethylene Terephthalate Nanoparticles by a Polyester Hydrolase from Thermobifida fusca. Biochem. Eng. J. 2015, 93, 222–228. 10.1016/j.bej.2014.10.012. [DOI] [Google Scholar]
- Carniel A.; Waldow V. d. A.; Castro A. M. d. A Comprehensive and Critical Review on Key Elements to Implement Enzymatic PET Depolymerization for Recycling Purposes. Biotechnol. Adv. 2021, 52, 107811. 10.1016/j.biotechadv.2021.107811. [DOI] [PubMed] [Google Scholar]
- Sulaiman S.; You D. J.; Kanaya E.; Koga Y.; Kanaya S. Crystal Structure and Thermodynamic and Kinetic Stability of Metagenome-Derived LC-Cutinase. Biochemistry 2014, 53, 1858–1869. 10.1021/bi401561p. [DOI] [PubMed] [Google Scholar]
- Wei R.; Breite D.; Song C.; Gräsing D.; Ploss T.; Hille P.; Schwerdtfeger R.; Matysik J.; Schulze A.; Zimmermann W. Biocatalytic Degradation Efficiency of Postconsumer Polyethylene Terephthalate Packaging Determined by Their Polymer Microstructures. Adv. Sci. 2019, 6, 1900491. 10.1002/advs.201900491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H. F.; Cho I. J.; Joo S.; Seo H.; Sagong H.-Y.; Choi S. Y.; Lee S. Y.; Kim K.-J. Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019, 9, 3519–3526. 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- Cui Y.; Chen Y.; Liu X.; Dong S.; Tian Y. e.; Qiao Y.; Mitra R.; Han J.; Li C.; Han X.; Liu W.; Chen Q.; Wei W.; Wang X.; Du W.; Tang S.; Xiang H.; Liu H.; Liang Y.; Houk K. N.; Wu B. Computational Redesign of a PETase for Plastic Biodegradation under Ambient Condition by the GRAPE Strategy. ACS Catal. 2021, 11, 1340–1350. 10.1021/acscatal.0c05126. [DOI] [Google Scholar]
- Brott S.; Pfaff L.; Schuricht J.; Schwarz J.-N.; Böttcher D.; Badenhorst C. P. S.; Wei R.; Bornscheuer U. T. Engineering and Evaluation of Thermostable IsPETase Variants for PET Degradation. Eng. Life Sci. 2021, 1. 10.1002/elsc.202100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt U.; Yamamoto M.; Seeliger U.; Müller R.-J.; Warzelhan V. Biodegradable Polymeric Materials—Not the Origin but the Chemical Structure Determines Biodegradability. Angew. Chem., Int. Ed. 1999, 38, 1438–1442. . [DOI] [PubMed] [Google Scholar]
- Marten E.; Müller R.-J.; Deckwer W.-D. Studies on the Enzymatic Hydrolysis of Polyesters. II. Aliphatic-Aromatic Copolyesters. Polym. Degrad. Stab. 2005, 88, 371–381. 10.1016/j.polymdegradstab.2004.12.001. [DOI] [Google Scholar]
- Keller A.; Lester G. R.; Morgan L. B. Crystallization Phenomena in Polymers. I. Preliminary Investigation of the Crystallization Characteristics of Polyethylene Terephthalate. Philos. Trans. Royal Soc. A 1954, 247, 1–12. [Google Scholar]
- Langevin D.; Grenet J.; Saiter J. M. Moisture Sorption in PET Influence on the Thermokinetic Parameters. Eur. Polym. J. 1994, 30, 339–345. 10.1016/0014-3057(94)90297-6. [DOI] [Google Scholar]
- Alves N. M.; Mano J. F.; Balaguer E.; Meseguer Dueñas J. M.; Gómez Ribelles J. L. Glass Transition and Structural Relaxation in Semi-Crystalline Poly(Ethylene Terephthalate): A DSC Study. Polymer 2002, 43, 4111–4122. 10.1016/S0032-3861(02)00236-7. [DOI] [Google Scholar]
- Wellen R. M. R.; Canedo E.; Rabello M. S. Nonisothermal Cold Crystallization of Poly(Ethylene Terephthalate). J. Mater. Res. 2011, 26, 1107–1115. 10.1557/jmr.2011.44. [DOI] [Google Scholar]
- Wei R.; Oeser T.; Barth M.; Weigl N.; Lübs A.; Schulz-Siegmund M.; Hacker M.; Zimmermann W. Turbidimetric Analysis of the Enzymatic Hydrolysis of Polyethylene Terephthalate Nanoparticles. J. Mol. Catal. B-Enzym. 2014, 103, 72–78. 10.1016/j.molcatb.2013.08.010. [DOI] [Google Scholar]
- Gamerith C.; Zartl B.; Pellis A.; Guillamot F.; Marty A.; Acero E. H.; Guebitz G. M. Enzymatic Recovery of Polyester Building Blocks from Polymer Blends. Process Biochem. 2017, 59, 58–64. 10.1016/j.procbio.2017.01.004. [DOI] [Google Scholar]
- Austin H. P.; Allen M. D.; Donohoe B. S.; Rorrer N. A.; Kearns F. L.; Silveira R. L.; Pollard B. C.; Dominick G.; Duman R.; El Omari K.; Mykhaylyk V.; Wagner A.; Michener W. E.; Amore A.; Skaf M. S.; Crowley M. F.; Thorne A. W.; Johnson C. W.; Woodcock H. L.; McGeehan J. E.; Beckham G. T. Characterization and Engineering of a Plastic-Degrading Aromatic Polyesterase. Proc. Nat. Acad. Sci. U.S.A. 2018, 115, E4350–E4357. 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E.; Kobayashi Y. Effects of Absorbed Water on Physical Properties of Polyesters. J. Appl. Polym. Sci. 1980, 25, 2145–2157. 10.1002/app.1980.070251001. [DOI] [Google Scholar]
- Jabarin S. A.; Lofgren E. A. Effects of Water Absorption on Physical Properties and Degree of Molecular Orientation of Poly (Ethylene Terephthalate). Polym. Eng. Sci. 1986, 26, 620–625. 10.1002/pen.760260907. [DOI] [Google Scholar]
- Bianchi R.; Chiavacci P.; Vosa R.; Guerra G. Effect of Moisture on the Crystallization Behavior of PET from the Quenched Amorphous Phase. J. Appl. Polym. Sci. 1991, 43, 1087–1089. 10.1002/app.1991.070430608. [DOI] [Google Scholar]
- Launay A.; Thominette F.; Verdu J. Water Sorption in Amorphous Poly(Ethylene Terephthalate). J. Appl. Polym. Sci. 1999, 73, 1131–1137. . [DOI] [Google Scholar]
- Shinotsuka K.; Bliznyuk V. N.; Assender H. E. Near-Surface Crystallization of PET. Polymer 2012, 53, 5554–5559. 10.1016/j.polymer.2012.09.048. [DOI] [Google Scholar]
- Zuo B.; Liu Y.; Liang Y.; Kawaguchi D.; Tanaka K.; Wang X. Glass Transition Behavior in Thin Polymer Films Covered with a Surface Crystalline Layer. Macromolecules 2017, 50, 2061–2068. 10.1021/acs.macromol.6b02740. [DOI] [Google Scholar]
- Falkenstein P.; Gräsing D.; Bielytskyi P.; Zimmermann W.; Matysik J.; Wei R.; Song C. UV Pretreatment Impairs the Enzymatic Degradation of Polyethylene Terephthalate. Front. Microbiol. 2020, 11, 1. 10.3389/fmicb.2020.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J.; Poultney C.; Liu Z.; Gross R.; Montclare J. K. Identification and Comparison of Cutinases for Synthetic Polyester Degradation. Appl. Microbiol. Biotechnol. 2012, 93, 229–240. 10.1007/s00253-011-3402-4. [DOI] [PubMed] [Google Scholar]
- Kawai F. The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts 2021, 11, 206. 10.3390/catal11020206. [DOI] [Google Scholar]
- Danso D.; Schmeisser C.; Chow J.; Zimmermann W.; Wei R.; Leggewie C.; Li X.; Hazen T.; Streit W. R. New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-02717 10.1128/AEM.02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T.; Mizushima H.; Ohtsuka J.; Oda M.; Kawai F.; Tanokura M. Structural Basis for the Ca2+-Enhanced Thermostability and Activity of PET-Degrading Cutinase-Like Enzyme from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2015, 99, 4297–4307. 10.1007/s00253-014-6272-8. [DOI] [PubMed] [Google Scholar]
- Then J.; Wei R.; Oeser T.; Gerdts A.; Schmidt J.; Barth M.; Zimmermann W. A Disulfide Bridge in the Calcium Binding Site of a Polyester Hydrolase Increases Its Thermal Stability and Activity against Polyethylene Terephthalate. FEBS Open Bio 2016, 6, 425–432. 10.1002/2211-5463.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M.; Yamagami Y.; Inaba S.; Oida T.; Yamamoto M.; Kitajima S.; Kawai F. Enzymatic Hydrolysis of PET: Functional Roles of Three Ca2+ Ions Bound to a Cutinase-Like Enzyme, Cut190*, and Its Engineering for Improved Activity. Appl. Microbiol. Biotechnol. 2018, 102, 10067–10077. 10.1007/s00253-018-9374-x. [DOI] [PubMed] [Google Scholar]
- Then J.; Wei R.; Oeser T.; Barth M.; Belisário-Ferrari M. R.; Schmidt J.; Zimmermann W. Ca2+ and Mg2+ Binding Site Engineering Increases the Degradation of Polyethylene Terephthalate Films by Polyester Hydrolases from Thermobifida fusca. Biotechnol. J. 2015, 10, 592–598. 10.1002/biot.201400620. [DOI] [PubMed] [Google Scholar]
- Numoto N.; Kamiya N.; Bekker G.-J.; Yamagami Y.; Inaba S.; Ishii K.; Uchiyama S.; Kawai F.; Ito N.; Oda M. Structural Dynamics of the PET-Degrading Cutinase-Like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle. Biochemistry 2018, 57, 5289–5300. 10.1021/acs.biochem.8b00624. [DOI] [PubMed] [Google Scholar]
- Emori M.; Numoto N.; Senga A.; Bekker G.-J.; Kamiya N.; Kobayashi Y.; Ito N.; Kawai F.; Oda M. Structural Basis of Mutants of PET-Degrading Enzyme from Saccharomonospora viridis AHK190 with High Activity and Thermal Stability. Proteins 2021, 89, 502–511. 10.1002/prot.26034. [DOI] [PubMed] [Google Scholar]
- Baker P.; Grossman R. F. Properties and Reactions of Metal Terephthalates. J. Vinyl Technol. 1989, 11, 59–61. 10.1002/vnl.730110204. [DOI] [Google Scholar]
- Zhong-Johnson E. Z. L.; Voigt C. A.; Sinskey A. J. An Absorbance Method for Analysis of Enzymatic Degradation Kinetics of Poly(Ethylene Terephthalate) Films. Sci. Rep. 2021, 11, 928. 10.1038/s41598-020-79031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A.; Kobayashi N.; Koga N.; Iino R. Positive Charge Introduction on the Surface of Thermostabilized PET Hydrolase Facilitates PET Binding and Degradation. ACS Catal. 2021, 11, 8550–8564. 10.1021/acscatal.1c01204. [DOI] [Google Scholar]
- Xi X.; Ni K.; Hao H.; Shang Y.; Zhao B.; Qian Z. Secretory Expression in Bacillus subtilis and Biochemical Characterization of a Highly Thermostable Polyethylene Terephthalate Hydrolase from Bacterium HR29. Enzyme Microb. Technol. 2021, 143, 109715. 10.1016/j.enzmictec.2020.109715. [DOI] [PubMed] [Google Scholar]
- Shirke A. N.; White C.; Englaender J. A.; Zwarycz A.; Butterfoss G. L.; Linhardt R. J.; Gross R. A. Stabilizing Leaf and Branch Compost Cutinase (LCC) with Glycosylation: Mechanism and Effect on PET Hydrolysis. Biochemistry 2018, 57, 1190–1200. 10.1021/acs.biochem.7b01189. [DOI] [PubMed] [Google Scholar]
- Fecker T.; Galaz-Davison P.; Engelberger F.; Narui Y.; Sotomayor M.; Parra L. P.; Ramírez-Sarmiento C. A. Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensisPETase. Biophys. J. 2018, 114, 1302–1312. 10.1016/j.bpj.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.; Diaz D. J.; Czarnecki N. J.; Zhu C.; Kim W.; Shroff R.; Acosta D. J.; Alexander B.; Cole H.; Zhang Y. J.; Lynd N.; Ellington A. D.; Alper H. S.. Deep Learning Redesign of PETase for Practical PET Degrading Applications. bioRxiv, 2021, 10.1101/2021.10.10.463845. [DOI] [Google Scholar]
- Bell E.; Smithson R.; Kilbride S.; Foster J.; Hardy F.; Ramachandran S.; Tedstone A.; Haigh S.; Garforth A.; Day P.; Levy C.; Shaver M.; Green A.. Directed Evolution of an Efficient and Thermostable PET Depolymerase. ChemRxiv, 2021, 10.26434/chemrxiv-2021-mcjh6. [DOI] [Google Scholar]
- Erickson E.; Shakespeare T. J.; Bratti F.; Buss B. L.; Graham R.; Hawkins M. A.; König G.; Michener W. E.; Miscall J.; Ramirez K. J.; Rorrer N. A.; Zahn M.; Pickford A. R.; McGeehan J. E.; Beckham G. T. Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation. ChemSusChem 2022, 15, e202101932 10.1002/cssc.202101932. [DOI] [PubMed] [Google Scholar]
- Meng X.; Yang L.; Liu H.; Li Q.; Xu G.; Zhang Y.; Guan F.; Zhang Y.; Zhang W.; Wu N.; Tian J. Protein Engineering of Stable IsPETase for PET Plastic Degradation by Premuse. Int. J. Biol. Macromol. 2021, 180, 667–676. 10.1016/j.ijbiomac.2021.03.058. [DOI] [PubMed] [Google Scholar]
- Singh A.; Rorrer N. A.; Nicholson S. R.; Erickson E.; DesVeaux J. S.; Avelino A. F. T.; Lamers P.; Bhatt A.; Zhang Y.; Avery G.; Tao L.; Pickford A. R.; Carpenter A. C.; McGeehan J. E.; Beckham G. T. Techno-Economic, Life-Cycle, and Socioeconomic Impact Analysis of Enzymatic Recycling of Poly(Ethylene Terephthalate). Joule 2021, 5, 2479–2503. 10.1016/j.joule.2021.06.015. [DOI] [Google Scholar]
- Woodard L. N.; Grunlan M. A. Hydrolytic Degradation and Erosion of Polyester Biomaterials. ACS Macro Lett. 2018, 7, 976–982. 10.1021/acsmacrolett.8b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari J.; Andersen M.; Borch K.; Westh P. An Inverse Michaelis–Menten Approach for Interfacial Enzyme Kinetics. ACS Catal. 2017, 7, 4904–4914. 10.1021/acscatal.7b00838. [DOI] [Google Scholar]
- Vogel K.; Wei R.; Pfaff L.; Breite D.; Al-Fathi H.; Ortmann C.; Estrela-Lopis I.; Venus T.; Schulze A.; Harms H.; Bornscheuer U. T.; Maskow T. Enzymatic Degradation of Polyethylene Terephthalate Nanoplastics Analyzed in Real Time by Isothermal Titration Calorimetry. Sci. Total Environ. 2021, 773, 145111. 10.1016/j.scitotenv.2021.145111. [DOI] [PubMed] [Google Scholar]
- Bååth J. A.; Borch K.; Jensen K.; Brask J.; Westh P. Comparative Biochemistry of Four Polyester (PET) Hydrolases. ChemBioChem. 2021, 22, 1627–1637. 10.1002/cbic.202000793. [DOI] [PubMed] [Google Scholar]
- Silva C.; Da S.; Silva N.; Matama T.; Araujo R.; Martins M.; Chen S.; Chen J.; Wu J.; Casal M.; Cavaco-Paulo A. Engineered Thermobifida fusca Cutinase with Increased Activity on Polyester Substrates. Biotechnol. J. 2011, 6, 1230–1239. 10.1002/biot.201000391. [DOI] [PubMed] [Google Scholar]
- Wei R.; Oeser T.; Then J.; Kühn N.; Barth M.; Schmidt J.; Zimmermann W. Functional Characterization and Structural Modeling of Synthetic Polyester-Degrading Hydrolases from Thermomonospora curvata. AMB Express 2014, 4, 44. 10.1186/s13568-014-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.; Oeser T.; Schmidt J.; Meier R.; Barth M.; Then J.; Zimmermann W. Engineered Bacterial Polyester Hydrolases Efficiently Degrade Polyethylene Terephthalate Due to Relieved Product Inhibition. Biotechnol. Bioeng. 2016, 113, 1658–1665. 10.1002/bit.25941. [DOI] [PubMed] [Google Scholar]
- Ribitsch D.; Yebra A. O.; Zitzenbacher S.; Wu J.; Nowitsch S.; Steinkellner G.; Greimel K.; Doliska A.; Oberdorfer G.; Gruber C. C.; Gruber K.; Schwab H.; Stana-Kleinschek K.; Acero E. H.; Guebitz G. M. Fusion of Binding Domains to Thermobifida cellulosilytica Cutinase to Tune Sorption Characteristics and Enhancing PET Hydrolysis. Biomacromolecules 2013, 14, 1769–1776. 10.1021/bm400140u. [DOI] [PubMed] [Google Scholar]
- Zumstein M. T.; Kohler H.-P. E.; McNeill K.; Sander M. Enzymatic Hydrolysis of Polyester Thin Films: Real-Time Analysis of Film Mass Changes and Dissipation Dynamics. Environ. Sci. Technol. 2016, 50, 197–206. 10.1021/acs.est.5b04103. [DOI] [PubMed] [Google Scholar]
- Weinberger S.; Haernvall K.; Scaini D.; Ghazaryan G.; Zumstein M. T.; Sander M.; Pellis A.; Guebitz G. M. Enzymatic Surface Hydrolysis of Poly(Ethylene Furanoate) Thin Films of Various Crystallinities. Green Chem. 2017, 19, 5381–5384. 10.1039/C7GC02905E. [DOI] [Google Scholar]
- Xue R.; Chen Y.; Rong H.; Wei R.; Cui Z.; Zhou J.; Dong W.; Jiang M. Fusion of Chitin-Binding Domain from Chitinolyticbacter meiyuanensis SYBC-H1 to the Leaf-Branch Compost Cutinase for Enhanced PET Hydrolysis. Front. Bioeng. Biotechnol. 2021, 9, 762854. 10.3389/fbioe.2021.762854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wang L.; Chen J.; Wu J. Enhanced Activity toward PET by Site-Directed Mutagenesis of Thermobifida fusca Cutinase-CBM Fusion Protein. Carbohydr. Polym. 2013, 97, 124–129. 10.1016/j.carbpol.2013.04.042. [DOI] [PubMed] [Google Scholar]
- Bååth J. A.; Novy V.; Carneiro L. V.; Guebitz G. M.; Olsson L.; Westh P.; Ribitsch D. Structure-Function Analysis of Two Closely Related Cutinases from Thermobifida cellulosilytica. Biotechnol. Bioeng. 2022, 119, 470–481. 10.1002/bit.27984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badino S. F.; Bååth J. A.; Borch K.; Jensen K.; Westh P. Adsorption of Enzymes with Hydrolytic Activity on Polyethylene Terephthalate. Enzyme Microb. Technol. 2021, 152, 109937. 10.1016/j.enzmictec.2021.109937. [DOI] [PubMed] [Google Scholar]
- Boneta S.; Arafet K.; Moliner V. QM/MM Study of the Enzymatic Biodegradation Mechanism of Polyethylene Terephthalate. J. Chem. Inf. Model. 2021, 61, 3041–3051. 10.1021/acs.jcim.1c00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.; Yue Y.; Zheng M.; Li Y.; Zhang Q.; Wang W. IsPETase- and IsMHETase-Catalyzed Cascade Degradation Mechanism toward Polyethylene Terephthalate. ACS Sustain. Chem. Eng. 2021, 9, 9823–9832. 10.1021/acssuschemeng.1c02420. [DOI] [Google Scholar]
- Jerves C.; Neves R. P. P.; Ramos M. J.; da Silva S.; Fernandes P. A. Reaction Mechanism of the PET Degrading Enzyme PETase Studied with DFT/MM Molecular Dynamics Simulations. ACS Catal. 2021, 11, 11626–11638. 10.1021/acscatal.1c03700. [DOI] [Google Scholar]
- Zheng M.; Li Y.; Dong W.; Feng S.; Zhang Q.; Wang W. Computational Biotransformation of Polyethylene Terephthalate by Depolymerase: A QM/MM Approach. J. Hazard. Mater. 2022, 423, 127017. 10.1016/j.jhazmat.2021.127017. [DOI] [PubMed] [Google Scholar]
- Roth C.; Wei R.; Oeser T.; Then J.; Foellner C.; Zimmermann W.; Sträter N. Structural and Functional Studies on a Thermostable Polyethylene Therephtalate Degrading Hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. 10.1007/s00253-014-5672-0. [DOI] [PubMed] [Google Scholar]
- Joo S.; Cho I. J.; Seo H.; Son H. F.; Sagong H.-Y.; Shin T. J.; Choi S. Y.; Lee S. Y.; Kim K.-J. Structural Insight into Molecular Mechanism of Poly(Ethylene Terephthalate) Degradation. Nat. Commun. 2018, 9, 382. 10.1038/s41467-018-02881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa C. H. S.; dos Santos A. M.; Alves C. N.; Martí S.; Moliner V.; Santana K.; Lameira J. Assessment of the PETase Conformational Changes Induced by Poly(Ethylene Terephthalate) Binding. Proteins 2021, 89, 1340–1352. 10.1002/prot.26155. [DOI] [PubMed] [Google Scholar]
- Taniguchi I.; Yoshida S.; Hiraga K.; Miyamoto K.; Kimura Y.; Oda K. Biodegradation of PET: Current Status and Application Aspects. ACS Catal. 2019, 9, 4089–4105. 10.1021/acscatal.8b05171. [DOI] [Google Scholar]
- Wei R.; Song C.; Gräsing D.; Schneider T.; Bielytskyi P.; Böttcher D.; Matysik J.; Bornscheuer U. T.; Zimmermann W. Conformational Fitting of a Flexible Oligomeric Substrate Does Not Explain the Enzymatic PET Degradation. Nat. Commun. 2019, 10, 5581. 10.1038/s41467-019-13492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Liu W.; Huang J.-W.; Ma J.; Zheng Y.; Ko T.-P.; Xu L.; Cheng Y.-S.; Chen C.-C.; Guo R.-T. Structural Insight into Catalytic Mechanism of PET Hydrolase. Nat. Commun. 2017, 8, 2106. 10.1038/s41467-017-02255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-C.; Han X.; Ko T.-P.; Liu W.; Guo R.-T. Structural Studies Reveal the Molecular Mechanism of PETase. FEBS J. 2018, 285, 3717–3723. 10.1111/febs.14612. [DOI] [PubMed] [Google Scholar]
- Liu B.; He L.; Wang L.; Li T.; Li C.; Liu H.; Luo Y.; Bao R. Protein Crystallography and Site-Direct Mutagenesis Analysis of the Poly(Ethylene Terephthalate) Hydrolase PETase from Ideonella sakaiensis. ChemBioChem. 2018, 19, 1471–1475. 10.1002/cbic.201800097. [DOI] [PubMed] [Google Scholar]
- Chen C.-C.; Han X.; Li X.; Jiang P.; Niu D.; Ma L.; Liu W.; Li S.; Qu Y.; Hu H.; Min J.; Yang Y.; Zhang L.; Zeng W.; Huang J.-W.; Dai L.; Guo R.-T. General Features to Enhance Enzymatic Activity of Poly(Ethylene Terephthalate) Hydrolysis. Nat. Catal. 2021, 4, 425–430. 10.1038/s41929-021-00616-y. [DOI] [Google Scholar]
- Araujo R.; Silva C.; O’Neill A.; Micaelo N.; Guebitz G.; Soares C. M.; Casal M.; Cavaco-Paulo A. Tailoring Cutinase Activity Towards Polyethylene Terephthalate and Polyamide 6,6 Fibers. J. Biotechnol. 2007, 128, 849–857. 10.1016/j.jbiotec.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Falkenstein P.; Wei R.; Matysik J.; Song C.. Mechanistic Investigation of Enzymatic Degradation of Polyethylene Terephthalate by Nuclear Magnetic Resonance. In Methods in Enzymology, Weber G.; Bornscheuer U. T.; Wei R., Eds.; Academic Press, 2021; Vol. 648, pp 231–252. [DOI] [PubMed] [Google Scholar]
- Furukawa M.; Kawakami N.; Tomizawa A.; Miyamoto K. Efficient Degradation of Poly(Ethylene Terephthalate) with Thermobifida fusca Cutinase Exhibiting Improved Catalytic Activity Generated Using Mutagenesis and Additive-Based Approaches. Sci. Rep. 2019, 9, 16038. 10.1038/s41598-019-52379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth M.; Honak A.; Oeser T.; Wei R.; Belisário-Ferrari M. R.; Then J.; Schmidt J.; Zimmermann W. A Dual Enzyme System Composed of a Polyester Hydrolase and a Carboxylesterase Enhances the Biocatalytic Degradation of Polyethylene Terephthalate Films. Biotechnol. J. 2016, 11, 1082–1087. 10.1002/biot.201600008. [DOI] [PubMed] [Google Scholar]
- Eugenio E. d. Q.; Campisano I. S. P.; de Castro A. M.; Coelho M. A. Z.; Langone M. A. P. Kinetic Modeling of the Post-Consumer Poly(Ethylene Terephthalate) Hydrolysis Catalyzed by Cutinase from Humicola insolens. J. Polym. Environ. 2021, 1. 10.1007/s10924-021-02301-4. [DOI] [PubMed] [Google Scholar]
- Knott B. C.; Erickson E.; Allen M. D.; Gado J. E.; Graham R.; Kearns F. L.; Pardo I.; Topuzlu E.; Anderson J. J.; Austin H. P.; Dominick G.; Johnson C. W.; Rorrer N. A.; Szostkiewicz C. J.; Copié V.; Payne C. M.; Woodcock H. L.; Donohoe B. S.; Beckham G. T.; McGeehan J. E. Characterization and Engineering of a Two-Enzyme System for Plastics Depolymerization. Proc. Nat. Acad. Sci. U.S.A. 2020, 117, 25476–25485. 10.1073/pnas.2006753117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Cifuentes I. E.; Öztürk B. Mle046 Is a Marine Mesophilic MHETase-Like Enzyme. Front. Microbiol. 2021, 12, 693985. 10.3389/fmicb.2021.693985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm G. J.; Reisky L.; Böttcher D.; Müller H.; Michels E. A. P.; Walczak M. C.; Berndt L.; Weiss M. S.; Bornscheuer U. T.; Weber G. Structure of the Plastic-Degrading Ideonella sakaiensis MHETase Bound to a Substrate. Nat. Commun. 2019, 10, 1717. 10.1038/s41467-019-09326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake L.; Jayakody L. N. Engineering Microbes to Bio-Upcycle Polyethylene Terephthalate. Front. Bioeng. Biotechnol. 2021, 9, 656465. 10.3389/fbioe.2021.656465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachisuka S.-i.; Nishii T.; Yoshida S. Development of a Targeted Gene Disruption System in the Poly(Ethylene Terephthalate)-Degrading Bacterium Ideonella sakaiensis and Its Applications to PETase and MHETase Genes. Appl. Environ. Microbiol. 2021, 87, e00020 10.1128/AEM.00020-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth M.; Wei R.; Oeser T.; Then J.; Schmidt J.; Wohlgemuth F.; Zimmermann W. Enzymatic Hydrolysis of Polyethylene Terephthalate Films in an Ultrafiltration Membrane Reactor. J. Membr. Sci. 2015, 494, 182–187. 10.1016/j.memsci.2015.07.030. [DOI] [Google Scholar]
- Dai L.; Qu Y.; Huang J.-W.; Hu Y.; Hu H.; Li S.; Chen C.-C.; Guo R.-T. Enhancing PET Hydrolytic Enzyme Activity by Fusion of the Cellulose–Binding Domain of Cellobiohydrolase I from Trichoderma reesei. J. Biotechnol. 2021, 334, 47–50. 10.1016/j.jbiotec.2021.05.006. [DOI] [PubMed] [Google Scholar]
- Weber J.; Petrović D.; Strodel B.; Smits S. H. J.; Kolkenbrock S.; Leggewie C.; Jaeger K.-E. Interaction of Carbohydrate-Binding Modules with Poly(Ethylene Terephthalate). Appl. Microbiol. Biotechnol. 2019, 103, 4801–4812. 10.1007/s00253-019-09760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B. W.; Sallada N. D. Hydrophobins: Multifunctional Biosurfactants for Interface Engineering. J. Biol. Eng. 2019, 13, 10. 10.1186/s13036-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino-Rammer L.; Ribitsch D.; Przylucka A.; Marold A.; Greimel K. J.; Herrero Acero E.; Guebitz G. M.; Kubicek C. P.; Druzhinina I. S. Two Novel Class II Hydrophobins from Trichoderma spp. Stimulate Enzymatic Hydrolysis of Poly(Ethylene Terephthalate) When Expressed as Fusion Proteins. Appl. Environ. Microbiol. 2013, 79, 4230–4238. 10.1128/AEM.01132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribitsch D.; Herrero Acero E.; Przylucka A.; Zitzenbacher S.; Marold A.; Gamerith C.; Tscheliessnig R.; Jungbauer A.; Rennhofer H.; Lichtenegger H.; Amenitsch H.; Bonazza K.; Kubicek C. P.; Druzhinina I. S.; Guebitz G. M. Enhanced Cutinase-Catalyzed Hydrolysis of Polyethylene Terephthalate by Covalent Fusion to Hydrophobins. Appl. Environ. Microbiol. 2015, 81, 3586–3592. 10.1128/AEM.04111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puspitasari N.; Tsai S.-L.; Lee C.-K. Class I Hydrophobins Pretreatment Stimulates PETase for Monomers Recycling of Waste PET. Int. J. Biol. Macromol. 2021, 176, 157–164. 10.1016/j.ijbiomac.2021.02.026. [DOI] [PubMed] [Google Scholar]
- Dai L.; Qu Y.; Hu Y.; Min J.; Yu X.; Chen C.-C.; Huang J.-W.; Guo R.-T. Catalytically Inactive Lytic Polysaccharide Monooxygenase PCAA14A Enhances the Enzyme-Mediated Hydrolysis of Polyethylene Terephthalate. Int. J. Biol. Macromol. 2021, 190, 456–462. 10.1016/j.ijbiomac.2021.09.005. [DOI] [PubMed] [Google Scholar]
- Chen K.; Hu Y.; Dong X.; Sun Y. Molecular Insights into the Enhanced Performance of EKylated PETase toward PET Degradation. ACS Catal. 2021, 11, 7358–7370. 10.1021/acscatal.1c01062. [DOI] [Google Scholar]
- Várnai A.; Siika-Aho M.; Viikari L. Carbohydrate-Binding Modules (CBMs) Revisited: Reduced Amount of Water Counterbalances the Need for CBMs. Biotechnol. Biofuels 2013, 6, 30. 10.1186/1754-6834-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari J.; Olsen J. P.; Jensen K.; Badino S. F.; Krogh K. B. R. M.; Borch K.; Westh P. Sabatier Principle for Interfacial (Heterogeneous) Enzyme Catalysis. ACS Catal. 2018, 8, 11966–11972. 10.1021/acscatal.8b03547. [DOI] [Google Scholar]
- Herrero Acero E.; Ribitsch D.; Steinkellner G.; Gruber K.; Greimel K.; Eiteljoerg I.; Trotscha E.; Wei R.; Zimmermann W.; Zinn M.; Cavaco-Paulo A.; Freddi G.; Schwab H.; Guebitz G. Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. 10.1021/ma200949p. [DOI] [Google Scholar]
- Schmidt J.; Wei R.; Oeser T.; Belisário-Ferrari M. R.; Barth M.; Then J.; Zimmermann W. Effect of Tris, MOPS, and Phosphate Buffers on the Hydrolysis of Polyethylene Terephthalate Films by Polyester Hydrolases. FEBS Open Bio 2016, 6, 919–927. 10.1002/2211-5463.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. T.; Hee Ryu M.; Jung Y. J.; Lim S.; Song H. M.; Park J.; Hwang S. Y.; Lee H.-S.; Yeon Y. J.; Sung B. H.; Bornscheuer U. T.; Park S. J.; Joo J. C.; Oh D. X. Chemo-Biological Upcycling of Poly(Ethylene Terephthalate) to Multifunctional Coating Materials. ChemSusChem 2021, 14, 4251–4259. 10.1002/cssc.202100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. A.; Okur H. I.; Yan C.; Yang T.; Heyda J.; Cremer P. S. Weakly Hydrated Anions Bind to Polymers but Not Monomers in Aqueous Solutions. Nat. Chem. 2022, 14, 40–45. 10.1038/s41557-021-00805-z. [DOI] [PubMed] [Google Scholar]
- Sonnendecker C.; Oeser J.; Richter P. K.; Hille P.; Zhao Z.; Fischer C.; Lippold H.; Blázquez-Sánchez P.; Engelberger F.; Ramírez-Sarmiento C. A.; Oeser T.; Lihanova Y.; Frank R.; Jahnke H.-G.; Billig S.; Abel B.; Sträter N.; Matysik J.; Zimmermann W. Low Carbon Footprint Recycling of Post-Consumer PET Plastic with a Metagenomic Polyester Hydrolase. ChemSusChem 2021, 1. 10.1002/cssc.202101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundupalli S. P.; Hait S.; Thakur A. A Review on Automated Sorting of Source-Separated Municipal Solid Waste for Recycling. Waste Manage. 2017, 60, 56–74. 10.1016/j.wasman.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Hahladakis J. N.; Iacovidou E. An Overview of the Challenges and Trade-Offs in Closing the Loop of Post-Consumer Plastic Waste (PCPW): Focus on Recycling. J. Hazard. Mater. 2019, 380, 120887. 10.1016/j.jhazmat.2019.120887. [DOI] [PubMed] [Google Scholar]
- Tsochatzis E. D.; Lopes J. A.; Corredig M. Chemical Testing of Mechanically Recycled Polyethylene Terephthalate for Food Packaging in the European Union. Resour. Conserv. Recycl. 2022, 179, 106096. 10.1016/j.resconrec.2021.106096. [DOI] [Google Scholar]
- Beckham G. T.; Matthews J. F.; Peters B.; Bomble Y. J.; Himmel M. E.; Crowley M. F. Molecular-Level Origins of Biomass Recalcitrance: Decrystallization Free Energies for Four Common Cellulose Polymorphs. J. Phys. Chem. B 2011, 115, 4118–4127. 10.1021/jp1106394. [DOI] [PubMed] [Google Scholar]
- Hon J.; Borko S.; Stourac J.; Prokop Z.; Zendulka J.; Bednar D.; Martinek T.; Damborsky J. EnzymeMiner: Automated Mining of Soluble Enzymes with Diverse Structures, Catalytic Properties and Stabilities. Nucleic Acids Res. 2020, 48, W104–W109. 10.1093/nar/gkaa372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumulya Y.; Baek J.-M.; Wun S.-J.; Thomson R. E. S.; Harris K. L.; Hunter D. J. B.; Behrendorff J. B. Y. H.; Kulig J.; Zheng S.; Wu X.; Wu B.; Stok J. E.; De Voss J. J.; Schenk G.; Jurva U.; Andersson S.; Isin E. M.; Bodén M.; Guddat L.; Gillam E. M. J. Engineering Highly Functional Thermostable Proteins Using Ancestral Sequence Reconstruction. Nat. Catal. 2018, 1, 878–888. 10.1038/s41929-018-0159-5. [DOI] [Google Scholar]
- Vanacek P.; Sebestova E.; Babkova P.; Bidmanova S.; Daniel L.; Dvorak P.; Stepankova V.; Chaloupkova R.; Brezovsky J.; Prokop Z.; Damborsky J. Exploration of Enzyme Diversity by Integrating Bioinformatics with Expression Analysis and Biochemical Characterization. ACS Catal. 2018, 8, 2402–2412. 10.1021/acscatal.7b03523. [DOI] [Google Scholar]
- Mazurenko S.; Prokop Z.; Damborsky J. Machine Learning in Enzyme Engineering. ACS Catal. 2020, 10, 1210–1223. 10.1021/acscatal.9b04321. [DOI] [Google Scholar]
- Musil M.; Khan R. T.; Beier A.; Stourac J.; Konegger H.; Damborsky J.; Bednar D. FireProtASR: A Web Server for Fully Automated Ancestral Sequence Reconstruction. Brief. Bioinformatics 2021, 22, bbaa337 10.1093/bib/bbaa337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M.; DiMaio F.; Anishchenko I.; Dauparas J.; Ovchinnikov S.; Lee G. R.; Wang J.; Cong Q.; Kinch L. N.; Schaeffer R. D.; Millán C.; Park H.; Adams C.; Glassman C. R.; DeGiovanni A.; Pereira J. H.; Rodrigues A. V.; Dijk A. A. v.; Ebrecht A. C.; Opperman D. J.; Sagmeister T.; Buhlheller C.; Pavkov-Keller T.; Rathinaswamy M. K.; Dalwadi U.; Yip C. K.; Burke J. E.; Garcia K. C.; Grishin N. V.; Adams P. D.; Read R. J.; Baker D. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe D. D.; Praetorius F.; Courbet A.; Hsia Y.; Wicky B. I. M.; Edman N. I.; Miller L. M.; Timmermans B. J. R.; Decarreau J.; Morris H. M.; Kang A.; Bera A. K.; Baker D. Reconfigurable Asymmetric Protein Assemblies through Implicit Negative Design. Science 2022, 375, eabj7662 10.1126/science.abj7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas J. V.; Crowley M. F.; Beckham G. T.; Payne C. M. Effects of Lytic Polysaccharide Monooxygenase Oxidation on Cellulose Structure and Binding of Oxidized Cellulose Oligomers to Cellulases. J. Phys. Chem. B 2015, 119, 6129–6143. 10.1021/acs.jpcb.5b00778. [DOI] [PubMed] [Google Scholar]
- Heinz H.; Lin T.-J.; Kishore Mishra R.; Emami F. S. Thermodynamically Consistent Force Fields for the Assembly of Inorganic, Organic, and Biological Nanostructures: The Interface Force Field. Langmuir 2013, 29, 1754–1765. 10.1021/la3038846. [DOI] [PubMed] [Google Scholar]
- Tiso T.; Narancic T.; Wei R.; Pollet E.; Beagan N.; Schröder K.; Honak A.; Jiang M.; Kenny S. T.; Wierckx N.; Perrin R.; Avérous L.; Zimmermann W.; O’Connor K.; Blank L. M. Towards Bio-Upcycling of Polyethylene Terephthalate. Metab. Eng. 2021, 66, 167–178. 10.1016/j.ymben.2021.03.011. [DOI] [PubMed] [Google Scholar]
- Werner A. Z.; Clare R.; Mand T. D.; Pardo I.; Ramirez K. J.; Haugen S. J.; Bratti F.; Dexter G. N.; Elmore J. R.; Huenemann J. D.; Peabody G. L.; Johnson C. W.; Rorrer N. A.; Salvachúa D.; Guss A. M.; Beckham G. T. Tandem Chemical Deconstruction and Biological Upcycling of Poly(Ethylene Terephthalate) to β-Ketoadipic Acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261. 10.1016/j.ymben.2021.07.005. [DOI] [PubMed] [Google Scholar]
- Kim H. T.; Kim J. K.; Cha H. G.; Kang M. J.; Lee H. S.; Khang T. U.; Yun E. J.; Lee D.-H.; Song B. K.; Park S. J.; Joo J. C.; Kim K. H. Biological Valorization of Poly(Ethylene Terephthalate) Monomers for Upcycling Waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. 10.1021/acssuschemeng.9b03908. [DOI] [Google Scholar]
- Heyde S. A. H.; Arnling Bååth J.; Westh P.; Nørholm M. H. H.; Jensen K. Surface Display as a Functional Screening Platform for Detecting Enzymes Active on PET. Microb. Cell Fact. 2021, 20, 93. 10.1186/s12934-021-01582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Xiao Y.; Weber G.; Wei R.; Wang Z.. Yeast Cell Surface Display of Bacterial PET Hydrolase as a Sustainable Biocatalyst for the Degradation of Polyethylene Terephthalate. In Methods in Enzymology, Weber G.; Bornscheuer U. T.; Wei R., Eds.; Academic Press, 2021; Vol. 648, pp 457–477. [DOI] [PubMed] [Google Scholar]
- Yan F.; Wei R.; Cui Q.; Bornscheuer U. T.; Liu Y.-J. Thermophilic Whole-Cell Degradation of Polyethylene Terephthalate Using Engineered Clostridium thermocellum. Microb. Biotechnol. 2021, 14, 374–385. 10.1111/1751-7915.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog D.; Schmitt J.; Senger J.; Zarzycki J.; Rexer K.-H.; Linne U.; Erb T.; Maier U. G. Using a Marine Microalga as a Chassis for Polyethylene Terephthalate (PET) Degradation. Microb. Cell Fact. 2019, 18, 171. 10.1186/s12934-019-1220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R.; Sun C. A Marine Bacterial Community Capable of Degrading Poly(Ethylene Terephthalate) and Polyethylene. J. Hazard. Mater. 2021, 416, 125928. 10.1016/j.jhazmat.2021.125928. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann S.; Labrenz M. Marine Microbial Assemblages on Microplastics: Diversity, Adaptation, and Role in Degradation. Annu. Rev. Mar. Sci. 2020, 12, 209. 10.1146/annurev-marine-010419-010633. [DOI] [PubMed] [Google Scholar]
- Liu J.; He J.; Xue R.; Xu B.; Qian X.; Xin F.; Blank L. M.; Zhou J.; Wei R.; Dong W.; Jiang M. Biodegradation and Up-Cycling of Polyurethanes: Progress, Challenges, and Prospects. Biotechnol. Adv. 2021, 48, 107730. 10.1016/j.biotechadv.2021.107730. [DOI] [PubMed] [Google Scholar]
- Knyazev V. D. Effects of Chain Length on the Rates of C-C Bond Dissociation in Linear Alkanes and Polyethylene. J. Phys. Chem. A 2007, 111, 3875–3883. 10.1021/jp066419e. [DOI] [PubMed] [Google Scholar]
- Jiang Q.; Li Z.; Cui Z.; Wei R.; Nie K.; Xu H.; Liu L. Quantum Mechanical Investigation of the Oxidative Cleavage of the C–C Backbone Bonds in Polyethylene Model Molecules. Polymers 2021, 13, 2730. 10.3390/polym13162730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik M. W.; Kenny S. T.; Duane G. F.; Casey E.; Woods T.; Babu R. P.; Nikodinovic-Runic J.; Murray M.; O’Connor K. E. Conversion of Post Consumer Polyethylene to the Biodegradable Polymer Polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2014, 98, 4223–4232. 10.1007/s00253-013-5489-2. [DOI] [PubMed] [Google Scholar]
- Ghatge S.; Yang Y.; Ko Y.; Yoon Y.; Ahn J.-H.; Kim J. J.; Hur H.-G. Degradation of Sulfonated Polyethylene by a Bio-Photo-Fenton Approach Using Glucose Oxidase Immobilized on Titanium Dioxide. J. Hazard. Mater. 2022, 423, 127067. 10.1016/j.jhazmat.2021.127067. [DOI] [PubMed] [Google Scholar]
- Ballerstedt H.; Tiso T.; Wierckx N.; Wei R.; Averous L.; Bornscheuer U.; O’Connor K.; Floehr T.; Jupke A.; Klankermayer J.; Liu L.; de Lorenzo V.; Narancic T.; Nogales J.; Perrin R.; Pollet E.; Prieto A.; Casey W.; Haarmann T.; Sarbu A.; Schwaneberg U.; Xin F.; Dong W.; Xing J.; Chen G.-Q.; Tan T.; Jiang M.; Blank L. M. Mixed Plastics Biodegradation and Upcycling Using Microbial Communities: EU Horizon 2020 Project MIX-UP Started January 2020. Environ. Sci. Europe 2021, 33, 99. 10.1186/s12302-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y.; Watanabe T.; Nakajima-Kambe T.; Kitamoto H. K. Rapid and Simple Colorimetric Assay for Detecting the Enzymatic Degradation of Biodegradable Plastic Films. J. Biosci. Bioeng. 2013, 115, 111–114. 10.1016/j.jbiosc.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Zumstein M. T.; Kohler H.-P. E.; McNeill K.; Sander M. High-Throughput Analysis of Enzymatic Hydrolysis of Biodegradable Polyesters by Monitoring Cohydrolysis of a Polyester-Embedded Fluorogenic Probe. Environ. Sci. Technol. 2017, 51, 4358–4367. 10.1021/acs.est.6b06060. [DOI] [PubMed] [Google Scholar]
- Wei R.; Oeser T.; Billig S.; Zimmermann W. A High-Throughput Assay for Enzymatic Polyester Hydrolysis Activity by Fluorimetric Detection. Biotechnol. J. 2012, 7, 1517–1521. 10.1002/biot.201200119. [DOI] [PubMed] [Google Scholar]
- Pfaff L.; Breite D.; Badenhorst C. P. S.; Bornscheuer U. T.; Wei R.. Fluorimetric High-Throughput Screening Method for Polyester Hydrolase Activity Using Polyethylene Terephthalate Nanoparticles. In Methods in Enzymology, Weber G.; Bornscheuer U. T.; Wei R., Eds.; Academic Press, 2021; Vol. 648, pp 253–270. [DOI] [PubMed] [Google Scholar]
- Weigert S.; Gagsteiger A.; Menzel T.; Höcker B. A Versatile Assay Platform for Enzymatic Poly(Ethylene-Terephthalate) Degradation. Protein Eng. Des. Sel. 2021, 34, gzab022. [DOI] [PubMed] [Google Scholar]
- Molitor R.; Bollinger A.; Kubicki S.; Loeschcke A.; Jaeger K.-E.; Thies S. Agar Plate-Based Screening Methods for the Identification of Polyester Hydrolysis by Pseudomonas Species. Microb. Biotechnol. 2020, 13, 274–284. 10.1111/1751-7915.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock C. A Simple and Novel Method for the Production of Polyethylene Terephthalate Containing Agar Plates for the Growth and Detection of Bacteria Able to Hydrolyze This Plastic. J. Microbiol. Methods 2021, 185, 106222. 10.1016/j.mimet.2021.106222. [DOI] [PubMed] [Google Scholar]
- Qiao Y.; Hu R.; Chen D.; Wang L.; Wang Z.; Yu H.; Fu Y.; Li C.; Dong Z.; Weng Y.-X.; Du W. Fluorescence-Activated Droplet Sorting of PET Degrading Microorganisms. J. Hazard. Mater. 2022, 424, 127417. 10.1016/j.jhazmat.2021.127417. [DOI] [PubMed] [Google Scholar]
- Pardo I.; Jha R. K.; Bermel R. E.; Bratti F.; Gaddis M.; McIntyre E.; Michener W.; Neidle E. L.; Dale T.; Beckham G. T.; Johnson C. W. Gene Amplification, Laboratory Evolution, and Biosensor Screening Reveal Muck as a Terephthalic Acid Transporter in Acinetobacter baylyi ADP1. Metab. Eng. 2020, 62, 260–274. 10.1016/j.ymben.2020.09.009. [DOI] [PubMed] [Google Scholar]
- Meyer A.; Pellaux R.; Potot S.; Becker K.; Hohmann H.-P.; Panke S.; Held M. Optimization of a Whole-Cell Biocatalyst by Employing Genetically Encoded Product Sensors inside Nanolitre Reactors. Nat. Chem. 2015, 7, 673–678. 10.1038/nchem.2301. [DOI] [PubMed] [Google Scholar]
- Clarkson S. M.; Giannone R. J.; Kridelbaugh D. M.; Elkins J. G.; Guss A. M.; Michener J. K.; Vieille C. Construction and Optimization of a Heterologous Pathway for Protocatechuate Catabolism in Escherichia coli Enables Bioconversion of Model Aromatic Compounds. Appl. Environ. Microbiol. 2017, 83, e01313 10.1128/AEM.01313-17. [DOI] [PMC free article] [PubMed] [Google Scholar]