ABSTRACT
Fish-pathogenic bacteria of the Tenacibaculum genus are a serious emerging concern in modern aquaculture, causing tenacibaculosis in a broad selection of cultured finfish. Data describing their virulence mechanisms are scarce and few means, antibiotic treatment aside, are available to control their proliferation in aquaculture systems. We genome sequenced a collection of 19 putative Tenacibaculum isolates from outbreaks at two aquaculture facilities and tested their susceptibility to treatment with tropodithietic acid (TDA)-producing Roseobacter group probiotics. We found that local outbreaks of Tenacibaculum can involve heterogeneous assemblages of species and strains with the capacity to produce multiple different virulence factors related to host invasion and infection. The probiotic Phaeobacter piscinae S26 proved efficient in killing pathogenic Tenacibaculum species such as T. maritimum, T. soleae, and some T. discolor strains. However, the T. mesophilum and T. gallaicum species exhibit natural tolerance toward TDA and are hence not likely to be easily killed by TDA-producing probiotics. Tolerance toward TDA in Tenacibaculum is likely involving multiple inherent physiological features pertaining to electron and proton transport, iron sequestration, and potentially also drug efflux mechanisms, since genetic determinants encoding such features were significantly associated with TDA tolerance. Collectively, our results support the use of TDA producers to prevent tenacibaculosis; however, their efficacy is likely limited to some Tenacibaculum species.
IMPORTANCE A productive and sustainable aquaculture sector is needed to meet the UN sustainable development goals and supply the growing world population with high-protein food sources. A sustainable way to prevent disease outbreaks in the industry is the application of probiotic bacteria that can antagonize fish pathogens in the aquaculture systems. TDA-producing Roseobacter group probiotics have proven efficient in killing important vibrio pathogens and protecting fish larvae against infection, and yet their efficacy against different fish pathogenic species of the Tenacibaculum genus has not been explored. Therefore, we tested the efficacy of such potential probiotics against a collection of different Tenacibaculum isolates and found the probiotic to efficiently kill a subset of relevant strains and species, supporting their use as sustainable disease control measure in aquaculture.
KEYWORDS: Tenacibaculum, flavobacteria, roseobacters, probiotics, aquaculture, tropodithietic acid, Phaeobacter
INTRODUCTION
The continuous growth of the human population poses a series of challenges in terms of efficient and sustainable production of high-quality foods. Fish and seafood constitute high-quality protein, and since the 1960s the seafood consumption per capita has doubled, and it is expected to increase even further in the coming years (1). To sustain this development, the aquaculture industry has grown concurrently, producing ∼115 million tons of seafood, exceeding the production from wild catches (1, 2).
Bacterial infections caused by fish pathogenic bacteria are some of the most critical challenges in the efficient production of fish, which are often reared at high densities. Members of the genera Vibrio, Aeromonas, and Yersinia are common pathogens in aquaculture, but, recently, flavobacteria from the Tenacibaculum genus have been observed as emerging fish pathogens in several fish farms (1, 3, 4). Here, Tenacibaculum causes tenacibaculosis in multiple species of finfish, including, but not limited to, red and black sea bream, trout, yellowtail, seabass, turbot, salmon, sole, and seabream (5–11). This broad-host-range pathogen represents a serious threat to a wide selection of marine aquaculture facilities worldwide. Our understanding of virulence factors involved in pathogenesis of Tenacibaculum spp. is limited and has been focused on T. maritimum (previously Flexibacter maritimus [12]), for which multiple genetic factors involved in the infection of fish have been identified through whole-genome sequencing (13). Within this species, factors relating to iron acquisition, degradation and utilization of host tissue, immune system evasion, and toxin production and excretion seem to be important. Furthermore, early symptoms of tenacibaculosis include scale loss, skin ulcerations, and inflammation where the formation of biofilms on the skin and gills is believed to be a prerequisite for infection (14). This is substantiated by the fact that the T. maritimum genome comprises a series of genetic determinants relating to host surface colonization such as adhesins, exopolysaccharide synthesis, and lectin or carbohydrate-binding motifs (13).
T. maritimum is susceptible to multiple antibiotics (11, 15, 16) and their application is a possible treatment of tenacibaculosis. However, the use of antibiotics poses a series of challenges, including poor efficacy when administered in situ (17), but also in terms of sustainable rearing practices. Extensive use of antibiotics, especially in smaller aquaculture facilities, has led to the spread of antibiotic-resistant bacteria in the environment and the spread of resistance determinants between aquatic bacteria and human pathogens (18–21). Therefore, sustainable alternatives to antibiotics are needed to ensure continued efficacy in the treatment of bacterial infections in humans and animals. One sustainable alternative is the development of vaccines. This, however, represents multiple caveats, since fish at the larval stage do not have adaptive immune systems, and vaccines exhibit high specificity to selected target serotypes. Vaccination attempts targeting Tenacibaculum spp. have only had limited success (22, 23) and, to our knowledge, the control of tenacibaculosis in the fish farming industry is currently limited to the administration of antibiotics with the exception of turbot, for which a vaccine is commercially available (16, 24). Another promising alternative disease control strategy is the use of probiotic bacteria that benefit the host health, often by antagonizing fish pathogens. One such group of potential probiotic bacteria is the tropodithietic acid (TDA)-producing roseobacters. These can kill or reduce the growth of several pathogens from the Vibrio genus in multiple different aquaculture relevant systems (25–28). Furthermore, they can improve survival of fish larvae and live feed challenged with pathogenic vibrios (29, 30). The antimicrobial compound TDA acts as a proton antiporter (31) and likely interacts with multiple targets in the cell, ultimately rendering fast-growing pathogens such as vibrios iron- and energy-depleted upon exposure (32). The numerous targets of TDA may be a contributing factor to the observation that the development of resistance is not readily inducible in susceptible organisms (33, 34). However, a substantial number of marine bacteria coinhabiting niches alongside TDA-producing roseobacters have a natural tolerance to TDA (35) and may even exhibit increased abundances in the presence of TDA producers in some cases (36). It is currently unknown to what extent the control of tenacibaculosis with roseobacter probiotics is a viable alternative to treatment with antibiotics and, hence, the aim of the present study is to determine to what extent TDA-producing roseobacters are capable of antagonizing different species of the emerging fish pathogenic Tenacibaculum genus.
RESULTS
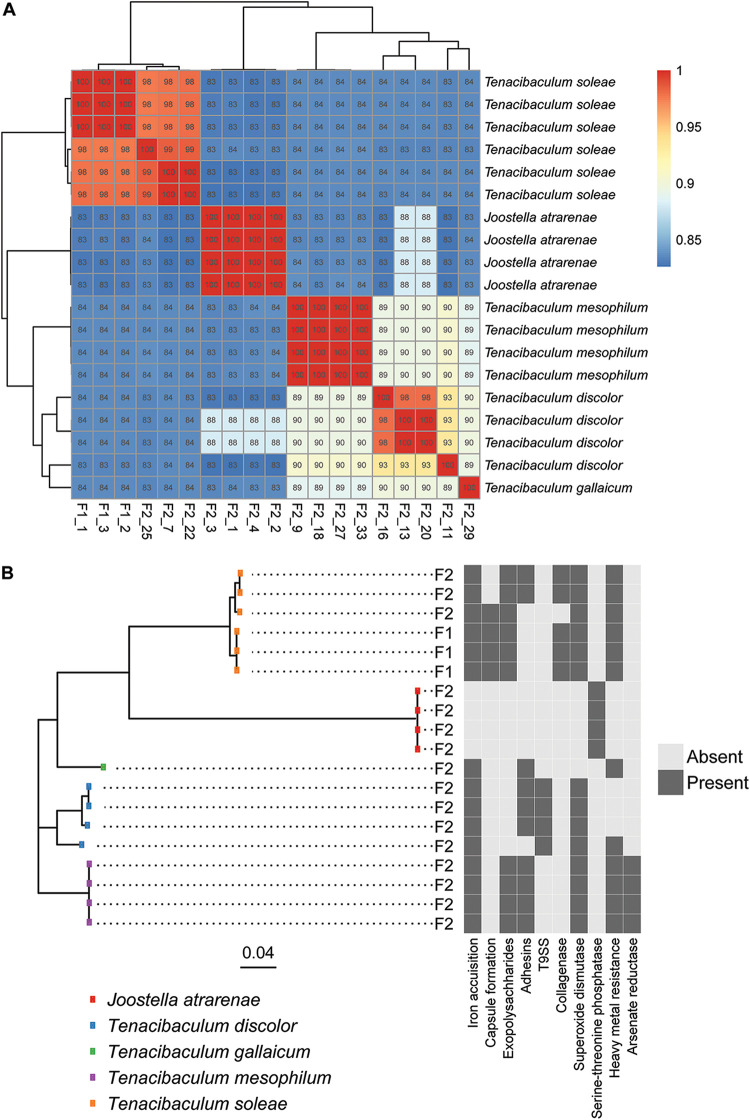
A total of 48 putative pathogenic Tenacibaculum isolates was obtained from disease outbreaks in two aquaculture facilities (F1 and F2) rearing Senegalese sole and seabass/seabream, respectively. An initial screening based on full-length 16S rRNA gene sequences indicated that 19 were flavobacterial isolates (three from F1 and 16 from F2). Fifteen belonged to the Tenacibaculum genus, and four belonged to the closely related Joostella genus (see Table S1 in the supplemental material). The remaining isolates comprised members of the Vibrio, Shewanella, Litoreibacter, Pseudoalteromonas, Bacillus, and Epibacterium genera. Whole-genome sequencing of the 19 flavobacteria and subsequent extraction and BLAST-analyses of the 16S rRNA genes confirmed that from F1, all three flavobacterial isolates belonged to the T. soleae species (Fig. 1A). From F2, the isolates represented T. soleae (three isolates), T. mesophilum (four clonal isolates), T. discolor (four isolates), T. gallaicum (one isolate), and Joostella atrarenae (four clonal isolates). Strains within the T. soleae and T. discolor species exhibited substantial genomic heterogeneity (Fig. 1A), and while the closest known relative to one of the T. discolor isolates, F2_11, was a T. discolor type strain according to the TYGS database, it was below the species-level cutoff in both an average nucleotide identity analysis (ANI; Fig. 1A) and in the TYGS comparisons, suggesting that this could be an undescribed species closely related to T. discolor. Due to the lack of representative sequences in the TYGS database, the identity of the T. gallaicum and the J. atrarenae could not be established beyond 16S rRNA gene phylogeny. Overall, the outbreak at the F2 facility involved a more diverse collection of potential flavobacterial pathogens.
FIG 1.
(A) Genomic heterogeneity among the 19 flavobacterial genomes depicted as the average nucleotide identity (ANI). The assigned taxonomy on the right is based on 16S rRNA gene sequence similarity. (B) Phylogenetic tree based on the alignment of 411 concatenated core genes with the virulence factor profile (a minimum of one gene per category) of the individual isolates on the right. F1 and F2 refers to the two aquaculture facilities, and the scale bar represents substitutions per nucleotide.
Virulence factors and biofilm formation.
Multiple genes encoding putative virulence factors were observed in the genomes of members of the Tenacibaculum genus, corroborating the assumption that these flavobacteria were involved in the disease outbreaks, however, a homolog to the stp gene, encoding a serine-threonine phosphatase (STP) presumed to be important for survival in the infected host, was the only virulence factor identified in the Joostella isolates from F2 (Fig. 1B). In contrast, this gene was not observed in any of the Tenacibaculum isolates. The most commonly observed virulence factors among Tenacibaculum isolates related to iron acquisition factors, manganese-dependent superoxide dismutases (SodA), heavy metal resistance factors, and exopolysaccharides and adhesins involved in biofilm formation (Fig. 1B).
Isolates of the T. soleae species (F1_1, F1_2, F1_3, F2_7, F2_22, and F2_25) represented three different clades (Fig. 1), all carrying virulence genes involved in iron acquisition, exopolysaccharide production, superoxide dismutase production, and heavy metal resistance. In addition, genes encoding collagenases for the degradation of host tissues were unique to the T. soleae species, and yet it was only observed in two of the three clades (Fig. 1B). In contrast to the other members of the species, one clade represented by two strains, F2_7 and F2_22, lacked genetic determinants involved in capsule formation, while they, on the other hand, carried genes relating to adhesin production, a trait not observed in the other clades within the species (Fig. 1B). Similarly, the T. discolor isolates (F2_11, F2_13, F2_16, and F2_20) represented three different clades. Members of all three clades harbored genes relating to iron acquisition and superoxide dismutase production, and in contrast to other members of the genus, all T. discolor genomes encoded a secretion system homologous to the Por system (type IX secretion system; T9SS) from the fish pathogen Flavobacterium psychrophilum. The strain F2_20 lacked the adhesin homologs present in the other T. discolor isolates and carried heavy metal resistance determinants (Fig. 1B). T. mesophilum isolates were highly genetically homogeneous, and all carried virulence factors relating to iron acquisition, exopolysaccharide and adhesin production, superoxide dismutase production, heavy metal resistance, and arsenate reductase production.
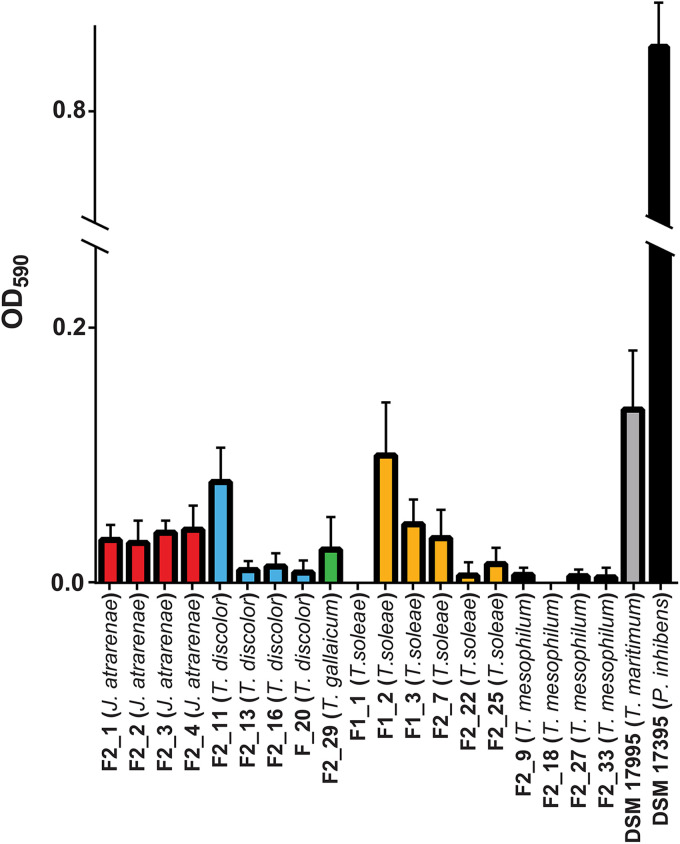
Biofilm formation is believed to be a key virulence factor in the early stages of infection by Tenacibaculum spp. Furthermore, biofilm formation in T. maritimum may facilitate persistence and reinfection in aquaculture facilities, and hence we sought to determine whether the isolates were proficient biofilm formers in vitro compared to the probiotic strain using a modified O’Toole-Kolter biofilm assay. Biofilm formation was observed for several isolates, although the ability to form biofilm was highly species and/or strain-specific (Fig. 2). The type strain T. maritimum DSM 17995 was the most proficient biofilm former of the tested flavobacteria, but none of the isolated pathogenic species, nor the included Tenacibaculum type strains, were as efficient in forming biofilm on abiotic surfaces as the potential probiotic roseobacter control strain Phaeobacter inhibens DSM 17395, which exhibited significantly higher optical density at 590 nm (OD590) values (one-way analysis of variance [ANOVA], Tukey’s post hoc, P < 0.05; Fig. 2).
FIG 2.
Biofilm formation by the 19 flavobacterial isolates, the Tenacibaculum type strains T. maritimum DSM 17995 and T. mesophilum DSM 17364, and the roseobacter Phaeobacter inhibens DSM 17395 (positive control). The OD590 values from the negative control (MB medium) were subtracted from the measurements. Error bars represent the standard deviations between biological triplicates.
Killing of flavobacterial pathogens by Phaeobacter piscinae S26.
TDA-producing roseobacters are proficient biofilm formers on biotic and abiotic surfaces, and the production of TDA (per cell) is enhanced at the biofilm stage (37). We therefore envision that the roseobacters will be introduced, e.g., as biofilm on algal cultures that are used as live feed in marine fish larval rearing. We therefore applied a combined surface-liquid system in the investigation of the potential antagonism of a TDA-producing Phaeobacter piscinae S26 (38) against selected representatives of the flavobacterial isolates, as well as the type strain T. maritimum DSM 17995. P. piscinae S26 biofilms were established on the pegs of minimum biofilm eradication concentration (MBEC) assay plates with a density of log10 6.52 ± 0.147 CFU peg−1. Subsequently, planktonic cultures of T. soleae F1_2, J. atrarenae F2_1, T. mesophilum F2_9, T. discolor F2_11, T. gallaicum F2_29, and T. maritimum DSM 17995 were exposed to pegs colonized with S26 for 4 days. With this approach, substantial killing (>1 log10-fold reduction) of flavobacterial strains was observed for four of the six tested strains and, as for biofilm formation, susceptibility to the probiotic treatment was highly species specific. The pathogenic T. maritimum DSM 17995 type strain was the most susceptible, exhibiting a >5-fold log10 reduction compared to the untreated control (P < 0.001; Table 1). For T. gallaicum F2_29 and T. mesophilum F2_9, little to no effect of the treatment was observed, whereas pronounced inhibition was observed for Tenacibaculum soleae F1_2, J. atrarenae F2_1, and T. discolor F2_11 with abundances 1.78 to 2.44 orders of magnitude lower in the presence of the probiotic strain. The observed differences in efficacy of the treatment were due to variations in TDA sensitivity between flavobacterial species as sizeable zones of inhibition (ZOIs) were observed when the TDA-producing P. piscinae S26 and P. inhibens DSM 17395 strains were spotted onto lawns of T. soleae F1_2, J. atrarenae F2_1, T. discolor F2_11, and T. maritimum DSM 17995, whereas little to no ZOIs were observed on T. mesophilum F2_9 and T. gallaicum F2_29 lawns. The ZOIs observed were similar to—and in the case of T. maritimum DSM 17995 larger than—the zones produced on the highly susceptible control strain Vibrio anguillarum 90-11-286. ZOIs were not observed when spotting a P. inhibens DSM 17395 ΔtdaB mutant against any of the strains (Fig. 3). No homologs to the known TDA resistance genes present in the genomes of TDA-producing roseobacters were found in the genomes of the flavobacterial isolates, and hence these strains draw on alternative means to evade the antibacterial effects of TDA.
TABLE 1.
Killing of representative flavobacterial isolates and the T. maritimum DSM 17995 type strain by Phaeobacter piscinae 26 biofilms
| Strain (species) | Mean ± SD |
Log10 reduction | P a | |
|---|---|---|---|---|
| Log10 CFU mL−1 after exposure to S26 | Control log10 CFU mL−1 | |||
| F1_2 (T. soleae) | 6.17 ± 0.28 | 8.45 ± 0.04 | 2.28 | 0.031 |
| F2_1 (J. atrarenae) | 6.83 ± 0.38 | 9.27 ± 0.03 | 2.44 | 0.0018 |
| F2_9 (T. mesophilum) | 8.39 ± 0.04 | 9.04 ± 0.08 | 0.65 | 0.017 |
| F2_11 (T. discolor) | 7.24 ± 0.20 | 9.02 ± 0.13 | 1.78 | <0.001 |
| F2_29 (T. gallaicum) | 8.84 ± 0.30 | 8.63 ± 0.23 | –0.21 | 0.39 |
| DSM 17995 (T. maritimum) | <3b | 8.40 ± 0.14 | >5.40 | <0.001 |
As determined by a paired t test.
Below the detection limit of 3.
FIG 3.
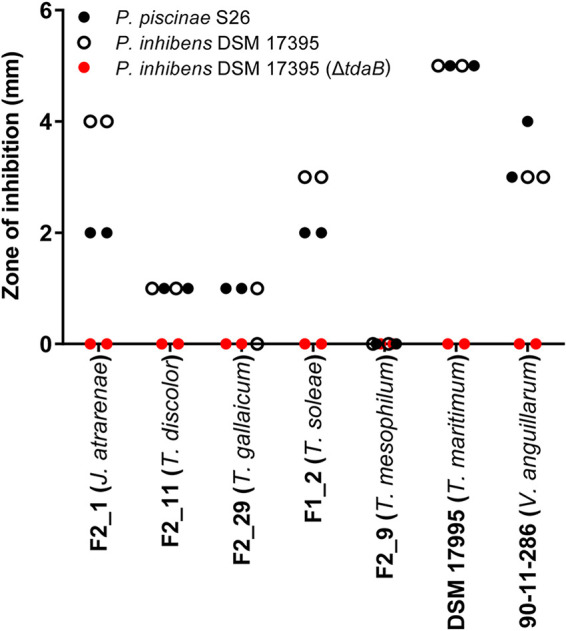
Zones of inhibition around roseobacter colonies spotted onto lawns of Joostella atrarenae F2_1, Tenacibaculum discolor F2_11, Tenacibaculum gallaicum F2_29, Tenacibaculum soleae F1_2, Tenacibaculum mesophilum F2_9, the Tenacibaculum maritimum type strain DSM 17955, and the TDA-sensitive Vibrio anguillarum 90-11-286. Zones were measured from the outside of the spot to correct for differences in roseobacter colony sizes on duplicate plates.
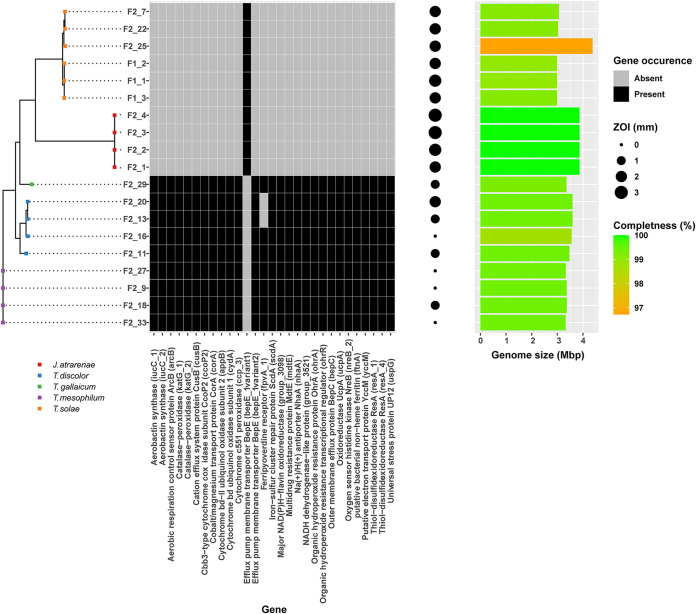
While TDA is known to interfere with ATP generation through disruption of the proton motive force in E. coli (31), gene expression data suggest that TDA may have multiple targets in marine fish- and human-pathogenic Vibrio vulnificus (32). To investigate whether the observed divergence in susceptibility among species and strains could be explained by the presence or absence of specific genes, per-gene Fisher exact tests using Scoary were performed using ZOIs as a proxy for susceptibility. The observed ZOIs were significantly associated with the presence of a total of 219 annotated consensus genes and the absence of nine genes (see Table S2). Among others, the presence of genes involved in putative drug efflux, electron transport and ATP generation, universal stress, oxidative stress, metal ion stress, and iron acquisition were identified as being significantly associated with TDA tolerance (Fig. 4). Species exhibiting mean ZOIs of ≤1 mm all possessed three putative drug efflux-encoding genes, i.e., the TolC-related tripartite efflux-encoding gene mdtE and the efflux pump membrane transporter-encoding genes bepC and bepE, which were absent in susceptible species (mean ZOIs > 1 mm), with the exception of F2_20, which was the only T. discolor strain exhibiting some susceptibility. Generally, susceptible strains harbored a divergent homolog to one of the efflux genes, bepE, yet it exhibited <70% sequence similarity to the gene found in tolerant strains. In addition, tolerant species possessed the gene encoding the Na+/H+ antiporter, NhaA, which facilitates the exchange of protons for sodium ions across the membrane. The same species harbored a series of genes related to electron transport, including the putative electron transport protein YccM, several cytochrome oxidases, oxidoreductases, and regulatory genes involved in oxygen sensing and respiration control (Fig. 4). Except for T. discolor F2_20, multiple stress-related genes were observed in species exhibiting tolerance toward TDA, including a universal stress protein-encoding gene homologous to uspG in E. coli, which is involved in regulation of cell motility in response to starvation, heat shock, and exposure to toxic substances. Furthermore, genes related to oxidative stress (peroxidase-encoding genes) and metal ion stress/efflux (cusB and corA) associated with TDA tolerance. In contrast to the susceptible strains, tolerant isolates also carried the capacity to synthesize a putative aerobactin-like siderophore and the iron storage protein ferritin. However, confirmation of the ability to produce siderophores under the tested laboratory conditions was not possible for any of the strains (see Table S3). All aforementioned traits were conserved at the species level; however, the T. discolor isolates represented more pronounced overall genomic heterogeneity within the species (Fig. 1), which also came across as a relatively high degree of variation in the ZOI sizes among isolates (x- = 1 ± 0.7 mm). Two T. discolor isolates exhibiting smaller ZOIs of 0 mm (F2_16) and 1 mm (F2_11) both harbored a gene homologous to a ferripyoverdine receptor, while the other two isolates did not (F2_20; 2 mm ZOI, F2_13; 1 mm ZOI).
FIG 4.
Gene occurrence profile of selected genes significantly associated with tolerance toward TDA in the 19 flavobacterial isolates. The sizes of the zones of inhibition are used as proxies for TDA susceptibility in the analysis. The genome completeness and size of the assembled genomes are presented as a bar chart on the right.
DISCUSSION
Cost-efficient rearing of fish at high densities is challenged by outbreaks of opportunistic pathogenic bacteria. To mitigate this issue while ensuring sustainability, probiotics have been suggested as an alternative to the prophylactic use of antibiotics to control pathogens and prevent disease outbreaks (2, 28). Currently, little is known about the virulence factors of the emerging broad-host-range pathogenic flavobacterial genus Tenacibaculum, let alone its susceptibility to probiotic treatment, and the results reported here demonstrate that outbreaks of flavobacterial fish pathogens may involve heterogeneous assemblages of strains that likely draw upon multiple different virulence factors. Furthermore, these diverse pathogenic strains exhibit different levels of susceptibility to the promising TDA-producing probiotic candidate P. piscinae S26 (28, 29). However, several strains were highly sensitive, indicating that TDA-producing roseobacters could hold potential in control of tenacibaculosis.
The 15 Tenacibaculum isolates obtained were distributed among four different species, all previously shown to be associated with disease in cultured finfish (5, 39–41). At F1, only three clonal isolates were recovered, whereas the outbreak at F2 exhibited significant heterogeneity within the genus and within T. discolor isolates, corroborating the notion, that such local Tenacibaculum epidemics can involve multiple different strains, hence complicating efforts to produce effective vaccines (42). Interestingly, four clonal isolates of a closely related genus, Joostella, was also recovered from F2, although isolates of this genus have previously only been recovered from environmental sources (43, 44) and has to our knowledge not been associated with disease outbreaks in aquaculture. To what extent these bacteria are actively involved in infection remains to be determined, but the four Joostella strains did carry the genetic capacity to produce a single, though unique potential virulence factor, namely, an STP, which have been reported to be important for the regulation of virulence expression and survival within infected mammalian hosts (45, 46).
Within the genus Tenacibaculum, fish-pathogenic lineages are dispersed among lineages of environmental strains across clades, suggesting parallel evolution of traits involved in pathogenicity (39). While all 15 Tenacibaculum strains in this study possessed genetic determinants relevant for iron acquisition, our results corroborate the observation of parallel evolution as no single virulence gene was conserved across all 15 genomes. In addition to iron acquisition factors, which may be broadly employed as a means to sequester iron from host proteins in flavobacterial fish pathogens (47), mechanisms to evade the reactive oxygen species (ROS) from host macrophages were widely distributed among the isolates as well. This suggests that mechanisms involved in survival within the host are essential and similar across Tenacibaculum species, while the mechanisms by which they gain entry into the host, e.g., through adhesion, collagenase production, or secretion systems, deviate.
Tenacibaculosis is generally assumed to be preceded by biofilm formation on skin and gills (14). T. maritimum has recently been shown to form biofilms on inert surfaces as well, allowing it to persist in the aquaculture environment (48). We observed biofilm formation by T. maritimum DSM 17995 and a subset of the T. soleae and T. discolor strains on inert surfaces. However, the TDA-producing roseobacter P. inhibens DSM 17395 control strain was much more proficient in biofilm formation. The closely related species Phaeobacter gallaeciensis 2.10 is capable of successfully invading and dominating marine epiphytic biofilms (49), and as we imagine probiotic TDA-producing Phaeobacter spp. (49), being administered as biofilms on live feed organisms, or as biofilter constituents (2), the pronounced difference in biofilm production between the pathogens and the roseobacter is a promising observation. Pursuing this line of thinking further, we investigated the ability of pregrown biofilms of the indigenous aquaculture probiotic P. piscinae S26 (29) to kill selected representative pathogenic strains.
While TDA can inhibit or kill a wide range of Gram-positive and Gram-negative bacteria, including fish and human pathogens (28, 29, 33, 50–55), a high number of naturally tolerant strains have been observed in marine microbial communities. Furthermore, the impact of TDA or TDA-producing microorganisms on microbiomes associated with eukaryotes, including aquaculture relevant live feed organisms, fish eggs, and larvae, is subtle and may only affect specific species within a genus (36, 56). Accordingly, we observed widely different killing effects of P. piscinae S26 biofilms on the different Tenacibaculum species. The fish pathogenic type strain T. maritimum DSM 17995, and the T. soleae, and T. discolor representative strains were efficiently killed, whereas the selected T. mesophilum and T. gallaicum strains were largely unaffected. We could confirm that tolerance toward TDA was the cause of the differential killing efficacy observed between the species, so TDA-producing probiotics may only provide efficient protection against a subset of Tenacibaculum pathogens. Still, more extensive testing in complex nonaxenic live feed systems and challenge trials are needed to ascertain the efficacy of P. piscinae S26 against these pathogens in situ.
TDA tolerance in T. discolor and T. mesophilum was not associated with identified TDA resistance genes (tdaR1-3 [31]) but rather with hitherto-undescribed mechanisms. We identified the presence of no less than 228 annotated genes, which were significantly associated with tolerance, and yet the tolerant phenotype also followed the overall phylogenetic relationship between strains; hence, causality could not be inferred directly from our analysis. Nonetheless, we did observe that the presence of a multidrug efflux pump was significantly associated with tolerance and similar systems have previously been demonstrated to confer drug resistance in fish pathogenic flavobacteria (57). Hence, chromosomally encoded efflux systems may be important for the efficacy of TDA against Tenacibaculum pathogens.
TDA inhibits target bacteria through acidification of the cytosol by import of protons and concurrent export of metal ions. As a result, the proton motive force is disrupted, ATP is depleted, and cells are killed (31). In the human and fish pathogen Vibrio vulnificus, the effects of TDA exposure results in reduced expression of the final parts of the electron transport chain (32), likely resulting in energy depletion and accumulation of intracellular superoxide radicals (58). The tolerant strains all harbored 15 genes involved in oxidative phosphorylation, in its regulation, and in oxidative stress relief. These genes were absent in susceptible strains and, accordingly, differences in TDA susceptibility across the Tenacibaculum genus may be a result of inherent differences in their physiology, specifically pertaining to energy generation and stress responses.
TDA has weak iron chelating properties, and yet its production in laboratory cultures is dependent on the presence of iron. Hence, iron sequestration is likely not the main function of TDA (59). However, when exposed to subinhibitory concentrations of TDA, V. vulnificus responds by upregulating the expression of genes involved in iron sequestration, including siderophore-encoding genes (32). Our findings substantiate the observation that iron is important for the efficacy of TDA since we found the tolerant strains to possess genetic determinants involved in iron sequestration and storage, e.g., through putative siderophores and siderophore receptors. Within the heterogeneous group of T. discolor isolates, the two strains exhibiting highest tolerance to TDA also possessed a ferripyoverdine receptor not recovered in the genomes of the two more susceptible strains.
In conclusion, local outbreaks of flavobacterial fish pathogens may involve heterogeneous groups of strains with the genetic capacity to produce multiple different virulence factors related to host invasion but with largely similar mechanisms of host immune evasion. The probiotic P. piscinae S26 is promising against Tenacibaculum pathogens such as T. maritimum and T. soleae. However, it may not prevent tenacibaculosis altogether, since strains of the T. discolor species, as well as the T. gallaicum and T. mesophilum species, exhibit a natural tolerance toward TDA. While we cannot determine which of the specific traits identified to correlate with tolerance are directly causative, the data suggest that multiple inherent physiological features contribute to TDA tolerance in concert, likely including multidrug efflux systems, electron and proton transport mechanisms, stress response mechanisms, and features involved in iron sequestration and storage.
MATERIALS AND METHODS
Strains and media.
Except for the type strains T. maritimum DSM 17995, which was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DMSZ), and the Vibrio anguillarum 90-11-286 control strain (60, 61), all pathogenic strains were supplied by aquaculture facilities (F1 and F2). Roseobacter group bacteria used in this study included P. piscinae S26 originally isolated from F2 (29, 38), as well as P. inhibens DSM 17395 and its corresponding TDA-deficient mutant (62). All bacteria were grown on marine agar (MA) or in marine broth (MB; Difco 2216) at 25°C for 48 h (with shaking at 200 rpm) unless otherwise stated.
16S rRNA gene sequencing.
For identification of bacterial isolates, colony mass from a single colony was transferred to 50 μL of 0.05 M NaOH in 1.5-mL microcentrifuge tubes. Tubes were incubated at 100°C for 10 min and centrifuged at 11,000 × g for 120 s. One microliter of supernatant was used as the template in a 25-μL PCR containing 2.5 μL of 10 × Hot Start PCR buffer, 2.5 μL of 2 mM dNTPs, 0.8 μL of 12.5 μM 27F primer, 0.8 μL of 12.5 μM 1492R primer (63), 4 μL of 25 mM MgCl2, and 0.125 μL of Maxima Hot Start polymerase in ddH2O (63). The reaction was performed in a Bio-Rad T100 thermal cycler as follows: 1 cycle at 95°C for 300 s; followed by 35 cycles at 95°C for 45 s, 51°C for 60 s, and 72°C for 120 s; followed finally by a cycle at 72°C for 420 s. The PCR product was cleaned by the addition of 0.5 μL of Exonuclease I (EN0581; Thermo Scientific) and 0.5 μL of Fast AP (EF0651, Thermo Scientific), followed by consecutive 15-min incubations at 37°C and 85°C. Amplicons were sequenced at Macrogen Europe (Amsterdam, Netherlands), and forward and reverse reads were trimmed (quality score of 0.05) and assembled (minimum aligned length, 20 nucleotides) using CLC Main Workbench 8.1 (Qiagen Bioinformatics). Some sequences could not be assembled using these criteria, and in these cases the forward and reverse reads were analyzed separately (see Table S1). Identities were assigned by BLASTn against the NCBI database.
DNA extraction and whole-genome sequencing of flavobacteria.
Cells from 2 mL of liquid culture were pelleted by centrifugation at 5,000 × g for 5 min, and DNA was extracted using a NucleoSpin tissue kit (Macherey-Nagel) according to the manufacturer’s instructions, using 10 mM Tris-HCl (pH 8.5) as the elution buffer. DNA concentrations and purity were determined using a Qubit 2.0 fluorometer (Invitrogen) and a DS-11+ NanoDrop spectrophotometer (DeNovix), respectively. Paired-end (2 × 300 PE) sequencing was performed using an Illumina MiSeq platform (V3 chemistry), aiming at a 100× coverage. De novo assembly was performed with SPAdes 3.13.0 using a Shovill pipeline (64, 65). 16S rRNA genes were extracted with Barrnap and aligned to the NCBI database for species verification (66), and the average nucleotide identity (ANI) between the genomes was calculated with MUMmer 3.0 using the Pyani module (67, 68). Genomes were compared to all type strain genomes available in the TYGS database via the MASH algorithm (69) to confirm 16S species verification where possible. Using BLASTn, the genomes were aligned to the “VFDB” virulence factor database (VFDB, 2019), as well as to a custom-made database consisting of the tdaR1-tdaR3 genes encoding the TDA resistance factors in P. inhibens (31), and virulence genes were identified in T. maritimum DSM 17995 (13) and F. psychrophilum (70). Homologs were determined as open reading frames with a minimum sequence similarity of 70% and a minimum length of 30 bp. Genome sizes were determined using Quast, including contigs of >1,000 bp, and genome completeness was estimated with Busco v4.1.4 using the lineage data set flavobacteriales_odb10 (71, 72).
Core genes were identified and aligned using Roary with a blastp cutoff of 70% and if they were present in at least 95% of the sequence isolates (73). The core genes at a nucleotide level were then aligned with MAFFT, and the resulting alignment was used to generate a phylogenetic tree using Fasttree (74).
Biofilm formation.
The ability of the isolates to produce biofilms on abiotic surfaces was determined using an O’Toole-Kolter assay (75) with modifications as described elsewhere (76). All flavobacterial isolates, the type strains T. maritimum DSM 17995 and T. mesophilum DSM 13764, and the positive-control P. inhibens DSM 17395 were grown in 10 mL of MB at 25°C overnight. All bacterial cultures were diluted to an OD600 of 0.01, and 100 μL was transferred in triplicate to a 96-well microtiter plate (Thermo Scientific Nucleon Delta Surface). Sterile medium was used for negative controls. Subsequently, the plate was incubated for 21 h at 25°C, followed by 10 s of shaking. The OD600 was measured using a SpectraMax i3 spectrophotometer. Next, the liquid was removed, the wells were washed three times with 100 μL of dH2O, and the plate was left to dry for 5 min. After drying, 125 μL of a 1% crystal violet (Sigma-Aldrich) solution was added to each well, followed by incubation for 15 min. The crystal violet solution was removed, and the wells were washed three times with 200 μL of dH2O. After the wells were dried for 15 min, 200 μL of 96% ethanol was added, and the plate was incubated for 30 min. Then, 100 μL of the ethanol-crystal violet mixture was added to a new microtiter plate. The plate was shaken for 10 s, followed by measurement of the optical density (OD590). To determine significant differences in biofilm formation by the pathogenic species compared to the probiotic P. inhibens DSM 17395 positive control, one-way ANOVA, followed by Tukey’s test, was performed.
Inhibition of pathogens by P. piscinae S26 biofilms.
The assay was performed following procedures described by D’Alvise et al. (77). Briefly, P. piscinae S26 was incubated in 10 mL of MB overnight at 25°C under stagnant growth conditions. Subsequently, the culture was diluted 40-fold in 200 μL of MB in wells of a 96-well Innovotech MBEC P&G assay plate. The plate was incubated stagnant for 4 days at 25°C. After incubation, duplicate pegs were removed and transferred to 2 mL of 3% Instant Ocean (Instant Ocean sea salts; AquariumSystems, Inc., Sarrebourg, France) and sonicated for 4 min at 28 kHz, followed by vortexing for 30 s to detach bacterial cells from the peg surfaces. A dilution series was plated on MA plates for enumeration. After the removal of excess liquid on the remaining pegs (not sonicated, vortexed) carrying P. piscinae S26 biofilms, the pegs were transferred directly to wells containing MB cultures of the strains F1_2, F2_1, F2_9, F2_11, F2_29, and T. maritimum DSM 17995 in triplicates. Sterile pegs and MB were used for negative controls. The MBEC plates were incubated for 4 days at 25°C under stagnant conditions after which bacterial abundances were determined on MA plates supplemented with 25 μg mL−1 streptomycin. Colony counts were log transformed, and significant changes were determined using F tests for variance, followed by two-tailed t tests (α = 0.05).
TDA susceptibility.
An agar diffusion (spot inhibition) assay was performed to determine the sensitivity of F1_2, F2_1, F2_9, F2_11, F2_29, and T. maritimum DSM 17995 toward TDA. The optical densities of flavobacterial cultures were adjusted to an OD600 of 0.5, and a sterile cotton swab was used to plate culture material onto MA plates to form bacterial lawns. Then, 5 μL of ON culture of the two wild-type TDA producers and the TDA-deficient mutant were spotted onto the surfaces of the lawns in duplicates. The TDA-sensitive V. anguillarum 90-11-286 strain was included as a positive control. ZOIs were measured from the edge of the spot to adjust for the slight variations in roseobacter colony sizes.
Associations of TDA susceptibility and gene occurrence.
Tolerance was assessed with an approach identical to the TDA susceptibility test described above, including all the flavobacterial isolates, but only including P. piscinae S26 as the spotted culture. The resulting ZOIs were used as a proxy for TDA susceptibility. A gene presence/absence matrix was generated based on the de novo-assembled genomes using Roary v3.13.0 with a minimum blastp percentage of 70% (73). Scoary v.1.6.16 was then run on the gene presence absence matrix, and the ZOIs were taken as a trait, using a cutoff of >1 mm (78). Genes with a Bonferroni-corrected P value of <0.05 in the Fisher exact test in Scoary were deemed significantly associated and annotated.
Determination of siderophore production.
Precultures of the representative strains F1_2, F2_1, F2_9, F2_11, F2_29, the type strain T. maritimum DSM 17995, and a siderophore-producing positive-control strain, Pseudoalteromonas piscicida S2040 (79), were diluted 100-fold into triplicate tubes of three different liquid growth substrates: 1.5% SSCAS+GLU (1.5% Sigma Sea Salts, 0.3% Casamino Acids, 0.4% glucose) 1.5% SSCAS+MAN (1.5% Sigma Sea Salts, 0.3% Casamino Acids, 0.4% mannose), and 1/2YTSS (2 g of Bacto yeast extract, 1.25 g of Bacto tryptone, 20 g of Sigma Sea Salts L−1). The cultures were incubated for 7 days at 15°C and 25°C, and sterile-filtered supernatant was subsequently mixed 1:1 with CAS solution (80). The assay reaction was inspected at 2 h, 24 h, and 5 days. Positive reactions were only observed for the positive-control strain, and the growth of the Tenacibaculum strains in the substrates was limited overall.
Data availability.
Partial 16S rRNA gene sequences obtained from the isolates were deposited at the National Center for Biotechnology Information (NCBI) under the accession numbers MN481000 to MN481046, and whole-genome sequencing reads were deposited in the Sequencing Read Archive at the NCBI under accession number PRJNA566079.
ACKNOWLEDGMENTS
We thank Jette Melchiorsen for technical assistance.
This study was supported by the Danish National Research Foundation (DNRF137) and by a grant from The Danish Council for Strategic Research Program Commission on Health, Food, and Welfare (12-132390; ProAqua).
We declare there are no conflicts of interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Mikkel Bentzon-Tilia, Email: mibti@bio.dtu.dk.
Laura Villanueva, Royal Netherlands Institute for Sea Research.
REFERENCES
- 1.FAO. 2020. The state of world fisheries and aquaculture: sustainability in action. FAO, Rome, Italy. [Google Scholar]
- 2.Bentzon-Tilia M, Sonnenschein EC, Gram L. 2016. Monitoring and managing microbes in aquaculture: towards a sustainable industry. Microb Biotechnol 9:576–584. doi: 10.1111/1751-7915.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Done HY, Venkatesan AK, Halden RU. 2015. Does the recent growth of aquaculture create antibiotic resistance threats different from those associated with land animal production in agriculture? AAPS J 17:513–524. doi: 10.1208/s12248-015-9722-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avendaño-Herrera R, Irgang R, Núñez S, Romalde JL, Toranzo AE. 2005. Recommendation of an appropriate medium for in vitro drug susceptibility testing of the fish pathogen Tenacibaculum maritimum. Antimicrob Agents Chemother 49:82–87. doi: 10.1128/AAC.49.1.82-87.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pineiro-Vidal M, Carballas CG, Gomez-Barreiro O, Riaza A, Santos Y. 2008. Tenacibaculum soleae sp. nov., isolated from diseased sole (Solea senegalensis Kaup). Int J Syst Evol Microbiol 58:881–885. doi: 10.1099/ijs.0.65539-0. [DOI] [PubMed] [Google Scholar]
- 6.Avendaño-Herrera R, Magariños B, López-Romalde S, Romalde J, Toranzo A. 2004. Phenotypic characterization and description of two major O-serotypes in Tenacibaculum maritimum strains from marine fishes. Dis Aquat Organ 58:1–8. doi: 10.3354/dao058001. [DOI] [PubMed] [Google Scholar]
- 7.Bernardet J-F, Kerouault B, Michel C. 1994. Comparative study on Flexibacter maritimus strains isolated from farmed sea bass (Dicentrarchus labrax) in France. Fish Pathol 29:105–111. doi: 10.3147/jsfp.29.105. [DOI] [Google Scholar]
- 8.Castro NF, Balboa S, Nuacute S, Toranzo AE, Magarintilde B. 2014. First isolation and characterization of Tenacibaculum soleae from sea bass Dicentrarchus labrax. Fish Pathol 49:16–22. doi: 10.3147/jsfp.49.16. [DOI] [Google Scholar]
- 9.Chen ME, Henry-Ford D, Groff JM. 1995. Isolation and characterization of Flexibacter maritimus from marine fishes of California. J Aquat Anim Health 7:318–326. doi:. [DOI] [Google Scholar]
- 10.Avendaño-Herrera R, Toranzo A, Magariños B. 2006. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis Aquat Organ 71:255–266. doi: 10.3354/dao071255. [DOI] [PubMed] [Google Scholar]
- 11.Handlinger J, Soltani M, Percival S. 1997. The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J Fish Dis 20:159–168. doi: 10.1046/j.1365-2761.1997.00288.x. [DOI] [Google Scholar]
- 12.Suzuki M, Nakagawa Y, Harayama S, Yamamoto S. 2001. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov. Int J Syst Evol Microbiol 51:1639–1652. doi: 10.1099/00207713-51-5-1639. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Pascual D, Lunazzi A, Magdelenat G, Rouy Z, Roulet A, Lopez-Roques C, Larocque R, Barbeyron T, Gobet A, Michel G, Bernardet J-F, Duchaud E. 2017. The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front Microbiol 8:1542. doi: 10.3389/fmicb.2017.01542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avendano-Herrera R, Toranzo AE, Magarinos B. 2006. A challenge model for Tenacibaculum maritimum infection in turbot, Scophthalmus maximus (L.). J Fish Dis 29:371–374. doi: 10.1111/j.1365-2761.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- 15.Florio D, Gridelli S, Fioravanti ML, Zanoni RG. 2016. First isolation of Tenacibaculum maritimum in a captive sand tiger shark (Carcharias taurus). J Zoo Wildl Med 47:351–353. doi: 10.1638/2015-0064.1. [DOI] [PubMed] [Google Scholar]
- 16.Avendaño-Herrera R, Núñez S, Barja JL, Toranzo AE. 2008. Evolution of drug resistance and minimum inhibitory concentration to enrofloxacin in Tenacibaculum maritimum strains isolated in fish farms. Aquacult Int 16:1–11. doi: 10.1007/s10499-007-9117-y. [DOI] [Google Scholar]
- 17.Cepeda C, Santos Y. 2002. First isolation of Flexibacter maritimus from farmed Senegalese sole (Solea senegalensis, Kaup) in Spain. Bull Eur Assoc Fish Pathol 22:388–392. [Google Scholar]
- 18.Cabello FC. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Pickup RW. 2000. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl Environ Microbiol 66:3883–3890. doi: 10.1128/AEM.66.9.3883-3890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda CD, Godoy FA, Lee MR. 2018. Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front Microbiol 9:1284. doi: 10.3389/fmicb.2018.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angulo FJ, Griffin PM. 2000. Changes in antimicrobial resistance in Salmonella enterica serovar typhimurium. Emerg Infect Dis 6:436–438. doi: 10.3201/eid0604.000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Gelderen R, Carson J, Nowak B. 2009. Experimental vaccination of Atlantic salmon (Salmo salar L.) against marine flexibacteriosis. Aquaculture 288:7–13. doi: 10.1016/j.aquaculture.2008.11.012. [DOI] [Google Scholar]
- 23.Småge SB, Frisch K, Vold V, Duesund H, Brevik ØJ, Olsen RH, Sjaatil ST, Klevan A, Brudeseth B, Watanabe K, Nylund A. 2018. Induction of tenacibaculosis in Atlantic salmon smolts using Tenacibaculum finnmarkense and the evaluation of a whole cell inactivated vaccine. Aquaculture 495:858–864. doi: 10.1016/j.aquaculture.2018.06.063. [DOI] [Google Scholar]
- 24.Toranzo AE, Magariños B, Romalde JL. 2005. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246:37–61. doi: 10.1016/j.aquaculture.2005.01.002. [DOI] [Google Scholar]
- 25.D’Alvise PW, Lillebø S, Prol-Garcia MJ, Wergeland HI, Nielsen KF, Bergh Ø, Gram L. 2012. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS One 7:e43996. doi: 10.1371/journal.pone.0043996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grotkjær T, Bentzon-Tilia M, D’Alvise P, Dierckens K, Bossier P, Gram L. 2016. Phaeobacter inhibens as probiotic bacteria in non-axenic Artemia and algae cultures. Aquaculture 462:64–69. doi: 10.1016/j.aquaculture.2016.05.001. [DOI] [Google Scholar]
- 27.Rasmussen BB, Erner KE, Bentzon-Tilia M, Gram L. 2018. Effect of TDA-producing Phaeobacter inhibens on the fish pathogen Vibrio anguillarum in non-axenic algae and copepod systems. Microb Biotechnol 11:1070–1079. doi: 10.1111/1751-7915.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenschein EC, Jimenez G, Castex M, Gram L. 2021. The Roseobacter-group bacterium Phaeobacter as a safe probiotic solution for aquaculture. Appl Environ Microbiol 87:1–15. doi: 10.1128/AEM.02581-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grotkjær T, Bentzon-Tilia M, D’Alvise P, Dourala N, Nielsen KF, Gram L. 2016. Isolation of TDA-producing Phaeobacter strains from sea bass larval rearing units and their probiotic effect against pathogenic Vibrio spp. in Artemia cultures. Syst Appl Microbiol 39:180–188. doi: 10.1016/j.syapm.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 30.D’Alvise PW, Lillebø S, Wergeland HI, Gram L, Bergh Ø. 2013. Protection of cod larvae from vibriosis by Phaeobacter spp.: a comparison of strains and introduction times. Aquaculture 384–387:82–86. doi: 10.1016/j.aquaculture.2012.12.013. [DOI] [Google Scholar]
- 31.Wilson MZ, Wang R, Gitai Z, Seyedsayamdost MR. 2016. Mode of action and resistance studies unveil new roles for tropodithietic acid as an anticancer agent and the γ-glutamyl cycle as a proton sink. Proc Natl Acad Sci USA 113:1630–1635. doi: 10.1073/pnas.1518034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dittmann KK, Porsby CH, Goncalves P, Mateiu RV, Sonnenschein EC, Bentzon-Tilia M, Egan S, Gram L. 2019. Tropodithietic acid induces oxidative stress response, cell envelope biogenesis and iron uptake in Vibrio vulnificus. Environ Microbiol Rep 11:581–588. doi: 10.1111/1758-2229.12771. [DOI] [PubMed] [Google Scholar]
- 33.Porsby CH, Webber MA, Nielsen KF, Piddock LV, Gram L. 2011. Resistance and tolerance to tropodithietic acid, an antimicrobial in aquaculture, is hard to select. Antimicrob Agents Chemother 55:1332–1337. doi: 10.1128/AAC.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen BB, Grotkjær T, D’Alvise PW, Yin G, Zhang F, Bunk B, Spröer C, Bentzon-Tilia M, Gram L. 2016. Vibrio anguillarum is genetically and phenotypically unaffected by long-term continuous exposure to the antibacterial compound tropodithietic acid. Appl Environ Microbiol 82:4802–4810. doi: 10.1128/AEM.01047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington C, Reen FJ, Mooij MJ, Stewart FA, Chabot J-B, Guerra AF, Glöckner FO, Nielsen KF, Gram L, Dobson ADW, Adams C, O’Gara F. 2014. Characterization of non-autoinducing tropodithietic acid (TDA) production from marine sponge Pseudovibrio species. Mar Drugs 12:5960–5978. doi: 10.3390/md12125960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dittmann KK, Sonnenschein EC, Egan S, Gram L, Bentzon-Tilia M. 2019. Impact of Phaeobacter inhibens on marine eukaryote-associated microbial communities. Environ Microbiol Rep 11:401–413. doi: 10.1111/1758-2229.12698. [DOI] [PubMed] [Google Scholar]
- 37.D’Alvise PW, Magdenoska O, Melchiorsen J, Nielsen KF, Gram L. 2014. Biofilm formation and antibiotic production in Ruegeria mobilis are influenced by intracellular concentrations of cyclic dimeric guanosinmonophosphate. Environ Microbiol 16:1252–1266. doi: 10.1111/1462-2920.12265. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenschein EC, Phippen CBW, Nielsen KF, Mateiu RV, Melchiorsen J, Gram L, Overmann J, Freese HM. 2017. Phaeobacter piscinae sp. nov., a species of the Roseobacter group and potential aquaculture probiont. Int J Syst Evol Microbiol 67:4559–4564. doi: 10.1099/ijsem.0.002331. [DOI] [PubMed] [Google Scholar]
- 39.Habib C, Houel A, Lunazzi A, Bernardet J-F, Olsen AB, Nilsen H, Toranzo AE, Castro N, Nicolas P, Duchaud E. 2014. Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl Environ Microbiol 80:5503–5514. doi: 10.1128/AEM.01177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López JR, Piñeiro-Vidal M, García-Lamas N, de la Herran R, Navas JI, Hachero-Cruzado I, Santos Y. 2010. First isolation of Tenacibaculum soleae from diseased cultured wedge sole, Dicologoglossa cuneata (Moreau), and brill, Scophthalmus rhombus (L.). J Fish Dis 33:273–278. doi: 10.1111/j.1365-2761.2009.01105.x. [DOI] [PubMed] [Google Scholar]
- 41.Pineiro-Vidal M, Riaza A, Santos Y. 2008. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int J Syst Evol Microbiol 58:21–25. doi: 10.1099/ijs.0.65397-0. [DOI] [PubMed] [Google Scholar]
- 42.Olsen AB, Gulla S, Steinum T, Colquhoun DJ, Nilsen HK, Duchaud E. 2017. Multilocus sequence analysis reveals extensive genetic variety within Tenacibaculum spp. associated with ulcers in sea-farmed fish in Norway. Vet Microbiol 205:39–45. doi: 10.1016/j.vetmic.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 43.Quan ZX, Xiao YP, Roh SW, Do Nam Y, Chang HW, Shin KS, Rhee SK, Park YH, Bae JW. 2008. Joostella marina gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from then East Sea. Int J Syst Evol Microbiol 58:1388–1392. doi: 10.1099/ijs.0.65611-0. [DOI] [PubMed] [Google Scholar]
- 44.Kim B-C, Lee KH, Kim MN, Jung MY, Chang Y-H, Lee J, Shin K-S. 2011. Joostella atrarenae sp. nov., a novel member of the Flavobacteriaceae originating from the black sea sand of Jeju Island. Curr Microbiol 62:606–611. doi: 10.1007/s00284-010-9750-y. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal S, Agarwal S, Pancholi P, Pancholi V. 2011. Role of serine/threonine phosphatase (SP-STP) in Streptococcus pyogenes physiology and virulence. J Biol Chem 286:41368–41380. doi: 10.1074/jbc.M111.286690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shakir SM, Bryant KM, Larabee JL, Hamm EE, Lovchik J, Lyons CR, Ballard JD. 2010. Regulatory interactions of a virulence-associated serine/threonine phosphatase-kinase pair in Bacillus anthracis. J Bacteriol 192:400–409. doi: 10.1128/JB.01221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck BH, Li C, Farmer BD, Barnett LM, Lange MD, Peatman E. 2016. A comparison of high- and low-virulence Flavobacterium columnare strains reveals differences in iron acquisition components and responses to iron restriction. J Fish Dis 39:259–268. doi: 10.1111/jfd.12343. [DOI] [PubMed] [Google Scholar]
- 48.Levipan HA, Tapia-Cammas D, Molina V, Irgang R, Toranzo AE, Magariños B, Avendaño-Herrera R. 2019. Biofilm development and cell viability: an undervalued mechanism in the persistence of the fish pathogen Tenacibaculum maritimum. Aquaculture 511:734267. doi: 10.1016/j.aquaculture.2019.734267. [DOI] [Google Scholar]
- 49.Rao D, Skovhus T, Tujula N, Holmström C, Dahllöf I, Webb JS, Kjelleberg S. 2010. Ability of Pseudoalteromonas tunicata to colonize natural biofilms and its effect on microbial community structure. FEMS Microbiol Ecol 73:450–457. doi: 10.1111/j.1574-6941.2010.00917.x. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Ponte C, Cilia V, Lambert C, Nicolas JL. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int J Syst Bacteriol 48:537–542. doi: 10.1099/00207713-48-2-537. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Ponte C, Samain JF, Sánchez JL, Nicolas JL. 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar Biotechnol 1:52–59. doi: 10.1007/pl00011751. [DOI] [PubMed] [Google Scholar]
- 52.Hjelm M, Riaza A, Formoso F, Melchiorsen J, Gram L. 2004. Seasonal incidence of autochthonous antagonistic Roseobacter spp. and Vibrionaceae strains in a turbot larva (Scophthalmus maximus) rearing system. Appl Environ Microbiol 70:7288–7294. doi: 10.1128/AEM.70.12.7288-7294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planas M, Pérez-Lorenzo M, Hjelm M, Gram L, Uglenes Fiksdal I, Bergh Ø, Pintado J. 2006. Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255:323–333. doi: 10.1016/j.aquaculture.2005.11.039. [DOI] [Google Scholar]
- 54.Porsby CH, Nielsen KF, Gram L. 2008. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl Environ Microbiol 74:7356–7364. doi: 10.1128/AEM.01738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W, Dao C, Karim M, Gomez-Chiarri M, Rowley D, Nelson DR. 2016. Contributions of tropodithietic acid and biofilm formation to the probiotic activity of Phaeobacter inhibens. BMC Microbiol 16:1–17. doi: 10.1186/s12866-015-0617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dittmann KK, Rasmussen BB, Melchiorsen J, Sonnenschein EC, Gram L, Bentzon-Tilia M. 2020. Changes in the microbiome of mariculture feed organisms after treatment with a potentially probiotic strain of Phaeobacter inhibens. Appl Environ Microbiol 86:1–17. doi: 10.1128/AEM.00499-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark SE, Jude BA, Danner GR, Fekete FA. 2009. Identification of a multidrug efflux pump in Flavobacterium johnsoniae. Vet Res 40:55. doi: 10.1051/vetres/2009038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poole RK, Cook GM. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol 43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 59.D’Alvise PW, Phippen CBW, Nielsen KF, Gram L. 2016. Influence of iron on production of the antibacterial compound tropodithietic acid and its noninhibitory analog in Phaeobacter inhibens. Appl Environ Microbiol 82:502–509. doi: 10.1128/AEM.02992-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedersen K, Larsen JL. 1993. rRNA gene restriction patterns of Vibrio anguillarum serogroup O1. Dis Aquat Org 16:121–126. doi: 10.3354/dao016121. [DOI] [Google Scholar]
- 61.Skov MN, Pedersen K, Larsen JL. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl Environ Microbiol 61:1540–1545. doi: 10.1128/aem.61.4.1540-1545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, Gallant É, Seyedsayamdost MR. 2016. Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. mBio 7:e02118. doi: 10.1128/mBio.02118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lane D. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. J Wiley & Sons, Amsterdam, Netherlands. [Google Scholar]
- 64.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, p 158–170. In Research in computational molecular biology. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shovill. 2019. GitHub–tseemann/shovill: faster SPAdes assembly of Illumina reads. https://github.com/tseemann/shovill.
- 66.Barrnap. 2019. GitHub–tseemann/barrnap: bacterial ribosomal RNA predictor. https://github.com/tseemann/barrnap.
- 67.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pyani. 2019. Python module for average nucleotide identity analyses. https://github.com/widdowquinn/pyani.
- 69.Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:1–10. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castillo D, Christiansen RH, Dalsgaard I, Madsen L, Espejo R, Middelboe M. 2016. Comparative genome analysis provides insights into the pathogenicity of Flavobacterium psychrophilum. PLoS One 11:e0152515. doi: 10.1371/journal.pone.0152515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 73.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 76.Jensen A, Larsen MH, Ingmer H, Fonnesbech Vogel B, Gram L. 2007. Sodium chloride enhances adherence and aggregation and strain variation influences invasiveness of Listeria monocytogenes strains. J Food Prot 70:592–599. doi: 10.4315/0362-028x-70.3.592. [DOI] [PubMed] [Google Scholar]
- 77.D’Alvise PW, Melchiorsen J, Porsby CH, Nielsen KF, Gram L. 2010. Inactivation of Vibrio anguillarum by attached and planktonic Roseobacter cells. Appl Environ Microbiol 76:2366–2370. doi: 10.1128/AEM.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:1–9. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nielsen A, Mansson M, Wietz M, Varming AN, Phipps RK, Larsen TO, Gram L, Ingmer H. 2012. Nigribactin, a novel siderophore from Vibrio nigripulchritudo, modulates Staphylococcus aureus virulence gene expression. Mar Drugs 10:2584–2595. doi: 10.3390/md10112584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download aem.02418-21-s0001.pdf, PDF file, 0.7 MB (744.6KB, pdf)
Data Availability Statement
Partial 16S rRNA gene sequences obtained from the isolates were deposited at the National Center for Biotechnology Information (NCBI) under the accession numbers MN481000 to MN481046, and whole-genome sequencing reads were deposited in the Sequencing Read Archive at the NCBI under accession number PRJNA566079.