ABSTRACT
The family “Candidatus Midichloriaceae” constitutes the most diverse but least studied lineage within the important order of intracellular bacteria Rickettsiales. “Candidatus Midichloriaceae” endosymbionts are found in many hosts, including terrestrial arthropods, aquatic invertebrates, and protists. Representatives of the family are not documented to be pathogenic, but some are associated with diseased fish or corals. Different genera display a range of unusual features, such as full sets of flagellar genes without visible flagella or the ability to invade host mitochondria. Since studies on “Ca. Midichloriaceae” tend to focus on the host, the family is rarely addressed as a unit, and we therefore lack a coherent picture of its diversity. Here, we provide four new midichloriaceae genomes, and we survey molecular and ecological data from the entire family. Features like genome size, ecological context, and host transitions vary considerably even among closely related midichloriaceae, suggesting a high frequency of such shifts, incomplete sampling, or both. Important functional traits involved in energy metabolism, flagella, and secretion systems were independently reduced multiple times with no obvious correspondence to host or habitat, corroborating the idea that many features of these “professional symbionts” are largely independent of host identity. Finally, despite “Ca. Midichloriaceae” being predominantly studied in ticks, our analyses show that the clade is mainly aquatic, with a few terrestrial offshoots. This highlights the importance of considering aquatic hosts, and protists in particular, when reconstructing the evolution of these endosymbionts and by extension all Rickettsiales.
IMPORTANCE Among endosymbiotic bacterial lineages, few are as intensely studied as Rickettsiales, which include the causative agents of spotted fever, typhus, and anaplasmosis. However, an important subgroup called “Candidatus Midichloriaceae” receives little attention despite accounting for a third of the diversity of Rickettsiales and harboring a wide range of bacteria with unique features, like the ability to infect mitochondria. Midichloriaceae are found in many hosts, from ticks to corals to unicellular protozoa, and studies on them tend to focus on the host groups. Here, for the first time since the establishment of this clade, we address the genomics, evolution, and ecology of “Ca. Midichloriaceae” as a whole, highlighting trends and patterns, the remaining gaps in our knowledge, and its importance for the understanding of symbiotic processes in intracellular bacteria.
KEYWORDS: “Candidatus Anadelfobacter”, “Candidatus Bandiella”, “Candidatus Cyrtobacter”, Euplotes, “Candidatus Grellia”, “Candidatus Midichloriaceae”, Rickettsiales, endosymbionts
INTRODUCTION
The alphaproteobacterial order Rickettsiales receives intense scrutiny for two reasons: several of its representatives are important intracellular pathogens of humans and livestock (1), and it has long been considered to be closely related to the ancestor of mitochondria (2, 3, but see also 4). Rickettsiales includes three main lineages with roughly similar observed molecular diversity: Rickettsiaceae, Anaplasmataceae, and “Candidatus Midichloriaceae” (5, 6). Of the three, “Ca. Midichloriaceae” is the most recently established as a family (5) as well as the least studied by far. None of its representatives have ever been cultivated in isolation, which explains the “Candidatus” status shared by the family and most of its representatives (7). A literature search yielded more than a hundred papers on midichloriaceae in the last decade (see Table S1 in the supplemental material), with a trend toward increasing numbers of publications over the years, and yet the group is often overlooked in broader analyses of Rickettsiales. It is common for nonpathogenic bacteria to receive less attention, especially when associated with unusual hosts; even protist-harbored representatives of the otherwise well-known Rickettsiaceae are severely understudied (8, 9). However, it is actually not clear if midichloriaceae are indeed all nonpathogenic, and they do occur in thoroughly studied hosts, such as ticks (10, 11) and corals (12).
The first-described member of the family, “Candidatus Midichloria mitochondrii,” is the only known bacterium capable of entering the mitochondria of its hosts, mainly in the ovarian cells of female ticks, such as the Eurasian Ixodes ricinus (13, 14). Molecular characterization (15) showed “Candidatus Midichloria” to be a “stray” member of Rickettsiales, belonging to neither of the two families known at the time. Relatives of “Ca. Midichloria” were then identified within amoebas (“Candidatus Jidaibacter”) (16, 17), ciliates (several bacterial genera) (18–20), and other protists (21), as well as insects (“Candidatus Lariskella”) (22) and marine metazoans, including corals (“Candidatus Aquarickettsia”) (12) and placozoans (“Candidatus Grellia”) (23, 24). High-throughput sequencing and quantitative PCR studies have now reliably detected environmental DNA belonging to “Ca. Midichloriaceae” in many aquatic habitats (for example, see reference 12) as well as the blood and tissues of vertebrates (25, 26).
Once “Ca. Midichloria” became recognized as a major symbiont of pathogen-carrying ticks, much of the literature revolved around microbiome surveys of such ticks from around the globe (for example, see references 11, 27, and 28) (Table S1). However, in addition to biogeographic accounts, midichloriaceae have been the subjects of various lines of enquiry addressing, broadly, (i) the phylogenetic relationships with other bacteria and their degree of horizontal gene transfer (16, 17, 29); (ii) their capabilities to be horizontally transmitted, infect different hosts, and possibly cause or facilitate diseases (26, 30–32); and (iii) unexpected genomic features, in particular the presence and expression of flagellar genes (16, 29, 33), whose absence was considered a defining feature of Rickettsiales not long ago (8).
The relative obscurity midichloriaceae endure despite their intriguing traits might be partially due to the fact that different genera are studied in isolation by communities of researchers interested in different hosts (ticks, corals, ciliates, etc.), instead of being treated as a coherent, and likely quite ancient, bacterial lineage. Here, we report four additional genomes of midichloriaceae from ciliates, significantly increasing the genomic coverage of the clade, and also take the opportunity to comparatively consider the characteristics of the family as a whole, summarizing the current state of knowledge and highlighting the most promising lines of research going forward.
RESULTS
General genomic features of “Ca. Midichloriaceae.”
We were able to assemble and annotate the genomes of three midichloriaceae symbionts of ciliates, namely, “Candidatus Cyrtobacter zanobii” from Euplotes aediculatus Eae1 (k-mer coverage, 12×), “Candidatus Anadelfobacter sociabilis” from Euplotes octocarinatus FL(12)-VI (k-mer coverage, 28×), and “Candidatus Grellia numerosa” from Euplotes woodruffi Ewo1 (k-mer coverage, 371×), formerly known as “Candidatus Bandiella numerosa” (see below). Reads from a “Candidatus Cyrtobacter sp.” symbiont of Euplotes eurystomus EM could not be confidently assembled into a complete genome due to binning ambivalence, and the partial assembly was not included in the functional analyses; however, enough individual genes could be recovered for the phylogenomic analysis (see below). Eight available genomes of other midichloriaceae were also comparatively analyzed in this study. The main features of each are reported in Table 1.
TABLE 1.
Main features of “Ca. Midichloriaceae” genomes analyzed
| Symbiont species | Host species | Size (kbp) | No. of contigs | G+C (%) | Coding (%) | Accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|
| “Ca. Anadelfobacter sociabilis” | Euplotes octocarinatus (ciliate) | 1,890 | 817 | 31.08 | 61.1 | JAJBJC000000000 | 20; this study |
| “Ca. Aquarickettsia rohweri” | Acropora cervicornis (coral) | 1,285 | 155 | 27.59 | 84.2 | GCA_003953955.1 | 12 |
| “Ca. Cyrtobacter zanobii” | Euplotes aediculatus (ciliate) | 1,326 | 350 | 37.01 | 77.1 | JAJBJB000000000 | 56; this study |
| “Ca. Fokinia solitaria” | Paramecium sp. (ciliate) | 837 | 1 | 35.79 | 88.9 | GCA_003072485.1 | 19 |
| “Ca. Grellia incantans” | Trichoplax sp. H2 (placozoan) | 1,258 | 74 | 30.04 | 88.9 | PRJEB30343 | 23 |
| “Ca. Grellia (formerly Bandiella) numerosa” | Euplotes woodruffi (ciliate) | 1,067 | 315 | 31.40 | 82.3 | JAJBJD000000000 | 20; this study |
| “Ca. Jidaibacter acanthamoeba” | Acanthamoeba sp. (amoebozoan) | 2,371 | 249 | 33.65 | 82.8 | GCA_000815465.1 | 16 |
| “Ca. Midichloria mitochondrii” | Ixodes ricinus (tick) | 1,184 | 1 | 36.55 | 78.7 | GCA_000219355.1 | 29 |
| Symbiont of Acanthamoeba sp. UWC8 | Acanthamoeba sp. (amoebozoan) | 1,615 | 1 | 34.76 | 89.2 | GCA_000730245.1 | 17 |
| Symbiont of Peranema trichophorum | Peranema trichophorum (euglenozoan) | 1,376 | 125 | 40.84 | 82.8 | GCA_004210275.1 | 21 |
| Symbiont of Trichoplax sp. H2 'Panama' | Trichoplax sp. H2 (placozoan) | 1,474 | 25 | 27.29 | 88.0 | GCA_009690945.1 | 24 |
Genome sizes among midichloriaceae range from 0.8 Mbp in “Candidatus Fokinia solitaria” to 2.4 Mbp in “Ca. Jidaibacter acanthamoeba” (average ± standard deviation [SD] = 1.4 Mbp ± 0.4 Mbp), with no clear trend matching relatedness (Fig. 1). The two closely related symbionts from different Acanthamoeba strains, for example, differ by almost 0.8 Mbp (half the size of the smaller). The number of coding DNA sequences in each genome ranged from 719 in “Ca. Fokinia solitaria” to 2,035 in “Ca. Jidaibacter acanthamoeba” (average ± SD = 1,341 ± 343), positively correlating with genome size (R2 = 0.89, P = 1.3 × 10−5; Student’s two-tailed t distribution). Coding genome percentage was lowest in “Ca. Anadelfobacter sociabilis” (61%) and highest in the symbiont of Acanthamoeba UWC8 (89%), spanning a relatively small range of values (mean ± SD = 82% ± 8%) not significantly correlated with genome size (R2 = 0.12, P = 0.29; Student’s two-tailed t distribution). G+C content ranged from 27% in the symbiont of Trichoplax (Placozoa) strain ‘Panama’ to 41% in the symbiont of Peranema trichophorum (mean ± SD = 33% ± 4%).
FIG 1.
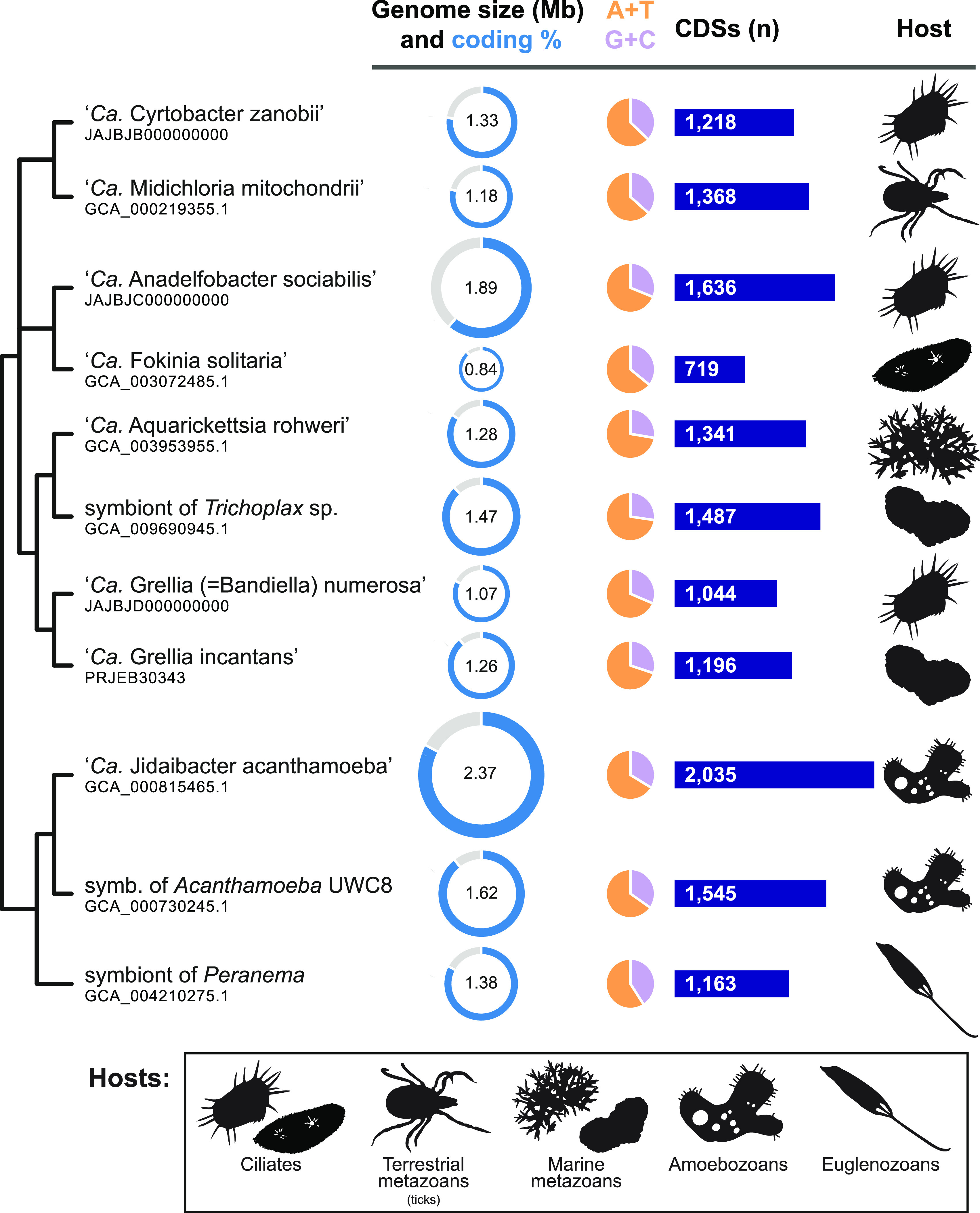
Hosts and genomic traits of 11 strains of “Ca. Midichloriaceae.” Bacteria are arranged according to their phylogenetic relationships (see Fig. 2). The genomes of “Ca. Cyrtobacter zanobii,” “Ca. Anadelfobacter sociabilis,” and “Ca. Grellia numerosa” (formerly “Ca. Bandiella numerosa”) are new in this study. CDS, coding DNA sequences.
Evolutionary relationships within “Ca. Midichloriaceae.”
Both models employed in the maximum-likelihood (ML) phylogenomic analysis outlined the same four well-supported clades, here called CM, AF, ABG, and J based on the initial of their named representatives (Fig. 2). Within clades, “Candidatus Aquarickettsia rohweri” is extremely closely related to the unnamed symbiont of Trichoplax strain ‘Panama’ (average nucleotide identity [ANI], 98.6%; average amino acid identity [AAI], 97.3%), which was putatively assigned to the genus “Ca. Grellia” in the original publication because it shared the same host species as “Candidatus Grellia incantans” (24). Conversely, the Euplotes symbiont described as “Ca. Bandiella numerosa” is actually a close sister group of “Ca. Grellia incantans” (ANI, 81.6%; AAI, 80.0%). The two symbionts of Acanthamoeba initially described by Fritsche et al. (34) but investigated since by different authors are also very similar at the molecular level (ANI, 92.8%; AAI, 91.9%).
FIG 2.
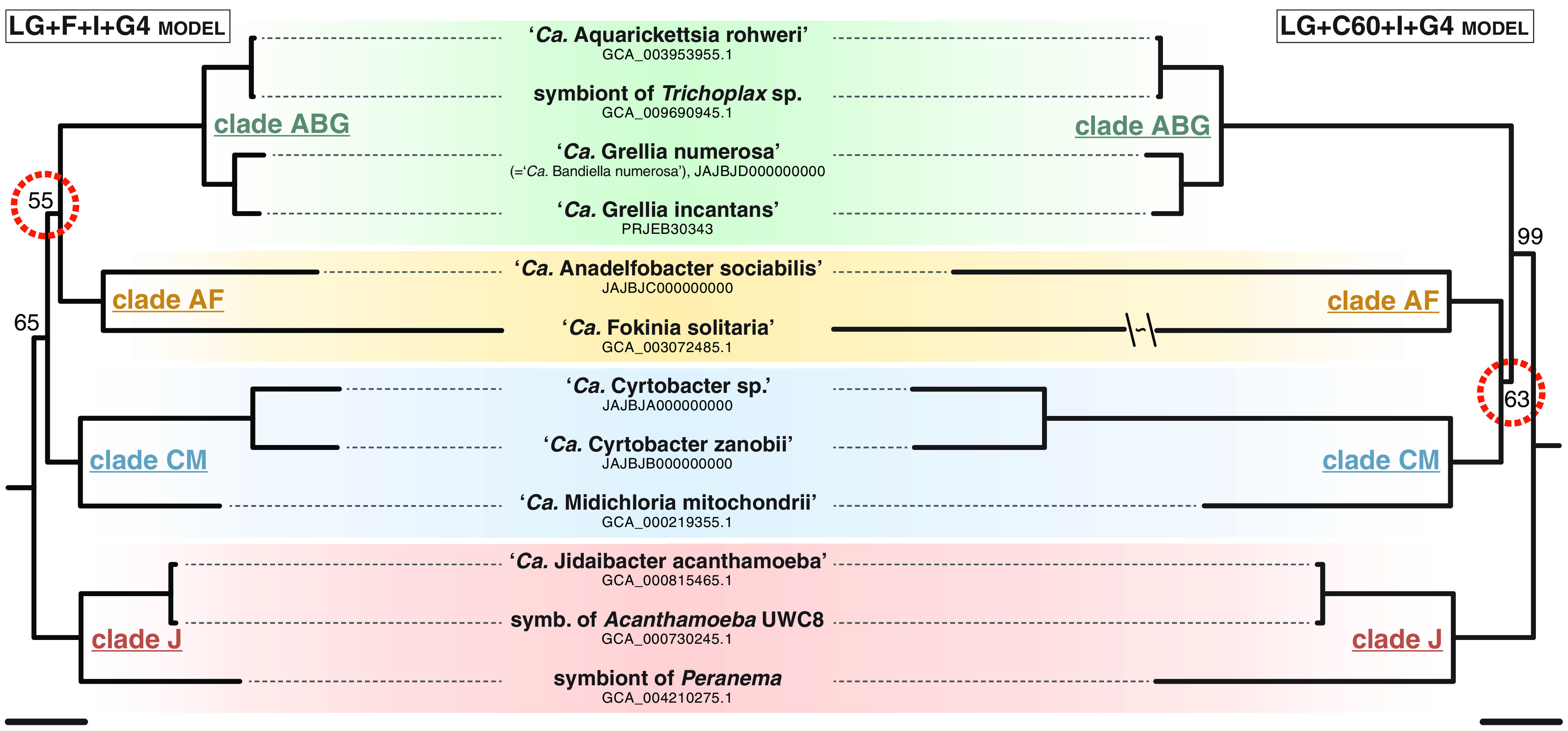
Phylogenomics of “Ca. Midichloriaceae.” Maximum-likelihood phylogenomic trees based on 179 concatenated genes (47,692 characters), obtained under two substitution models. Twelve ingroup plus 10 outgroup (Anaplasmataceae and Rickettsiaceae; not shown) sequences were included; only bootstrap values below 100% are shown for each node, and nodes underlining the difference between the two topologies are circled in red. Bars correspond to an estimated sequence divergence of 0.2. The branch leading to “Ca. Fokinia solitaria” in the LG+C60+I+G4 tree was shortened because of space constraints.
The branching order of the four clades differs between models. Clade J is consistently recovered as a sister group of the other midichloriaceae, but clades ABG and AF are sister groups under the LG+F+I+G4 model, whereas clades CM and AF are sister groups instead under the LG+C60+I+G4 model. Low bootstrap support (55% and 63%, respectively) underlined the different nodes, and approximately unbiased (AU) tests did not provide statistical support to reject either topology. Therefore, that part of the tree is treated as an unresolved polytomy here.
Trends of ecological distribution.
Using the consensus topology resulting from the phylogenomic analyses as a constraint, a phylogenetic tree based on the 16S rRNA gene sequence was also obtained, including described species and environmental sequences clustering within the family “Ca. Midichloriaceae.” Ecological metadata, namely, sampling environment and host taxonomy, were then mapped to observe possible evolutionary patterns (Fig. 3). Ecological transitions seem to be numerous and trends complex to identify, but it appears clear that terrestrial lineages, mainly the tick-associated “Ca. Midichloria” and the arthropod-associated “Ca. Lariskella,” are nested within aquatic clades. Similarly, metazoan-associated midichloriaceae are either interspersed or nested within clades of environmental (unknown host) or protist-harbored bacteria. In particular, “Ca. Midichloria” stems within a group of symbionts of various ciliates.
FIG 3.
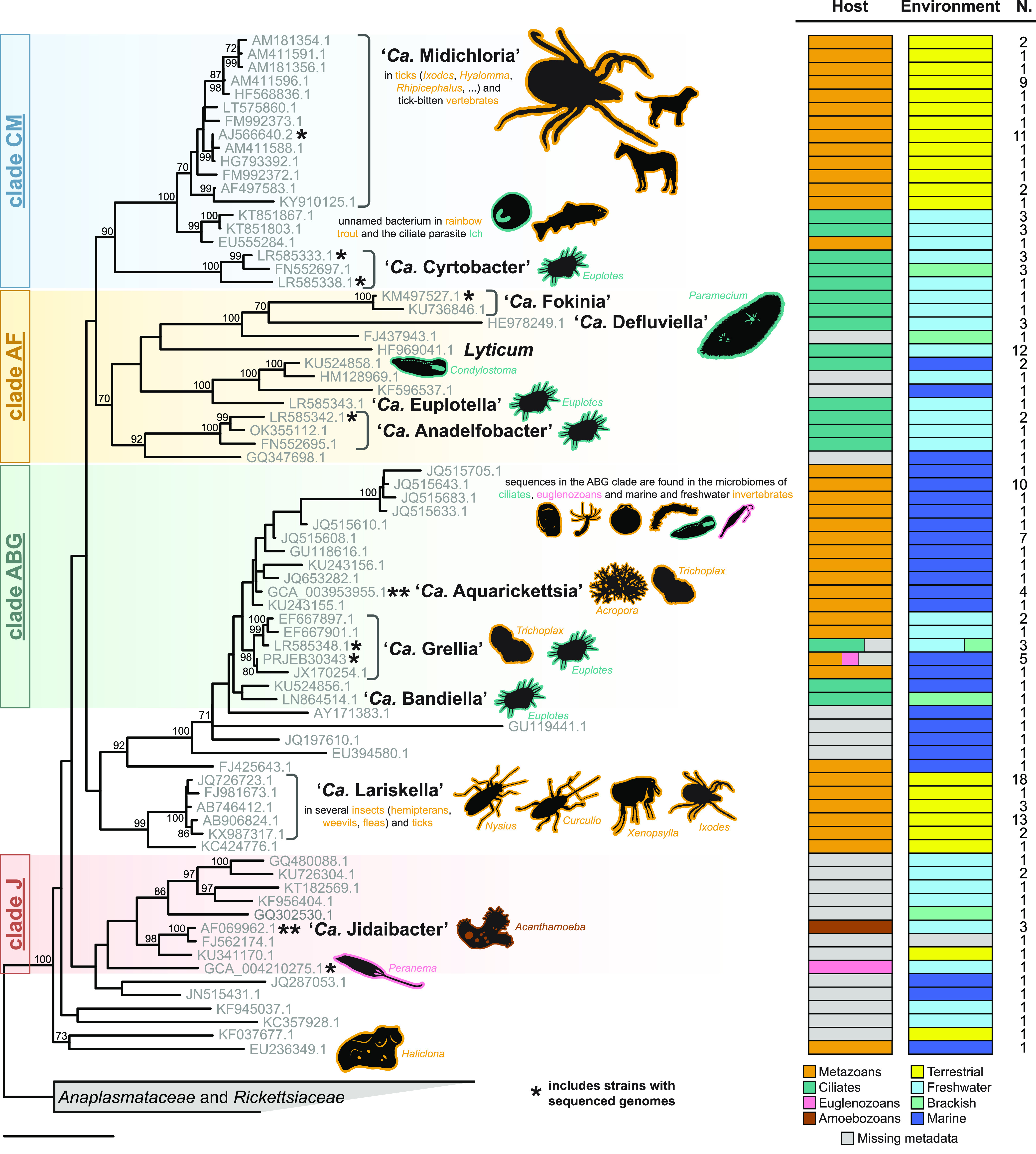
16S rRNA gene phylogeny and ecological traits of “Ca. Midichloriaceae.” Maximum-likelihood single-gene inference obtained after constraining the four main clades recovered by the phylogenomic analysis (labeled on the left). Each sequence in the tree represents a cluster of ≥99% similar sequences, whose numbers are shown in the rightmost column (N.). Host and environment bar plots depict trait frequencies in each cluster. Named and otherwise relevant bacterial taxa are highlighted, and the main hosts are shown as silhouettes with lines colored corresponding to their taxonomy (metazoans, ciliates, amoebozoans, and euglenozoans). Note that for most marine invertebrate hosts, the presence of “Ca. Midichloriaceae” was detected only by high-throughput sequencing, without histological confirmation that the bacterium infects animal tissues (instead of, for example, protist symbionts in the animal). Asterisks mark clusters that include one or more strains with a sequenced genome (Fig. 1 and 2). Bootstrap values below 70% are not shown. The bar corresponds to an estimated sequence divergence of 0.1.
Reconstructed metabolic capabilities.
Overall, the inferred metabolic capabilities of symbionts belonging to “Ca. Midichloriaceae” appear to be heavily depleted (Table S2), a noticeable feature being the lack of an intact Embden-Meyerhof-Parnas glycolytic pathway due to the absence of the phosphofructokinase-1 gene (Fig. 4A). In at least three cases (“Ca. Anadelfobacter sociabilis,” “Ca. Fokinia solitaria,” and the symbiont of Peranema), the energy metabolism appears to be further reduced due to the loss of a large number of genes involved in the tricarboxylic acid cycle and in oxidative phosphorylation. None of the analyzed genomes seem to possess an aa3 cytochrome c oxidase in their electron transport chain, replaced by the cbb3 type as originally reported for “Ca. Midichloria” (29). Genes coding for the ATP synthase subunits were mostly present, except for four occurrences in which a single gene remained undetected. Finally, in no case could a complete pentose phosphate oxidative pathway be observed.
FIG 4.
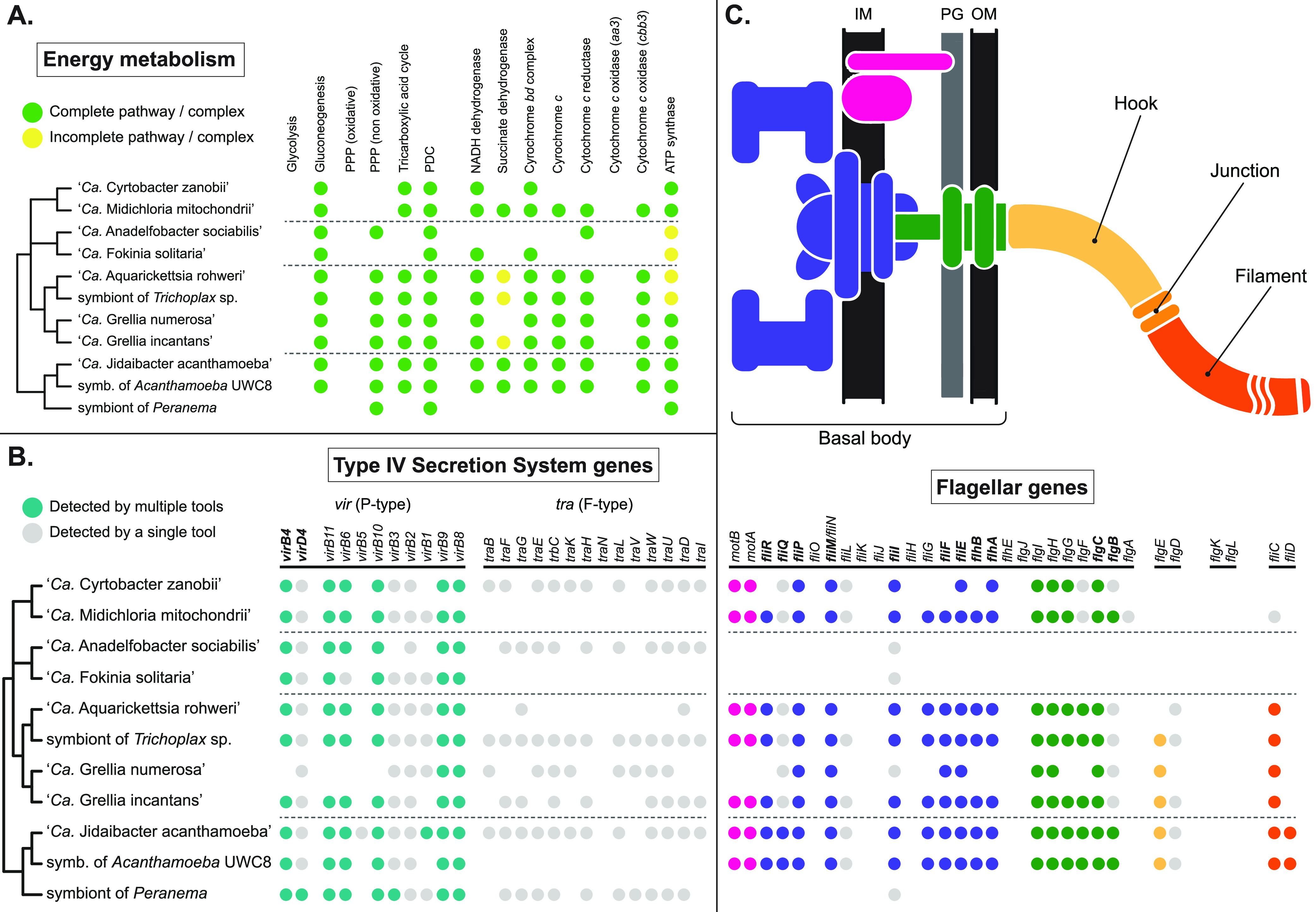
Selected functional traits mapped on the consensus phylogeny of “Ca. Midichloriaceae.” Dashed lines separate the four clades identified in Fig. 2. (A) Energy metabolism-related genes, complexes, and pathways, as annotated by the KEGG Automatic Annotation System (KAAS) and, for oxidative phosphorylation, Prokka. Pathways and complexes were considered complete (green circles) when all essential genes known to be exclusively involved in that pathway/complex were detected, possibly incomplete (yellow circles) when at least half but not all such genes were detected, and likely absent (no circle) otherwise. PPP, pentose phosphate pathway; PDC, pyruvate dehydrogenase complex. (B) Genes involved in T4SS, as annotated by KAAS, Prokka, and TXSScan. Teal circles mark genes detected by at least two analytical tools; gray circles mark genes detected by only one of the three (usually TXSScan, which is optimized for the detection of T4SS genes; see also Fig. S1). Names of genes considered mandatory (73) are in bold. (C) Flagellum and flagellar assembly-related genes, annotated and displayed as in panel B, but with different colors used for different parts of the flagellum, as shown in the schematic drawing above the table. IM, internal membrane; PG, peptidoglycan layer; OM, outer membrane. More details on the functional analysis are shown in Table S2.
Regarding biosynthetic capabilities, only phosphatidyl-l-serine and phosphatidylethanolamine autonomous synthetic pathways could be identified among all possible membrane glycerophospholipid anabolic pathways. Genes involved in cell envelope (peptidoglycan and outer membrane lipopolysaccharides [LPS]) biosynthesis are mostly present across the analyzed genomes, except for the lack of genes for LPS synthesis in “Ca. Fokinia solitaria” and “Ca. Cyrtobacter zanobii.” Both purine and pyrimidine pathways are considerably depleted across the analyzed genomes, to the point where no standard nucleotide can be autonomously produced by any of the considered species. Few amino acid synthetic pathways are intact, while the vast majority appear to be completely absent: indeed, complete biosynthetic pathways were consistently identified only for l-glutamate, l-glutamine, l-aspartate, l-lysine, and glycine (starting from l-serine). Vitamin production also appears very depleted, with complete biosynthetic pathways found only for lipoic acid and, in some species, biotin. Coenzyme A (CoA) can likely be produced in at least five of the 11 analyzed species starting from pantothenate, but in none of the examined genomes was an intact pathway for the synthesis of pantothenate detected. Genes for NAD+ production from nicotinamide-d-ribonucleotide were found in seven genomes, but no surveyed midichloriacea seems able to synthesize nicotinamide-d-ribonucleotide. The pathway from GTP to riboflavin is incomplete in all examined species, whereas the pathway from riboflavin to FAD is present in most cases.
Transport, secretion, and flagellar assemblies.
The assortment of transporters from each genome represents a truly diverse pattern with few regularities (Table S2), the most remarkable common feature being the ubiquitous occurrence of the tlcA gene, coding for the ATP/ADP translocase.
Sets of genes coding for type IV secretion systems (T4SS) could be found in most species considered in this study (Table S2 and Fig. 4B). In particular, two different kinds of T4SS were retrieved: the vir secretion system (type IVa/P type; in Rickettsiales also called rvh, for “Rickettsiales vir homologue”) and the tra conjugative system (type IVa/F type). The genome of “Ca. Grellia numerosa” has the fewest genes for vir secretion system biosynthesis. Genes coding for the tra system subunits are more patchily distributed and are largely missing in “Ca. Midichloria mitochondrii,” “Ca. Fokinia solitaria,” “Ca. Aquarickettsia rohweri,” and the symbiont of Acanthamoeba UWC8. However, their synteny is quite conserved, and open reading frames with sequences homologous to those of the missing genes, possibly pseudogenes or more divergent gene variants, are often present, as are mobile element-related genes flanking regions with tra genes (Fig. S1).
An extensive collection of flagellar genes was retrieved in all but three analyzed genomes (Table S2 and Fig. 4C), namely, those of “Ca. Anadelfobacter sociabilis,” “Ca. Fokinia solitaria,” and the symbiont of Peranema, the species with especially reduced energy metabolism. However, no evidence for genes involved in the hook-filament junction (flgL and flgK) could be retrieved from any genome, whereas fliD, which codes for the flagellar cap, was observed only in the two genomes of Acanthamoeba symbionts. Finally, “Ca. Midichloria mitochondrii” and “Ca. Cyrtobacter zanobii” showed no gene involved in hook biosynthesis.
DISCUSSION
Evolutionary trends in “Ca. Midichloriaceae.”
Phylogenetic analyses of Rickettsiales usually recover “Ca. Midichloriaceae” and Anaplasmataceae as sister groups, with Rickettsiaceae more distantly related (19, 20, 29). Within “Ca. Midichloriaceae,” relationships based on single-gene trees have instead been conflicting and poorly supported (for example, see references 5 and 6). Even the phylogenomic analyses conducted here, despite adding about 50% more data to what was previously available, are not conclusive: some clades including two or more genera each can now be reliably defined (Fig. 2), but relationships among them remain uncertain. However, some general observations can be made regarding the evolutionary history of the family. First, despite being originally described in terrestrial arthropods (15, 35), the current distribution suggests that midichloriaceae are ancestrally aquatic, and most of their diversity still lies in freshwater and marine environments. Transitions to terrestrial hosts, probably overrepresented due to arthropod symbionts being more frequently investigated, happened multiple times independently (Fig. 3) and at least twice in ticks (11, 22). Second, most of the molecular diversity in “Ca. Midichloriaceae” is found in unicellular protists, which are the most likely hosts for the ancestor of the family; current knowledge suggests that the same is true also of Rickettsiaceae (36), but interestingly not Anaplasmataceae, which have never been found in protists (37). Obviously the “protist” label is very broad, since it encompasses most of the diversity of eukaryotes, and the current sampling is insufficient to speculate on the identity of the ancestral midichloriaceae host in more detail; the abundance of ciliate hosts is compatible with a corresponding bias in symbiosis studies within protists (37).
The dominant feature of “Ca. Midichloriaceae” as a whole is their diversity. All known representatives are intracellular symbionts, but they vastly differ in genomic and ecological properties. Strikingly, most of the differences do not seem to be tied to their phylogeny, even within well-supported clades. For example, large reductions in energy production- and flagellum-related genes occurred in clade AF as well as the distantly related symbiont of the euglenozoan Peranema. On the other hand, two very closely related Acanthamoeba symbionts (“Ca. Jidaibacter”) differ considerably in genome size and gene content. Finally, bacterial taxa in the more genetically homogeneous clade ABG can be found in hosts as different as marine corals and freshwater ciliates. All of this suggests that either large and discontinuous evolutionary transitions happen frequently among midichloriaceae or we have uncovered only a tiny fraction of the actual diversity required to reveal more gradual changes. Both explanations might be true, but some discontinuities are not sampling dependent, as genome size in “Ca. Jidaibacter” exemplifies. Irregular trends in genome size within clades of intracellular bacteria are actually more puzzling than they appear at first: we usually rely on models for genome evolution of intracellular symbionts (38) that have proved to be extremely successful but that account only for genome erosion and reduction. However, the ancestor of midichloriaceae, as well as the ancestor of all Rickettsiales, was already a specialized intracellular bacterium, arguably subjected to genome streamlining for a long time. A purely reductive ratchet-like model cannot explain the large genome size range in midichloriaceae, even admitting differential evolutionary rates; other mechanisms must be invoked to account for the expansion of coding and noncoding genomic regions, for example lateral gene transfers among co-occurring symbionts (17, 39).
It is also clear that the diversity of midichloriaceae has to be explored more before we can fully interpret the complex evolution of this clade. Most studies tend to compare individual midichloriaceae with distantly related bacteria, but the evolution of the group would be better understood by examining the forces and processes acting within the family, since such signals are likely obfuscated in comparisons to distant relatives. Protists are the most promising hosts in which to search for diverse symbionts, but analyses of high-throughput sequencing environmental data suggest that the most abundant and ecologically relevant midichloriaceae might be the (mostly) marine representatives of clade ABG, which are mainly associated with invertebrates (12, 20, 23, 24, 40). A more unified approach to studying midichloriaceae is also needed to prevent taxonomic confusion. For example, taxa in clade ABG are very closely related to each other (minimum ANI, 76.6%; minimum AAI, 69.9%; minimum 16S rRNA gene sequence similarity, 95.8%), and several of them were described independently and without following taxonomic guidelines, eventually making the earliest established genus, “Ca. Bandiella,” nonmonophyletic. According to commonly used molecular thresholds, all “Ca. Bandiella,” “Ca. Grellia,” and “Ca. Aquarickettsia” strains should have been assigned to the same genus, but we opted here for the least disruptive solution of the issue, and simply reassigned “Ca. Bandiella numerosa” to the genus “Ca. Grellia” (as “Ca. Grellia numerosa”).
Ecological insights and host-symbiont relationship.
Midichloriaceae are “professional symbionts” (37), i.e., bacteria that have long become adapted to the intracellular lifestyle, are not trapped within a specific host (at least over evolutionary time), and do not seem to be subject to ongoing massive genome decay. Contrary to general expectations, symbionts of this kind are rarely seriously harmful for their hosts. Horizontal (infectious) transmission is inferred in “Ca. Midichloriaceae” by the discrepancy between bacterial and host phylogenies but has not yet been definitively proven by direct observation. Experimental setups involving ciliates (31), ciliates and fish (41), and ticks and mammals (42) have shown that some midichloriaceae (“Ca. Bandiella woodruffi,” an unnamed “Ca. Midichloria”-like symbiont, and “Ca. Midichloria mitochondrii,” respectively) can be transmitted between hosts in experimental contexts, but the infection often does not persist upon removal of the original source. Vertical transmission has instead been proven in protists and Ixodes ricinus ticks (43) but challenged in corals, where eggs and larvae from adults heavily infected by “Ca. Aquarickettsia” seem to harbor no bacteria (44). Midichloriaceae vary in their tissue and subcellular compartment specificity, with “Ca. Midichloria” and “Ca. Grellia incantans” showing a preference for specific organelles—mitochondria and the rough endoplasmic reticulum, respectively (14, 23)—and most protist symbionts occupying the host cytoplasm, either in free form or surrounded by vacuoles (16, 20). The symbionts of terrestrial arthropods, “Ca. Midichloria” and “Ca. Lariskella,” are usually concentrated in specialized organs, the symbiosomes (14, 22).
We can only speculate as to the roles different midichloriaceae play in their hosts, which might vary with species and even species-host combination. “Ca. Midichloria,” for example, is sometimes assumed to be on the beneficial end of the symbiosis spectrum, possibly a nutritional mutualist in Ixodes ricinus (45), like bacterial symbionts of arthropods with restrictive diets (46). However, long-term laboratory-maintained tick populations can lose the symbionts, proving that they are not strictly essential (43). Integrated symbiont-host metabolic pathways, known from ancient essential symbionts of insects (47), have not been characterized in midichloriaceae. On the other end of the spectrum, “Ca. Aquarickettsia” and an unnamed midichloriacea in rainbow trout are consistently associated with diseased individuals, susceptible to white-band disease (32) and suffering from red mark syndrome (26), respectively. However, the possibility that the debilitated state of the host is a cause rather than a consequence of the infection has not been ruled out. In most cases, including the ciliate hosts examined here, no obvious effect on host fitness can be observed (14, 20), which might be a mark of successful long-term commensalism.
Opportunistic commensalism would indeed fit many of the observed features of midichloriacea distribution, which, admittedly, are only partially and patchily characterized. Some taxa, chiefly “Ca. Aquarickettsia” and “Ca. Lariskella,” have very broad host ranges (12, 22). Midichloriaceae are usually only one component of more complex microbiomes, in both metazoan (44, 48) and protist (20, 49) hosts. Their frequency is usually far lower than 100%, the exceptions being “Ca. Grellia numerosa” in Euplotes woodruffi (49) and “Ca. Midichloria” in Ixodes ricinus, where it is found in every adult female as the predominant symbiont. Interestingly, this is not true for the other ticks “Ca. Midichloria” infects (48). Although more data are clearly needed for each of these patterns, we can speculate that most, but not necessarily all, midichloriaceae are probably commensals that neither hurt nor particularly benefit their hosts.
Functional genomics. (i) Metabolism and energetics.
It should be noted that inferences based on gene annotations, while useful and routinely performed, can rarely provide definitive proof of the metabolic capabilities of an organism. There are even small discrepancies between annotations obtained here and previous analyses on the same published genomes, to be ascribed to differences in the annotation strategy. Regardless of individual details, the general picture of severely reduced anabolism and catabolism has now been consistently reported for all “Ca. Midichloriaceae” (19, 23, 29), in line with other representatives of Rickettsiales. These bacteria depend on their hosts for most carbon compounds and energy, although it is not clear if any of them is a true “energy parasite,” a term usually restricted to intracellular symbionts that cannot produce ATP in any way and have to import it from their host, such as chlamydiae (50) and some microsporidia (51). All “Ca. Midichloriaceae” possess both an ATP/ADP transporter as well as a (mostly) complete ATP synthase complex. Hence, they either both produce and steal ATP, possibly alternating between the two strategies depending on specific conditions, or use the synthase for purposes other than energy production, like pH homeostasis, and rely on their hosts for energy.
(ii) Secretion systems.
T4SS are a peculiar topic in studies of symbiotic bacteria, because they are consistently invoked as essential tools for host interactions, but abundant experimental evidence is only available for some gene clusters and/or in some model systems. None has been produced yet for “Ca. Midichloriaceae.” Our comparative analysis confirms, however, that genes involved in T4SS are widespread in the family and belong to two clusters already observed to be vertically inherited in Rickettsiales: vir and tra genes. Schulz et al. (16) reported a third cluster in “Ca. Jidaibacter acanthamoeba,” trb, which is not usually found in Rickettsiales and was apparently horizontally acquired by “Ca. Jidaibacter” from outside the alphaproteobacteria. The vir cluster is the most homogeneously distributed in “Ca. Midichloriaceae,” supporting the hypothesis that it plays a key role in host interactions. The tra cluster, whose phylogeny and function are more uncertain, is considerably variable both in terms of gene content (Fig. 4) and number of gene copies (Fig. S1) and appears to be highly depleted in four unrelated species that do not share other obvious unifying traits. The conserved synteny of tra genes suggests that many of them are probably functional, and their association with mobile element-related genes corroborates the phylogenetic-based conclusion of Schulz et al. (16) that tra operons in “Ca. Midichloriaceae” belong to the RAGEs (Rickettsiales amplified genetic elements), conjugative transposons that might be involved in horizontal gene transfers.
(iii) Flagellar apparatus.
The presence, expression, and function of flagellar genes in symbiotic bacteria is a topic that does not receive enough attention (52, 53). Some symbionts rely on a motile stage to infect their hosts (54) but Rickettsiales, traditionally studied in arthropods and transmitted through fluid exchanges between metazoans, were supposed to always lack a flagellum. Representatives of Rickettsiaceae have recently been discovered not just with flagella but with extensive flagellar movement inside their hosts (8). In contrast, flagellar genes without visible corresponding structures have been reported in most “Ca. Midichloriaceae” (12, 16, 24, 29); the opposite is true for Lyticum, which possesses many flagella but whose genome has not been yet sequenced (55). Flagellar genes are an ancestral, inherited feature of Rickettsiales (29), and we observed here only two independent major losses (Fig. 4C), not clearly tied to host identity or overall genome reduction. These genes must be functional to be preserved by selection. One hypothesis is that they still express a complete flagellar apparatus, possibly used only during horizontal transmission in the aquatic environment, but this is unlikely considering the apparent infrequency of this transmission mode and the depleted energy metabolism that may be insufficient to support locomotion outside the host. Alternatively, these genes might have been repurposed and might now be involved in interactions with the host. FliD, a filament protein whose gene was not even detected in “Ca. Midichloria” in our analyses, has been found to be expressed on the outer membrane of bacteria in Ixodes ticks, but it is not part of a classical flagellar structure and instead seems to be situated at the interface with the host cytoplasm in an unknown conformation (33).
MATERIALS AND METHODS
Genome assembly, binning, and annotation.
Analyses were performed on high-throughput metagenomic sequencing data from four strains/populations of the ciliate Euplotes, obtained as described by Boscaro et al. (20): FL(12)-VI (E. octocarinatus), Ewo1 (E. woodruffi), Eae1 (E. aediculatus), and EM (E. eurystomus). Each Euplotes culture harbors a different “Ca. Midichloriaceae” species (Table 1) formally described in previous papers (20, 56). Raw reads were assembled using SPAdes v3.13.0 (57). BLASTn, Diamond BLASTx, and the “taxify” blobtools v1.0 command (58) were used to assign assembled sequences to bacterial taxa. Hits were then organized into a database using the blobtools “create” command, and contigs assigned to “Ca. Midichloriaceae” (minimum size, 200 bp) were collected and further processed. To recover taxonomically unassigned contigs belonging to the genomes of interest, sequences from the previous step were subjected to BLAST search against the assembled metagenome assembly graph using Bandage v0.8.1 (59), adding contigs with significant similarity (>97%) and those clearly connected to the assembly subgraph representing the specific symbiont. Prokka v1.12 (60) was used for the functional annotation of all genomes.
Functional analysis.
Eight available genomes from “Ca. Midichloriaceae” symbionts (Table 1) were analyzed together with the newly obtained genomes for consistency. The partial genome of “Ca. Cyrtobacter sp.” from E. eurystomus EM was not included in functional analyses. Artemis v18.1.0 (61) was used to visualize and navigate through the annotations. Annotated genomes were submitted to the KEGG Automatic Annotation Server (KAAS) (62) using the bidirectional best-hit BLAST method on prokaryotic references in order to reconstruct putative functions and metabolic pathways. The same data were also uploaded on Galaxy (63) at the Pasteur web platform (galaxy.pasteur.fr) (64) to further identify bacterial secretion system and flagellar genes using the MacsyFinder-based detection tool TXSScan (65). The annotation of the patchily distributed tra genes was also manually verified using BLASTp and OrthoFinder v2.3.3 (66).
Phylogenomic analyses.
Twelve “Ca. Midichloriaceae” genomes, including four obtained here, were used for phylogenetic inference, in addition to 10 genomes from Anaplasmataceae and Rickettsiaceae as outgroups (outgroup accession numbers: GCA_000018225.1, GCA_000020305.1, GCA_000309075.1, GCA_000367405.1, GCA_000632985.1, GCA_000964685.1, GCA_002879995.1, GCA_003015145.1, GCA_004171285.1, GCA_900327275.1). OrthoFinder was used for clustering homologous genes and amino acid sequence alignment. 179 single-copy orthologs, missing at most from one genome, were used in the analysis. Amino acid sequences were trimmed using trimAl v1.2 (67) choosing the “-automated1” option, concatenated, and then submitted to IQ-TREE v.1.5.4 (68) for maximum-likelihood (ML) tree reconstruction. The best-fitting model selected by the Akaike and Bayesian information criteria (AIC and BIC), i.e., LG+F+I+G4, and the mixed model LG+C60+I+G4 were employed, running 1,000 ultrafast bootstraps (69) for topology support. Approximately unbiased (AU) statistical tests (70) were then performed to evaluate the statistical significance of the differences between topologies.
16S rRNA gene-based phylogenetic analyses.
Fifty-two full-length 16S rRNA gene sequences associated with described species of “Ca. Midichloriaceae,” Anaplasmataceae, and Rickettsiaceae were collected from GenBank, from the genomes assembled in this study, and from a novel “Ca. Anadelfobacter” symbiont of Euplotes strain THA1 characterized using methods described by Boscaro et al. (20) (16S rRNA gene sequence accession number OK355112). Sequences were aligned using MAFFT v7.222 (71) and used for a series of preliminary ML phylogenetic analyses with IQ-TREE, setting nodes strongly supported (100% bootstrap values) by the phylogenomic analyses as topological constraints. Nine sequences evenly spread across the tree were individually submitted as queries on BLASTn to recover a larger diversity of “Ca. Midichloriaceae.” Matching 16S rRNA gene sequences (≥85% identical to the query and ≥1,000 bp long) from bacterial species or environmental samples were collected and added to the phylogenetic analysis. Environmental sequences clustering outside the clade of known midichloriaceae were discarded. Sequences on long branches were manually inspected to confirm that none were of chimeric origin. Ecological and environmental metadata associated with the remaining 206 sequences were downloaded from GenBank. To improve legibility with limited loss of information, the sequences were clustered using the “cluster_smallmem” UCLAST algorithm in USEARCH (72), setting 99% as identity threshold, thus obtaining 102 representative sequences. A final ML phylogenetic analysis was then performed using these representatives as input for IQ-TREE under the GTR+I+G4 model, selected by AIC and BIC, and running 100 standard nonparametric bootstraps with the topological constraints described earlier.
Data availability.
Genome sequences were deposited in the DDBJ/ENA/GenBank database with accession numbers JAJBJA000000000 to JAJBJD000000000.
ACKNOWLEDGMENTS
We thank Sergei Fokin for his assistance in sampling Euplotes strain THA1.
This work was supported by grants from the University of Pisa (565-60% 2018 and 565-60% 2020 to C.V.; Mobility Grant 2019 to D.G.) and the Gordon and Betty Moore Foundation (grant GBMF9201 [https://doi.org/10.37807/GBMF9201] to P.J.K.).
Footnotes
Supplemental material is available online only.
Contributor Information
Vittorio Boscaro, Email: vittorio.boscaro@botany.ubc.ca.
Karyn N. Johnson, University of Queensland
REFERENCES
- 1.Salje J. 2021. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat Rev Microbiol 19:375–390. 10.1038/s41579-020-00507-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, Näslund AK, Eriksson AS, Winkler HH, Kurland CG. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133–140. 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Fitzpatrick DA, Creevey CJ, McInerney JO. 2006. Genome phylogeny indicate a meaningful α-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol Biol Evol 23:74–85. 10.1093/molbev/msj009. [DOI] [PubMed] [Google Scholar]
- 4.Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. 2018. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557:101–105. 10.1038/s41586-018-0059-5. [DOI] [PubMed] [Google Scholar]
- 5.Montagna M, Sassera D, Epis S, Bazzocchi C, Vannini C, Lo N, Sacchi L, Fukatsu T, Petroni G, Bandi C. 2013. “Candidatus Midichloriaceae” fam. nov. (Rickettsiales), an ecologically widespread clade of intracellular alphaproteobacteria. Appl Environ Microbiol 79:3241–3248. 10.1128/AEM.03971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szokoli F, Castelli M, Sabaneyeva E, Schrallhammer M, Krenek S, Doak TG, Berendonk TU, Petroni G. 2016. Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia. Appl Environ Microbiol 82:7236–7247. 10.1128/AEM.02284-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray RGE, Stackebrandt E. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45:186–187. 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 8.Vannini C, Boscaro V, Ferrantini F, Benken KA, Mironov TI, Schweikert M, Görtz H, Fokin SI, Sabaneyeva EV, Petroni G. 2014. Flagellar movement in two bacteria of the family Rickettsiaceae: a re-evaluation of motility in an evolutionary perspective. PLoS One 9:e87718. 10.1371/journal.pone.0087718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanzoni O, Sabaneyeva E, Modeo L, Castelli M, Lebedeva N, Verni F, Schrallhammer M, Potekhin A, Petroni G. 2019. Diversity and environmental distribution of the cosmopolitan endosymbiont “Candidatus Megaira”. Sci Rep 9:1179. 10.1038/s41598-018-37629-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epis S, Sassera D, Beninati T, Lo N, Beati L, Piesman J, Rinaldi L, McCoy KD, Torina A, Sacchi L, Clementi E, Genchi M, Magnino S, Bandi C. 2008. Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 135:485–494. 10.1017/S0031182007004052. [DOI] [PubMed] [Google Scholar]
- 11.Lejal E, Chiquet J, Aubert J, Robin S, Estrada-Peña A, Rue O, Midoux C, Mariadassou M, Bailly X, Cougoul A, Gasqui P, Cosson JF, Chalvet-Monfray K, Vayssier-Taussat M, Pollet T. 2021. Temporal patterns in Ixodes ricinus microbial communities: an insight into tick-borne microbe interactions. Microbiome 9:153. 10.1186/s40168-021-01051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klinges JG, Rosales SM, McMinds R, Shaver EC, Shantz AA, Peters EC, Eitel M, Wörheide G, Sharp KH, Burkepile DE, Silliman BR, Vega Thurber RL. 2019. Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J 13:2938–2953. 10.1038/s41396-019-0482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Aeschlimann A, Gern L. 1992. Rickettsia-like microorganisms in the ovarian primordia of molting Ixodes ricinus (Acari: Ixodidae) larvae and nymphs. Ann Parasitol Hum Comp 67:99–110. 10.1051/parasite/199267499. [DOI] [Google Scholar]
- 14.Comandatore F, Radaelli G, Montante S, Sacchi L, Clementi E, Epis S, Cafiso A, Serra V, Pajoro M, Di Carlo D, Floriano AM, Stavru F, Bandi C, Sassera D. 2021. Modelling the life cycle of the intramitochondrial bacterium “Candidatus Midichloria mitochondrii” using electron microscopy data. mBio 12:e00574-21. 10.1128/mBio.00574-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beninati T, Lo N, Sacchi L, Genchi C, Noda H, Bandi C. 2004. A novel alphaproteobacterium resides in the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl Environ Microbiol 70:2596–2602. 10.1128/AEM.70.5.2596-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz F, Martijn J, Wascher F, Lagkouvardos I, Kostanjšek R, Ettema TJ, Horn M. 2016. A Rickettsiales symbiont of amoebae with ancient features. Environ Microbiol 18:2326–2342. 10.1111/1462-2920.12881. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Wu M. 2017. Comparative genomic analysis of Acanthamoeba endosymbionts highlights the role of amoebae as a “melting pot” shaping the Rickettsiales evolution. Genome Biol Evol 9:3214–3224. 10.1093/gbe/evx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vannini C, Ferrantini F, Schleifer K, Ludwig W, Verni F, Petroni G. 2010. “Candidatus Anadelfobacter veles” and “Candidatus Cyrtobacter comes,” two new Rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Appl Environ Microbiol 76:4047–4054. 10.1128/AEM.03105-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floriano AM, Castelli M, Krenek S, Berendonk TU, Bazzocchi C, Petroni G, Sassera D. 2018. The genome sequence of “Candidatus Fokinia solitaria”: insights on reductive evolution in Rickettsiales. Genome Biol Evol 10:1120–1126. 10.1093/gbe/evy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boscaro V, Husnik F, Vannini C, Keeling PJ. 2019. Symbionts of the ciliate Euplotes: diversity, patterns and potential as models for bacteria-eukaryote endosymbioses. Proc Biol Sci 286:20190693. 10.1098/rspb.2019.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Gómez SA, Hess S, Burger G, Lang BF, Susko E, Slamovits CH, Roger AJ. 2019. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. Elife 8:e42535. 10.7554/eLife.42535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura Y, Kikuchi Y, Meng XY, Koga E, Fukatsu T. 2012. Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl Environ Microbiol 78:4149–4156. 10.1128/AEM.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber-Vodicka HR, Leisch N, Kleiner M, Hinzke T, Liebeke M, McFall-Ngai M, Hadfield MG, Dubilier N. 2019. Two intracellular and cell type-specific bacterial symbionts in the placozoan Trichoplax H2. Nat Microbiol 4:1465–1474. 10.1038/s41564-019-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamm K, Osigus H-J, Stadler PF, DeSalle R, Schierwater B. 2019. Genome analyses of a placozoan rickettsial endosymbiont show a combination of mutualistic and parasitic traits. Sci Rep 9:17561. 10.1038/s41598-019-54037-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Lecce I, Bazzocchi C, Cecere JG, Epis S, Sassera D, Villani BM, Bazzi G, Negri A, Saino N, Spina F, Bandi C, Rubolini D. 2018. Patterns of Midichloria infection in avian-borne African ticks and their trans-Saharan migratory hosts. Paras Vect 11:106. 10.1186/s13071-018-2669-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metselaar M, Thompson KD, Paley R, Green DM, Verner-Jeffreys D, Feist S, Adams A. 2020. Investigating the involvement of a Midichloria-like organism (MLO) in red mark syndrome in rainbow trout Oncorhynchus mykiss. Aquaculture 528:735485. 10.1016/j.aquaculture.2020.735485. [DOI] [Google Scholar]
- 27.Fischer T, Myalkhaa M, Krücken J, Battsetseg G, Batsukh Z, Baumann MPO, Clausen P-H, Nijhof AM. 2020. Molecular detection of tick-borne pathogens in bovine blood and ticks from Khentii, Mongolia. Transbound Emerg Dis 67:111–118. 10.1111/tbed.13315. [DOI] [PubMed] [Google Scholar]
- 28.Arrais RC, Paula RC, Martins TF, Nieri-Bastos FA, Marcili A, Labruna MB. 2021. Survey of ticks and tick-borne agents in maned wolves (Chrysocyon brachyurus) from a natural landscape in Brazil. Ticks Tick Borne Dis 12:1011639. 10.1016/j.ttbdis.2020.101639. [DOI] [PubMed] [Google Scholar]
- 29.Sassera D, Lo N, Epis S, D'Auria G, Montagna M, Comandatore F, Horner D, Peretó J, Luciano AM, Franciosi F, Ferri E, Crotti E, Bazzocchi C, Daffonchio D, Sacchi L, Moya A, Latorre A, Bandi C. 2011. Phylogenomic evidence for the presence of a flagellum and cbb3 oxidase in the free-living mitochondrial ancestor. Mol Biol Evol 28:3285–3296. 10.1093/molbev/msr159. [DOI] [PubMed] [Google Scholar]
- 30.Mariconti M, Epis S, Gaibani P, Dalla Valle C, Sassera D, Tomao P, Fabbi M, Castelli F, Marone P, Sambri V, Bazzocchi C, Bandi C. 2012. Humans parasitized by the hard tick Ixodes ricinus are seropositive to Midichloria mitochondrii: is Midichloria a novel pathogen, or just a marker of tick bite? Pathog Glob Health 106:391–396. 10.1179/2047773212Y.0000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senra MVX, Dias RJP, Castelli M, Silva-Neto ID, Verni F, Soares CAG, Petroni G. 2016. A house for two—double bacterial infection in Euplotes woodruffi Sq1 (Ciliophora, Euplotia) sampled in Southeastern Brazil. Microb Ecol 71:505–517. 10.1007/s00248-015-0668-6. [DOI] [PubMed] [Google Scholar]
- 32.Klinges G, Maher RL, Vega Thurber R, Muller EM. 2020. Parasitic “Candidatus Aquarickettsia rohweri” is a marker of disease susceptibility in Acropora cervicornis but is lost during thermal stress. Environ Microbiol 22:5341–5355. 10.1111/1462-2920.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariconti M, Epis S, Sacchi L, Biggiogera M, Sassera D, Genchi M, Alberti E, Montagna M, Bandi C, Bazzocchi C. 2012. A study on the presence of flagella in the order Rickettsiales: the case of “Candidatus Midichloria mitochondrii”. Microbiology 158:1677–1683. 10.1099/mic.0.057174-0. [DOI] [PubMed] [Google Scholar]
- 34.Fritsche TR, Horn M, Seyedirashti S, Gautom RK, Schleifer KH, Wagner M. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl Environ Microbiol 65:206–212. 10.1128/AEM.65.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassera D, Beninati T, Bandi C, Bouman EAP, Sacchi L, Fabbi M, Lo N. 2006. “Candidatus Midichloria mitochondrii,” an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol 56:2535–2540. 10.1099/ijs.0.64386-0. [DOI] [PubMed] [Google Scholar]
- 36.Castelli M, Lanzoni O, Nardi T, Lometto S, Modeo L, Potekhin A, Sassera D, Petroni G. 2021. ‘Candidatus Sarmatiella mevalonica’ endosymbiont of the ciliate Paramecium provides insights on evolutionary plasticity among Rickettsiales. Environ Microbiol 23:1684–1701. 10.1111/1462-2920.15396. [DOI] [PubMed] [Google Scholar]
- 37.Husnik F, Tashyreva D, Boscaro V, George EE, Lukeš J, Keeling PJ. 2021. Bacterial and archaeal symbioses with protists. Curr Biol 31:R862–R877. 10.1016/j.cub.2021.05.049. [DOI] [PubMed] [Google Scholar]
- 38.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 39.Tsai Y-M, Chang A, Kuo CH. 2018. Horizontal gene acquisitions contributed to genome expansion in insect-symbiotic Spiroplasma clarkii. Genome Biol Evol 10:1526–1532. 10.1093/gbe/evy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraune S, Bosch T. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA 104:13146–13151. 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasqualetti C, Schmidt JG, Cafiso A, Gammuto L, Lanzoni O, Sepulveda D, Manfrin A, Benedetti Cecchi L, Olesen NJ, Bazzocchi C, Petroni G. 2021. Double trouble: could Ichthyophthirius multifiliis be a vehicle for the bacterium associated with red mark syndrome in rainbow trout, Oncorhynchus mykiss? Aquaculture 533:736230. 10.1016/j.aquaculture.2020.736230. [DOI] [Google Scholar]
- 42.Cafiso A, Sassera D, Romeo C, Serra V, Caroline H, Bandi C, Plantard O, Bazzocchi C. 2019. Midichloria mitochondrii, endosymbiont of Ixodes ricinus: evidence for the transmission to the vertebrate host during the tick blood meal. Ticks Tick Borne Dis 10:5–12. 10.1016/j.ttbdis.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Lo N, Beninati T, Sassera D, Bouman EAP, Santagati S, Gern L, Sambri V, Masuzawa T, Gray JS, Jaenson TGT, Bouattour A, Kenny MJ, Guner ES, Kharitonenkov IG, Bitam I, Bandi C. 2006. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ Microbiol 8:1280–1287. 10.1111/j.1462-2920.2006.01024.x. [DOI] [PubMed] [Google Scholar]
- 44.Baker LJ, Reich HG, Kitchen SA, Klinges JG, Koch HR, Baums IB, Muller EM, Vega Thurber R. 2022. The coral symbiont Candidatus Aquarickettsia is variably abundant in threatened Caribbean acroporids and transmitted horizontally. ISME J 16:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duron O, Binetruy F, Noël V, Cremaschi J, McCoy KD, Arnathau C, Plantard O, Goolsby J, Pérez de León AA, Heylen DJA, Van Oosten AR, Gottlieb Y, Baneth G, Guglielmone AA, Estrada-Peña A, Opara MN, Zenner L, Vavre F, Chevillon C. 2017. Evolutionary changes in symbiont community structure in ticks. Mol Ecol 26:2905–2921. 10.1111/mec.14094. [DOI] [PubMed] [Google Scholar]
- 46.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 47.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson ACC, von Dohlen C, Fukatsu T, McCutcheon JP. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 48.Cafiso A, Bazzocchi C, De Marco L, Opara MN, Sassera D, Plantard O. 2016. Molecular screening for Midichloria in hard and soft ticks reveals variable prevalence levels and bacterial loads in different tick species. Ticks Tick Borne Dis 7:1186–1192. 10.1016/j.ttbdis.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 49.Boscaro V, Manassero V, Keeling PJ, Vannini C. Single-cell microbiomics unveils distribution and patterns of microbial symbioses in the natural environment. Microb Ecol, in press. [DOI] [PubMed] [Google Scholar]
- 50.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 51.Keeling PJ, Corradi N, Morrison HG, Haag KL, Ebert D, Weiss LM, Akiyoshi DE, Tzipori S. 2010. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol Evol 2:304–309. 10.1093/gbe/evq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toft C, Fares MA. 2008. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol Biol Evol 25:2069–2076. 10.1093/molbev/msn153. [DOI] [PubMed] [Google Scholar]
- 53.Hendry TA, de Wet JR, Dougan KE, Dunlap PV. 2016. Genome evolution in the obligate but environmentally active luminous symbionts of flashlight fish. Genome Biol Evol 8:2203–2213. 10.1093/gbe/evw161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visick KL, Stabb EV, Ruby EG. 2021. A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host. Nat Rev Microbiol 19:654–665. 10.1038/s41579-021-00557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boscaro V, Schrallhammer M, Benken KA, Krenek S, Szokoli F, Berendonk TU, Schweikert M, Verni F, Sabaneyeva EV, Petroni G. 2013. Rediscovering the genus Lyticum, multiflagellated symbionts of the order Rickettsiales. Sci Rep 3:3305. 10.1038/srep03305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boscaro V, Petroni G, Ristori A, Verni F, Vannini C. 2013. “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii”, two novel ciliate endosymbionts belonging to the “Midichloria clade”. Microb Ecol 65:302–310. 10.1007/s00248-012-0170-3. [DOI] [PubMed] [Google Scholar]
- 57.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinf 70:e102. 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 58.Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Res 6:1287. 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- 59.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 61.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mareuil F, Doppelt-Azeroual O, Ménager H. 2017. A public Galaxy platform at Pasteur used as an execution engine for web services. F1000Research 6:1020. [Google Scholar]
- 65.Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. 2014. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9:e110726. 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emms D, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Phylogenetics trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508. 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 73.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha EP. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aem.02432-21-s0001.xlsx, XLSX file, 0.05 MB (53KB, xlsx)
Table S2. Download aem.02432-21-s0002.xlsx, XLSX file, 0.03 MB (27.7KB, xlsx)
Fig. S1. Download aem.02432-21-s0003.pdf, PDF file, 0.6 MB (649.8KB, pdf)
Data Availability Statement
Genome sequences were deposited in the DDBJ/ENA/GenBank database with accession numbers JAJBJA000000000 to JAJBJD000000000.


