ABSTRACT
Spermidine, a kind of polycation and one important member of the polyamine family, is essential for survival in many kinds of organisms and participates in the regulation of cell growth and metabolism. To explore the mechanism by which spermidine regulates ganoderic acid (GA) biosynthesis in Ganoderma lucidum, the effects of spermidine on GA and reactive oxygen species (ROS) contents were examined. Our data suggested that spermidine promoted the production of mitochondrial ROS and positively regulated GA biosynthesis. Further research revealed that spermidine promoted the translation of mitochondrial complexes I and II and subsequently influenced their activity. With a reduction in eukaryotic translation initiation factor 5A (eIF5A) hypusination by over 50% in spermidine synthase gene (spds) knockdown strains, the activities of mitochondrial complexes I and II were reduced by nearly 60% and 80%, respectively, and the protein contents were reduced by over 50%, suggesting that the effect of spermidine on mitochondrial complexes I and II was mediated through its influence on eIF5A hypusination. Furthermore, after knocking down eIF5A, the deoxyhypusine synthase gene (dhs), and the deoxyhypusine hydroxylase gene (dohh), the mitochondrial ROS level was reduced by nearly 50%, and the GA content was reduced by over 40%, suggesting that eIF5A hypusination contributed to mitochondrial ROS production and GA biosynthesis. In summary, spermidine maintains mitochondrial ROS homeostasis by regulating the translation and subsequent activity of complexes I and II via eIF5A hypusination and promotes GA biosynthesis via mitochondrial ROS signaling. The present findings provide new insight into the spermidine-mediated biosynthesis of secondary metabolites.
IMPORTANCE Spermidine is necessary for organism survival and is involved in the regulation of various biological processes. However, the specific mechanisms underlying the various physiological functions of spermidine are poorly understood, especially in microorganisms. In this study, we found that spermidine hypusinates eIF5A to promote the production of mitochondrial ROS and subsequently regulate secondary metabolism in microorganisms. Our study provides a better understanding of the mechanism by which spermidine regulates mitochondrial function and provides new insight into the spermidine-mediated biosynthesis of secondary metabolites.
KEYWORDS: polyamine, mitochondrial complexes, ROS signaling, secondary metabolism
INTRODUCTION
Polyamines are a class of polycationic compounds that are ubiquitous and indispensable in both prokaryotic and eukaryotic cells (1). Spermidine (Spd), an important member of the polyamine family, is necessary for cell survival and participates in the regulation of cell proliferation and metabolism in all kinds of organisms (2). Under normal physiological conditions, protonated spermidine, as a polycation, can interact with some biomacromolecules with negative charges, such as DNA, RNA, ATP, proteins, and phospholipids (3–5). Therefore, spermidine plays important roles in the regulation of transcription, translation, signal transduction, and other biological processes and is involved in the responses to multiple stresses and diseases as well as the regulation of growth, development, and secondary metabolism.
The functions of spermidine and the underlying mechanisms have been studied extensively in animals and plants. In animals, spermidine extends the life span of aging organisms by exhibiting anti-inflammatory and antioxidant properties and enhancing mitochondrial metabolic function and respiration, etc. (6, 7). In addition, spermidine participates in the regulation of a variety of biological processes by acting on different targets. In plants, spermidine regulates root growth, tissue differentiation, photosynthesis, and pollen development, etc. (1, 8, 9). Moreover, spermidine increases plant tolerance to various biotic and abiotic stresses (9–11). There are fewer studies on the functions of spermidine in microorganisms than in animals and plants, and most in the former have been focused on the roles of spermidine in the regulation of secondary metabolite biosynthesis and the maintenance of cell viability. In fungi, spermidine promotes the biosynthesis of 2-acetyl-1-pyrroline, a common food-flavoring compound, in Aspergillus awamori (12); upregulates lovastatin production in Aspergillus terreus (13); and increases the production of penicillin in Penicillium chrysogenum (14). In addition, spermidine plays important roles in mycelial growth, conidiation, the synthesis of secondary metabolites, and infection viability in Aspergillus flavus (15). These observations prove that spermidine has a variety of physiological functions in microorganisms. However, the mechanisms by which spermidine regulates various biological processes remain unclear. Therefore, it is of great significance to perform in-depth studies on the functions of spermidine in microorganisms.
Ganoderma lucidum, a well-known large basidiomycete fungus, is highly valued in Asia due to not only its ornamental value but also its medicinal efficacy (16, 17). Modern medical studies have shown that G. lucidum has significant antiaging, antioxidant, antiviral, and immunomodulatory therapeutic effects, which are thought to be mainly due to its secondary metabolites ganoderic acids (GAs) (16, 18). Therefore, it is of great significance to study the regulatory mechanism of GA biosynthesis in G. lucidum. Our previous study revealed that there are two kinds of polyamines, putrescine and spermidine, in G. lucidum (19). Putrescine negatively regulates GA biosynthesis by reducing cellular reactive oxygen species (ROS) contents (20). Heat stress also induces GA biosynthesis (21). As polyamines are important heat stress response factors, we studied the functions of polyamines in the regulation of GA biosynthesis. We found that heat stress induced the conversion of putrescine to spermidine, which was accompanied by an increase in the GA content, and that spermidine synthase (Spds) played an important role in this process (19). However, the function of spermidine in GA biosynthesis and the underlying mechanism remain unclear. In this study, we found that spermidine positively regulated GA biosynthesis by promoting the production of cellular ROS. Further study revealed that spermidine regulated the activities of mitochondrial complex I (NADH dehydrogenase [NDH]) and complex II (succinate dehydrogenase [SDH]) by hypusinating eukaryotic translation initiation factor 5A (eIF5A), resulting in the positive regulation of ROS production and GA biosynthesis. This study exposed the mechanism by which spermidine regulates the biosynthesis of GAs and reveals the important roles of spermidine in regulating mitochondrial redox homeostasis and function as well as secondary metabolism in fungi.
RESULTS
Spermidine positively regulates GA biosynthesis in G. lucidum.
Spds is a key enzyme for spermidine biosynthesis in G. lucidum. As Spds converts putrescine to spermidine, the accumulation of putrescine and the reduction of spermidine were observed after Spds gene (spds) knockdown (see Fig. S1 in the supplemental material). In strains Spds si 40 and Spds si 55, compared with the wild type (WT), the contents of putrescine increased by 52.16% and 40.68%, respectively, and those of spermidine decreased by 56.37% and 48.9%, respectively. To explore the effect of spermidine on the biosynthesis of GAs, the GA contents were analyzed in two spds gene knockdown strains. The contents of GAs were decreased by 44.34% and 39.32% in the two spds knockdown strains compared with the WT (Fig. 1). After supplementation with 1 mM spermidine, the GA contents of the spds knockdown strains were restored to levels consistent with that of the WT (Fig. 1), revealing that spermidine plays an important role in the regulation of GA biosynthesis in G. lucidum.
FIG 1.
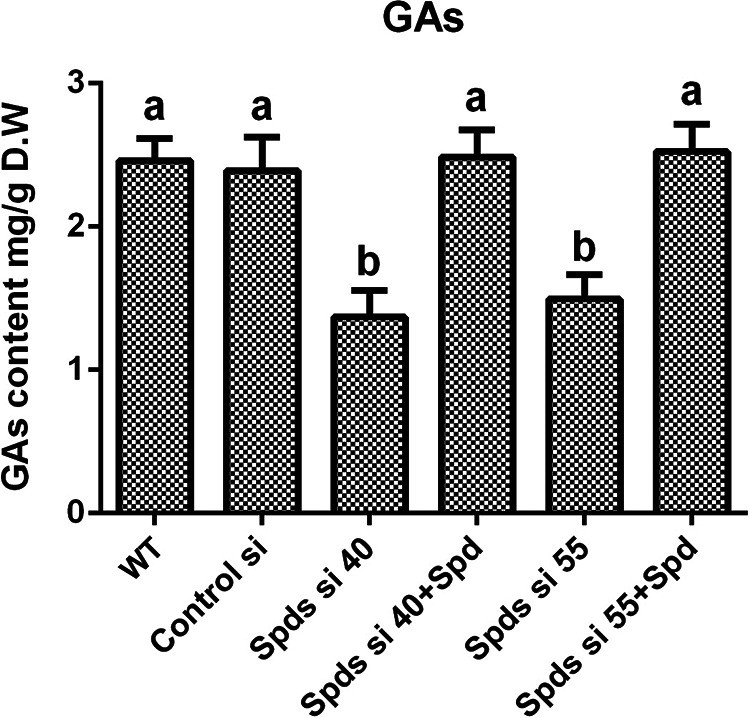
Knockdown of the spds gene decreases the content of GAs. GA contents in the WT, the empty vector control (control silence (Control si)), two spds knockdown strains, and strains supplied with 1 mM spermidine (Spd) from 7-day-old liquid mycelia were determined. The values presented are the means ± standard deviations (SD) of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test). D.W, dry weight.
Spermidine regulates GA biosynthesis by ROS.
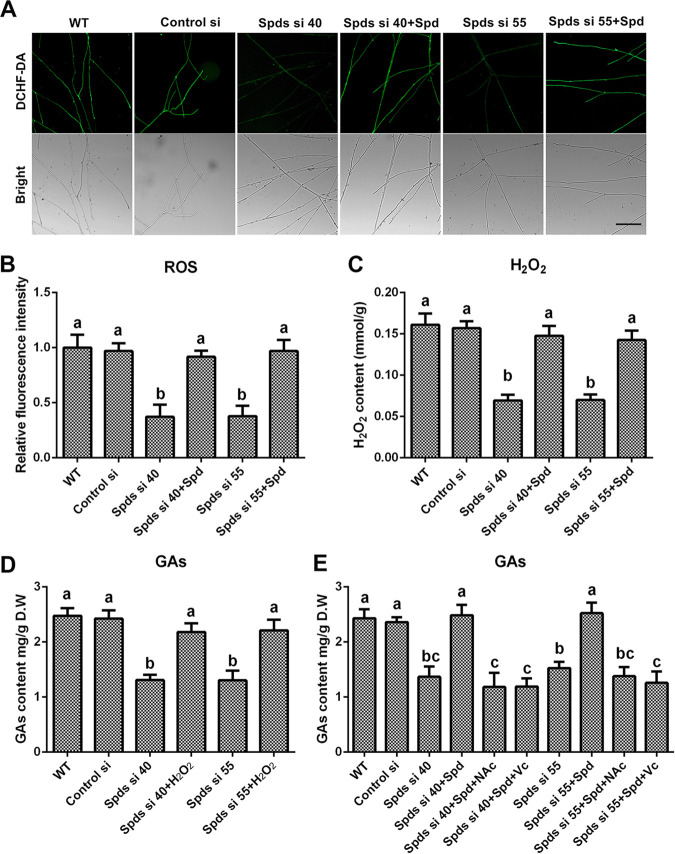
ROS signaling induces GA biosynthesis in G. lucidum (21, 22), and polyamine regulates redox homeostasis in multiple species (23). The ROS levels in the spds knockdown strains were determined to explore whether spermidine regulates GA biosynthesis via ROS signaling. In strains Spds si 40 and Spds si 55, compared with the WT, the ROS levels were decreased by 62.86% and 62.23%, respectively (Fig. 2A and B), and the H2O2 contents were decreased by 59.6% and 56.59%, respectively (Fig. 2C). After supplementation with 1 mM spermidine, the ROS levels and H2O2 contents in the spds knockdown strains were restored to levels consistent with those of the WT, exhibiting trends similar to those for GA contents. To further explore the relationships among spermidine, ROS signaling, and GA biosynthesis, H2O2 (5 mM), the ROS scavenger N-acetyl-l-cysteine (NAc) (2 mM), and ascorbic acid vitamin C (Vc) (1 mM) were used. The GA contents were restored to 88.06% and 89.31% of the WT level in Spds si 40 and Spds si 55, respectively, after supplementation with H2O2 (Fig. 2D). However, even with spermidine supplementation, after treatment with NAc, the GA contents in Spds si 40 and Spds si 55 were decreased by 51.27% and 43.31%, respectively, compared with that in the WT, and a similar result was observed after treatment with Vc (Fig. 2E). These data revealed that spermidine regulated GA biosynthesis mainly via ROS signaling in G. lucidum. Taken together, these results indicate that spermidine deficiency disrupted cellular redox homeostasis and decreased cellular ROS production and subsequent GA biosynthesis.
FIG 2.
Knockdown of the spds gene decreases the content of GAs by decreasing ROS production. (A) ROS levels determined by DCHF-DA staining in the WT, Control si, two spds knockdown strains, and strains supplied with 1 mM spermidine (Spd) from 5-day-old plate mycelia. Bar = 50 μm. (B) Relative fluorescence intensities from panel A. (C) H2O2 contents in the WT, Control si, two spds knockdown strains, and strains supplied with 1 mM Spd from 5-day-old plate mycelia. (D) GA contents in the WT, Control si, two spds knockdown strains, and strains supplied with 5 mM H2O2 from 7-day-old liquid mycelia. (E) GA contents in the WT, Control si, two spds knockdown strains, and strains supplied with 1 mM Spd and treated with 2 mM ascorbic acid (Vc) or 1 mM N-acetyl cysteine (NAc) from 7-day-old liquid mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
Spermidine regulates cellular redox homeostasis mainly by promoting mitochondrial ROS production.
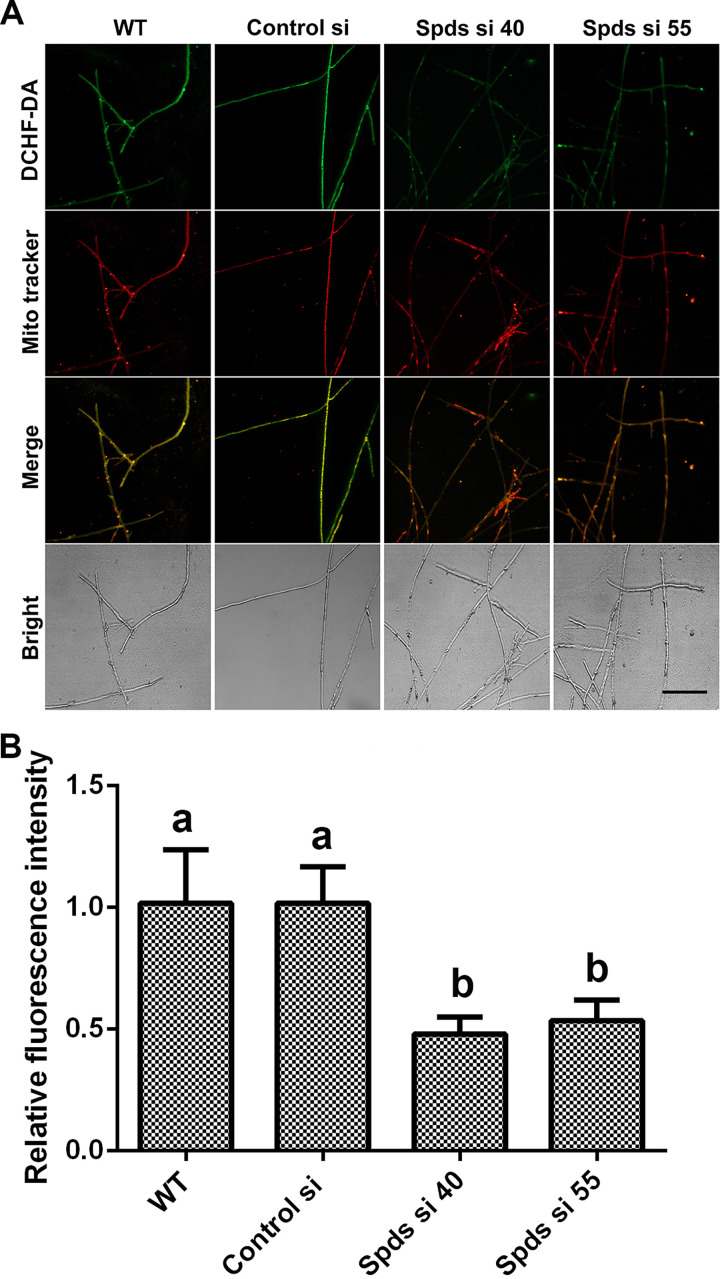
To further explore the mechanism by which spermidine regulates the redox system, we first tested the activities of the antioxidant enzymes ascorbate peroxidase (APX), catalase (CAT), glutathione peroxidase (GPX), and superoxide dismutase (SOD), in addition to NADPH oxidase (NOX), in G. lucidum. Our results showed no significant difference between either of the spds knockdown strains and the WT (Fig. S2A to E), revealing that spermidine regulates ROS potentially without affecting the oxidoreductase system. Spermidine is oxidized by polyamine oxidase (Pao) to produce H2O2 (23). Subsequently, ROS levels were analyzed in two polyamine oxidase gene (pao) knockdown strains. Inexplicably, there was also no significant difference between the pao knockdown strains and the WT (Fig. S2F), revealing that the catabolism of spermidine has no significant effect on ROS homeostasis. An additional major source of ROS is mitochondria. Next, we tested the mitochondrial ROS (mtROS) levels in the spds knockdown strains. The mitochondrial ROS levels were decreased by 53.66% and 48.24% in Spds si 40 and Spds si 55, respectively, compared with the WT, as shown in Fig. 3A and B. Collectively, these data indicated that spermidine regulated cellular redox homeostasis mainly by regulating mitochondrial ROS production.
FIG 3.
Knockdown of the spds gene decreases mitochondrial ROS (mtROS) production. (A) Mitochondrial ROS levels determined by DCHF-DA and MitoTracker Red double staining in the WT, Control si, and two spds knockdown strains from 5-day-old plate mycelia. Bar = 50 μm. (B) Relative fluorescence intensities from panel A. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
Spermidine regulates mitochondrial ROS by regulating mitochondrial complexes I and II.
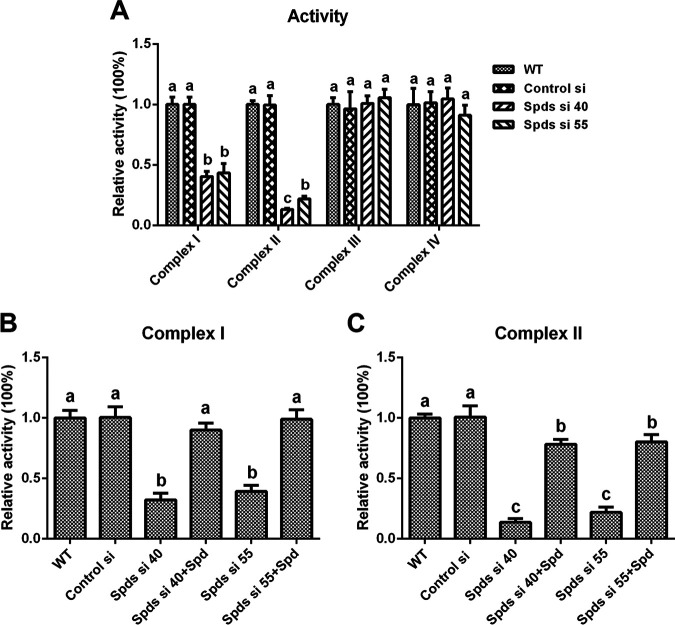
The mitochondrial electron transport chain (ETC) is a major place where mitochondrial ROS are produced. The enzymatic activities of mitochondrial complexes I (NADH dehydrogenase), II (succinate dehydrogenase), III (CoQ-cytochrome c reductase), and IV (cytochrome c oxidase) in the spds knockdown strains were assessed. The activities of complex I were decreased by 67.74% and 60.74% in Spds si 40 and Spds si 55 compared with the WT, respectively, and they were restored to levels consistent with that in the WT after supplementation with spermidine (Fig. 4B). The activities of complex II were decreased by 86.26% and 78.03% in Spds si 40 and Spds si 55 compared with the WT, respectively, and were restored to 78.36% and 80.36% of the activity in the WT, respectively, after supplementation with spermidine (Fig. 4C). However, there was no significant difference in the activities of complex III or IV between Spds si 40 or Spds si 55 and the WT (Fig. 4A). These data demonstrated that the inhibition of spermidine biosynthesis reduced the activities of mitochondrial complexes I and II.
FIG 4.
Knockdown of the spds gene leads to the loss of activity of mitochondrial complexes I and II. (A) Relative activities of mitochondrial complexes I, II, III, and IV in the WT, Control si, and two spds knockdown strains from 5-day-old plate mycelia. (B and C) Relative activities of mitochondrial complexes I and II in the WT, Control si, two spds knockdown strains, and strains supplied with 1 mM spermidine (Spd) from 5-day-old plate mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
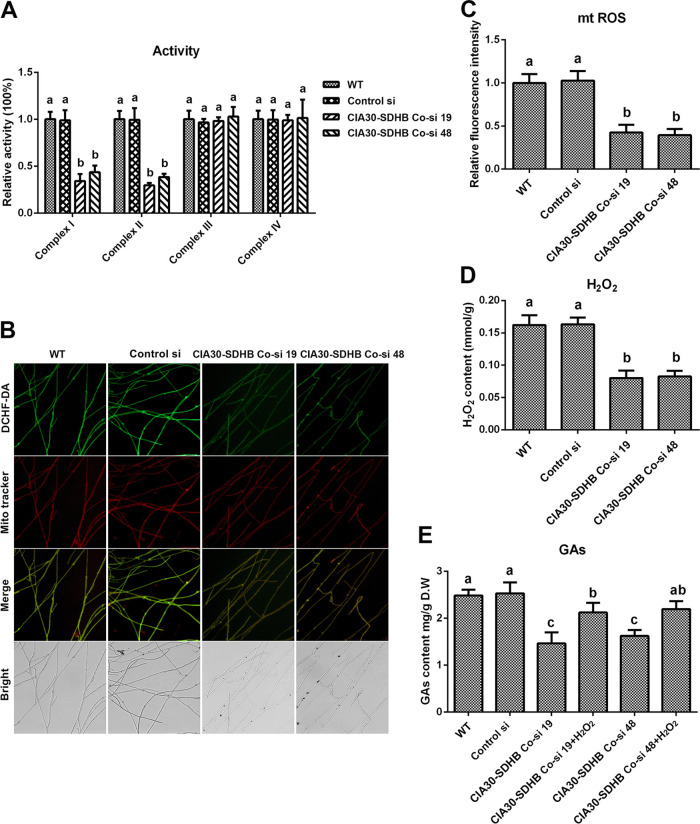
To achieve a simultaneous reduction in mitochondrial complex I and II activities, complex I and II coknockdown strains were constructed and analyzed. The complex I assembly factor NDUFAF1 (complex I intermediate-associated protein 30 [CIA30]) and complex II functional subunit B (SDHB) are key subunits of mitochondrial complexes I and II, respectively, and the CIA30 and SDHB genes were used as target genes for establishing complex I and II coknockdown strains (24). Two complex I and II coknockdown strains were screened from 52 presumptive transformants, named CIA30-SDHB Co-si 19 and CIA30-SDHB Co-si 48, respectively. The gene expression levels of CIA30 were decreased by 79.05% and 72.77% and those of SDHB were decreased by 84.91% and 77.24% in CIA30-SDHB Co-si 19 and CIA30-SDHB Co-si 48, respectively, compared with the WT (Fig. S3A). Furthermore, in CIA30-SDHB Co-si 19 and CIA30-SDHB Co-si 48, compared with the WT, the protein contents of CIA30 were decreased by 52.13% and 45.42%, respectively, and those of SDHB were decreased by 62.46% and 53.68%, respectively (Fig. S3B and C). The activities of mitochondrial complex I were decreased by 65.7% and 56.29% and those of complex II were decreased by 65.7% and 56.29% in the two complex I and II coknockdown strains compared with the WT (Fig. 5A). There was no significant difference between the WT and the coknockdown strains in the activities of complexes III and IV. The constructed mitochondrial complex I and II coknockdown strains were used for the next study.
FIG 5.
Loss of activity of mitochondrial complexes I and II leads to decreases in mitochondrial ROS (mtROS) production and GA contents. (A) Relative activities of mitochondrial complexes I, II, III, and IV in the WT, Control si, and two complex I (CIA30) and complex II (SDHB) coknockdown strains from 5-day-old plate mycelia. (B) Mitochondrial ROS levels determined by DCHF-DA and MitoTracker Red double staining in the WT, Control si, and two CIA30 and SDHB coknockdown strains from 5-day-old plate mycelia. Bar = 50 μm. (C) Relative fluorescence intensities from panel B. (D) H2O2 contents in the WT, Control si, and two CIA30 and SDHB coknockdown mutants from 5-day-old plate mycelia. (E) GA contents in the WT, Control si, two CIA30 and SDHB coknockdown strains, and strains supplied with 5 mM H2O2 from 7-day-old liquid mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
Next, the ROS levels and GA contents were analyzed in the two complex I and II coknockdown strains. The levels of mitochondrial ROS were decreased by 57.47% and 60.41% (Fig. 5B and C) and the contents of cellular H2O2 were decreased by 50.64% and 49.09% in the two complex I and II coknockdown strains compared with the WT (Fig. 5D). The GA contents were reduced by 41.13% and 34.67% in the two complex I and II coknockdown strains compared with the WT and were restored to 85.46% and 88.31% of the WT level in the coknockdown strains after supplementation with 5 mM H2O2 (Fig. 5E). These results suggested that the inhibition of complexes I and II reduced the production of ROS and the subsequent biosynthesis of GAs. Taken together, these findings indicate that the deficiency of spermidine reduced the activity of mitochondrial complexes I and II and led to a decrease in ROS production by the ETC and a subsequent decrease in the biosynthesis of GAs.
Spermidine promotes the translation of mitochondrial complexes I and II.
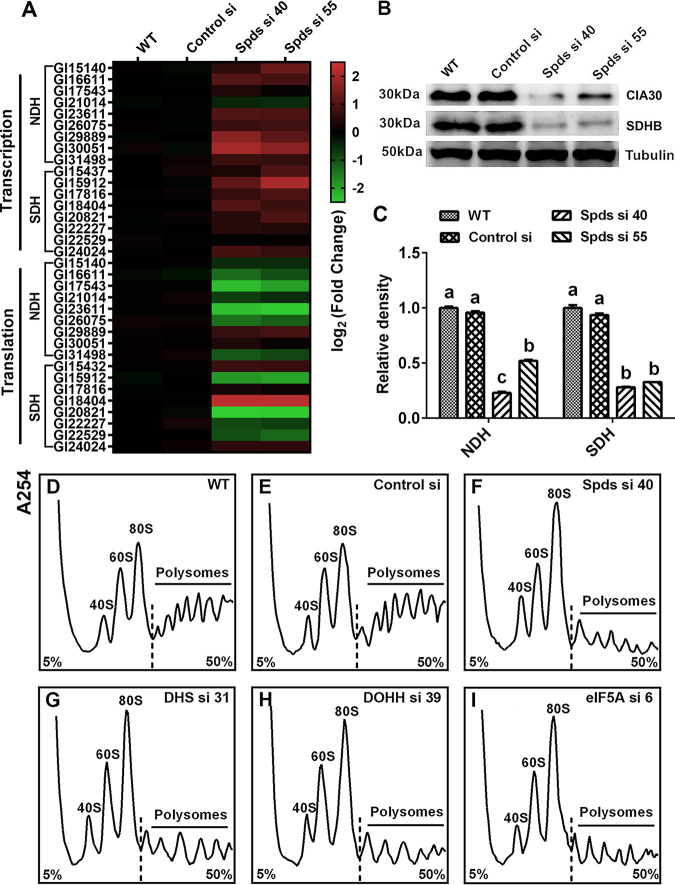
To explore the mechanism by which spermidine regulates the activities of mitochondrial complexes I and II, a gene expression analysis was first performed to assess the transcription levels of genes that encode the mitochondrial complex I and II subunits. The gene transcription levels of most subunits of mitochondrial complexes I and II were increased in the spds knockdown strains, as shown in Fig. 6A. However, this result was not consistent with the activities of mitochondrial complexes I and II after spds knockdown. Next, Western blotting was performed to determine the protein levels of complexes I and II. As shown in Fig. 6B and C, the contents of complex I and II proteins were reduced by over 50% in the spds knockdown strains compared with the WT, consistent with the activities of mitochondrial complexes I and II. Next, we investigated the translation status of the mitochondrial complex I and II subunits by determining their mRNA levels combined with polysomes and 80S monosomes. As shown in Fig. 6A, the translation levels of most subunits were reduced in the spds knockdown strains. Based on the above-described results, we speculated that the decreases in protein levels were due to abnormal translation and that the increase in the transcription of mitochondrial complexes I and II was a response to repair the damage to mitochondrial function in G. lucidum.
FIG 6.
Knockdown of the spds gene blocks the translation of complexes I and II. (A) Protein contents of complex I (anti-CIA30 [assembly factor NDUFAF1]) and complex II (anti-SDHB [succinate dehydrogenase subunit B]) determined by Western blotting in the WT, Control si, and two spds knockdown strains from 5-day-old plate mycelia. (B) Relative densities from panel A. (C) Transcription and translation of complex I and II subunits assessed by qRT-PCR in the WT, Control si, and two spds knockdown strains from 5-day-old plate mycelia. (D to I) Polyribosome profiles of the WT, Control si, and spds/eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
To explore the mechanism of translation regulation by spermidine, a polyribosome profile analysis was performed. As shown in Fig. 6D to F, in the spds knockdown strains, a loss of polysomes but an increase in 80S monosomes was observed, suggesting that the 80S monosomes cannot bind normally to mRNA in these strains, resulting in a blockade of translation initiation. These results reveal an important role of cellular spermidine in the regulation of translation. Taken together, these results indicate that spermidine regulates the activities of mitochondrial complexes I and II by promoting the translation of subunits of mitochondrial complexes I and II in G. lucidum.
Spermidine regulated the translation of mitochondrial complexes I and II by hypusinating eIF5A.
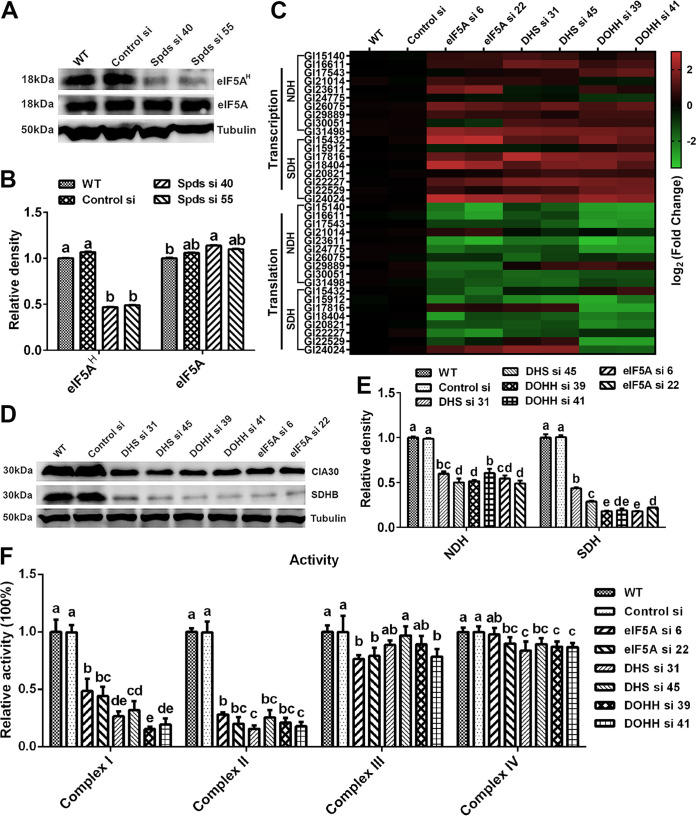
eIF5A is hypusinated with spermidine as the substrate and participates in the regulation of eukaryotic protein translation (25–27). The blockade of translation initiation observed after knocking down the spds gene was suspected to be the result of deficient eIF5A hypusination in G. lucidum. To test this hypothesis, the level of hypusinated eIF5A was determined by Western blotting. The level of eIF5A hypusination was decreased by more than 50% in the two spds knockdown strains compared with the WT, but there was a slight increase in the eIF5A protein level in the knockdown strains, as shown in Fig. 7A and B.
FIG 7.
Blockade of translation of complexes I and II after spds knockdown due to a decrease in eIF5A hypusination. (A) eIF5A protein contents and eIF5A hypusination levels determined by Western blotting in the WT, Control si, and two spds knockdown strains from 5-day-old plate mycelia. (B) Relative densities from panel A. (C) Transcription and translation of complex I and II subunits assessed by qRT-PCR in the WT, Control si, and eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. (D) Protein contents of complex I (CIA30) and complex II (SDHB) determined by Western blotting in the WT, Control si, and eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. (E) Relative densities from panel D. (F) Relative activities of mitochondrial complexes I, II, III, and IV in the WT, Control si, and eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
To determine whether the deficiency in hypusinated eIF5A was related to the blockade of mitochondrial complex I and II translation, we constructed knockdown strains for three genes: the eIF5A gene and two genes necessary for eIF5A hypusination, the deoxyhypusine synthase gene (dhs) and the deoxyhypusine hydroxylase gene (dohh) (28). Two knockdown strains with >65% silencing efficiency were screened for each gene (Fig. S4A to C). Next, we investigated the hypusination and protein levels of eIF5A in the eIF5A, dhs, and dohh knockdown strains by Western blotting. The eIF5A hypusination levels were decreased by more than 50% in the eIF5A, dhs, and dohh knockdown strains compared with the WT. In addition, the protein contents of eIF5A were decreased by more than 70% in the eIF5A knockdown strains, whereas there was little difference in eIF5A protein contents between either the dhs or dohh knockdown strain and the WT (Fig. S4D and E). Furthermore, we determined the transcription, translation, and protein contents of mitochondrial complexes I and II in the above-described six strains. Consistent with the findings for the spds knockdown strains, the transcription levels of most subunits of mitochondrial complexes I and II were upregulated and the translation levels were downregulated in the six knockdown strains compared with the WT (Fig. 7C). In the six knockdown strains, the protein content of mitochondrial complex I was decreased by more than 40%, and that of complex II was decreased by more than 60% (Fig. 7D and E). In addition, the polyribosome profile analysis revealed that translation was blocked during initiation due to a decrease in polysomes and an increase in 80S monosomes in the eIF5A, dhs, and dohh knockdown strains (Fig. 6G to I). Next, the activities of mitochondrial complexes I, II, III, and IV were tested. Similar to the trends observed in the spds knockdown strains, the activity of mitochondrial complex I was decreased by more than 50% and that of complex II was decreased by more than 70% in the six knockdown strains compared with the WT (Fig. 7F). In contrast, although the activities of mitochondrial complexes III and IV were decreased, the decreases did not exceed 25% (Fig. 7F). Therefore, the decrease in eIF5A hypusination mainly blocked the translation of mitochondrial complex I and II proteins, resulting in the loss of activity of mitochondrial complexes I and II in G. lucidum. Considering all of these results together, we concluded that after the knockdown of the spds gene in G. lucidum, the decrease in eIF5A hypusination was the immediate cause of the blockage of translation initiation and thus caused decreases in the activities of mitochondrial complexes I and II.
eIF5A hypusination modulates the production of ROS and GAs in G. lucidum.
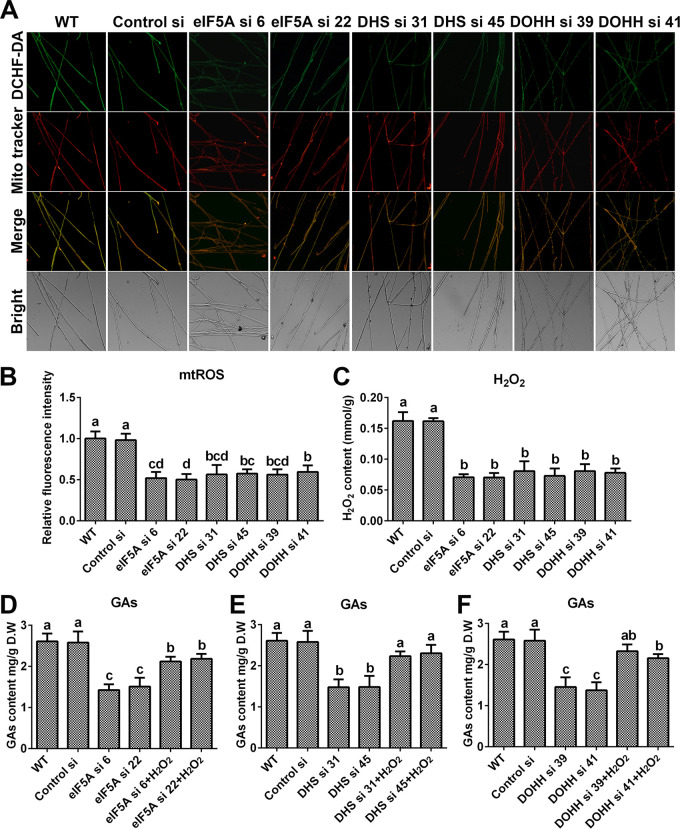
To determine whether spermidine affects mitochondrial ROS homeostasis and GA biosynthesis by hypusinating eIF5A, the mitochondrial ROS levels and GA contents were determined in the eIF5A, dhs, and dohh knockdown strains. As shown in Fig. 8A and B, the mitochondrial ROS levels were decreased by nearly 50% in all six strains compared with the WT, and H2O2 had trends similar to those of mitochondrial ROS (Fig. 8C). In addition, the content of GAs decreased significantly after knocking down the eIF5A, dhs, or dohh gene. Compared with that in the WT, the GA content decreased by more than 40% in the eIF5A and dhs knockdown strains and by more than 50% in the dohh knockdown strain. However, after supplementation with 5 mM H2O2, the GA contents were restored to more than 80% of the WT level in the six knockdown strains (Fig. 8D to F). Both the ROS levels and GA contents in the eIF5A, dhs, and dohh knockdown strains had trends similar to those for the spds knockdown strains. These results showed that eIF5A hypusination promoted mitochondrial ROS production and GA biosynthesis in G. lucidum. In summary, these results suggest that spermidine regulates mitochondrial ROS production and GA biosynthesis by hypusinating eIF5A.
FIG 8.
Knockdown of the eIF5A gene or blockade of eIF5A hypusination leads to decreases in mitochondrial ROS production and GA contents. (A) Mitochondrial ROS levels determined by DCHF-DA and MitoTracker Red double staining in the WT, Control si, and eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. Bar = 50 μm. (B) Relative fluorescence intensities from panel A. (C) H2O2 contents in the WT, Control si, and eIF5A/dhs/dohh knockdown strains from 5-day-old plate mycelia. (D to F) GA contents in the WT, Control si, eIF5A/dhs/dohh knockdown strains, and strains supplied with 5 mM H2O2 from 7-day-old liquid mycelia. The values presented are the means ± SD of data from three independent experiments. Different letters indicate significant differences between the lines (P < 0.05, according to Duncan’s multiple-range test).
DISCUSSION
As a natural polyamine, spermidine is necessary for the survival of organisms and is involved in the regulation of various biological processes. However, the mechanisms by which spermidine affects various physiological functions are poorly understood, especially in microorganisms. In this study, we found that spermidine hypusinates eIF5A to promote the production of mitochondrial ROS and subsequently regulate secondary metabolism in microorganisms. Our study provides a better understanding of the mechanism by which spermidine regulates mitochondrial function and provides new insight into the spermidine-mediated biosynthesis of secondary metabolites.
Our previous studies indicated that the regulation of GA biosynthesis in G. lucidum involves various signals, such as ROS, nitric oxide (NO), and Ca2+ signals, and complex cross talk among them (29, 30). Putrescine and spermidine regulate GA biosynthesis mainly through ROS signaling. However, putrescine and spermidine play different roles in the regulation of ROS production and GA biosynthesis. A previous study demonstrated that putrescine negatively regulates ROS signaling by enhancing the antioxidant enzyme system and subsequently reducing the accumulation of GAs (20). In this research, we found that spermidine positively regulates ROS signaling by enhancing mitochondrial function and subsequently increasing the accumulation of GAs. Importantly, the balance of putrescine and spermidine is an important regulatory mechanism to stabilize redox homeostasis in G. lucidum and to stabilize cellular physiology and metabolism by ROS signaling. In contrast to putrescine, spermidine has little effect on the redox system. The H2O2 produced by spermidine catabolism does not affect redox homeostasis, possibly due to its low content. Moreover, NO signaling is involved in the polyamine regulation of GA biosynthesis and negatively regulates the biosynthesis of GAs in G. lucidum (31, 32). Putrescine promotes the accumulation of NO signaling by enhancing the activity of nitrate reductase and reduces the accumulation of GAs (31). Similarly, in animals, spermidine induces the accumulation of NO signaling by its metabolite γ-aminobutyric acid (GABA) (7). However, the global promotion of GA accumulation by spermidine indicates that NO signaling is not a dominant pathway by which spermidine regulates GA biosynthesis. Furthermore, a previous study demonstrated that polyamine activates Ca2+ signaling in plants (33). Further research is needed to determine whether Ca2+ is also involved in polyamine regulation of GA biosynthesis in G. lucidum. In contrast to their roles in ROS signaling, putrescine and spermidine play similar roles in the regulation of NO and Ca2+ signaling. Notably, environmental stress can create an imbalance of putrescine and spermidine, destroy redox homeostasis, and alter biological processes. In G. lucidum, heat stress induces the conversion of putrescine to spermidine, which promotes the accumulation of ROS signals and the biosynthesis of GAs (19, 21). Spds plays a crucial role in this process. However, the mechanism underlying the heat stress-induced conversion of putrescine to spermidine remains unclear and deserves further attention.
eIF5A is a small, highly expressed, and conserved protein in all eukaryotes, and its activity is strictly dependent on the spermidine level (34). Both eIF5A and its hypusination are essential for cell viability (35, 36). A proteomics analysis in animals showed that 153 proteins were altered significantly after treatment with N1-guanyl-1,7-diaminoheptane (GC7) (an inhibitor of deoxyhypusine synthase), including 63 proteins from mitochondria, such as tricarboxylic acid (TCA) cycle enzymes and ETC proteins, revealing that eIF5A and its hypusination are closely related to mitochondrial function (28). The mitochondrial respiration rate is decreased after inhibition of eIF5A hypusination and results in a decrease in ROS production under anoxic conditions (37). In this work, blockage of eIF5A hypusination blocked the translation of complexes I and II and reduced the number of electrons entering the ETC. Subsequently, the production of mitochondrial ROS was reduced and negatively regulated the biosynthesis of GAs. In mouse kidney cells, the inhibition of eIF5A hypusination by GC7 has been shown to reduce the entry of pyruvate into mitochondria for aerobic respiration, resulting in the accumulation of cytoplasmic pyruvate (38). In Metarhizium robertsii, the accumulation of cytoplasmic pyruvate eliminates ROS (39). These observations reveal that the blockade of eIF5A hypusination regulates ROS signaling through multiple pathways. However, after blocking eIF5A hypusination, the GA contents did not completely recover to levels equal to those of the WT, even after supplementation with H2O2. We speculate that there are some other factors involved in this process. A metabolome analysis in animals showed that the content of acetyl coenzyme A (Ac-CoA) is significantly reduced after the blockade of eIF5A hypusination (28). Ac-CoA serves as the precursor for GA biosynthesis in G. lucidum. Therefore, we speculate that both signal and substrate regulation after the inhibition of eIF5A hypusination are simultaneously involved in GA biosynthesis. In addition, blockage of eIF5A hypusination reprograms metabolism, promoting the transformation from aerobic respiration to anaerobic glycolysis (38), and the enhancement of anaerobic glycolysis is also involved in the regulation of GA biosynthesis in G. lucidum (40). How metabolic reprogramming regulates secondary metabolism attracts our attention and is an urgent question to be answered.
In conclusion, our data suggested that a lack of spermidine led to a decrease in mitochondrial ROS production and a decrease in GA biosynthesis. Further experiments revealed that the decrease in mitochondrial ROS production was due to the loss of complex I and II activities. In this process, eIF5A and its hypusination played important roles. The lack of spermidine led to deficient eIF5A hypusination, resulting in the blockade of complex I and II protein translation and a subsequent loss of activity. Based on these findings, a potential cascade of cellular events constituting the spermidine-mediated regulation of mitochondrial function is proposed (Fig. 9). Spermidine maintains mitochondrial ROS homeostasis through the regulation of translation and the subsequent activity of complexes I and II by hypusinating eIF5A and regulates GA biosynthesis via mitochondrial ROS signaling. Our work is the first to report a detailed mechanism by which spermidine regulates mitochondrial ROS homeostasis in microorganisms. This research can facilitate studies of the function of spermidine in regulating signal transduction and secondary metabolism in other species.
FIG 9.

Schematic describing a model by which spermidine regulates mitochondrial ROS production and GA biosynthesis by hypusinating eIF5A in Ganoderma lucidum. Spermidine maintains mitochondrial ROS homeostasis through the regulation of translation and subsequent activity of complexes I and II by hypusinating eIF5A and regulates GA biosynthesis by mitochondrial ROS signaling. OXPHOS, oxidative phosphorylation; eIF5AD, deoxyhypusinated eIF5A; eIF5AH, hypusinated eIF5A.
MATERIALS AND METHODS
Strains and culture conditions.
A G. lucidum strain (ACCC53264, provided by the Agricultural Culture Collection of China) was used as the WT. spds and pao knockdown strains and empty vector controls (Control silence (Control si)) were established previously by Tao et al. (19). The strains were grown in CYM (1% maltose, 2% glucose, 0.2% yeast extract, 0.2% tryptone, 0.05% MgSO4·7H2O, and 0.46% KH2PO4 [in the absence of agar for fermentation and in the presence of agar for growth]) for 7 days at 28°C in a shaker incubator at 150 rpm for fermentation. H2O2 (5 mM), N-acetyl-l-cysteine (NAc) (2 mM), and ascorbic acid (Vc) (1 mM) were added to media according to a previously described method (24).
Extraction and detection of GAs.
The GAs were extracted and detected using previously described methods (40). Two hundred milligrams of dried mycelial powder was added to 10 mL of ethanol (95%) for a 2-h sonication. After the mixture was centrifuged at 1,520 × g for 10 min, the supernatant was dried with rotary evaporation. Next, the crude extract was resuspended in methanol (0.5 mL) and analyzed by ultraperformance liquid chromatography (UPLC) (Agilent Technologies, Santa Clara, CA, USA) (32).
Detection of ROS and H2O2.
The level of ROS was assessed by fluorescence analysis according to a previously described method (24). The mycelia were stained with 2′,7′-dichlorodihydro-fluorescein diacetate (DCHF-DA) for 20 min, fluorescence was detected with a fluorescence microscope (Zeiss Axio Imager A1), and the fluorescence intensity was analyzed with ZEN lite software (Zeiss). Similarly, the mycelia were doubly stained with DCHF-DA and MitoTracker Red to detect mitochondrial ROS, as described previously by Liu et al. (24). The mitochondria were stained with MitoTracker Red, and ROS were stained with DCHF-DA for 20 min. Fluorescence was detected with a fluorescence microscope, and the fluorescence intensity was analyzed with ZEN lite software. The content of intracellular H2O2 was determined using a hydrogen peroxide assay kit (Beyotime, China) according to the manufacturer’s protocol.
Enzymatic activity assay.
The activities of APX, CAT, GPX, SOD, and NOX were determined using an ascorbate peroxidase assay kit (Solarbio, China), a catalase assay kit (Solarbio, China), a glutathione peroxidase assay kit (Solarbio, China), a superoxide dismutase assay kit (Solarbio, China), and an NADPH oxidase assay kit (Beyotime, China), respectively, according to the manufacturers’ protocols. Respiratory chain complex enzyme activity assays were performed using mitochondrial complex I, II, III, and IV assay kits (Solarbio, China) according to the manufacturer’s protocols, as described previously by Liu et al. (24).
Construction of RNA interference plasmids and gene knockdown strains.
The G. lucidum RNA interference (RNAi) knockdown vector was constructed as previously described (41). The complex I assembly factor NDUFAF1 (CIA30) gene and the complex II succinate dehydrogenase subunit B (SDHB) gene were amplified by PCR using G. lucidum cDNA as the template with primers CIA30-F/R and SDHB-F/R (Table 1), respectively (24), to construct the complex I and complex II RNA cointerference plasmid. The eIF5A, dhs, and dohh genes were amplified by PCR using G. lucidum cDNA as a template with primers eIF5A-F/R, DHS-F/R, and DOHH-F/R (Table 1), respectively, to construct eIF5A, dhs, and dohh RNA interference plasmids. RNA interference plasmids were transferred into G. lucidum according to a previously described method (32). Two independent knockdown strains with the highest efficiency for each interference plasmid were selected for subsequent experiments.
TABLE 1.
Primers used for the construction of gene knockdown strainsa
| Primer | Sequence (5′–3′) | Description |
|---|---|---|
| CIA30 si-F | GGTACCAAACCTCAGTCAACCCTCTT | Gl26075; CIA30 |
| CIA30 si-R | ACTAGTAGTCAATGCCCAGCTCATA | |
| RT-CIA30-F | TCCCATCACAACCGACCTA | |
| RT-CIA30-R | TGTTTCCGCCCAACAGAG | |
| SDHB si-F | ACTAGTTTGGAACCCAGATGAGCC | Gl15912; complex II succinate dehydrogenase subunit B |
| SDHB si-R | TCTAGATCCTGTCCTCCTGCGACT | |
| RT-SDHB-F | TGCGGACCCATGATTCTC | |
| RT-SDHB-R | ATCCTTGCTCGCGTTTGT | |
| eIF5A si-F | GGTACCGTCCCTGCAAGATCGTCG | Gl21362; eukaryotic translation initiation factor 5A |
| eIF5A si-R | ACTAGTGCCTGCTCCTCACCCATA | |
| RT-eIF5A-F | CGATGAGCAGCACAACCA | |
| RT-eIF5A-R | GCCCTTGATGACGACGTG | |
| DHS si-F | GGTACCCTGGGCGTACAAAGTGAA | Gl31472; deoxyhypusine synthase |
| DHS si-R | ACTAGTCAAAGGTGGCTGCTACAA | |
| RT-DHS-F | CCTCGGCTACACCTCCAA | |
| RT-DHS-R | CGATGCGGTTCATTCCAC | |
| DOHH si-F | GGTACCGGCATCTCCTTCCTCGTT | Gl23607; deoxyhypusine hydroxylase |
| DOHH si-R | ACTAGTGCCGCTTCATCCACTCCT | |
| RT-DOHH-F | ATCATCTCGAAGGTGCCTAT | |
| RT-DOHH-R | CGAGGACGGACTCCAGTA | |
The sequences with underline were restriction sites that were used for plasmid construction. Gl, Ganoderma lucidum.
Gene expression analysis.
Gene expression analysis was performed according to a previously reported method (40). Total RNA was isolated from 50 mg fresh mycelia with 1 mL RNAiso Plus (TaKaRa) and reverse transcribed to cDNA with a PrimeScript RT reagent kit (TaKaRa). For quantitative real-time PCR (qRT-PCR) analysis, a SYBR kit (TaKaRa) was used according to the manufacturer’s instructions. The relative expression levels were calculated with the 2−ΔΔCT method (primers are listed in Tables 1 to 3).
TABLE 2.
Primers used for transcription and translation analyses of mitochondrial complex Ia
| Primer | Sequence (5′–3′) | Description |
|---|---|---|
| Gl23611-RT-F | ACCTTGTTGGTCGGGAGT | NDUFA9 |
| Gl23611-RT-R | TGTGGGATGCGTTGAGAT | |
| Gl26075-RT-F | TCCCATCACAACCGACCTA | CIA30 |
| Gl26075-RT-R | TGTTTCCGCCCAACAGAG | |
| Gl21014-RT-F | AGTGGAACGGTTTCAGCATC | NDUFS1 |
| Gl21014-RT-R | CAGGGATCTCCTTCGTATTGAC | |
| Gl16611-RT-F | GCCCAACAACTGTCGCTAA | NDUFV1 |
| Gl16611-RT-R | GGATGCTCATCTCCTCCTCTAC | |
| GL15140-RT-F | CCAACCTCCTTGAGATGC | NDUFV2 |
| GL15140-RT-R | GTCCTTGGTGGTTTGACC | |
| GL30051-RT-F | CGCCTTCACGCCGCATAT | NDUFS2 |
| GL30051-RT-R | GGTCCGCTCCTTCCAGATA | |
| Gl29889-RT-F | CAACCTTATTCATTTGCGTCTG | NDUFS3 |
| Gl29889-RT-R | CTTCGCCAATCTGCTCCC | |
| GL17543-RT-F | CTGGGCGTCGTGTTTCGT | NDUFS7 |
| GL17543-RT-R | GGTAGTAACCTCCTCCATTCG | |
| GL31498-RT-F | CTGTTTCTCGCTCCTTCG | NDUFS8 |
| GL31498-RT-R | CACTTGGACCATCCTTGTAG | |
| 18S-RT-F | TCGAGTTCTGACTGGGTTGT | 18S rRNA gene as a reference gene |
| 18S-RT-R | TCCGTTGCTGAAAGTTGTAT | |
NDUFA9, NADH:ubiquinone oxidoreductase alpha subunit 9; CIA30, complex I intermediate-associated protein 30; NDUFS1, NADH:ubiquinone oxidoreductase core subunit S1; NDUFV1, NADH:ubiquinone oxidoreductase core subunit V1.
TABLE 3.
Primers used for transcription and translation analyses of mitochondrial complex IIa
| Primer | Sequence (5′–3′) | Description |
|---|---|---|
| Gl20821-RT-F | TCGGTGGTCAGTCCCTCA | SdhA1 |
| Gl20821-RT-R | CGACGCACTCGCCATTTT | |
| Gl18404-RT-F | AGTCAGGTGCTGTCGTTGGC | SdhA2 |
| Gl18404-RT-R | ACAGTGCTCGCCGTTGGT | |
| Gl17816-RT-F | GACTACTTCAACGTCGCCATCA | SdhA3 |
| Gl17816-RT-R | ACCACGCAGCCGAGCAAA | |
| Gl22529-RT-F | AGAGCGACCTGCTGCTGAG | SdhE |
| Gl22529-RT-R | CCTTGCCCTCGTTCTTCG | |
| Gl15437-RT-F | TCAAGTACGCAGGCAAGGC | SdhC1 |
| Gl15437-RT-R | AACGCAATCCACGAAGGT | |
| Gl24204-RT-F | GCCCGAATCCGTCAAGTA | SdhC2 |
| Gl24204-RT-R | GCAATAATCACAGCAGCACAA | |
| Gl15912-RT-F | TGCGGACCCATGATTCTC | SdhB |
| Gl15912-RT-R | ATCCTTGCTCGCGTTTGT | |
| Gl22227-RT-F | TTCCCTGCTCCCTCCAAA | CybS |
| Gl22227-RT-R | GGCTCAACAACCCACAAAA | |
SdhA1, succinate dehydrogenase subunit A1; CybS, succinate dehydrogenase cytochrome b small subunit.
Western blotting analysis.
Extraction of G. lucidum proteins and Western blotting were performed according to previously described methods (42). Twenty-five micrograms of protein from each sample was separated in a 12% (wt/vol) SDS-PAGE gel, transferred to polyvinylidene difluoride membranes (Bio-Rad), and incubated with the primary antibody anti-eIF5A (1:2,000, rabbit; BBI Solutions), antihypusine (1:2,000, rabbit; Merck), anti-CIA30 (1:2,000, rabbit; CMCTAG), anti-SDHB (1:2,000, rabbit; CMCTAG), or anti-β-tubulin (1:2,000, mouse; CMCTAG). Next, the polyvinylidene difluoride membranes were incubated with goat anti-rabbit IgG or goat anti-mouse IgG (1:5,000) (horseradish peroxidase [HRP]-conjugated) secondary antibodies and observed with an ECL Western blot detection system (Amersham Bioscience). The densities of the bands were assessed using ImageJ 1.8.0 software.
Polyribosome profile analysis.
Polyribosome profile analysis was performed according to previously reported methods with slight modifications (43, 44). One gram of 5-day-old fresh mycelia was pulverized in liquid nitrogen and homogenized in 2 mL of ribosome extraction buffer (50 mM Tris-HCl [pH 7.5], 400 mM KCl, 30 mM MgCl2, 5 mM dithiothreitol, 100 μg·mL−1 cycloheximide, 100 μg·mL−1 chloramphenicol, 1% Triton X-100). The mixture was centrifuged at 2,380 × g for 10 min at 4°C to obtain the supernatant. Two hundred microliters of 20% Triton X-100 was added to the supernatant, and the mixture was centrifuged at 18,630 × g for 20 min at 4°C to obtain the crude extract (supernatant). Five hundred microliters of the ribosome crude extract was layered onto a 10-mL 5 to 50% linear sucrose gradient and centrifuged at 170,000 × g for 2.5 h at 4°C, and the absorbance of each fraction at 254 nm was measured with a fraction collection system (Biocomp).
Mitochondrial complex I and II subunit translation analysis.
According to previously reported methods (43, 45), polysome fractions were collected and precipitated in 2 volumes of ice-chilled ethanol at 4°C for 12 h. After centrifugation at 18,630 × g for 10 min at 4°C, the pellet was collected and resuspended in 50 μL of RNase-free water. Polysomal and subpolysomal RNAs were reverse transcribed to cDNA with a PrimeScript RT reagent kit (TaKaRa). Subsequently, qRT-PCR was performed to analyze the translation levels of complex I and II partial subunits. The relative translation levels were calculated with the 2−ΔΔCT method (primers are listed in Tables 2 and 3).
Statistical analysis.
Every experimental data point was obtained from three independent experiments to ensure reproducible trends and relationships. Each error bar indicates the standard deviation (SD) from the mean obtained from triplicate samples. Differences in mean values between groups were analyzed by one-way analysis of variance followed by Duncan’s multiple-range test. Different letters indicate significant differences between groups (P < 0.05).
Data availability.
The nucleotide sequences used in this study from G. lucidum strain ACCC53264 have been deposited in GenBank under accession numbers OK666533 (eIF5A), OK666534 (dhs), OK666535 (dohh), OK666536 (CIA30), and OK666537 (SDHB).
ACKNOWLEDGMENTS
This work was supported by China Agriculture Research System of MOF and MARA (project number CARS20), the National Natural Science Foundation of China (project number 31972059), the Fundamental Research Funds for the Central Universities (project number KYYZ202002), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (project number KYCX20_0563).
Footnotes
Supplemental material is available online only.
Contributor Information
Mingwen Zhao, Email: mwzhao@njau.edu.cn.
Nicole R. Buan, University of Nebraska—Lincoln
REFERENCES
- 1.Tassoni A, van Buuren M, Franceschetti M, Fornale S, Bagni N. 2000. Polyamine content and metabolism in Arabidopsis thaliana and effect of spermidine on plant development. Plant Physiol Biochem 38:383–393. 10.1016/S0981-9428(00)00757-9. [DOI] [Google Scholar]
- 2.Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. 2013. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci USA 110:2169–2174. 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe S, Kusama-Eguchi K, Kobayashi H, Igarashi K. 1991. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J Biol Chem 266:20803–20809. 10.1016/S0021-9258(18)54780-3. [DOI] [PubMed] [Google Scholar]
- 4.Lee C-Y, Su G-C, Huang W-Y, Ko M-Y, Yeh H-Y, Chang G-D, Lin S-J, Chi P. 2019. Promotion of homology-directed DNA repair by polyamines. Nat Commun 10:65. 10.1038/s41467-018-08011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Igarashi K, Kashiwagi K. 2010. Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42:39–51. 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Madeo F, Bauer MA, Carmona-Gutierrez D, Kroemer G. 2019. Spermidine: a physiological autophagy inducer acting as an anti-aging vitamin in humans? Autophagy 15:165–168. 10.1080/15548627.2018.1530929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. 2018. Spermidine in health and disease. Science 359:eaan2788. 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 8.Hussain A, Nazir F, Fariduddin Q. 2019. Polyamines (spermidine and putrescine) mitigate the adverse effects of manganese induced toxicity through improved antioxidant system and photosynthetic attributes in Brassica juncea. Chemosphere 236:124830. 10.1016/j.chemosphere.2019.124830. [DOI] [PubMed] [Google Scholar]
- 9.Tiburcio AF, Altabella T, Bitrian M, Alcazar R. 2014. The roles of polyamines during the lifespan of plants: from development to stress. Planta 240:1–18. 10.1007/s00425-014-2055-9. [DOI] [PubMed] [Google Scholar]
- 10.Alcazar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF. 2010. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231:1237–1249. 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- 11.Wimalasekera R, Tebartz F, Scherer GF. 2011. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci 181:593–603. 10.1016/j.plantsci.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Wongsadee T, Vatanyoopaisarn S, Rungsardthong V, Thumthanaruk B, Puttanlek C, Uttapap D, Wetprasit N. 2021. Effect of polyamine on growth, intermediates and 2-acetyl-1-pyrroline formation by Aspergillus awamori. Flavour Fragr J 36:395–403. 10.1002/ffj.3651. [DOI] [Google Scholar]
- 13.Zhgun AA, Nuraeva GK, Dumina MV, Voinova TM, Dzhavakhiya VV, Eldarov MA. 2019. 1,3-Diaminopropane and spermidine upregulate lovastatin production and expression of lovastatin biosynthetic genes in Aspergillus terreus via LaeA regulation. Appl Biochem Microbiol 55:243–254. 10.1134/S0003683819020170. [DOI] [Google Scholar]
- 14.Martin J, Garcia-Estrada C, Rumbero A, Recio E, Albillos SM, Ullan RV, Martin JF. 2011. Characterization of an autoinducer of penicillin biosynthesis in Penicillium chrysogenum. Appl Environ Microbiol 77:5688–5696. 10.1128/AEM.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majumdar R, Lebar M, Mack B, Minocha R, Minocha S, Carter-Wientjes C, Sickler C, Rajasekaran K, Cary JW. 2018. The Aspergillus flavus spermidine synthase (spds) gene, is required for normal development, aflatoxin production, and pathogenesis during infection of maize kernels. Front Plant Sci 9:317. 10.3389/fpls.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiao MS. 2003. Natural products of the medicinal fungus Ganoderma lucidum: occurrence, biological activities, and pharmacological functions. Chem Rec 3:172–180. 10.1002/tcr.10058. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, Ren A, Mu DS, Zhao MW. 2010. Current progress in the study on biosynthesis and regulation of ganoderic acids. Appl Microbiol Biotechnol 88:1243–1251. 10.1007/s00253-010-2871-1. [DOI] [PubMed] [Google Scholar]
- 18.Sanodiya BS, Thakur GS, Baghel RK, Prasad GBKS, Bisen PS. 2009. Ganoderma lucidum: a potent pharmacological macrofungus. Curr Pharm Biotechnol 10:717–742. 10.2174/138920109789978757. [DOI] [PubMed] [Google Scholar]
- 19.Tao YX, Han XF, Ren A, Li J, Song HB, Xie BG, Zhao MW. 2021. Heat stress promotes the conversion of putrescine to spermidine and plays an important role in regulating ganoderic acid biosynthesis in Ganoderma lucidum. Appl Microbiol Biotechnol 105:5039–5051. 10.1007/s00253-021-11373-0. [DOI] [PubMed] [Google Scholar]
- 20.Wu C-G, Tian J-L, Liu R, Cao P-F, Zhang T-J, Ren A, Shi L, Zhao M-W. 2017. Ornithine decarboxylase-mediated production of putrescine influences ganoderic acid biosynthesis by regulating reactive oxygen species in Ganoderma lucidum. Appl Environ Microbiol 83:e01289-17. 10.1128/AEM.01289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, Zhang X, Ren A, Shi DK, Shi L, Zhu J, Yu HS, Zhao MW. 2018. Heat stress-induced reactive oxygen species participate in the regulation of HSP expression, hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum. Microbiol Res 209:43–54. 10.1016/j.micres.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Mu DS, Li CY, Zhang XC, Li XB, Shi L, Ren A, Zhao MW. 2014. Functions of the nicotinamide adenine dinucleotide phosphate oxidase family in Ganoderma lucidum: an essential role in ganoderic acid biosynthesis regulation, hyphal branching, fruiting body development, and oxidative-stress resistance. Environ Microbiol 16:1709–1728. 10.1111/1462-2920.12326. [DOI] [PubMed] [Google Scholar]
- 23.Murray Stewart T, Dunston TT, Woster PM, Casero RA, Jr.. 2018. Polyamine catabolism and oxidative damage. J Biol Chem 293:18736–18745. 10.1074/jbc.TM118.003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Cao PF, Ren A, Wang SL, Yang T, Zhu T, Shi L, Zhu J, Jiang AL, Zhao MW. 2018. SA inhibits complex III activity to generate reactive oxygen species and thereby induces GA overproduction in Ganoderma lucidum. Redox Biol 16:388–400. 10.1016/j.redox.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson A, Hershey JW. 2011. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 108:6415–6419. 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuller AP, Wu CC-C, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66:194–205.e5. 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park MHC, Cooper HL, Folk JE. 1981. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA 78:2869–2873. 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AM, Zhang Y, Flachsmann LJ, Field CS, Patterson AE, Scherer S, Alfei F, Baixauli F, Austin SK, Kelly B, Matsushita M, Curtis JD, Grzes KM, Villa M, Corrado M, Sanin DE, Qiu J, Pallman N, Paz K, Maccari ME, Blazar BR, Mittler G, Buescher JM, Zehn D, Rospert S, Pearce EJ, Balabanov S, Pearce EL. 2019. Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab 30:352–363.e8. 10.1016/j.cmet.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao T, Shi L, Zhang TJ, Ren A, Jiang AL, Yu HS, Zhao MW. 2018. Cross talk between calcium and reactive oxygen species regulates hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum under copper stress. Appl Environ Microbiol 84:e00438-18. 10.1128/AEM.00438-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Shi L, Zhu T, Yang T, Ren A, Zhu J, Zhao MW. 2018. Cross talk between nitric oxide and calcium-calmodulin regulates ganoderic acid biosynthesis in Ganoderma lucidum under heat stress. Appl Environ Microbiol 84:e00043-18. 10.1128/AEM.00043-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia JL, Wu CG, Ren A, Hu YR, Wang SL, Han XF, Shi L, Zhu J, Zhao MW. 2020. Putrescine regulates nitric oxide accumulation in Ganoderma lucidum partly by influencing cellular glutamine levels under heat stress. Microbiol Res 239:126521. 10.1016/j.micres.2020.126521. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Zhu T, Yang T, Yang ZY, Ren A, Shi L, Zhu J, Yu HS, Zhao MW. 2021. Nitric oxide regulates ganoderic acid biosynthesis by the S-nitrosylation of aconitase under heat stress in Ganoderma lucidum. Environ Microbiol 23:682–695. 10.1111/1462-2920.15109. [DOI] [PubMed] [Google Scholar]
- 33.Pottosin I, Velarde-Buendia AM, Bose J, Fuglsang AT, Shabala S. 2014. Polyamines cause plasma membrane depolarization, activate Ca2+-, and modulate H+-ATPase pump activity in pea roots. J Exp Bot 65:2463–2472. 10.1093/jxb/eru133. [DOI] [PubMed] [Google Scholar]
- 34.Coni S, Serrao SM, Yurtsever ZN, Di Magno L, Bordone R, Bertani C, Licursi V, Ianniello Z, Infante P, Moretti M, Petroni M, Guerrieri F, Fatica A, Macone A, De Smaele E, Di Marcotullio L, Giannini G, Maroder M, Agostinelli E, Canettieri G. 2020. Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis 11:1045. 10.1038/s41419-020-03174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JWB. 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol 11:3105–3114. 10.1128/mcb.11.6.3105-3114.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dever TE, Ivanov IP. 2018. Roles of polyamines in translation. J Biol Chem 293:18719–18729. 10.1074/jbc.TM118.003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melis N, Rubera I, Cougnon M, Giraud S, Mograbi B, Belaid A, Pisani DF, Huber SM, Lacas-Gervais S, Fragaki K, Blondeau N, Vigne P, Frelin C, Hauet T, Duranton C, Tauc M. 2017. Targeting eIF5A hypusination prevents anoxic cell death through mitochondrial silencing and improves kidney transplant outcome. J Am Soc Nephrol 28:811–822. 10.1681/ASN.2016010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cougnon M, Carcy R, Melis N, Rubera I, Duranton C, Dumas K, Tanti JF, Pons C, Soubeiran N, Shkreli M, Hauet T, Pellerin L, Giraud S, Blondeau N, Tauc M, Pisani DF. 2021. Inhibition of eIF5A hypusination reprogrammes metabolism and glucose handling in mouse kidney. Cell Death Dis 12:283. 10.1038/s41419-021-03577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, St Leger RJ, Fang WG. 2017. Pyruvate accumulation is the first line of cell defense against heat stress in a fungus. mBio 8:e01284-17. 10.1128/mBio.01284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu YR, Xu WZ, Hu SS, Lian LD, Zhu J, Ren A, Shi L, Zhao MW. 2020. Glsnf1-mediated metabolic rearrangement participates in coping with heat stress and influencing secondary metabolism in Ganoderma lucidum. Free Radic Biol Med 147:220–230. 10.1016/j.freeradbiomed.2019.12.041. [DOI] [PubMed] [Google Scholar]
- 41.Mu DS, Shi L, Ren A, Li MJ, Wu FL, Jiang AL, Zhao MW. 2012. The development and application of a multiple gene co-silencing system using endogenous URA3 as a reporter gene in Ganoderma lucidum. PLoS One 7:e43737. 10.1371/journal.pone.0043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu YR, Xu WZ, Hu SS, Lian LD, Zhu J, Shi L, Ren A, Zhao MW. 2020. In Ganoderma lucidum, Glsnf1 regulates cellulose degradation by inhibiting GlCreA during the utilization of cellulose. Environ Microbiol 22:107–121. 10.1111/1462-2920.14826. [DOI] [PubMed] [Google Scholar]
- 43.Zheng M, Wang YH, Liu X, Sun J, Wang YL, Xu Y, Lv J, Long WH, Zhu XP, Guo XP, Jiang L, Wang CM, Wan JM. 2016. The RICE MINUTE-LIKE1 (RML1) gene, encoding a ribosomal large subunit protein L3B, regulates leaf morphology and plant architecture in rice. J Exp Bot 67:3457–3469. 10.1093/jxb/erw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adjibade P, Grenier St-Sauveur V, Droit A, Khandjian EW, Toren P, Mazroui R. 2016. Analysis of the translatome in solid tumors using polyribosome profiling/RNA-Seq. J Biol Methods 3:e59. 10.14440/jbm.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan TCJ, Knight J, Sbarrato T, Dudek K, Willis AE, Zamoyska R. 2017. Suboptimal T-cell receptor signaling compromises protein translation, ribosome biogenesis, and proliferation of mouse CD8 T cells. Proc Natl Acad Sci USA 114:E6117–E6126. 10.1073/pnas.1700939114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download aem.02037-21-s0001.pdf, PDF file, 0.7 MB (691KB, pdf)
Data Availability Statement
The nucleotide sequences used in this study from G. lucidum strain ACCC53264 have been deposited in GenBank under accession numbers OK666533 (eIF5A), OK666534 (dhs), OK666535 (dohh), OK666536 (CIA30), and OK666537 (SDHB).