Abstract
The COVID-19 pandemic has brought to light the need for fast and sensitive detection methods to prevent the spread of pathogens. The scientific community is making a great effort to design new molecular detection methods suitable for fast point-of-care applications. In this regard, a variety of approaches have been developed or optimized, including isothermal amplification of viral nucleic acids, CRISPR-mediated target recognition, and read-out systems based on nanomaterials.
Herein, we present CASCADE (CRISPR/CAS-based Colorimetric nucleic Acid DEtection), a sensing system for fast and specific naked-eye detection of SARS-CoV-2 RNA. In this approach, viral RNA is recognized by the LwaCas13a CRISPR protein, which activates its collateral RNase activity. Upon target recognition, Cas13a cleaves ssRNA oligonucleotides conjugated to gold nanoparticles (AuNPs), thus inducing their colloidal aggregation, which can be easily visualized. After an exhaustive optimization of functionalized AuNPs, CASCADE can detect picomolar concentrations of SARS-CoV-2 RNA. This sensitivity is further increased to low femtomolar (3 fM) and even attomolar (40 aM) ranges when CASCADE is coupled to RPA or NASBA isothermal nucleic acid amplification, respectively. We finally demonstrate that CASCADE succeeds in detecting SARS-CoV-2 in clinical samples from nasopharyngeal swabs.
In conclusion, CASCADE is a fast and versatile RNA biosensor that can be coupled to different isothermal nucleic acid amplification methods for naked-eye diagnosis of infectious diseases.
Keywords: CRISPR/Cas13, Point-of-Care, SARS-CoV-2, Diagnostics, Gold nanoparticles, Naked-eye
Abbreviations: CASCADE, CRISPR/CAS-based colorimetric nucleic acid detection; AuNPs, gold nanoparticles; ssRNA, single-stranded RNA; PoC, point-of-care; RNP, ribonucleoprotein; RPA, Recombinase Polymerase Amplification; NASBA, Nucleic Acid Sequence Based Amplification; SSB, single-stranded DNA binding protein; dsDNA, double-stranded DNA; Ct, cycle threshold; PAM, protospacer adjacent motif
Graphical abstract
1. Introduction
In March 2020, WHO declared COVID-19 a global pandemic. Twenty months later (December 20th, 2021), 275,265,910 confirmed cases and 5,373,186 COVID-19 related deaths had been reported. This severe acute respiratory syndrome is caused by the β-coronavirus SARS-CoV-2, a single-stranded RNA (ssRNA) virus whose genome includes at least 5 genes: ORF1ab, N, M, E and S [1]. Since SARS-CoV-2 is easily transmitted [2], general population screening and rapid isolation of infected individuals are crucial to control its spread. Consequently, there is an urgent need to develop rapid, sensitive, specific, and affordable PoC devices to cover the global demand required to tackle the current and future pandemics.
RT-qPCR is the gold standard method for SARS-CoV-2 detection. It is a specific, sensitive, and reliable technique for viral nucleic acid sensing. However, it requires specialized technicians and expensive machinery and reagents, and is thus unsuitable for fast PoC detection. The most widespread SARS-CoV-2 PoC tests nowadays are serological and antigen lateral flow sensors. Both are fast (15 min) and easy to use, but they have some limitations. Serological tests detect the anti-SARS-CoV-2 antibodies, produced 3–7 days after infection onset, making early detection challenging. Antigen tests are reliable devices to identify symptomatic COVID-19 patients, but their sensitivity drops for low viral load samples compared to RT-qPCR [3]. Due to their extreme importance and high demand, a variety of PoC COVID-19 sensors are currently under development [4]. Most of these are based on viral RNA detection using isothermal amplification techniques such as LAMP, RPA, RCA, EXPAR or NASBA, which simplify the procedure by eliminating the need for thermal cycling. Following this step, amplification products must be specifically detected. Some of the most successful detection methods are based on the CRISPR/Cas system, which can be easily coupled to isothermal amplification strategies [5]. The Class 2 CRISPR/Cas system consists of a Cas nuclease forming a ribonucleoprotein (RNP) complex with a guide RNA (crRNA) which is complementary to a target nucleic acid (DNA/RNA). crRNA-target interaction through base pair complementarity activates the nuclease activity of the Cas protein. Thus, cleavage specificity can be easily modulated through the crRNA sequence. Two types of CRISPR/Cas nucleases, Cas12 and Cas13, possess an additional multiple-turnover trans-cleavage activity that is specifically activated by target recognition and has been successfully exploited in SARS-CoV-2 nucleic acid PoC detectors [6]. Cas12 recognizes dsDNA, and is then able to cleave unspecific ssDNA, while Cas13 recognizes and cleaves ssRNA. Since this trans-cleavage activity is multiple-turnover, the signal resulting from target recognition can be amplified using a large amount of unspecific ssDNA or ssRNA oligonucleotides that act as reporters which produce a detectable signal once cleaved by the CRISPR collateral activity. The ability to amplify the signal is the basis for the highly sensitive CRISPR-mediated nucleic acid detection. The Sherlock™ CRISPR SARS-CoV-2 kit, which combines isothermal amplification, Cas13 recognition, and lateral flow read-out [7], is the first FDA-authorized CRISPR-based diagnostic test for viral RNA detection. This illustrates how CRISPR/Cas technology is becoming increasingly relevant for SARS-CoV-2 diagnosis [6]. In this context, the development of CRISPR-based PoC devices with naked-eye read-out is of great interest. Gold nanoparticles (AuNPs) are perfectly suited for this purpose due to their high color contrast and the dependency of their colorimetric properties on their aggregation state [8]. A target-triggered change in AuNPs' colloidal aggregation can be easily detected by the naked eye. To this end, our group has developed an RNA sensor using disperse AuNPs functionalized with oligonucleotides that aggregate in the presence of a target nucleic acid [9]. The combination of CRISPR-based systems with AuNPs to develop naked-eye sensors for SARS-CoV-2 constitutes a promising field. However, to date, it has been explored in just five scientific works [[10], [11], [12], [13], [14]], of which only two are related to SARS-CoV-2 detection [13,14]. In most of these projects [[10], [11], [12]] Cas12a's trans activity cleaves a ssDNA linker, which avoids the aggregation of AuNPs functionalized with a ssDNA complementary to the linker (Fig. 1 A). In this approach, the negative signal (the absence of target) drives the color change, and therefore carefully selected negative controls are needed to avoid false positives. This problem was solved by Cao et al., who developed a detection method based on the cleavage of a DNA/RNA hybrid hairpin transducer, which releases a RNA crosslinker that promotes the aggregation of AuNPs functionalized with oligonucleotides complementary to the linker [13] (Fig. 1B). In this design the presence of the target molecule induces the color change of the AuNP solution. Zhang et al. used a more direct approach where the target activation of Cas12a results in the cleavage of ssDNA oligonucleotides present on the AuNPs surface, which promote their destabilization and their color shift from red to purple [14] (Fig. 1C). However, in these methods, centrifugation is needed to accelerate detection and increase sensitivity (Fig. 1). Moreover, Cao et al.’s system also requires the addition of high salt concentrations to destabilize the AuNPs (Fig. 1B) [13]. New approaches are thus needed to improve the coupling of CRISPR detection and naked-eye readouts based on AuNP aggregation. So far, the combination of Cas13 and AuNPs for the naked-eye detection of SARS-CoV-2 RNA remains an unexplored field. Cas13a has been successfully used to detect viral nucleic acids using fluorimetric and lateral flow read-outs [5] and it is a good candidate for naked-eye sensing. While Cas12a target recognition relies on the presence of a protospacer adjacent motif (PAM), Cas13 lacks this kind of sequence requirements, which makes it more versatile. Finally, the urgent global need for fast sensing methods to fight viral outbreaks encourages the development of as many new detection systems as possible.
Fig. 1.
Schematic representation of the main AuNPs-CRISPR based nucleic acid detection methods. (A) Cas12a trans-activity cleaves an ssDNA linker, reverting AuNP aggregation. (B) Cas12a trans-activity cleaves a DNA/RNA hybrid hairpin transducer, promoting the aggregation of functionalized AuNPs. (C) Cas12a trans-activity promotes AuNP aggregation by cleaving ssDNA oligonucleotides present on the AuNP's surface.
In this work, we present CASCADE (CRISPR/CAS-based Colorimetric nucleic Acid DEtection), a CRISPR/Cas13-based sensor for SARS-CoV-2 RNA detection using AuNPs. A thorough AuNP optimization provides a fast read-out in 15–30 min. Furthermore, its combination with isothermal RNA amplification allows the detection of SARS-CoV-2 RNA in clinical samples.
2. Material and methods
2.1. AuNP synthesis
Gold nanoparticles were synthesized based on the Turkevich method [[15], [16], [17]]. Briefly, an aqueous sodium citrate solution was added to a HAuCl4 solution in water under reflux. The reaction was stirred for 15 min at a constant temperature. The mixture was cooled down at room temperature and filtered using a 0.3 μm frit. Gold nanoparticle concentration was determined by UV/VIS spectroscopy using Beer's law [16]. By adjusting the reaction conditions, AuNPs of 3 different sizes were prepared: 12–15 nm/22–25 nm/30–34 nm [16]. AuNP sizes were measured by TEM.
2.2. Oligonucleotide synthesis
The RNA2, RNA3, RNA3-medium, RNA3-short, RNA3-FAM, RNA3-7 nt oligonucleotides (Table S1) were prepared using a H6/H8 DNA/RNA Synthesizer (K&A). After solid-phase synthesis, the solid support was transferred to a screw-cap glass vial and incubated at room temperature for 16 h with 2 mL of ammonia/ethanol solution (3:1). Then, the samples were dried and purified using standard DNA/RNA purification columns (LCG Biosearch Micropure purification columns). Oligonucleotide purity was confirmed by polyacrylamide gel electrophoresis. RNA1 was purchased from Integrated DNA Technologies (IDT).
2.3. AuNP functionalization
1 mL of 23.4 nM gold nanoparticles was incubated with modified oligonucleotides (2.2 pmol μL−1) and Tris(2-carboxyethyl) phosphine (TCEP, 100 equivalents per oligo) for 30 min. Then, 12 μL of a 5 M NaCl solution were added every 20 min (5 times) until 0.3 M salt concentration was reached in the solution. The mixture was incubated for 16 h in the dark at room temperature. The gold nanoparticles were then centrifuged at 13200 rpm at 4 °C for 35 min, the supernatant was removed, and the pellet was resuspended in water. This process was repeated twice to remove the unbound oligonucleotides. To obtain the 0.7 and 2.3 pmol μL−1 AuNP loadings we employed 1.1 pmol μL−1 and 4.5 pmol μL−1 oligonucleotide initial concentration, respectively.
To quantify the attached oligonucleotide, the supernatants were collected, concentrated, and quantified by spectroscopy at 260 nm.
2.4. Cas13a purification
LwaCas13a was purified following Gootenberg et al. purification protocol with some variations [18]. LwaCas13a bacterial expression plasmid (p2CT-His-MBP-Lwa_Cas13a_WT, Addgene) was transformed into Escherichia coli (E. coli) BL21 competent cells. A 25 mL starter culture was grown overnight in Luria-Bertani (LB) broth. The starter culture was inoculated in 3L of LB media with Ampicillin and grown at 37 °C, 200 rpm until OD600 = 0.3, then changed to a 16 °C incubator and MBP-LwaCas13a expression was induced with 0.5 mM IPTG when OD600 = 0.5–0.6. Following overnight growth, cells were centrifuged and pellets were harvested, sonicated and lysed in a buffer containing 50 mM Tris-Cl pH 7.5, 500 mM NaCl, 5% Glycerol, 5 mM β-mercaptoethanol, 0.5 mM PMSF, lysozyme and EDTA-free protease inhibitor. Then the lysate was centrifuged at 10000 rpm for 1 h at 4 °C. Soluble His6-MBP-TEV-Cas13a was purified through affinity chromatography using a HisTrap HP column, (GE Healthcare). Fractions containing the protein were pooled and concentrated by centrifugation. TEV protease was added to the protein and dialyzed overnight at 4 °C in dialysis buffer containing 20 mM Tris-Cl pH 7.0, 500 mM NaCl, 5% Glycerol and 1 mM DTT. Following overnight dialysis, the protein was loaded on a size exclusion column (Superdex 200, GE Healthcare). Fractions containing the protein were pooled and the buffer solution containing the protein was exchanged using a Nap-10 column (GE Healthcare) and a buffer containing 20 mM Tris-Cl pH 7.0, 500 mM NaCl and 5% Glycerol to eliminate the DTT. Protein was concentrated and stored at −80 °C. All protein purification steps were performed at 4 °C.
2.5. crRNA design
crRNAs R2, S2 and NASBA for LwaCas13a were designed to target regions with minimal secondary structure. UNAFold (IDT) and RNAfold (Vienna) software was used for predicting the secondary structure of crRNAs and their targets. crRNAs R1 and S1 were described by Zhang et al. [19]. All crRNAs were purchased from IDT.
2.6. Target preparation
Cas13 RNA Orf1ab and S targets were obtained by RT-PCR from total RNA extracted from SARS-CoV-2 infected cells (donated by Isabel Sola and Sonia Zúñiga, CNB) using the SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Thermo Fisher). Primers used here containing a 5′ T7 promoter sequence were described by Zhang et al. [19] and ordered to IDT. RNA template NASBA was obtained by PCR from pSL_WH_SARS2_S plasmid (donated by Isabel Sola and Sonia Zúñiga, CNB). Later, a T7 RNA transcription was performed using the MEGASCRIPT Kit (Thermo Fisher) for 16 h at 37 °C following the manufacturer's instructions.
After T7 RNA transcription, the samples were treated with TurboDNase from MEGASCRIPT Kit for 25 min at 37 °C. Next, a phenol-chloroform RNA extraction was performed following the kit indications with some variations. 115 μL RNase-free water and 15 μL Ammonium Acetate stop solution were added to the samples and mixed well. Next, samples were extracted with phenol and then with chloroform. Then, RNA was precipitated by adding ethanol. Samples were cooled for 2 h at −20 °C. Then, samples were centrifuged for 20 min at 4 °C and at maximum speed. The supernatant was decanted until ethanol was completely removed. The RNAs obtained were diluted in RNase-free water.
2.7. Isothermal amplification
RPA was performed using the TwistAmp® Basic Kit (TwistDx) following manufacturer's instructions. Since the template was RNA, ProtoScript® II Reverse Transcriptase (New England Biolabs) was added to the reaction. The reaction was performed in a total volume of 10 μL. Samples were incubated at 42 °C for 50 min. Next, RNA transcription was performed for 1 h at 37 °C and samples were kept at 4 °C or −20 °C for short- and long-term storage, respectively. Only 10% of the volume of an RPA reaction is required for a CASCADE assay.
Nucleic Acid Sequence Based Amplification (NASBA) was carried out using NASBA Lyophilized Kit (Asmbio) following manufacturer's instructions. NASBA primer FW and RV (Table S1), designed by Wu et al. [20], were used. Samples were incubated at 65 °C for 2 min first and then at 41 °C for 90 min. Amplified samples were stored at 4 °C or −20 °C for short- and long-term usage, respectively. 25% of the total volume of a NASBA reaction is used for a CASCADE assay.
2.8. CASCADE sensing assay
For ribonucleoprotein (RNP) formation, LwaCas13a was incubated with its crRNA in a 1:1 M ratio for 30 min at 37 °C. Following incubation, 20 units of Murine RNase inhibitor (New England Biolabs) per CASCADE reaction were added.
Cas13a RNP (final concentration of 50 nM) was mixed with 50 μL of RNA-functionalized AuNPs (12–48 nM) in cleavage buffer containing 60 mM Tris pH 7.5, 60 mM NaCl, 6 mM MgCl2. Next, the corresponding target was added, and samples were incubated at 37 °C. The total volume of the reaction was 100 μl. Absorbance measurements and pictures were recorded at 15, 30 and 60 min time points. 80 μL from each sample were carefully transferred to a 96-well UV/Vis plate at the indicated time points. UV/Vis absorbance spectrums were measured in a plate reader (Plate reader Synergy H4, Biotek) and then the samples were returned to their tubes to continue the incubation at 37 °C.
2.9. RNA extraction from clinical samples
RNA from inactivated clinical remnant nasopharyngeal swab samples obtained from patients suspected of SARS-CoV-2 infection and donated by Hospital Ramón y Cajal (Madrid) was extracted using the QIAamp Viral RNA kit (Qiagen). 300 μL of each sample, in transport media (containing guanidine isothiocyanate as viral inactivating agent), were processed in each case. After cellular and viral lysis, total RNA was purified by sequential centrifugation through silica-based membranes using ethanol-containing washing buffers. Total RNA was eluted in RNase-free water. Its concentration was measured using a Nanodrop and it was stored at – 80 °C. Samples were anonymized and results were not used for patient care.
3. Results and discussion
3.1. CASCADE design
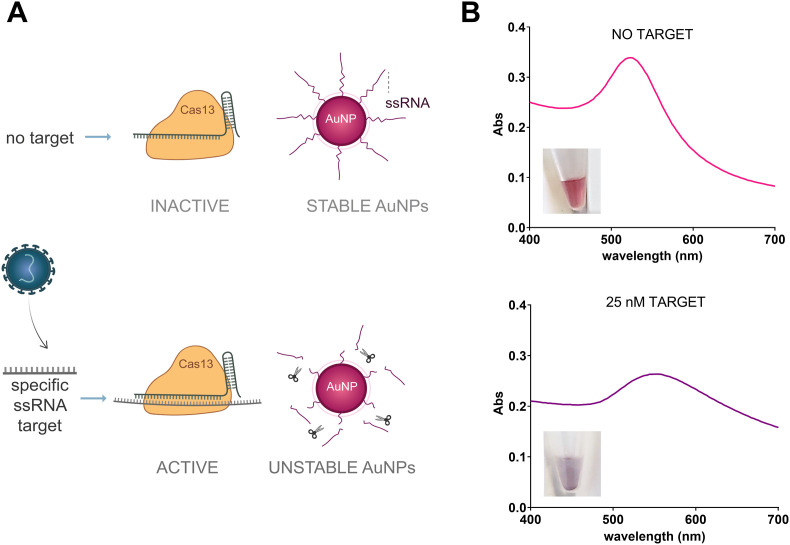
The optical properties of AuNPs can be modulated through their interaction with different molecules. These properties have been extensively investigated for the development of multiple sensors [9,[21], [22], [23], [24]]. Some of these approaches exploit the destabilization of AuNPs by a ligand exchange process, where the incoming ligand induces the aggregation of the AuNPs. Our strategy instead exploits the enzymatic degradation of the stabilizing ligands. We used AuNPs stabilized with ssRNAs, which can be cleaved by Cas13a's trans-activity nuclease upon specific target recognition. Cas13a's activation results in the shortening of these AuNP-bound ssRNAs. Since short oligonucleotides cannot cover the entire AuNP's surface, they cannot exert their stabilizing influence. This may result in target-triggered AuNP aggregation [25] (Fig. 2 A). To test this hypothesis, AuNPs were coated with different ssRNA oligonucleotides. Two target RNAs from SARS-CoV-2 genome were selected [19]: Orf1ab_ssRNAtarget (Table S1), corresponding to the region spanning nucleotides 6432–6582 (Orf1ab gene) of the SARS-CoV-2 genome; and S_ssRNAtarget (Table S1) comprising nucleotides 22271–22382 (S gene) of the SARS-CoV-2 genome. These RNA targets were amplified and in vitro transcribed from the total RNA extracted from SARS-CoV-2 infected cells. Specific crRNAs (R1, R2, S1 and S2) (Table S1) targeting these regions were used to assemble the following RNPs: RNP R1, R2, S1 and S2. The recognition of these target RNAs activated Cas13a's trans-cleavage. In the absence of target RNA, AuNPs remain stable in the aqueous media, with their characteristic reddish color and an absorption band centered at 523 nm. In contrast, when the target RNA is present, Cas13a's trans-cleavage activity promotes ssRNA degradation and subsequent AuNP aggregation, resulting in a substantial decrease of the absorbance together with a spectrum displacement to higher wavelengths, which can be seen by the naked eye (Fig. 2B). Crucially, this naked-eye detectable color change does not require additional steps such as centrifugation [13,14] or salt addition [13] (Table S2). The AuNP destabilization induced by Cas13a's target recognition is stronger than that of previous published works based on Cas12a and ssDNA-AuNPs, which constitutes a major advantage.
Fig. 2.
CASCADE description. (A) Schematic representation of the CASCADE sensing system. In the absence of SARS-CoV-2 RNA, Cas13a remains inactive, and the nanoparticles are stable. However, in the presence of SARS-CoV-2 RNA, the recognition of the target RNA complementary to the crRNA activates Cas13 nuclease activity, cleaving the RNAs stabilizing the gold nanoparticles. (B) Cas13 activity triggers the aggregation of gold nanoparticles and, consequently, a dramatic change in the absorption spectrum, visible to the naked eye. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We performed several control experiments to further confirm that the mechanism underlying AuNP aggregation is the degradation of the ssRNAs bound to the nanoparticle surface by Cas13a's nuclease activity. First, to evaluate whether these ssRNAs are actually cut by Cas13a, we used AuNPs functionalized with ssRNA oligonucleotides labelled with a fluorophore (FAM, Carboxyfluorescein). We observed that, in the presence of a specific target, Cas13a was able to release the FAM from the AuNPs, as shown by the increase of the fluorescence intensity in the supernatant. (Fig. S1A). This indicates that the specific activation of Cas13a results in the cleavage of the ssRNAs coating the AuNPs. Secondly, Cas13a's trans-activity presumably shortens the length of the ssRNAs attached to the nanoparticles but does not eliminate them completely from their surface. To demonstrate that the remaining short oligonucleotides are unable to stabilize the nanoparticles, we attempted to functionalize AuNPs with short ssRNAs. As previously shown by Storhoff et al. [25], we observed that oligonucleotides containing only 7 bases could not yield stable AuNPs (Fig. S1B). Finally, we investigated whether other RNA-degrading enzymes might exert a similar destabilizing effect on our ssRNA-AuNPs. The addition of RNase A induced fast AuNP aggregation (Fig. S1C), which further supports our hypothesis that ssRNA cleavage is the driving force behind AuNP destabilization.
3.2. Optimization of CASCADE assay conditions
ssRNA-functionalized AuNPs play an essential role in CASCADE, as they act as a substrate for Cas13a's trans-cleavage activity and provide its colorimetric read-out. Thus, we decided to assess the main critical parameters that might control this process. We particularly focused on the size and concentration of AuNPs as well as the sequence, length and loading of ssRNA. First, we tested three different RNA sequences for AuNP functionalization: RNA1, RNA2 and RNA3 (Table S1). All three resulted in AuNP aggregation in the presence of Cas13a in complex with crRNA_R1 (RNP R1) and a specific target (25 nM Orf1ab_ssRNAtarget), indicating that they were suitable for RNA sensing. However, in the case of AuNPs coated with RNA1 and RNA2 we observed slight aggregation even in the absence of the target (Fig. S2). Thus, RNA3 was selected for further experiments. We evaluated three different AuNP diameters (12, 22 and 34 nm) (Fig. 3 A) loaded with 1.5 pmol μl−1 RNA3 for the detection of Orf1ab. All samples presented similar AuNP aggregation in the presence of the target after 30 min of incubation (Fig. 3A). However, 12 nm AuNPs responded faster, which enabled target detection after 15 min of incubation and presented a more intense color change for naked-eye detection (Fig. S3). Since PoC detection must prioritize rapidity and specificity, we selected 12 nm AuNPs to develop our sensor. Furthermore, the length of the ssRNA oligonucleotide coating the AuNPs has a significant effect on their stability and aggregation dynamics. Specifically, long oligonucleotides increase the colloidal stability of the nanoparticles, while short ones cannot stabilize the AuNPs enough (Fig. S1B). Additionally, ssRNA length can influence Cas13a's collateral activity: if too short, the oligonucleotides might be inaccessible to the nuclease. Thus, we assessed three lengths of RNA3: long (33 nt), medium (23 nt) and short (13 nt). AuNPs coated with the 33 nt RNA3 yielded the best results for the detection of Orf1ab at the time points tested, showing greater absorbance reduction (Fig. 3B and Fig. S4). These AuNPs were therefore chosen for subsequent experiments. The amount of oligonucleotide coating the nanoparticle (AuNP loading) was also optimized. This parameter is crucial for AuNP stability and can also affect the interaction between the Cas13a and the bound RNAs. The three loadings tested (0.7, 1.5 and 2.3 pmol μL−1) were suitable for detecting the target sequence (Fig. 3C, Fig. S5). However, AuNPs loaded with 0.7 pmol μL−1 ssRNA did not contain enough oligonucleotide to stabilize the AuNPs (Fig. S5B). Even in the absence of target, they aggregated and showed a color shift, making them unsuitable for reliable detection. On the other hand, the 2.3 pmol μL−1 loading yielded the most stable nanoparticles, which required longer destabilization times. Therefore, the 1.5 pmol μL−1 loading was selected since it provided stable nanostructures and the fastest read-out. Finally, we evaluated the effect of AuNP concentration on detection. Three different AuNP concentrations were assessed: 11.7, 23.4 and 46.8 nM. At 11.7 nM the AuNP suspension was so diluted that changes in absorbance were hardly noticeable. At 46.8 nM, no significant differences could be observed between the samples with and without target. However, at 23.4 nM the decrease in absorbance was readily apparent at the three time points evaluated (Fig. 3D, Fig. S6). Furthermore, at this concentration, color changes were easily detectable by the naked eye (Fig. S6B).
Fig. 3.
Optimization of CASCADE detection conditions. (A–D) Normalized absorbance and naked-eye detectable color change of RNA-functionalized AuNPs and Cas13 RNP (50 nM) mixtures at 523 nm in the presence or absence of Orf1ab target after 30 min reaction. Several parameters were optimized: (A) AuNPs of different sizes modified with 1.5 pmol μL−1 of RNA3. (B) AuNPs functionalized with 1.5 pmol μL−1 of RNA3 (33 nt), RNA3-medium (23 nt) and RNA3-short (13 nt). (C) AuNPs functionalized with different concentrations of RNA3. (D) Different concentrations of 12 nm AuNPs functionalized with 1.5 pmol μL−1 of RNA3. Values represent the mean of replicate samples (n = 3) ± SD ∗P < 0 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant. (E) CASCADE's kinetic behavior with and without Orf1ab target (25 nM) from 0 to 30 min. RNP R1 was used for Orf1ab detection. UV–vis absorption spectra were registered at a 2 min interval. Error bars indicate the SD of replicate samples (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
After the optimization of the system, we evaluated the temporal response of our sensor to determine the time-span during which the first changes in absorbance occurred. AuNP aggregation in the presence of 25 nM of the Orf1ab target was measured every 2 min in a plate reader at 37 °C (Fig. 3E). Remarkably, in the samples containing the target sequence, the reduction in absorbance and the blue-shift were already detectable after 2-min incubation. These differences became increasingly evident throughout the rest of the experiment. In the absence of the target sequence, no changes were observed at any time. This indicates that CASCADE recognizes the target sequence and provides fast spectroscopic read-out in less than 10 min, and naked-eye detection in 15 min, making it suitable for fast PoC detection.
Specificity is an essential feature for diagnostic tools to be reliable. To determine the specificity of CASCADE we confronted RNP R1 with the S target and RNP S1 with the Orf1ab target (Fig. S7). In the presence of the unspecific target, there was no detection, demonstrating that the AuNP colloidal destabilization is only produced by specific target recognition. Furthermore, there was specific detection even when both target sequences were mixed, meaning that the presence of unspecific sequences did not hinder target recognition. This is a necessary feature for sensors, since they must recognize specific sequences in complex samples.
Finally, we tested the long-term colloidal stability of CASCADE's main components. We stored ssRNA3-AuNPs for 5 months at 4 °C and used them for CASCADE detection. They showed an adequate stability when the target was not present, and a detection performance comparable to that of freshly prepared AuNPs (Figs. S8A,B, C). Their colloidal stability was further tested by comparing the plasmon spectra of stored and freshly made ssRNA3-AuNPs (Fig. S8D). Their plasmon spectra were similar, meaning that they were the same size. In conclusion, our RNA-coated AuNPs can be safely kept at 4 °C for months. In addition, it has been previously observed that Cas13a-crRNA complexes can be lyophilized for portable deployment [26]. To further investigate Cas13 RNP stability, it was kept for 22 days at 4 °C. Figs. S8E and F show that stored RNP did not lose its target recognition capability. (Figs. S8E and F). The high stability of the CASCADE's components contributes to its portability.
3.3. Evaluation of CASCADE's sensitivity
Next, we studied CASCADE's sensitivity, another essential feature for PoC diagnosis. Guide RNA design crucially influences Cas13a's activity, and therefore its overall sensitivity. To assess the role of the crRNA sequences, two guides were used to recognize a specific region of the Orf1ab target RNA (R1 and R2) and another two to recognize the S target RNA (S1 and S2) (Table S1). R1 and S1 were described by Zhang et al. [19], while R2 and S2 were carefully designed using RNA secondary structure prediction software. R1, R2 and S1 yielded comparable results, enabling the detection of 0.5 nM of their respective target RNAs at 15–30 min (Fig. 4 , Figs. S9–S11), determined by the statistical significance of the absorbance measurements in comparison with the negative control. Moreover, R1 and S1 allowed the detection of 0.1 nM of their target after a 1-h incubation (Fig. S9, Fig. S11). The detection with S2, however, was notably less sensitive (Fig. S12). Although S1 and S2 target sites are not very distant, their behavior is markedly different. This highlights the importance of crRNA design. crRNAs should be carefully selected so that they target an accessible RNA region with minimal secondary structure. Furthermore, several guides should be tested to choose the one with optimal performance.
Fig. 4.
Evaluation of CASCADE's sensitivity for Orf1ab and S detection using different crRNAs. (A–D) Normalized sample absorbance at 523 nm at 30 min. 12 nm AuNPs functionalized with 1.5 pmol μL−1 of RNA3 were used. Orf1ab detection with 50 nM of (A) RNP R1 and (B) RNP R2. S detection with 50 nM of (C) RNP S1 and (D) RNP S2. Sample absorbance was normalized against the 0 nM target condition. Bar labels indicate target concentration. Values represent the mean of replicate samples (n = 3) ± SD ∗P < 0 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant.
The combination of two spectral changes, the blue-shift and the decrease in intensity due to nanoparticle aggregation (Fig. 3B), induces a strong reduction of the absorbance in presence of the target. This facilitates the detection of low-concentration samples without additional procedures such as centrifugation and/or salt addition (Table S2) [13],.
The sensitivity of CASCADE in the pM range (100 pM) is comparable to other CRISPR-based nucleic acid colorimetric sensors [10,12,14,24,25]. Although fluorimetric methods such as SHERLOCK [18] present fM sensitivity, they require a fluorimeter, which restricts their PoC applicability (Table S2).
3.4. CASCADE coupling to isothermal nucleic acid amplification
The optimized CASCADE sensor can detect SARS-CoV-2 RNA at pM concentrations. This, however, was insufficient for SARS-CoV-2 detection in clinical samples, and therefore we explored new approaches to increase sensitivity. One possible strategy was the coupling of CASCADE to isothermal nucleic acid amplification. Another option consisted of the combination of various RNPs targeting the same RNA region. Fozouni et al. used this approach to enhance Cas13 trans-cleavage, thus increasing the system's sensitivity [27]. We then evaluated whether using this strategy for CASCADE would improve the sensitivity of the sensor in comparison with single-guide targeting. We combined RNP R1 with RNP R2 and RNP S1 with RNP S2 to detect Orf1ab or S RNA targets, respectively. These targets have a size similar to that of amplicons resulting from isothermal amplification. However, the RNP combinations did not result in improved sensitivity (Fig. S13) as compared to the use of single RNPs (Fig. 4). It is likely that the binding sites in the target for the two guides tested were too close (Fig. S13A). This might induce competition between the RNPs in their interaction with the target RNA, which decreases sensitivity. In this regard, it is important to consider that isothermal amplifications, such as RPA and NASBA, produce amplicons of limited size and the design of multiple guides with such a restriction is challenging. Fozouni et al. designed and tested several crRNAs to target distant regions in viral RNA [27]. Their approach is therefore more suitable to detect viral RNA without previous amplification. Although it is a convenient and promising strategy, it would require extensive crRNA testing and optimization to detect new sequences of interest.
As mentioned above, isothermal nucleic acid amplification also constitutes a valuable tool to improve sensitivity. Furthermore, the combination of specific amplification primers and carefully designed crRNAs for Cas13a-mediated recognition results in enhanced specificity. We tested the coupling of two different isothermal amplification methods, RPA and NASBA, to the CASCADE sensor.
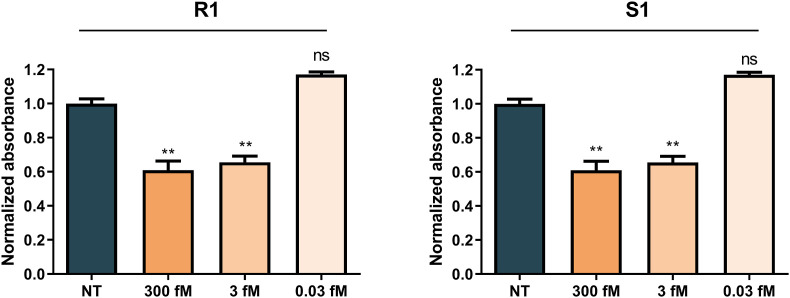
RPA is a fast and sensitive amplification method that has already been used in CRISPR-based sensors [18,26,28,29]. RPA combines the activity of a recombinase, a single-stranded DNA binding protein (SSB), and a strand-displacing polymerase to amplify dsDNA at a constant temperature of 37–42 °C in 30–60 min. Since SARS-CoV-2 is an RNA virus and RPA amplifies dsDNA, a previous retrotranscription step was required (RT-RPA). We generated two short RNA templates of the Orf1a and S regions by RT-PCR and subsequent T7 transcription using the RPA primers specified in Table S1. For the assay, different concentrations of these template RNAs (0.03–300 fM) were subjected to RT-RPA followed by T7 transcription. The total duration of the process was 80 min. The amplification products were then added to CASCADE using RNP R1 and S1 (Table S1) for the detection of Orf1ab and S, respectively. This addition induced a decrease in AuNP absorbance (Fig. 5 ) that, in the case of the highest concentration (300 fM), was already detectable after a 15-min incubation (Fig. S14). At 30 min, 3 fM samples showed decreased absorbance for both regions (Fig. 5, Fig. S14), although the difference was only detectable by the naked eye in the case of Orf1ab (RNP R1) (Fig. S14). After 60 min, RNP S1 allowed naked-eye detection of the S target at 3 fM target concentration (Fig. S14).
Fig. 5.
RPA coupling to CASCADE for S and Orf1ab colorimetric detection. Normalized sample absorbance at 523 nm at 30 min. 50 nM of RNP R1 and RNP S1 were used to detect Orf1ab (A) and S (B) regions, respectively. Sample absorbance was normalized against the no target (NT) control. Bar labels indicate target concentration before amplification. Values represent the mean of replicate samples (n = 9 for R1 and n = 3 for S1) ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant.
These results indicate that RPA-CASCADE can detect RNA sequences from SARS-CoV-2 with low femtomolar sensitivity (3 fM). The detection of Orf1ab using RNP R1 was faster and more sensitive, which highlights the importance of target site selection and crRNA design. Femtomolar target concentrations correspond to 106-104 viral copies μL−1, which resembles the number of copies present in clinical samples with cycle threshold (Ct) values ranging from 20 to 30 [30]. The additional amplification step extends the total detection time to approximately 2 h, which is comparable to that of other CRISPR-based sensors developed for PoC diagnosis, and even shorter than established methods like SHERLOCK (Table S2) [5].
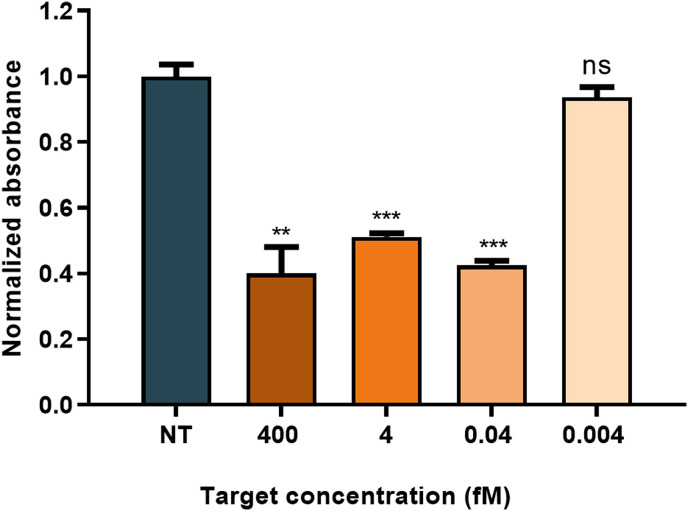
Nucleic Acid Sequence-Based Amplification (NASBA) constitutes a highly convenient tool for specific RNA amplification. It can directly amplify RNA, yielding complementary ssRNA amplicons at a constant temperature of 41 °C [31]. It is therefore a highly convenient method to be coupled with Cas13a, since it eliminates the need for both the retro- and T7 transcriptions. Strikingly, it has barely been applied before to CRISPR-based detection. To assess whether this amplification method could be coupled to CASCADE-based detection, an RNA template (RNA template NASBA, Table S1) was generated via RT-PCR and T7 transcription using T7 S_200 5′ PCR and S_200 3’ PCR primers (Table S1). Different concentrations of this RNA were subjected to NASBA amplification using NASBA FW and RV primers (Table S1) described by Wu et al. [20]. After a 90-min incubation at 41 °C, samples were added to CASCADE. 15 min later, we observed a significant decrease in absorbance for all the template concentrations tested (Fig. S15). After 30 min of incubation, absorbance continued to decrease, and samples also displayed a color change visible to the naked eye (Fig. 6 and Fig. S15). NASBA-CASCADE was able to detect RNA sequences from SARS-CoV-2 with attomolar sensitivity (40 aM) in 2 h.
Fig. 6.
NASBA coupling to CASCADE for colorimetric S detection. Normalized sample absorbance at 523 nm at 30 min 50 nM of RNP S were used to detect S. Sample absorbance was normalized against the no target (NT) control. Bar labels indicate target concentration before amplification. Values represent the mean of replicate samples (n = 3) ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant.
Taken together, these results suggest that our AuNP-Cas13a sensor is a versatile detection system that can be coupled to different isothermal amplification methods, which further highlights its potential for PoC applications. Remarkably, the sensitivity measured by the absorbance decrease correlates with naked-eye detection (Figs. S14 and S15), supporting the use of CASCADE as a reliable naked-eye sensor.
3.5. SARS-CoV-2 detection in clinical samples using CASCADE
Our RPA/NASBA-CASCADE results suggested that this system could be used for SARS-CoV-2 detection in clinical samples. For evaluating the clinical applicability of our sensing method, we used remnant genetic material obtained from patients’ nasopharyngeal swabs provided by Hospital Ramón y Cajal (Madrid), which had been previously analyzed by RT-qPCR (Table 1 ). The RNA from inactivated nasopharyngeal swab samples was extracted using the QIAamp Viral RNA kit (Qiagen). Patient RNA was first amplified by NASBA or RT-RPA-T7 transcription. In the case of RPA, as mentioned above, Orf1ab detection with RNP R1 was faster and more sensitive and was therefore chosen for the experiments with clinical samples. Next, amplification products were subjected to CASCADE detection. In total, 5 negative and 10 positive samples previously assessed by qRT-PCR (Table 1) were processed with CASCADE.
Table 1.
Comparison of RT-qPCR and CASCADE viral detection of clinical samples.
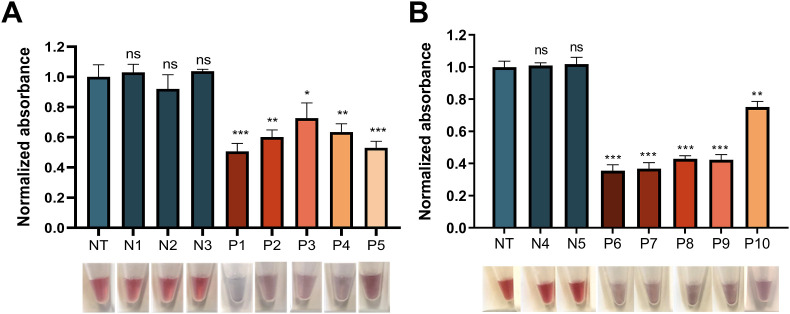
After only 15 min of incubation, four out of five samples subjected to RPA-CASCADE corresponding to positive patients (P1-4) already showed a significant decrease in absorbance and the color change of two of them (P1 and P4) was detectable by the naked eye (Table 1; Fig. S16). At 30 min, the decrease in absorbance of the positive samples was statistically significant (Fig. 7 A) and a perceivable color shift had taken place in all the samples from positive patients. These samples turned purple and could be clearly differentiated from the reddish negative samples (N1-3) (Fig. 7A). After 60 min, positive samples showed not only a different color, but also a dark AuNP precipitate (Fig. S16). In contrast, both the absorbance and the color of negative samples were always comparable to those of the no-target control (NT).
Fig. 7.
Isothermal amplification coupling to CASCADE for Orf1ab/S detection in clinical samples. Patient RNA samples amplified by RT-RPA(A)/NASBA(B) were subjected to CASCADE detection using 50 nM of RNP R1 (Orf1ab) or RNP S, respectively. Normalized sample absorbance at 523 nm (above) and naked-eye detectable color change upon Cas13a activation and subsequent AuNP aggregation (below) at 30 min. Sample absorbance was normalized against the no target (NT) control. N1, N2, N3 and N4 correspond to negative patient samples. P1– P10 correspond to positive patient samples. Values represent the mean of replicate samples (n = 3) ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns: non-significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In the case of NASBA-CASCADE, at 15 min all samples from positive patients already showed a statistically significant decrease in absorbance and the characteristic red to purple color shift (Table 1; Fig. S17). At 30 (Fig. 7B) and 60 (Fig. S17) minutes the absorbance continued to decrease, and the color change became increasingly evident. The absorbance and the color of negative samples remained unchanged and not significantly different from the no-target (NT) control.
To further investigate CASCADE's specificity, nasopharyngeal swabs from patients infected with other human coronaviruses (HCoV-NL63, HCoV-OC43 and HCoV-229E) were subjected to CASCADE detection using RPA amplification and RNP R1. The RNA extracted from these other coronaviruses did not induce AuNP aggregation, proving CASCADE's high specificity. Furthermore, we assessed if the simultaneous presence of SARS-CoV-2 and other human coronavirus in the same sample could affect SARS-CoV-2 detection. Thus, the RNA extracted from patients infected with other coronaviruses and the RNA from patients infected with SARS-CoV-2 were mixed and subjected to RPA-CASCADE. Fig. S18 shows that the presence of RNA from other coronaviruses did not impair SARS-CoV-2 detection by CASCADE. In conclusion, CASCADE is a highly specific detection method able to detect all positive samples tested (Cts 16–28), while showing no changes for negative (Ct > 35) (Table 1) and unspecific samples. These results suggest that our CASCADE sensor can be used for SARS-CoV-2 RNA detection in clinical samples as it provides reliable colorimetric and naked-eye sensing in less than two and a half hours.
4. Conclusions
In this work, we present CASCADE, a new SARS-CoV-2 detection method that combines isothermal amplification, specific CRISPR/Cas13a recognition and AuNP colloidal properties.
CASCADE is a highly versatile detection system. Thanks to the use of Cas13a instead of Cas12a, it can be easily customized to detect other pathogens and nucleic acids of interest. Although crRNA optimization is indispensable, Cas13a lacks target sequence requirements, in contrast to Cas12a and Cas9, which rely on the presence of a protospacer adjacent motif (PAM) flanking the region of interest. Furthermore, we have shown that CASCADE can be coupled to different isothermal nucleic acid amplification methods like RPA or NASBA. NASBA is especially suited for use with Cas13a, as it directly amplifies RNA with no need for additional retro- and T7 transcription reactions.
CASCADE is characterized by its inexpensive straightforward read-out, whether by using a simple spectrophotometer or even the naked eye. In contrast to other similar methods that require thermocyclers, centrifuges or fluorimeters, CASCADE only needs a heatblock (Table S2). CASCADE's sensing, which involves Cas13a target recognition and subsequent AuNP aggregation, requires very short times (15–30 min). Additionally, all its components are commercially available and it can be carried out by non-specialized staff. These features reinforce its feasibility and future practical application.
After a thorough optimization of AuNP features, CASCADE can detect SARS-CoV-2 RNA in nasopharyngeal swabs with good sensitivity, specificity and reliability.
In conclusion, CASCADE is a new and versatile molecular detection method with potential for PoC diagnosis of infectious diseases.
Authors' contributions
ML-V, CE-N and CR-D contributed equally to this work.
AS and BS have conceived the experiments, supervised the experimental results, and elaborated the manuscript.
ML-V, CE-N and CR-D have carried out the experiments and have elaborated the manuscript.
DP prepared gold nanoparticles.
MC performed NASBA experiments and extracted the RNA from patient samples.
PM extracted the RNA from patient samples.
CM-G has performed biosensing experiments.
RC designed NASBA experiments and prepared RNA targets.
MA, RC and JCG, obtained, classified, and analysed by RT-qPCR the patient samples.
RM designed the approach, coordinated the groups, and revised the manuscript.
All authors have given approval to the final version of the manuscript.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (SAF2017-87305-R, PID2020-119352RB-I00), Instituto de Salud Carlos III (FONDO-COVID19:COV20/00144 and COV20/00122) and Madrid Regional Government (NANOCOV-CM and IND2017/IND7809).
C.E-N, M. L-V and C.R-D thank Madrid Regional Government for the pre-doctoral Grants (PEJD-2017-PRE/BMD-3730, PEJD-2018-PRE/IND-9584 and PEJD-2017-PRE/IND-4438).
P.M.R thanks the Ministry of Economy, Industry and competitiveness of Spain for the FPI fellowship (BES-2017.082521).
IMDEA Nanociencia acknowledges support from the ‘Severo Ochoa’ Programme for Centres of Excellence in R&D (MINECO, Grant SEV-2016-0686, CEX2020-001039-S).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge Isabel Sola and Sonia Zúñiga for kindly providing the plasmids encoding the SARS-CoV-2 protein sequences and RNA from SARS-CoV-2 infected cells. We acknowledge Norman Feltz for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2022.339749.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wong N.A., Saier M.H. The SARS-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci. 2021;22(3):1308. doi: 10.3390/ijms22031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae S., Lim J.S., Kim J.Y., Jung J., Kim S.H. Transmission characteristics of SARS-CoV-2 that hinder effective control. Immune Netw. 2021;21(1):e9. doi: 10.4110/in.2021.21.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jääskeläinen A.E., Ahava M.J., Jokela P., Szirovicza L., Pohjala S., Vapalahti O., et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Virol. 2021;137:104785. doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., et al. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21(9):1634–1660. doi: 10.1039/d0lc01156h. [DOI] [PubMed] [Google Scholar]
- 5.Escalona-Noguero C., López-Valls M., Sot B. CRISPR/Cas technology as a promising weapon to combat viral infections. Bioessays. 2021;43(4) doi: 10.1002/bies.202000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 2021;178:113012. doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldewachi H., Chalati T., Woodroofe M.N., Bricklebank N., Sharrack B., Gardiner P. Gold nanoparticle-based colorimetric biosensors. Nanoscale. 2018;10(1):18–33. doi: 10.1039/c7nr06367a. [DOI] [PubMed] [Google Scholar]
- 9.Latorre A., Posch C., Garcimartín Y., Ortiz-Urda S., Somoza Á. Single-point mutation detection in RNA extracts using gold nanoparticles modified with hydrophobic molecular beacon-like structures. Chem. Commun. 2014;50(23):3018–3020. doi: 10.1039/c3cc47862a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan C., Tian T., Sun J., Hu M., Wang X., Xiong E., et al. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/cas12a/13a system. Anal. Chem. 2020;92(5):4029–4037. doi: 10.1021/acs.analchem.9b05597. [DOI] [PubMed] [Google Scholar]
- 11.Ma L., Peng L., Yin L., Liu G., Man S. CRISPR-Cas12a-Powered dual-mode biosensor for ultrasensitive and cross-validating detection of pathogenic bacteria. ACS Sens. 2021;6(8):2920–2927. doi: 10.1021/acssensors.1c00686. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Mansour H., Wang T., Poojari S., Li F. Naked-eye detection of grapevine red-blotch viral infection using a plasmonic CRISPR Cas12a assay. Anal. Chem. 2019;91(18):11510–11513. doi: 10.1021/acs.analchem.9b03545. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y., Wu J., Pang B., Zhang H., Le X.C. CRISPR/Cas12a-mediated gold nanoparticle aggregation for colorimetric detection of SARS-CoV-2. Chem. Commun. 2021;57(56):6871–6874. doi: 10.1039/d1cc02546e. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W.S., Pan J., Li F., Zhu M., Xu M., Zhu H., et al. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection. Anal. Chem. 2021;93(8):4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- 15.Kimling J., Maier M., Okenve B., Kotaidis V., Ballot H., Plech A. Turkevich method for gold nanoparticle synthesis revisited. J. Phys. Chem. B. 2006;110(32):15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D., Sarkar D., Girigoswami A., Chattopadhyay N. A fully standardized method of synthesis of gold nanoparticles of desired dimension in the range 15 nm-60 nm. J. Nanosci. Nanotechnol. 2011;11(2):1141–1146. doi: 10.1166/jnn.2011.3090. [DOI] [PubMed] [Google Scholar]
- 17.Turkevich J., Stevenson P.C., Hillier J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951;11:55–75. The Royal Society of Chemistry. [Google Scholar]
- 18.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F., Abudayyeh O.O., Gootenberg J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. Bioarchive. 2020 [Google Scholar]
- 20.Wu Q., Suo C., Brown T., Wang T., Teichmann S.A., Bassett A.R. INSIGHT: a population-scale COVID-19 testing strategy combining point-of-care diagnosis with centralized high-throughput sequencing. Sci. Adv. 2021;7(7) doi: 10.1126/sciadv.abe5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C.C., Chen C.P., Wu T.H., Yang C.H., Lin C.W., Chen C.Y. Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials. 2019;9(6):861. doi: 10.3390/nano9060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutinho C., Somoza Á. MicroRNA sensors based on gold nanoparticles. Anal. Bioanal. Chem. 2019;411(9):1807–1824. doi: 10.1007/s00216-018-1450-7. [DOI] [PubMed] [Google Scholar]
- 23.Mendez-Gonzalez D., Laurenti M., Latorre A., Somoza A., Vazquez A., Negredo A.I., et al. Oligonucleotide sensor based on selective capture of upconversion nanoparticles triggered by target-induced DNA interstrand ligand reaction. ACS Appl. Mater. Interfaces. 2017;9(14):12272–12281. doi: 10.1021/acsami.7b00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posch C., Latorre A., Crosby M.B., Celli A., Latorre A., Vujic I., et al. Detection of GNAQ mutations and reduction of cell viability in uveal melanoma cells with functionalized gold nanoparticles. Biomed. Microdevices. 2015;17(1):15. doi: 10.1007/s10544-014-9908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storhoff J.J., Elghanian R., Mirkin C.A., Letsinger R.L. Sequence-dependent stability of DNA-modified gold nanoparticles. Langmuir. 2002;18(17):6666–6670. [Google Scholar]
- 26.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D'Ambrosio M.V., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020;18(12) doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350(6313):91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.