Abstract
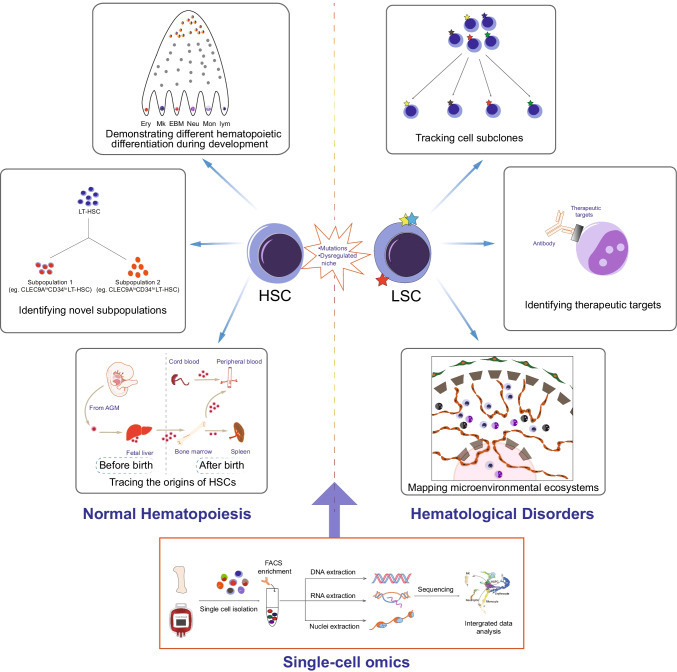
Residing at the apex of the hematopoietic hierarchy, hematopoietic stem and progenitor cells (HSPCs) give rise to all mature blood cells. In the last decade, significant progress has been made in single-cell RNA sequencing as well as multi-omics technologies that have facilitated elucidation of the heterogeneity of previously defined human HSPCs. From the embryonic stage through the adult stage to aging, single-cell studies have enabled us to trace the origins of hematopoietic stem cells (HSCs), demonstrating different hematopoietic differentiation during development, as well as identifying novel cell populations. In both hematological benign diseases and malignancies, single-cell omics technologies have begun to reveal tissue heterogeneity and have permitted mapping of microenvironmental ecosystems and tracking of cell subclones, thereby greatly broadening our understanding of disease development. Furthermore, advances have also been made in elucidating the molecular mechanisms for relapse and identifying therapeutic targets of hematological disorders and other non-hematological diseases. Extensive exploration of hematopoiesis at the single-cell level may thus have great potential for broad clinical applications of HSPCs, as well as disease prognosis.
Graphical abstract
Keywords: Hematopoietic stem and progenitor cells, Single-cell omics, Hematopoiesis, Hematopoietic malignancies
Introduction of human hematopoietic stem and progenitor cells
The hematopoietic system is one of the most highly regenerative tissues, which is composed of billions of erythrocytes, platelets, myeloid cells, innate and adaptive immune cells. It is involved in blood cell formation, coagulation function, immune response, and other physiological processes. Residing at the hierarchical top of hematopoiesis, hematopoietic stem and progenitor cells (HSPCs) give rise to all mature blood cells [1], and even a single hematopoietic stem cell (HSC) can produce long-term and multipotent reconstitution of the entire blood system [2, 3].
In 1961, Till and McCulloch first demonstrated the existence of multipotent HSCs through observing colony-forming units in the spleen following in vivo lethally irradiated transplantation [4]. The subsequent discovery of surface markers to purify HSCs, coupled with fluorescence-activated cell sorting (FACS), made it possible to isolate HSC and progenitor populations. In human hematopoiesis, CD34, which is expressed on a minority of blood cells, was the first human HSPC surface marker discovered in 1984 [5] and was widely used in clinical HSC transplantation [6, 7]. However, some studies showed that CD34− HSCs in human cord blood (CB) may exist [8–10]. To date, surface markers including CD38 [11, 12], CD45RA [13], CD90 [14], CD49f, rhodamine-123 [3], GPI80 [15], etc. have been applied to purify human HSCs in different laboratories. Similarly, multi-, oligo- and unipotent progenitor cells also have been separated by different surface markers [16]. These immunophenotype-defined assays have been commonly used and have contributed greatly to the research of hematopoiesis over the past decades. However, it is worth noting that due to alterations of surface marker expression after culture or transplantation [17], immunophenotype may not be an accurate representation of the true HSPCs in all scenarios.
Functional assays such as in vivo xenograft and in vitro colony formation are the “gold standard” to confirm the existence of human HSCs and progenitor cells [16]. HSCs are characterized by self-renewal and multipotency, while progenitor cells are oligo- or unipotent due to their insufficiency to differentiate into all blood lineages and lack of self-renewal ability [18, 19]. The classical human hematopoietic hierarchy is usually described as a tree-like model, which starts with HSCs, followed by multipotent progenitors (MPPs), downstream progenitors and mature cells [16]. The first lineage decision followed by MPPs to segregate myeloid/erythroid from myeloid/lymphoid fates coincides with the population of common myeloid progenitors (CMPs) and multi-lymphoid progenitors (MLPs) [20–22]. A second lineage decision occurs allowing CMPs to form megakaryocyte-erythroid progenitor progenitors (MEPs) and granulocyte-monocyte progenitors (GMPs). These progenitors differentiate into erythrocytes, megakaryocytes, granulocytes, monocytes, T, B, NK, and dendritic cells. Although the classical human hierarchical model has been a great tool to understand hematopoiesis, it should be noted that this model was established by immunophenotyped populations and functional assays of bulk cells.
The heterogeneity of immunophenotype-defined HSPC populations has restricted further understanding of human hematopoiesis in normal and disease states, thus, to overcome these limitations, researchers have turned to study HSPCs at more precise levels, even at the single-cell level. From a single-cell perspective, in vitro single-cell culture cannot fully display all blood lineages due to technical limitations and differentiation bias caused by different culture systems. Moreover, the insufficient cell numbers (or just one cell) make the in vivo xenograft experiments extremely difficult. Therefore, neither immunophenotype nor functional experiments can entirely define true HSPCs. With recent advances in single-cell technologies, single-cell transcriptomics became another powerful means to recognize hematopoiesis as well as HSPCs within the past few years. Through this new technology, new subpopulations of HSPCs [23], a revised roadmap of the hematopoietic system [24] and malignancy hierarchies relevant to disease progression [25] have been proposed by different groups. Our group has also detailed a single-cell landscape of the human blood cells for providing a comprehensive reference of hematopoiesis [26]. In conclusion, functional, immunophenotypic and transcriptomic (FIT)-defined HSPCs represent future directions for advancing the understanding of hematopoiesis [27].
Advances in single-cell technologies
Single-cell transcriptomics
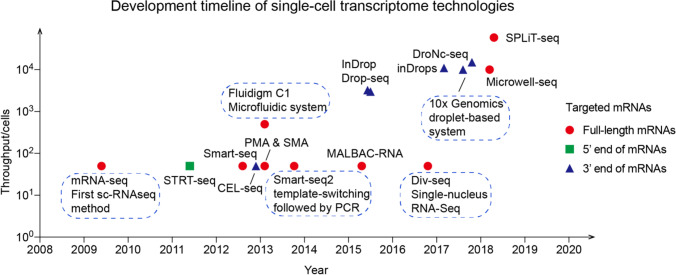
Since Tang et al. [28] first established the single-cell mRNA-Seq method in 2009, single-cell transcriptomic technologies have enabled great progress in the last ten years, owing to the capability of sequencing at increasing throughput and decreasing cost. Single-cell RNA sequencing (scRNA-seq) is composed of several steps: single-cell capture, RNA obtainment, cDNA amplification, and library construction. Wu et al. [29] used a microfluidic-based system for cell capture, lysis, and pre-amplification in one chip. In 2015, scientists developed two methods, separately termed Drop-seq [30] and InDrop [31], which combined barcoded primer beads and droplets to achieve throughput reaching thousands of individual cells. With the development of droplet-based assays such as 10 × Genomics [32] and inDrops [33], tens of thousands of single cells could be captured per sample. Subsequently, Guo’s laboratory developed Microwell-Seq [34] using microwell arrays constructed from agarose and barcoded beads, which drastically reduced the cost. SPLiT-seq [35] utilized four rounds of barcoding strategies to label the cellular origins of RNA. Each round of barcoding could append different labels to cDNA, ultimately labeling over 1 million cells. Figure 1 summarizes the recent advances in single-cell technologies.
Fig. 1.
Development timeline of single-cell transcriptome technologies
To achieve high efficiency and low bias of single-cell sequencing, researchers developed various assays: STRT-seq [36] and Smart-seq/Smart-seq2 [37–39] used template-switching technology followed by PCR for full-length cDNA; others like CEL-Seq [40], Phi29-mRNA amplification (PMA), SRP-mRNA amplification (SMA) [41] and Multiple Annealing and Looping-Based Amplification Cycles (MALBAC) [42] adopted linear amplification or other methods for cDNA. However, most of the aforementioned assays need freshly prepared and live single-cell suspensions to extract cytoplasmic RNA, hence Div-Seq [43] and DroNc-Seq [44] were developed to profile RNA from preserved or undissociated samples. Although there is still some room for improvement in single-cell transcriptomic technologies, it will have broad potential applications in biological and clinical research.
Single-cell multi-omics
Although single-cell transcriptomics is a great tool to quantify expression variability between individual cells, it is not possible to use it to explore the correlation of genotype, phenotype, gene expression, or even chromatin structure in single cells. It was a great challenge for scientists to integrate transcriptomics, genomics, proteomics and epigenomics in a single cell. However, in 2015, DR-Seq [45] and G&T-seq [46] made it possible to separate and sequence genomic DNA and full-length mRNA from single cells. Moreover, DNTR-seq [47] can combine whole genome DNA and mRNA sequencing at the single-cell level simultaneously. Tang’s laboratory described scTrio-seq [48], which can simultaneously obtain the single-cell transcriptome, DNA methylome, and genomic copy-number variations. In CITE-seq [49] and REAP-seq [50], cells labeled with antibodies conjugated to DNA barcodes and cellular protein and transcriptome can be measured simultaneously. In addition to revealing phenotypic differences and heterogeneity of cell populations, these methods enable large-scale immunophenotyping of dozens to hundreds of antibodies. Greenleaf's laboratory combined chromatin accessibility assays with gene expression data at the single-cell level to portray regulatory features in the human hematopoietic system [51]. Recently, his laboratory integrated protein quantification, gene expression, and chromatin accessibility to resolve molecular features of patients with mixed-phenotype acute leukemia (MPAL) [52]. Recently, a single-cell multi-omics assay has been developed by 10 × genomics in individual nucleus. Instead of using algorithmic integration method, this technology enables to perform RNA and ATAC sequencing simultaneously in single cells. This would be able to simultaneously investigate how chromatin accessibility and RNA expression determine cell fate in individual cells [53–55]. Single-cell multi-omics can provide sufficient information of cell heterogeneity, cellular subpopulations, epigenetic transitions, and the cell regulation state in the past and future. Thus, the correlation between genotype and phenotype could be captured by single-cell multi-omics. Methods for processing the single-cell multi-omics sequencing data usually include graph-based learning models and unsupervised learning methods [56]. Deep learning methods (e.g. Convolutional Neural Network (CNN) [57], Recurrent Neural Network (RNN) [58] and transformer model) have great potential in processing multi-omics sequencing which increased the data-processing performance. Recently, Simon Haas [59] et al. generated a single-cell proteo-genomic reference map, which linked the expression of 197 surface markers to cellular identities and biological processes across main hematopoietic cell types of BM and peripheral blood from human adults, aged and AML patients. Moreover, they developed computational tools that enable the automatic design of high-throughput cytometry schemes to isolate the molecularly defined cell state from blood and BM. However, most integrating data from DNA, RNA, or ATAC are not from the same cell. We suggest that the integration of these single-cell sequencing datasets should be used cautiously in hematological studies. Moreover, single-cell multi-omics sequencing usually generated a large number of heterogeneous data including different numbers of distributions, variables and diverse data modalities. Thus, developing a method to excavate data-specific information and to use different molecular layers information at the same time is crucial for data processing [56]. In conclusion, single-cell multi-omics could provide more insight into regulatory dynamics both in normal and disease conditions, although new technologies with low cost and high throughput are still needed.
Applications of single-cell omics in normal hematopoiesis and diseases
Embryonic hematopoiesis
The development of hematopoiesis is a complex process that switches from different organs in the embryonic period [60, 61]. During the prenatal stage, human HSCs first occur in the aorta-gonad-mesonephros (AGM), later in the yolk sac, fetal liver, and then finally migrate to bone marrow (BM). Additionally, some previously unappreciated sites like placenta [62, 63], umbilical arteries [64], and embryonic head [65] have been demonstrated to harbor HSCs in mice. Due to technical limitations and human sample rarity, as well as a rare number of HSC at the embryonic stage, it has been difficult to explore the precise origin of HSCs and the immune system. However, through scRNA-seq, Liu’s laboratory first established a gene expression atlas of HSC generation in the AGM region. HSC-primed hemogenic endothelial cells (HECs), the origin of HSPCs, were defined transcriptomically and could be enriched tenfold via the surface marker CD44. EMCN, PROCR, and RUNX1T1 were overexpressed at hemogenic fate choice from arterial endothelial cells via HSC-primed HECs to HSPCs [66]. To further identify the molecular mechanisms for promoting HSC emergence, Crosse et al. [67] applied spatial transcriptomics to compare the gene expression differences in dorsoventral polarized signaling in the aorta. A subpopulation of aortic endothelial cells, which had downregulated aortic signatures and may associate with HSPC emergence, was predicted. Also, endothelin 1 was found to be an important regulating factor of HSC development. Another study mainly focused on definitive hematopoiesis in fetal liver, in which over 200,000 single cells between 7 and 17 post-conception weeks (PCW) from liver, skin, kidney and yolk sac were sequenced to explore the human blood and immune cell developmental trajectory. Twenty-seven major cell clusters were transcriptomically defined in fetal liver and validated by morphology and imaging. Also, fetal skin was demonstrated as a physiological erythropoietic tissue and HSPCs in fetal liver showed decreased erythroid differentiation potential during gestation with functional validation [68]. Recently, hematopoietic development in human fetal BM was detailed from 12 to 19 PCW from nine normal fetuses. The whole blood and immune system were established during the early second trimester. Also, alterations of gene expression and cellular composition between fetal liver and fetal BM were revealed [69]. Integrative analysis of both scRNA-seq and scATAC-seq highlighted epigenetic priming of HSC/MPPs before lineage commitment [70], which was divergent from the conventional opinion that transcriptional priming emerged first. Although there have been many studies on the development of HSPCs during the fetal period, a more detailed molecular analysis of the dynamics of HSPCs as well as determining when transplantable HSCs emerge in fetal BM remains to be precisely investigated.
The development and origin of the immune system have also been revealed at the single-cell level in the human embryo. Macrophage is a regulator which is essential to tissue development and homeostasis. Bian et al. [71] comprehensively characterized macrophage development during embryogenesis and found a new cell population of yolk sac-derived myeloid-biased progenitors via functional validation. This could be crucial for the diagnosis and treatment of the disease associated with the new population. T and B cells are major components of human adaptive immunity. Thus, decoding their lymphopoiesis at the fetal stage not only provides insights into the physiological development of lymphopoiesis, but also helps to understand the etiology of related blood diseases. Zeng et al. [72] portrayed early T lymphopoiesis and thymus organogenesis at the human embryonic stage at single-cell resolution. A new subset of early thymic progenitors was defined, which shared transcriptional similarity with thymus-seeding progenitors in the fetal liver. Also, pre-thymic lymphoid progenitors were demonstrated in the AGM region. O’Byrne et al. demonstrated several B-cell progenitor populations by constructing the human fetal B-lymphocyte development hierarchy in fetal BM and liver. The PreProB-progenitor was confirmed as the first B-lymphoid–restricted progenitor and upstream of ProB-progenitors at the fetal stage [73]. Meanwhile, a series of studies focused on other cell population like megakaryocyte [74] has also been published.
In summary, single-cell technologies have greatly assisted the investigation of the detailed developmental processes underlying embryonic hematopoiesis and prenatal disease initiation. It may give us more information about hematopoietic development. In the future, we might be able to improve HSC reconstitution in the patient with delayed engraftment, and manipulate the lineage bias of progenitor cells for cell therapy.
Adult hematopoiesis
After the embryonic stage, BM is the most important microenvironment for maintaining hematopoiesis. As estimated by somatic clonal dynamics, there are approximately 50,000–200,000 HSPCs that contribute to white blood cells in adult BM [75]. Using scRNA-seq, several groups constructed the transcriptional landscape of human HSPCs in BM or CB [24, 76, 77]. A comprehensive human hematopoietic landscape constructed by single-cell transcriptome and chromatin accessibility revealed the differentiation trajectories of human hematopoiesis [51]. The lncRNAs expression profile of human HSPCs has also been generated [78]. Additionally, immunophenotype-defined stem and progenitor populations have been proven to be heterogeneous according to single-cell omics. Combining CD34 and CLEC9A expression, HSCs were divided into two groups: CLEC9AhighCD34low HSCs with multipotency and more quiescence and CLEC9AlowCD34high HSCs with myeloid and lymphoid potential [23]. Another study pointed out that compared to CD33− HSCs, CD33+ HSCs have more durable regenerative potential [79]. For the progenitors, the immunophenotypic CMPs and GMPs were reported to be heterogenous by an integrated analysis of scRNA-seq and scATAC-seq. GMPs were further divided into three sequential populations with distinct myeloid developmental levels [51]. Karamitros et al. showed via transcriptional and functional validation that lymphoid primed multi-potential progenitors (LMPPs), GMPs and MLPs were heterogeneous populations. Although most of them were unipotent, there were still a few of these progenitors that showed bi- and multipotency [21].
The molecular mechanisms of fate decision that play an important role in hematopoiesis have also been investigated. Recently, Lu et al. [80] demonstrated that the fate decision of MEPs was affected by cell cycle speed. When the cell cycle speed was increased, MEPs were biased towards erythrocyte specification, and when the cell cycle speed was decreased, MEPs were biased towards megakaryocyte differentiation. According to the upregulation of transcription factor GATA2, it is plausible that eosinophils/basophils/mast cells may have common ancestors with erythrocytes in human hematopoiesis, and this has been proved in the mouse model [81]. Taken together, more precise definition of subsets from HSPCs could be acquired through single-cell omics, thus leading to a better understanding of hematopoiesis.
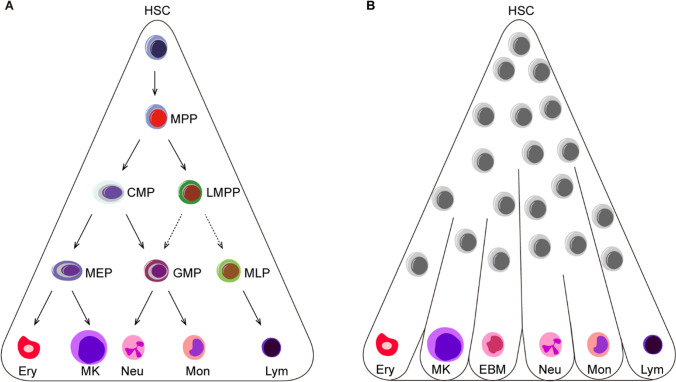
To date, several studies have challenged the classical tree-like hierarchy (Fig. 2) [21, 24, 76, 82]. Recent research provided a continuum differentiation model through a comprehensive overview of human BM HSPCs by single-cell technologies [24]. A continuum of low-primed undifferentiated HSPCs was at the hierarchical top of hematopoiesis, and there were no specific progenitor populations like CMPs during the period from HSCs to unilineage-restricted cells. This view was confirmed by another single-cell study performed in human CB HSPCs with unbiased computational analysis, in which there were intermediate stages closely related to stem cell populations without distinct fate choices [76]. Distinct differentiation potentials of LMPPs, MLPs, and GMPs also confirmed that a continuum of progenitors differentiate downstream of stem cells [21]. Additionally, the differentiation model of adult hematopoiesis is different from that at the fetal stage. Dick’s group demonstrated previously through in vivo and in vitro functional assays that immunophenotypic MPPs, CMPs and MEPs are heterogeneous populations, and that there is a differentiation shift from fetal to adult: many HSPCs were multipotent at the fetal stage, while stem cells show multipotency and progenitors were unipotent at the adult stage [83]. However, although this study focused on adult and fetal stage, alterations of human HSPCs in the period after birth to adult still requires further research.
Fig. 2.
Human hematopoietic hierarchy models. A The classical tree-like human hematopoietic hierarchy. HSCs differentiate into multi-, bi- and unipotent progenitor cells, and progenitors give rise to all mature blood cells. B The continuum human hematopoietic differentiation model. Continuum of low-primed undifferentiated HSPCs sit at the hierarchical top of hematopoiesis and differentiate into unilineage-restricted cells. Ery, erythrocyte; MK, megakaryocyte; EBM, eosinophil, basophil and mast cells; Neu, neutrophils; Mon, monocyte; Lym, lymphocyte
In summary, single-cell omics plays an important role in understanding human hematopoiesis. It is of critical importance to integrate the tools of immunophenotyping, functional validation, and transcriptomics to properly define HSPCs and gain a better understanding of the blood system.
HSPC niche
Single-cell omics are great tools to explore HSPC niche, which represents a three-dimensional space comprising several types of components interact with and regulate HSPCs [84, 85]. Recently, spatial transcriptomic technologies provided new perspectives in exploring HSPC niche both in human and mouse models [67, 86–88]. In short, there are four types of spatial transcriptomics, including computational methods for spatial reconstruction, laser capture microdissection (LCM)-based methods, in situ RNA imaging methods and in situ sequencing methods. Crosse et al. combined spatial, population, and single-cell transcriptomics to interrogate signaling in the human HSC embryonic niche. By using LCM coupled with RNA sequencing, molecular differences between orsal-ventral and the dorsal aorta were carefully explored. And it focused on cell layers close to intra-aortic hematopoietic cell clusters (IAHCs) formation. By analysis of the ventrally polarized molecular landscape, endothelin 1 was identified as an important secreted regulator for human HSC development. Interestingly, cardiac EGF pathway was enriched next to developing IAHCs/HSCs, and endothelin 1, secreted by ventral portions, was found to promote the development of HSCs. However, studies related to the niche of human adult hematopoiesis were still lacking [67]. The immune microenvironment in hematopoietic malignance and diseases could be the future directions of the spatial transcriptomics.
Aging and benign diseases
Hematopoietic aging in humans always manifests a high risk of myeloid malignancies and functional decline of HSCs. With an integrative characterization of epigenomic and transcriptomic changes, Adelman et al. [89] portrayed the map of regulatory elements in normal human HSC during aging. The epigenetic reprogramming of human HSCs occurred with age, which was particularly evident at active enhancers. For example, KLF6 was identified as not only the most downregulated transcription factor with aging, but also as aberrantly methylated in acute myeloid leukemia (AML). This indicated that age-related dysfunction of the human HSC may also be mediated by dysfunction of these regulatory elements, which may increase the risk of malignant transformation.
In benign diseases like BM failure (BMF) syndromes and aplastic anemia (AA), chromosomal abnormalities were frequently observed and monosomy 7 in BMF was associated with poor clinical outcomes. Young’s group [90] used scRNA-seq to distinguish aneuploid cells from diploid cells within the HSPCs of BMF patients. The aneuploid cells in this study were demonstrated to exhibit downregulation of genes involved in immune response and DNA stability. Recently, our group analyzed the relationship between HSPCs and T cells in AA patients through single-cell transcriptome. Cell-type-specific ligand-receptor interactions were revealed as potential factors for the continuous destruction of HSPCs by T cells [91].
Though single-cell omics have been applied to explore numerous hematopoietic disorders, a few studies related to rare disorders of the hematopoietic system have not been reported. Diamond-Blackfan anemia (DBA) is a rare ribosomopathy. Patients who develop DBA usually receive limited therapeutic options. Using single-cell transcriptomes, Deena et al. [92] presented an unbiased charting of erythropoiesis in RPS-DBA and RPL-DBA and defined genotype–phenotype correlations in DBA. Moreover, they found compensatory stress erythropoiesis in RPL-DBA exhibited altered glucocorticoid molecular signature, including reduced ZFP36L2 expression, leading to milder anemia and improved corticosteroid response. Therefore, ZFP36L2 may become candidate therapeutic targets for failing erythropoiesis.
Paroxysmal nocturnal hemoglobinuria (PNH), which is a rare clonal hematopoietic stem cell disorder that manifests with hemolytic anemia, thrombosis and peripheral blood cytopenia. PNH begins with the expansion of a HSC that has a severe deficiency or absence for GPI, a glycolipid moiety that anchors > 150 different proteins to the cell surface which deficiency in virtually all PNH cases is the result of a somatic mutation in PIGA [93]. The mechanisms leading to PNH stem cell clonal expansion and dominance remain unclear. Therefore, research into this disease at single cell level is needed.
On the other hand, although the increased throughput and decreased cost of single-cell omics sequencing emerged recently, it is still a problem how to change it from descriptive research towards mechanistic insights and/or precision medicine. Further research is needed to explore physiological regulation of human hematopoietic development and discover potential gene targets for directed therapy.
Malignancies
A series of mutations and/or epigenetic events in HSCs can lead to the occurrence of malignant hematopoiesis. In this section, we will discuss the application of single-cell omics to malignant hematopoiesis (Table 1), which has provided new insights into mechanisms of clonal evolution, drug resistance, and disease relapse.
Table 1.
Single-cell omics on malignant hematopoiesis
| Disease | Methodology | Sample input | Ref |
|---|---|---|---|
| AML | ScRNA-seq (Seq-Well) and single-cell genotyping | 30,712 cells from 16 AML patients and 7,698 cells from 5 healthy donors | [25] |
| ScATAC-seq |
71 LSCs and 42 blast cells from two AML patients 88 normal monocytes and 94 LMPPs isolated from healthy donors |
[99] | |
| ScRNA-seq (10 × Genomics) | Peripheral blood mononuclear cells from one older AML patient at baseline and after two and four days of therapy | [101] | |
| ScRNA-seq (MutaSeq) | 618 to 1,430 cells per patient from 4 AML patients | [96] | |
| ScDNA-seq | 735,483 cells from 154 AML samples (140 from BM and 14 from peripheral blood) of 123 patients | [102] | |
| ScDNA-seq | 740,529 cells from 146 samples of 123 patients with myeloid malignancies (clonal hematopoiesis, MPN or AML) | [103] | |
| ALL | ScDNA-seq and scRNA-seq (10 × Genomics) |
1,332 leukemic cells from 4 childhood T-ALL patients 8,296 cells from 4 childhood T-ALL |
[105] |
| Single-cell targeted DNA sequencing | 108,188 cells from 25 samples (12 from BM and 13 from peripheral blood) of 8 T-ALL patients | [106] | |
| Mass cytometry analysis | BM aspirates from 60 patients with BCP-ALL and five healthy donors | [104] | |
| ScRNA-seq (5′ 10 × Genomics and TCR V(D)J) | 25,386 CD19+ cells and 24,157 CD19−CD3+ cells from 4 samples from B-ALL patients before blinatumomab treatment (two responders and two non-responders) | [107] | |
| ScRNA-seq (10 × Genomics) | 53,447 cells from BM samples of 7 B-ALL patients (CD19+ cells: CD19−CD45+ cells, radio = 1:5) and 4 healthy controls (CD45+ cells) | [109] | |
| MPAL | ScRNA-seq (CITE-seq) and scATAC-seq |
CITE-seq: 35,882 cells from 6 healthy donors, 18,056 cells from 6 MPAL patients ScATAC-seq: 35,038 cells from 10 healthy donors, 35,423 cells from 6 MPAL patients |
[52] |
| CML | Sc-qPCR | 2,151 single LSCs from 22 CP-CML patients 5 age-matched healthy controls | [111] |
| ScRNA-seq (Smart-Seq2) | Over 2,000 stem cells from CML patients | [112] | |
| ScRNA-seq (Fluidigm C1) | 150 LSC-CD93+cells, 150 LSC-CD93− cells from 2 CP-CML patients | [113] | |
| ScRNA-seq (Smart-Seq2) | 144 CML-stem cells and 144 HSCs from 3 CML patients | [114] | |
| ScRNA-seq (Smart-Seq2) | 245 cells from 16 CML patients | [115] | |
| CLL |
MscRRBS ScRNA-seq (Smart-Seq2) |
831 normal B cells from six healthy donors, and 1,821 cells from 12 primary IGHV mutated and unmutated CLLs | [118] |
|
Single-cell targeted DNA sequencing ScRNA-seq (Smart-Seq) |
1,152 cells from the 5 CLL patients 96 cells from 4 CLL patients 384 cells from each of 7 CLL patients and from normal CD19+ B cells |
[119] | |
| ScRNA-seq (inDrops) | 1,035–3,751 cells per sample from 4 patients | [120] | |
| MDS | Single cell targeted sequencing | Sorted stem and blast populations with selected mutations in 7 MDS patients who had later progressed to AML | [122] |
| ScDNA-seq | Mononuclear cells in 21 BM samples from 8 patients with MDS and progression to AML | [123] | |
| MPN | scRNA-seq (3’-TARGET-seq) | 752 HSCs from 7 JAK2-V617F+ essential thrombocythemia patients, 359 HSCs from 6 JAK2-V617F+ patients posttreatment and 485 from 7 healthy controls | [124] |
| ScRNA-seq (10 × Genomics) | 93,157 lin−CD34+ HSPCs from 15 patients with myelofibrosis and 42,772 lin−CD34+ HSPCs from 6 healthy donors | [125] | |
| ScRNA-seq (10 × Genomics) | 52,127 HSPCs from 7 newly diagnosed patients with polycythemia vera (n = 3), essential thrombocythemia (n = 4) and healthy controls (n = 2) | [126] | |
| MM | ScRNA-seq (Mars-seq) | 20,586 single plasma cells from the BM and 3,540 single plasma cells from 11 control individuals and 29 newly diagnosed MM patients | [128] |
| Single-cell targeted qRT-PCR | 528 pre-treatment single cells from 11 myeloma cell lines and 418 single cells from 8 drug-naive MM patients | [129] | |
| ScRNA-seq (10 × Genomics) | 17,267 plasma cells and 57,719 immune cells from 29 samples with 14 MM patients at different disease stages | [130] |
Acute leukemia
AML is an aggressive hematological malignancy which leads to a poor clinical outcome. Most patients die from the disease relapse that is related to clonal evolution at the level of cytogenetics [94, 95]. ScRNA-seq is well-suited to characterize AML heterogeneity, illustrate AML tumor ecosystems, and validate subclones. Hence, many studies have focused on distinguishing AML blasts from normal cells [25, 96, 97]. Galen et al. [25] integrated single-cell transcriptomics and genomic data of AML samples and healthy donors to distinguish malignant from normal cells in AML samples. They identified six malignant cell types along the axis of HSC to myeloid differentiation and revealed a striking consistency between developmental hierarchy and tumor genetics. Gene expression analysis revealed that patients with higher HSC/progenitor-like signals exhibited significantly worse clinical outcomes than patients with higher expression of GMP-like genes. These results are in accordance with the existence of leukemia stem cells (LSCs), which have been proved to be capable of initiating and maintaining leukemia and linked to poor prognosis, therapy resistance and high rate of relapse in AML [98]. Velten et al. [96] revealed that LSCs, pre-LSCs and normal HSCs could be distinguished through single-cell transcriptomics due to genomic and mitochondrial mutations. Additionally, Corces et al. [99] demonstrated that Hox-mediated chromatin accessibility loss was the most common defect in pre-leukemic HSCs (pHSCs). Losing HOX factors may lead to differentiation defects like those observed in pHSCs and contribute to an evolutionary advantage. Xu et al. [100] also added the evidence of heterogeneity of the pHSCs population and revealed that the pHSCs burden may reflect the diversity of pHSCs and predict poor prognosis. Moreover, scRNA-seq studies revealed the disease dynamics on older AML patients before and after exposure to the B-cell lymphoma 2 inhibitor venetoclax and azacytidine, in which there were no changes in normal hematopoietic cells, whereas cell blasts were rapidly depleted [101]. These studies also showed that therapeutic interventions eradicate LSCs in AML patients by disrupting metabolic mechanisms that drive energy metabolism, providing insight into clinical use in patients with historically poor outcomes. Recently, clonal evolution in AML was revealed by different groups [102, 103]. Through single-cell DNA sequencing of 146 samples from 123 patients with myeloid malignancies, Miles et al. [103] found that AML was dominated by a few clones which mostly cover co-occurring epigenetic mutations. In contrast, signaling mutations often occurred in subclones more than one time. Such studies at the single-cell level could thus provide further crucial information on initiation and progression for AML.
To date, there have been limited such studies regarding acute lymphoblastic leukemia (ALL) initiation and progression in comparison with AML [104, 105]. Cools’ laboratory used targeted single-cell sequencing of total BM cells and CD34+CD38− multipotent progenitor cells to reveal the genetic basis of disease initiation in T-cell ALL (T-ALL). In half of the cases, mutations could be detected in CD34+CD38− cells, which proved that the order of mutation acquisition in T-ALL may initiate from the multipotent progenitor cells [105]. Another study described clonal evolution at diagnosis and during treatment in T-ALL patients in which a minor clone evolved to the major clone at the advanced stage of disease[106]. Additionally, through single-cell mass cytometry and machine learning, individual B cell precursor ALL (BCP-ALL) cells were mapped to normal B-cell trajectories and pre-pro-B cell to pre-BI cell transition was expanded. A new model, termed ‘developmentally dependent predictor of relapse’ was developed to predict the risk of relapse at diagnosis in BCP-ALL patients [104]. In addition, some studies have applied single-cell technologies to focus on effects of therapies for B-ALL, including blinatumomab [107] and chimeric antigen receptor T cell therapy [108]. Immune microenvironment re-modeling has also been characterized via single-cell technologies during B-ALL progression, in which low non-classical monocytes frequency implied a high survival rate in B-ALL [109]. Furthermore, single-cell omics has been used to reveal epigenetic alternations in mixed phenotype acute leukemia (MPAL), in which RUNX1 may act as a potential oncogene, resulting in poor survival of MPAL patients [52].
Taken together, such studies have offered new insights into heterogeneity, clonal evolution and cellular hierarchies during disease initiation and progression of acute leukemia at the single-cell level that could not be unraveled by bulk analysis. In future studies, determining the precise kinetics of clonal evolution from diagnosis and remission to relapse and response to new therapies would be promising avenues for the further application of single-cell methodologies.
Chronic leukemia
Chronic myeloid leukemia (CML) is primarily caused by the oncogenic fusion protein BCR-ABL, and tyrosine kinase inhibitors (TKIs) have shown potent efficacy in the treatment of CML. Nonetheless, many patients relapse after treatment, mainly due to selective resistance of CML stem cells (CML-SCs) to TKIs [110, 111]. Recently, a variety of studies focused on this issue have sought to identify the molecular mechanisms of relapse as well as therapeutic targets at the single-cell level [112–115]. Giustacchini et al. developed a new method, which combined high-sensitivity mutation detection with whole transcriptome analysis in the same cell, to analyze more than 2,000 CML-SCs from patients. Through this method, BCR-ABL+ SCs can be separated from BCR-ABL− SCs, and a subpopulation of BCR-ABL+ SCs resistant to TKIs was identified, which may become a putative therapeutic target. [112]. In addition, several surface markers were identified to enrich the TKIs-resistant CML-SCs, such as CD26 and CD93 [111, 113]. Another study confirmed that PIM2, a serine/threonine kinase, was required for imatinib mesylate (one of the TKIs) resistance in CML-SCs. A combination of imatinib mesylate with a PIM inhibitor can increase CML-SCs apoptosis, decrease colony formation, and prolong survival of the CML mouse model, without obvious side effects on HSCs [114]. Additionally, the bone morphogenetic protein receptor type-1B and Jak2/Stat3 signaling were activated in persisting and dormant SCs, and targeting these signals could affect CML-SCs in the BM niche [115].
Chronic lymphocytic leukemia (CLL) is a complex heterogeneous cancer with substantial genetic diversity and evolution during disease progression and treatment [116–118]. Combining whole transcriptome analysis with genomic information at the single-cell level could help to elucidate the underpinnings of CLL disease initiation and development. Wang et al. [119] utilized single-cell genomic and transcriptome analysis to reveal that mutations in LCP1 and WNK1 may be novel drivers of CLL, and that there was a high degree of genetic complexity in each CLL. This phenomenon was also observed epigenetically by another study from Landau’s laboratory, in which multiplexed single-cell reduced representation bisulfite sequencing was employed to identify the lineage history and evolution accompanying the therapy of CLL, demonstrating disease heterogeneity at the epigenetic level [118]. In addition, a study of the dynamics of relapse in CLL patients after allogeneic HSC transplantation revealed that later relapses showed accelerated epigenetic alterations in comparison to early relapses. These results provided new evidence of the molecular kinetics of relapse in CLL patients [120].
Other malignances
Myelodysplastic syndrome (MDS) progress to AML in approximately one-third of patients. A series of studies showed that MDS originated from a small group of disease-induced HSCs, which was sustained and expanded by conventional therapy and became a major factor in disease progression and relapse [121]. However, the cellular origins and mechanisms of malignant transformation from MDS to AML have not been clearly defined. Chen et al. [122] performed single-cell sequencing to identify stem cell and blast populations of MDS and matched AML, and found that the MDS stem cells had a higher subclonal mutation complexity than the blast cells. Also, a significant increase in phenotypic malignant stem cells in the overall HSPC population was observed during the development from MDS to AML. These results revealed a nonlinear, parallel clonal evolution in rare subclones in the progression of MDS to AML. Another study by Stosch et al. merged single-cell and bulk sample information to illustrate genetic aberrations, the pertinent clonal architectures, and DNA methylation patterns during the progression of MDS into AML [123]. Single-cell omics has thus matured into a valuable methodology to research disease progression and has provided evidence for subsequent therapy.
JAK2-V617F is the most common mutation in myeloproliferative neoplasm (MPN). Recently, combined with scRNA-seq and mutation detection, Tong et al. [124] revealed that JAK2-V617F+ HSCs exhibited a bias towards megakaryocyte differentiation. This finding was in accordance with other studies in which megakaryocyte differentiation bias in myelofibrosis [125] and increased frequency of erythroid-megakaryocyte progenitors in MPN [126] were observed. This differentiation bias in MPN indicates that the heterogeneity of stem cells in cancer could help to inform therapeutic guidelines.
Multiple myeloma (MM) is a neoplastic hematologic disorder manifested by a clonal proliferation of malignant plasma cells in the BM [127]. Single-cell transcriptome sequencing can not only study intra- and inter-tumor heterogeneity but also provide new ideas for clinical detection and screening of target drugs [128–130]. Ledergor et al. [128] performed scRNA-seq of BM and blood in diagnosed asymptomatic, symptomatic and control individuals to detail the molecular characteristics of MM plasma cells. CD52 was found to enrich circulating tumor cells from peripheral blood, which provided the same or more sensitive genetic information than BM plasma cells. In asymptomatic individuals with early disease, rare tumor plasma cells with molecular characteristics like those of active myeloma could be detected, suggesting that scRNA-seq of early MM may be applied for clinical use. Also, scRNA-seq was reported to detect gene fusions such as t(4;14) in MM [131], implying a new potential application for scRNA-seq.
In conclusion, single-cell omics, combined with integrative bioinformatic analysis, provides new insights into pHSCs, cancer stem cells in hematopoietic malignancies. In spite of this, we usually used mixed samples from different patients in the experiment, it is hard to apply it into individual level. If we want to target some subpopulations or clonal mutations, more meticulous sequencing that personalized to the individual patients is needed. Although it is difficult to apply it into clinical use in a short period of time, that’s making it possible to provide a novel targeted therapy aimed at complex heterogeneous subsets, identify new biomarkers for prediction of prognosis, refine personalized medicine for patients, MRD detection, therapeutic target discovery and even predict response to certain therapies.
Non-hematological diseases
In human cytomegalovirus (CMV) infection, the molecular mechanisms underlying the latent stage and reactivation still require further research. Through scRNA-seq, it was determined that a small group of CD34+ HSPCs expressed markers of Colony Forming Unit—Granulocyte, Erythrocyte, Monocyte, Megakaryocyte (CFU-GEMM) were infected for viral replication [132]. However, another study demonstrated that monocyte progenitors with repressed immune response were the only population in which viral transcripts could be detected in the latent HSPCs. The infection of CMV drives HSPCs towards the weaker immune stage of monocytes, which provides the optimal environment for viral replication [133]. Since 2019, severe coronavirus disease 2019 (COVID-19) became pandemic globally. Interestingly, HSPCs from COVID-19 patients were also impaired due to this highly contagious virus. Through single-cell transcriptome analysis, Wang et al. [134] demonstrated that in severe cases, immature myeloid progenitors accumulated and lymphoid progenitors were reduced, and also observed the upregulation of some transcriptome factors (SPI1, LMO4.etc.). Additionally, with the increasing severity of the COVID-19, monocytes showed decreased cell number and weakened response to this disease [135]. Thus, although HSPCs are the basis of hematopoiesis, perturbations in their numbers, differentiation, or temporal dynamics could partly reflect alterations by some non-hematopoietic diseases (e.g., viral infections) or other environmental factors.
Concluding Remarks
In summary, single-cell omics technologies present a powerful means to reveal the heterogeneity of human HSPCs. Functionally defined, immunophenotyped and transcriptomic-defined HSPCs represent fruitful avenues for future research. The application of single-cell omics to malignant hematopoiesis in recent years has provided new insights into the molecular mechanisms of clonal evolution, disease relapse, and the screening of targeted drugs. Gene and cell therapy areas including hematopoietic stem cell transplantation are important therapies in the treatment of hematologic diseases. However, the intricate operation and high detection cost limit the promotion of single-cell omics. In the future, new simplified single-cell technologies with low cost and high throughput may also lead the way in quality control in the gene and cell therapy arena.
Acknowledgements
We thank Hui Cheng for his helpful advice.
Abbreviations
- HSPCs
Hematopoietic stem and progenitor cells
- HSCs
Hematopoietic stem cells
- FACS
Fluorescence-activated cell sorting
- CB
Cord blood
- MPPs
Multipotent progenitors
- CMPs
Common myeloid progenitors
- MLPs
Multi-lymphoid progenitors
- MEPs
Megakaryocyte-erythroid progenitors
- GMPs
Granulocyte-monocyte progenitors
- scRNA-seq
Single-cell RNA sequencing
- MPAL
Mixed-phenotype acute leukemia
- AGM
Aorta-gonad-mesonephros
- PCW
Post-conception weeks
- BM
Bone marrow
- HECs
Hemogenic endothelial cells
- LMPPs
Lymphoid primed multi-potential progenitors
- BMF
Bone marrow failure
- AA
Aplastic anemia
- DBA
Diamond-Blackfan anemia
- PNH
Paroxysmal nocturnal hemoglobinuria
- AML
Acute myeloid leukemia
- LSCs
Leukemia stem cells
- pHSCs
Pre-leukemic HSCs
- ALL
Acute lymphoblastic leukemia
- T-ALL
T-cell acute lymphoblastic leukemia
- BCP-ALL
B cell precursor acute lymphoblastic leukemia
- CML
Chronic myeloid leukemia
- TKIs
Tyrosine kinase inhibitions
- CML-SCs
Chronic myeloid leukemia stem cells
- CLL
Chronic lymphocytic leukemia
- MDS
Myelodysplastic syndrome
- MM
Multiple myeloma
- CMV
Cytomegalovirus
Authors' contributions
YZ and YH wrote the manuscript. TC and LH gave suggestions and revisions. All authors contributed to this review and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81970104, 81730006, 81890990), National Key R&D Program of China (2021YFA1100900), CAMS Innovation Fund for Medical Sciences(CIFMS) (2021-I2M-1–040), and CAMS Fundamental Research Funds for Central Research Institutes (3332021093).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data Availability
Not applicable.
Competing interests
The authors declare that there is no conflict of interest regarding the publication of this paper.
Code availability
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yawen Zhang and Yaojin Huang are co-first author of this article.
Contributor Information
Linping Hu, Email: hulinping@ihcams.ac.cn.
Tao Cheng, Email: chengtao@ihcams.ac.cn.
References
- 1.Laurenti E, Gottgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553(7689):418–426. doi: 10.1038/nature25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ema, H., Morita, Y., Yamazaki, S., Matsubara, A., Seita, J., Tadokoro, Y., . . . Nakauchi, H. (2006). Adult mouse hematopoietic stem cells: purification and single-cell assays. Nature Protocols, 1(6), 2979–2987. [DOI] [PubMed]
- 3.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science (New York, NY) 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 4.Till JE, Mc CE. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiation research. 1961;14:213–222. doi: 10.2307/3570892. [DOI] [PubMed] [Google Scholar]
- 5.Civin CI, Strauss LC, Brovall C, Fackler MJ, Schwartz JF, Shaper JH. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J Immunol. 1984;133(1):157–165. [PubMed] [Google Scholar]
- 6.Platzbecker, U., Middeke, J. M., Sockel, K., Herbst, R., Wolf, D., Baldus, C. D., . . . Thiede, C. (2018). Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multicentre, phase 2 trial. The Lancet. Oncology, 19(12), 1668–1679. [DOI] [PubMed]
- 7.Cohen, S., Roy, J., Lachance, S., Delisle, J. S., Marinier, A., Busque, L., . . . Sauvageau, G. (2020). Hematopoietic stem cell transplantation using single UM171-expanded cord blood: a single-arm, phase 1-2 safety and feasibility study. The Lancet. Haematology, 7(2), e134–e145. [DOI] [PubMed]
- 8.Ishii, M., Matsuoka, Y., Sasaki, Y., Nakatsuka, R., Takahashi, M., Nakamoto, T., . . . Sonoda, Y. (2011). Development of a high-resolution purification method for precise functional characterization of primitive human cord blood-derived CD34-negative SCID-repopulating cells. Experimental Hematology, 39(2), 203–213 e201. [DOI] [PubMed]
- 9.Sumide, K., Matsuoka, Y., Kawamura, H., Nakatsuka, R., Fujioka, T., Asano, H., . . . Sonoda, Y. (2018). A revised road map for the commitment of human cord blood CD34-negative hematopoietic stem cells. Nature Communications, 9(1), 2202–2202. [DOI] [PMC free article] [PubMed]
- 10.Matsuoka Y, Takahashi M, Sumide K, Kawamura H, Nakatsuka R, Fujioka T, Sonoda Y. CD34 Antigen and the MPL Receptor Expression Defines a Novel Class of Human Cord Blood-Derived Primitive Hematopoietic Stem Cells. Cell Transplantation. 2017;26(6):1043–1058. doi: 10.3727/096368916X694201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(10):5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao QL, Shah AJ, Thiemann FT, Smogorzewska EM, Crooks GM. A functional comparison of CD34 + CD38- cells in cord blood and bone marrow. Blood. 1995;86(10):3745–3753. doi: 10.1182/blood.V86.10.3745.bloodjournal86103745. [DOI] [PubMed] [Google Scholar]
- 13.Mayani H, Dragowska W, Lansdorp PM. Characterization of functionally distinct subpopulations of CD34+ cord blood cells in serum-free long-term cultures supplemented with hematopoietic cytokines. Blood. 1993;82(9):2664–2672. doi: 10.1182/blood.V82.9.2664.bloodjournal8292664. [DOI] [PubMed] [Google Scholar]
- 14.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(7):2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prashad, S. L., Calvanese, V., Yao, C. Y., Kaiser, J., Wang, Y., Sasidharan, R., . . . Mikkola, H. K. (2015). GPI-80 defines self-renewal ability in hematopoietic stem cells during human development. Cell stem cell, 16(1), 80–87. [DOI] [PMC free article] [PubMed]
- 16.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: A human perspective. Cell Stem Cell. 2012;10(2):120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Chen, Y., Yao, C., Teng, Y., Jiang, R., Huang, X., Liu, S., . . . Guo, B. (2019). Phorbol ester induced ex vivo expansion of rigorously-defined phenotypic but not functional human cord blood hematopoietic stem cells: a cautionary tale demonstrating that phenotype does not always recapitulate stem cell function. Leukemia, 33(12), 2962–2966. [DOI] [PMC free article] [PubMed]
- 18.Dick JE. Stem cells: Self-renewal writ in blood. Nature. 2003;423(6937):231–233. doi: 10.1038/423231a. [DOI] [PubMed] [Google Scholar]
- 19.Yuan, S., Sun, G., Zhang, Y., Dong, F., Cheng, H., & Cheng, T. (2021). Understanding the "SMART" features of hematopoietic stem cells and beyond. Sci China Life Sci, 1–15. [DOI] [PMC free article] [PubMed]
- 20.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karamitros, D., Stoilova, B., Aboukhalil, Z., Hamey, F., Reinisch, A., Samitsch, M., . . . Vyas, P. (2018). Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nature immunology, 19(1), 85-97. [DOI] [PMC free article] [PubMed]
- 22.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature immunology. 2010;11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 23.Belluschi, S., Calderbank, E. F., Ciaurro, V., Pijuan-Sala, B., Santoro, A., Mende, N., . . . Laurenti, E. (2018). Myelo-lymphoid lineage restriction occurs in the human haematopoietic stem cell compartment before lymphoid-primed multipotent progenitors. Nature communications, 9(1), 4100–4100. [DOI] [PMC free article] [PubMed]
- 24.Velten, L., Haas, S. F., Raffel, S., Blaszkiewicz, S., Islam, S., Hennig, B. P., . . . Steinmetz, Lars M. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nature cell biology, 19(4), 271–281. [DOI] [PMC free article] [PubMed]
- 25.van Galen, P., Hovestadt, V., Wadsworth Ii, M. H., Hughes, T. K., Griffin, G. K., Battaglia, S., . . . Bernstein, B. E. (2019). Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell, 176(6), 1265-1281.e1224. [DOI] [PMC free article] [PubMed]
- 26.Xie, X., Liu, M., Zhang, Y., Wang, B., Zhu, C., Wang, C., . . . Cheng, T. (2021). Single-cell transcriptomic landscape of human blood cells. National Science Review, 8(3). [DOI] [PMC free article] [PubMed]
- 27.Dong F, Cheng H, Ema H, Cheng T. Probing the fate of transplanted hematopoietic stem cells: Is the combinational approach "FIT" for purpose? Sci China Life Sci. 2020;63(11):1755–1758. doi: 10.1007/s11427-020-1786-4. [DOI] [PubMed] [Google Scholar]
- 28.Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., . . . Surani, M. A. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods, 6(5), 377–382. [DOI] [PubMed]
- 29.Wu, A. R., Neff, N. F., Kalisky, T., Dalerba, P., Treutlein, B., Rothenberg, M. E., . . . Quake, S. R. (2014). Quantitative assessment of single-cell RNA-sequencing methods. Nature methods, 11(1), 41–46. [DOI] [PMC free article] [PubMed]
- 30.Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., . . . McCarroll, S. A. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 161(5), 1202–1214. [DOI] [PMC free article] [PubMed]
- 31.Klein, A. M., Mazutis, L., Akartuna, I., Tallapragada, N., Veres, A., Li, V., . . . Kirschner, M. W. (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell, 161(5), 1187–1201. [DOI] [PMC free article] [PubMed]
- 32.Zheng, G. X., Terry, J. M., Belgrader, P., Ryvkin, P., Bent, Z. W., Wilson, R., . . . Bielas, J. H. (2017). Massively parallel digital transcriptional profiling of single cells. Nature communications, 8, 14049. [DOI] [PMC free article] [PubMed]
- 33.Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L. Single-cell barcoding and sequencing using droplet microfluidics. Nature protocols. 2017;12(1):44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 34.Han, X., Wang, R., Zhou, Y., Fei, L., Sun, H., Lai, S., . . . Guo, G. (2018). Mapping the Mouse Cell Atlas by Microwell-Seq. Cell, 172(5), 1091–1107.e1017. [DOI] [PubMed]
- 35.Rosenberg, A. B., Roco, C. M., Muscat, R. A., Kuchina, A., Sample, P., Yao, Z., . . . Seelig, G. (2018). Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science (New York, N.Y.), 360(6385), 176–182. [DOI] [PMC free article] [PubMed]
- 36.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramskold, D., Luo, S., Wang, Y. C., Li, R., Deng, Q., Faridani, O. R., . . . Sandberg, R. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology, 30(8), 777–782. [DOI] [PMC free article] [PubMed]
- 38.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 39.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 40.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell reports. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Pan, X., Durrett, R. E., Zhu, H., Tanaka, Y., Li, Y., Zi, X., . . . Weissman, S. M. (2013). Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proceedings of the National Academy of Sciences of the United States of America, 110(2), 594–599. [DOI] [PMC free article] [PubMed]
- 42.Chapman, A. R., He, Z., Lu, S., Yong, J., Tan, L., Tang, F., & Xie, X. S. (2015). Single cell transcriptome amplification with MALBAC. PloS one, 10(3), e0120889. [DOI] [PMC free article] [PubMed]
- 43.Habib, N., Li, Y., Heidenreich, M., Swiech, L., Avraham-Davidi, I., Trombetta, J. J., . . . Regev, A. (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science (New York, N.Y.), 353(6302), 925–928. [DOI] [PMC free article] [PubMed]
- 44.Habib, N., Avraham-Davidi, I., Basu, A., Burks, T., Shekhar, K., Hofree, M., . . . Regev, A. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature methods, 14(10), 955-958. [DOI] [PMC free article] [PubMed]
- 45.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nature biotechnology. 2015;33(3):285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macaulay, I. C., Haerty, W., Kumar, P., Li, Y. I., Hu, T. X., Teng, M. J., . . . Voet, T. (2015). G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nature methods, 12(6), 519–522. [DOI] [PubMed]
- 47.Zachariadis, V., Cheng, H., Andrews, N., & Enge, M. (2020). A Highly Scalable Method for Joint Whole-Genome Sequencing and Gene-Expression Profiling of Single Cells. Mol Cell, 80(3), 541–553 e545. [DOI] [PubMed]
- 48.Hou, Y., Guo, H., Cao, C., Li, X., Hu, B., Zhu, P., . . . Peng, J. (2016). Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell research, 26(3), 304–319. [DOI] [PMC free article] [PubMed]
- 49.Stoeckius, M., Hafemeister, C., Stephenson, W., Houck-Loomis, B., Chattopadhyay, P. K., Swerdlow, H., . . . Smibert, P. (2017). Simultaneous epitope and transcriptome measurement in single cells. Nature methods, 14(9), 865–868. [DOI] [PMC free article] [PubMed]
- 50.Peterson, V. M., Zhang, K. X., Kumar, N., Wong, J., Li, L., Wilson, D. C., . . . Klappenbach, J. A. (2017). Multiplexed quantification of proteins and transcripts in single cells. Nature biotechnology, 35(10), 936–939. [DOI] [PubMed]
- 51.Buenrostro, J. D., Corces, M. R., Lareau, C. A., Wu, B., Schep, A. N., Aryee, M. J., . . . Greenleaf, W. J. (2018). Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell, 173(6), 1535–1548 e1516. [DOI] [PMC free article] [PubMed]
- 52.Granja, J. M., Klemm, S., McGinnis, L. M., Kathiria, A. S., Mezger, A., Corces, M. R., . . . Greenleaf, W. J. (2019). Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nature biotechnology, 37(12), 1458–1465. [DOI] [PMC free article] [PubMed]
- 53.Llorens-Bobadilla, E., Chell, J. M., Le Merre, P., Wu, Y., Zamboni, M., Bergenstråhle, J., . . . Frisén, J. (2020). A latent lineage potential in resident neural stem cells enables spinal cord repair. Science, 370(6512). [DOI] [PubMed]
- 54.Hung, K. L., Yost, K. E., Xie, L., Shi, Q., Helmsauer, K., Luebeck, J., . . . Chang, H. Y. (2021). ecDNA hubs drive cooperative intermolecular oncogene expression. Nature, 600(7890), 731–736. [DOI] [PMC free article] [PubMed]
- 55.Chen S, Lake BB, Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nature Biotechnology. 2019;37(12):1452–1457. doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huo, L., Jiao Li, J., Chen, L., Yu, Z., Hutvagner, G., & Li, J. (2021). Single-cell multi-omics sequencing: application trends, COVID-19, data analysis issues and prospects. Brief Bioinform, 22(6), bbab229. [DOI] [PMC free article] [PubMed]
- 57.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Communications of the ACM. 2017;60(6):84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 58.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533–536. doi: 10.1038/323533a0. [DOI] [Google Scholar]
- 59.Triana, S., Vonficht, D., Jopp-Saile, L., Raffel, S., Lutz, R., Leonce, D., . . . Haas, S. (2021). Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat Immunol, 22(12), 1577–1589. [DOI] [PMC free article] [PubMed]
- 60.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, Medvinsky A. Human haematopoietic stem cell development: From the embryo to the dish. Development (Cambridge, England) 2017;144(13):2323–2337. doi: 10.1242/dev.134866. [DOI] [PubMed] [Google Scholar]
- 62.Robin, C., Bollerot, K., Mendes, S., Haak, E., Crisan, M., Cerisoli, F., . . . Dzierzak, E. (2009). Human placenta is a potent hematopoietic niche containing hematopoietic stem and progenitor cells throughout development. Cell stem cell, 5(4), 385–395. [DOI] [PMC free article] [PubMed]
- 63.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Developmental cell. 2005;8(3):365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 64.Inman KE, Downs KM. The murine allantois: emerging paradigms in development of the mammalian umbilical cord and its relation to the fetus. Genesis (New York, NY 2000) 2007;45(5):237–258. doi: 10.1002/dvg.20281. [DOI] [PubMed] [Google Scholar]
- 65.Li, Z., Lan, Y., He, W., Chen, D., Wang, J., Zhou, F., . . . Liu, B. (2012). Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell, 11(5), 663–675. [DOI] [PubMed]
- 66.Zeng, Y., He, J., Bai, Z., Li, Z., Gong, Y., Liu, C., . . . Liu, B. (2019). Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell research, 29(11), 881–894. [DOI] [PMC free article] [PubMed]
- 67.Crosse, E. I., Gordon-Keylock, S., Rybtsov, S., Binagui-Casas, A., Felchle, H., Nnadi, N. C., . . . Medvinsky, A. (2020). Multi-layered Spatial Transcriptomics Identify Secretory Factors Promoting Human Hematopoietic Stem Cell Development. Cell Stem Cell, 27(5), 822–839 e828. [DOI] [PMC free article] [PubMed]
- 68.Popescu, D. M., Botting, R. A., Stephenson, E., Green, K., Webb, S., Jardine, L., . . . Haniffa, M. (2019). Decoding human fetal liver haematopoiesis. Nature, 574(7778), 365–371. [DOI] [PMC free article] [PubMed]
- 69.Jardine, L., Webb, S., Goh, I., Londoño, M. Q., Reynolds, G., Mather, M., . . . Haniffa, M. (2021). Intrinsic and extrinsic regulation of human fetal bone marrow haematopoiesis and perturbations in Down syndrome. bioRxiv.
- 70.Ranzoni, A. M., Tangherloni, A., Berest, I., Riva, S. G., Myers, B., Strzelecka, P. M., . . . Cvejic, A. (2021). Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Human Developmental Hematopoiesis. Cell stem cell, 28(3), 472–487 e477. [DOI] [PMC free article] [PubMed]
- 71.Bian, Z., Gong, Y., Huang, T., Lee, C. Z. W., Bian, L., Bai, Z., . . . Liu, B. (2020). Deciphering human macrophage development at single-cell resolution. Nature, 582(7813), 571–576. [DOI] [PubMed]
- 72.Zeng, Y., Liu, C., Gong, Y., Bai, Z., Hou, S., He, J., . . . Hu, H. (2019). Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos. Immunity, 51(5), 930–948.e936. [DOI] [PubMed]
- 73.O'Byrne, S., Elliott, N., Rice, S., Buck, G., Fordham, N., Garnett, C., . . . Roy, A. (2019). Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood, 134(13), 1059–1071. [DOI] [PubMed]
- 74.Wang, H., He, J., Xu, C., Chen, X., Yang, H., Shi, S., . . . Zhou, J. (2021). Decoding Human Megakaryocyte Development. Cell stem cell, 28(3), 535–549 e538. [DOI] [PubMed]
- 75.Lee-Six, H., Obro, N. F., Shepherd, M. S., Grossmann, S., Dawson, K., Belmonte, M., . . . Campbell, P. J. (2018). Population dynamics of normal human blood inferred from somatic mutations. Nature, 561(7724), 473–478. [DOI] [PMC free article] [PubMed]
- 76.Zheng, S., Papalexi, E., Butler, A., Stephenson, W., & Satija, R. (2018). Molecular transitions in early progenitors during human cord blood hematopoiesis. Molecular systems biology, 14(3), e8041. [DOI] [PMC free article] [PubMed]
- 77.Pellin, D., Loperfido, M., Baricordi, C., Wolock, S. L., Montepeloso, A., Weinberg, O. K., . . . Biasco, L. (2019). A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nature communications, 10(1), 2395. [DOI] [PMC free article] [PubMed]
- 78.Wu, Z., Gao, S., Zhao, X., Chen, J., Keyvanfar, K., Feng, X., . . . Young, N. S. (2019). Long noncoding RNAs of single hematopoietic stem and progenitor cells in healthy and dysplastic human bone marrow. Haematologica, 104(5), 894–906. [DOI] [PMC free article] [PubMed]
- 79.Knapp, D., Hammond, C. A., Hui, T., van Loenhout, M. T. J., Wang, F., Aghaeepour, N., . . . Eaves, C. J. (2018). Single-cell analysis identifies a CD33(+) subset of human cord blood cells with high regenerative potential. Nature cell biology, 20(6), 710–720. [DOI] [PubMed]
- 80.Lu, Y. C., Sanada, C., Xavier-Ferrucio, J., Wang, L., Zhang, P. X., Grimes, H. L., . . . Krause, D. S. (2018). The Molecular Signature of Megakaryocyte-Erythroid Progenitors Reveals a Role for the Cell Cycle in Fate Specification. Cell reports, 25(8), 2083–2093 e2084. [DOI] [PMC free article] [PubMed]
- 81.Drissen, R., Buza-Vidas, N., Woll, P., Thongjuea, S., Gambardella, A., Giustacchini, A., . . . Nerlov, C. (2016). Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nature immunology, 17(6), 666–676. [DOI] [PMC free article] [PubMed]
- 82.Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein & Cell. 2020;11(1):34–44. doi: 10.1007/s13238-019-0633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Notta, F., Zandi, S., Takayama, N., Dobson, S., Gan, O. I., Wilson, G., . . . Dick, J. E. (2016). Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science (New York, N.Y.), 351(6269), aab2116. [DOI] [PMC free article] [PubMed]
- 84.Tikhonova, A. N., Dolgalev, I., Hu, H., Sivaraj, K. K., Hoxha, E., Cuesta-Dominguez, A., . . . Aifantis, I. (2019). The bone marrow microenvironment at single-cell resolution. Nature, 569(7755), 222–228. [DOI] [PMC free article] [PubMed]
- 85.Hu L, Zhang Y, Miao W, Cheng T. Reactive Oxygen Species and Nrf2: Functional and Transcriptional Regulators of Hematopoiesis. Oxidative Medicine and Cellular Longevity. 2019;2019:5153268. doi: 10.1155/2019/5153268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao J, Lu X, Shao X, Zhu L, Fan X. Uncovering an Organ's Molecular Architecture at Single-Cell Resolution by Spatially Resolved Transcriptomics. Trends in Biotechnology. 2021;39(1):43–58. doi: 10.1016/j.tibtech.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Baccin, C., Al-Sabah, J., Velten, L., Helbling, P. M., Grünschläger, F., Hernández-Malmierca, P., . . . Haas, S. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol, 22(1), 38–48. [DOI] [PMC free article] [PubMed]
- 88.Gao, S., Shi, Q., Zhang, Y., Liang, G., Kang, Z., Huang, B., . . . Liu, F. (2022). Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res, 32(1), 38–53. [DOI] [PMC free article] [PubMed]
- 89.Adelman, E. R., Huang, H. T., Roisman, A., Olsson, A., Colaprico, A., Qin, T., . . . Figueroa, M. E. (2019). Aging Human Hematopoietic Stem Cells Manifest Profound Epigenetic Reprogramming of Enhancers That May Predispose to Leukemia. Cancer discovery, 9(8), 1080–1101. [DOI] [PMC free article] [PubMed]
- 90.Zhao, X., Gao, S., Wu, Z., Kajigaya, S., Feng, X., Liu, Q., . . . Young, N. S. (2017). Single-cell RNA-seq reveals a distinct transcriptome signature of aneuploid hematopoietic cells. Blood, 130(25), 2762–2773. [DOI] [PMC free article] [PubMed]
- 91.Zhu, C., Lian, Y., Wang, C., Wu, P., Li, X., Gao, Y., . . . Zhu, P. (2021). Single-cell transcriptomics dissects hematopoietic cell destruction and T-cell engagement in aplastic anemia. Blood, 138(1), 23–33. [DOI] [PMC free article] [PubMed]
- 92.Iskander, D., Wang, G., Heuston, E. F., Christodoulidou, C., Psaila, B., Ponnusamy, K., . . . Karadimitris, A. (2021). Single-cell profiling of human bone marrow progenitors reveals mechanisms of failing erythropoiesis in Diamond-Blackfan anemia. Sci Transl Med, 13(610), eabf0113. [DOI] [PubMed]
- 93.Brodsky RA. Paroxysmal nocturnal hemoglobinuria. Blood. 2014;124(18):2804–2811. doi: 10.1182/blood-2014-02-522128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riether C, Schurch CM, Ochsenbein AF. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell death and differentiation. 2015;22(2):187–198. doi: 10.1038/cdd.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ng, S. W., Mitchell, A., Kennedy, J. A., Chen, W. C., McLeod, J., Ibrahimova, N., . . . Wang, J. C. (2016). A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature, 540(7633), 433–437. [DOI] [PubMed]
- 96.Velten, L., Story, B. A., Hernandez-Malmierca, P., Raffel, S., Leonce, D. R., Milbank, J., . . . Steinmetz, L. M. (2021). Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nature communications, 12(1), 1366. [DOI] [PMC free article] [PubMed]
- 97.Petti, A. A., Williams, S. R., Miller, C. A., Fiddes, I. T., Srivatsan, S. N., Chen, D. Y., . . . Ley, T. J. (2019). A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun, 10(1), 3660. [DOI] [PMC free article] [PubMed]
- 98.Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: Similarities, differences and clinical prospects in CML and AML. Nature Reviews Cancer. 2020;20(3):158–173. doi: 10.1038/s41568-019-0230-9. [DOI] [PubMed] [Google Scholar]
- 99.Corces, M. R., Buenrostro, J. D., Wu, B., Greenside, P. G., Chan, S. M., Koenig, J. L., . . . Chang, H. Y. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature genetics, 48(10), 1193–1203. [DOI] [PMC free article] [PubMed]
- 100.Xu, J., Nuno, K., Litzenburger, U. M., Qi, Y., Corces, M. R., Majeti, R., & Chang, H. Y. (2019). Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. eLife, 8, e45105. [DOI] [PMC free article] [PubMed]
- 101.Pollyea, D. A., Stevens, B. M., Jones, C. L., Winters, A., Pei, S., Minhajuddin, M., . . . Jordan, C. T. (2018). Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nature medicine, 24(12), 1859–1866. [DOI] [PMC free article] [PubMed]
- 102.Morita, K., Wang, F., Jahn, K., Hu, T., Tanaka, T., Sasaki, Y., . . . Takahashi, K. (2020). Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nature communications, 11(1), 5327. [DOI] [PMC free article] [PubMed]
- 103.Miles, L. A., Bowman, R. L., Merlinsky, T. R., Csete, I. S., Ooi, A. T., Durruthy-Durruthy, R., . . . Levine, R. L. (2020). Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature, 587(7834), 477–482. [DOI] [PMC free article] [PubMed]
- 104.Good, Z., Sarno, J., Jager, A., Samusik, N., Aghaeepour, N., Simonds, E. F., . . . Davis, K. L. (2018). Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med, 24(4), 474–483. [DOI] [PMC free article] [PubMed]
- 105.De Bie, J., Demeyer, S., Alberti-Servera, L., Geerdens, E., Segers, H., Broux, M., . . . Cools, J. (2018). Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia, 32(6), 1358–1369. [DOI] [PMC free article] [PubMed]
- 106.Alberti-Servera, L., Demeyer, S., Govaerts, I., Swings, T., De Bie, J., Gielen, O., . . . Cools, J. (2021). Single-cell DNA amplicon sequencing reveals clonal heterogeneity and evolution in T-cell acute lymphoblastic leukemia. Blood, 137(6), 801–811. [DOI] [PMC free article] [PubMed]
- 107.Zhao, Y., Aldoss, I., Qu, C., Crawford, J. C., Gu, Z., Allen, E. K., . . . Roberts, K. G. (2021). Tumor-intrinsic and -extrinsic determinants of response to blinatumomab in adults with B-ALL. Blood, 137(4), 471–484. [DOI] [PMC free article] [PubMed]
- 108.Rabilloud T, Potier D, Pankaew S, Nozais M, Loosveld M, Payet-Bornet D. Single-cell profiling identifies pre-existing CD19-negative subclones in a B-ALL patient with CD19-negative relapse after CAR-T therapy. Nature communications. 2021;12(1):865. doi: 10.1038/s41467-021-21168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Witkowski, M. T., Dolgalev, I., Evensen, N. A., Ma, C., Chambers, T., Roberts, K. G., . . . Aifantis, I. (2020). Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia. Cancer Cell, 37(6), 867–882 e812. [DOI] [PMC free article] [PubMed]
- 110.Longo DL. Imatinib Changed Everything. New England Journal of Medicine. 2017;376(10):982–983. doi: 10.1056/NEJMe1700833. [DOI] [PubMed] [Google Scholar]
- 111.Warfvinge, R., Geironson, L., Sommarin, M. N. E., Lang, S., Karlsson, C., Roschupkina, T., . . . Karlsson, G. (2017). Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood, 129(17), 2384–2394. [DOI] [PMC free article] [PubMed]
- 112.Giustacchini, A., Thongjuea, S., Barkas, N., Woll, P. S., Povinelli, B. J., Booth, C. A. G., . . . Mead, A. J. (2017). Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nature medicine, 23(6), 692–702. [DOI] [PubMed]
- 113.Kinstrie, R., Horne, G. A., Morrison, H., Irvine, D., Munje, C., Castaneda, E. G., . . . Copland, M. (2020). CD93 is expressed on chronic myeloid leukemia stem cells and identifies a quiescent population which persists after tyrosine kinase inhibitor therapy. Leukemia, 34(6), 1613–1625. [DOI] [PMC free article] [PubMed]
- 114.Ma, L., Pak, M. L., Ou, J., Yu, J., St Louis, P., Shan, Y., . . . Green, M. R. (2019). Prosurvival kinase PIM2 is a therapeutic target for eradication of chronic myeloid leukemia stem cells. Proceedings of the National Academy of Sciences of the United States of America, 116(21), 10482–10487. [DOI] [PMC free article] [PubMed]
- 115.Jeanpierre, S., Arizkane, K., Thongjuea, S., Grockowiak, E., Geistlich, K., Barral, L., . . . Lefort, S. (2021). The quiescent fraction of chronic myeloid leukemic stem cells depends on BMPR1B, Stat3 and BMP4-niche signals to persist in patients in remission. Haematologica, 106(1), 111–122. [DOI] [PMC free article] [PubMed]
- 116.Landau, D. A., Tausch, E., Taylor-Weiner, A. N., Stewart, C., Reiter, J. G., Bahlo, J., . . . Wu, C. J. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature, 526(7574), 525–530. [DOI] [PMC free article] [PubMed]
- 117.Burger, J. A., Landau, D. A., Taylor-Weiner, A., Bozic, I., Zhang, H., Sarosiek, K., . . . Wu, C. J. (2016). Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun, 7, 11589. [DOI] [PMC free article] [PubMed]
- 118.Gaiti, F., Chaligne, R., Gu, H., Brand, R. M., Kothen-Hill, S., Schulman, R. C., . . . Landau, D. A. (2019). Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature, 569(7757), 576–580. [DOI] [PMC free article] [PubMed]
- 119.Wang, L., Fan, J., Francis, J. M., Georghiou, G., Hergert, S., Li, S., . . . Wu, C. J. (2017). Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res, 27(8), 1300–1311. [DOI] [PMC free article] [PubMed]
- 120.Bachireddy, P., Ennis, C., Nguyen, V. N., Gohil, S. H., Clement, K., Shukla, S. A., . . . Wu, C. J. (2020). Distinct evolutionary paths in chronic lymphocytic leukemia during resistance to the graft-versus-leukemia effect. Science translational medicine, 12(561). [DOI] [PMC free article] [PubMed]
- 121.Shastri A, Will B, Steidl U, Verma A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12):1586–1594. doi: 10.1182/blood-2016-10-696062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen, J., Kao, Y. R., Sun, D., Todorova, T. I., Reynolds, D., Narayanagari, S. R., . . . Steidl, U. (2019). Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med, 25(1), 103–110. [DOI] [PMC free article] [PubMed]
- 123.Stosch, J. M., Heumuller, A., Niemoller, C., Bleul, S., Rothenberg-Thurley, M., Riba, J., . . . Becker, H. (2018). Gene mutations and clonal architecture in myelodysplastic syndromes and changes upon progression to acute myeloid leukaemia and under treatment. Br J Haematol, 182(6), 830–842. [DOI] [PubMed]
- 124.Tong, J., Sun, T., Ma, S., Zhao, Y., Ju, M., Gao, Y., . . . Shi, L. (2021). Hematopoietic stem cell heterogeneity is linked to the initiation and therapeutic response of myeloproliferative neoplasms. Cell stem cell, 28(4), 780. [DOI] [PMC free article] [PubMed]
- 125.Psaila, B., Wang, G., Rodriguez-Meira, A., Li, R., Heuston, E. F., Murphy, L., . . . Mead, A. J. (2020). Single-Cell Analyses Reveal Megakaryocyte-Biased Hematopoiesis in Myelofibrosis and Identify Mutant Clone-Specific Targets. Mol Cell, 78(3), 477–492 e478. [DOI] [PMC free article] [PubMed]
- 126.Van Egeren, D., Escabi, J., Nguyen, M., Liu, S., Reilly, C. R., Patel, S., . . . Hormoz, S. (2021). Reconstructing the Lineage Histories and Differentiation Trajectories of Individual Cancer Cells in Myeloproliferative Neoplasms. Cell stem cell, 28(3), 514–523 e519. [DOI] [PMC free article] [PubMed]
- 127.Rajkumar SV. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. American Journal of Hematology. 2016;91(7):719–734. doi: 10.1002/ajh.24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ledergor, G., Weiner, A., Zada, M., Wang, S. Y., Cohen, Y. C., Gatt, M. E., . . . Amit, I. (2018). Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med, 24(12), 1867–1876. [DOI] [PubMed]
- 129.Mitra, A. K., Mukherjee, U. K., Harding, T., Jang, J. S., Stessman, H., Li, Y., . . . Van Ness, B. (2016). Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia, 30(5), 1094–1102. [DOI] [PubMed]
- 130.Liu, R., Gao, Q., Foltz, S. M., Fowles, J. S., Yao, L., Wang, J. T., . . . Ding, L. (2021). Co-evolution of tumor and immune cells during progression of multiple myeloma. Nature communications, 12(1), 2559. [DOI] [PMC free article] [PubMed]
- 131.Foltz, S. M., Gao, Q., Yoon, C. J., Sun, H., Yao, L., Li, Y., . . . Vij, R. (2020). Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nature communications, 11(1), 2666. [DOI] [PMC free article] [PubMed]
- 132.Galinato M, Shimoda K, Aguiar A, Hennig F, Boffelli D, McVoy MA, Hertel L. Single-Cell Transcriptome Analysis of CD34(+) Stem Cell-Derived Myeloid Cells Infected With Human Cytomegalovirus. Frontiers in Microbiology. 2019;10:577. doi: 10.3389/fmicb.2019.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shnayder, M., Nachshon, A., Rozman, B., Bernshtein, B., Lavi, M., Fein, N., . . . Schwartz, M. (2020). Single cell analysis reveals human cytomegalovirus drives latently infected cells towards an anergic-like monocyte state. eLife, 9, e52168. [DOI] [PMC free article] [PubMed]
- 134.Wang, X., Wen, Y., Xie, X., Liu, Y., Tan, X., Cai, Q., . . . Zhang, Z. (2021). Dysregulated hematopoiesis in bone marrow marks severe COVID-19. Cell Discov, 7(1), 60. [DOI] [PMC free article] [PubMed]
- 135.Xie, X., Cheng, X., Wang, G., Zhang, B., Liu, M., Chen, L., . . . Cheng, T. (2021). Single-cell transcriptomes of peripheral blood cells indicate and elucidate severity of COVID-19. Sci China Life Sci, 64(10), 1634–1644. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.