Objective:
Carotid artery intima-media thickness (cIMT) is a widely accepted marker of subclinical atherosclerosis. Twenty susceptibility loci for cIMT were previously identified and the identification of additional susceptibility loci furthers our knowledge on the genetic architecture underlying atherosclerosis.
Approach and Results:
We performed 3 genome-wide association studies in 45 185 participants from the UK Biobank study who underwent cIMT measurements and had data on minimum, mean, and maximum thickness. We replicated 15 known loci and identified 20 novel loci associated with cIMT at P<5×10−8. Seven novel loci (ZNF385D, ADAMTS9, EDNRA, HAND2, MYOCD, ITCH/EDEM2/MMP24, and MRTFA) were identified in all 3 phenotypes. An additional new locus (LOXL1) was identified in the meta-analysis of the 3 phenotypes. Sex interaction analysis revealed sex differences in 7 loci including a novel locus (SYNE3) in males. Meta-analysis of UK Biobank data with a previous meta-analysis led to identification of three novel loci (APOB, FIP1L1, and LOXL4). Transcriptome-wide association analyses implicated additional genes ARHGAP42, NDRG4, and KANK2. Gene set analysis showed an enrichment in extracellular organization and the PDGF (platelet-derived growth factor) signaling pathway. We found positive genetic correlations of cIMT with coronary artery disease rg=0.21 (P=1.4×10-7), peripheral artery disease rg=0.45 (P=5.3×10-5), and systolic blood pressure rg=0.30 (P=4.0×10-18). A negative genetic correlation between average of maximum cIMT and high-density lipoprotein was found rg=−0.12 (P=7.0×10-4).
Conclusions:
Genome-wide association meta-analyses in >100 000 individuals identified 25 novel loci associated with cIMT providing insights into genes and tissue-specific regulatory mechanisms of proatherosclerotic processes. We found evidence for shared biological mechanisms with cardiovascular diseases.
Keywords: carotid intima-media thickness, genetics, genome-wide association study, meta, analysis, population genetics
Highlights.
Leveraging data from over 45 000 UK Biobank participants, we discovered 22 novel genetic loci and replicated 15 known loci associated with carotid intima-media thickness from genome-wide association study.
Meta-analysis of >100k individuals combining UK Biobank data and a previous meta-analysis yielded three additional novel loci.
Interaction analysis revealed presence of sex-specific effects in one of the novel loci and six previously reported loci.
Expression analyses revealed many prioritized genes enriched in endothelial cells and smooth muscle cells.
Positive genetic correlations between carotid intima-media thickness and respectively peripheral artery disease, coronary artery disease, ischemic stroke and systolic blood pressure were observed; A negative genetic correlation was found between carotid intima-media thickness and high-density lipoprotein cholesterol.
See accompanying editorial on page 502
Cardiovascular disease is a leading cause of death worldwide with atherosclerosis as the underlying cause of most cases.1 Atherosclerosis is characterized by the accumulation of lipids in the subintimal space of medium-sized and large arteries. A fibrous cap is formed on the atherosclerotic plaque due to the proliferation of smooth muscle cells and matrix deposition.2 Plaques can gradually impede the blood flow, eventually leading to ischemia of end-organ tissues. Rupture of plaques due to unstable, thin caps and subsequent thrombosis can lead to abrupt occlusion of the artery, causing a stop of the blood flow and thereby ischemia and necrosis of the tissue following the occlusion.3 This process is the underlying pathophysiological mechanism of major cardiovascular events, such as myocardial infarction4 and ischemic stroke,5 which are both leading causes of mortality and disability in developed countries.6
Due to the long preceding phase of atherosclerosis, it can take decades before the clinical presentation of symptoms. An early detection of atherosclerotic plaque formation may aid in the risk stratification and identification of individuals at high risk of atherosclerotic cardiovascular diseases, such as myocardial infarction, coronary artery disease (CAD), and peripheral artery disease (PAD). Noninvasive imaging techniques such as ultrasound measurement of the carotid artery intima-media thickness (cIMT) can detect subclinical atherosclerosis and is widely accepted as an index of atherosclerosis severity.7 Increased cIMT has been associated with higher risk of cardiovascular diseases.8,9 Twin studies suggest a substantial genetic component in cIMT,10 and previous genome-wide association studies (GWAS) of cIMT have identified 20 genetic loci linked to known genes as well as novel candidate genes associated with atherosclerosis.11–13 In the current study, we aimed to identify additional genetic loci to further our knowledge on the genetic architecture of increased cIMT using a large population-based study.
Methods
Please see the Major Resources Table in the Supplemental Material. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Study Population
The UK Biobank study is a population-based prospective cohort in the United Kingdom in which ≈500 000 individuals aged between 40 and 69 years were included between 2006 and 2010. All participants have given informed consent for this study. The UK Biobank has ethical approval from North West–Haydock Research Ethics Committee (REC reference: 16/NW/0274). Details of the UK Biobank study have been described in detail previously.14 This research has been conducted using the UK Biobank Resource under Application Number 15031.
cIMT Measurements
At the time of analyses, cIMT measurements were available for 46 705 participants who had completed the second follow-up visit for the UK Biobank. The cIMT measurements were derived automatically from the 2-dimensional carotid scan using a CardioHealth Station (Panasonic Biomedical Sales Europe BV, Leicestershire, United Kingdom). A total of 4 cIMT measurements were taken, namely at 120° and 150° for the right carotid and 210° and 240° for the left carotid artery. For each angle, the minimum, mean and maximum values were recorded.15
Quality control of cIMT measurements was performed by the UK Biobank, validated both internally and externally using a set of predefined criteria.15,16 cIMT measurements that did not pass the quality control, either with cIMT values of zero or as indicated by the corresponding quality control flags (field IDs 22682, 22683, 22684, 22685) were excluded from the analyses. Measurements collected at the second follow-up visit (imaging visit) were considered for the current analysis. Average values of the cIMT of the four angles were taken as the final cIMT measurements, namely average of minimum cIMT (cIMTmin), average of mean cIMT (cIMTmean), and average of maximum cIMT (cIMTmax).
Genotyping and Imputation
Genotype data for 488 377 participants were available in the UK Biobank study. Two custom Affymetrix Axiom (UK Biobank Lung Exome Variant Evaluation or UK Biobank) genotyping arrays with >95% common content were used. Imputation was performed using Haplotype Reference Consortium as the primary reference panel with addition of merged UK10K and 1000 Genome phase 3 reference panels. Quality control of samples and variants, as well as of the imputation was performed by the Wellcome Trust Centre for Human Genetics.17 The current study was performed on the imputed data supplied by the UK Biobank. Individuals with overall missingness >5% or excessive heterozygosity, and individuals whose genetically inferred sex did not match with the reported sex, were excluded from the analyses. Additionally, variants with a minor allele frequency <0.5% or an INFO-score (imputation information score) smaller than 0.3 were excluded.
Genome-Wide Association Studies
A transethnic GWAS (demographics shown in Table S1 in the Supplemental Material) was performed on 3 averaged cIMT measurements; cIMTmin, cIMTmean, and cIMTmax, using BOLT-LMM v2.3.1.18 A linear mixed model was fitted for each of the inverse rank normalized cIMT measurements. GWAS analyses were adjusted for age at the time of the imaging visit, sex, genotyping array, and the first 30 principal components (provided by UK Biobank) to adjust for population stratification. Individuals with missing information on any of the covariates were excluded from the GWAS analyses. To obtain a set of independent single nucleotide polymorphisms (SNPs) associated with cIMT phenotypes, clumping was performed on variants (lead variants) that passed the genome-wide significance threshold of P<5×10-8 based on linkage disequilibrium (LD) r2>0.005 and 2.5-Mb distance using the clumping procedure in PLINK 1.9.19 A locus was defined as a 1-Mb region surrounding the most significant variant. The nearest protein-coding gene and any additional gene within 10 kb of each locus’ lead variant were annotated to the locus.
Statistical Fine-Mapping
To further refine the signals detected in GWAS, we applied statistical fine-mapping across a 1-MB region around the lead variant for each of the loci identified to prioritize putative causal variants. Bayesian fine-mapping was performed on summary statistics of cIMTmean using FINEMAP (v1.4).20 A shotgun stochastic search method was used to produce 95% credible sets under the assumption of several causal variants (k) from 1 up to 5, each with estimated posterior probabilities which jointly summed up to 1. We considered the variants in the top causal configuration under k with the highest posterior probabilities as the likely causal variants. For each likely causal variant, we searched for coding variants in high LD (R2 > 0.8) with dbNSFP (database for nonsynonymous SNPs’ functional predictions, v.4.2).21
Meta-Analysis of cIMT Measurements
Meta-analysis of the UK Biobank and the largest GWAS meta-analysis of cIMT previously published by the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology)/UCLEB (UCL-LSHTM-Edinburgh-Bristol) consortia (71 128 European participants from 31 studies)12 was performed for cIMTmax using Multi-Trait Analysis of GWAS (MTAG)22; MTAG is a tool for analysis of multiple GWAS summary statistics which applies a generalized inverse-variance-weighted meta-analysis that allows for sample overlap. Additionally, a second meta-analysis of the 3 cIMT measurements within the UK Biobank was performed with MTAG. All MTAG analyses were performed on common variants (minor allele frequency >0.01) with an INFO-score >0.3 under the assumption of perfect genetic correlation and equal heritability (with the options --perfect_gencov and --equal_h2).
Sex Interaction Analysis
To examine if there are sex-specific differences in the genetic loci associated with cIMT, we performed sex-stratified GWAS on all 3 cIMT traits. We systematically tested the difference between the effect size estimates obtained from the stratified GWAS. A P value (Psexdiff) was obtained using the following test statistics23:
where rsex was the Spearman rank correlation coefficient between sexes across all variants included in the sex-stratified GWAS. We examined the sex-specific effects on loci either identified in the main GWAS or sex-stratified GWAS, and a locus was considered to demonstrate sex difference if Psexdiff passed Bonferroni-corrected threshold (=0.05/number of loci tested).
Transcriptome-Wide Association Study
To prioritize genes identified in GWAS, we incorporated expression quantitative trait loci data and performed a transcriptome-wide association study (TWAS). Tissue-specific expression levels of the genes were predicted using a model trained on an external reference panel with both genotype and expression data available; associations between the predicted expression levels and cIMT phenotypes were then tested on a genic level. We performed the TWAS using the Unified Test for Molecular SignaTures method with imputation models pretrained on data from the Genotype-Tissue Expression project.24,25 Briefly, gene expression levels were imputed by training a multivariate lasso regression model taking all available tissues into considerations (separate penalty terms for within-tissue and cross-tissue effects which allow different effect directions in different tissues). Imputed gene expressions were then used for the association tests with the cIMT traits (TWAS). Association analyses were performed on three arterial tissues, namely aorta, coronary and tibial, to obtain tissue-specific result (arterial TWAS). Given that the relatively smaller sample size for arterial tissues may restrict the power to detect potential associations, a joint test combining the test statistics of 44 tissues was calculated using the Generalized Berk-Jones test (cross-tissue TWAS).24
Multi-Marker Analysis for Genomic Annotation
In addition to TWAS, we applied Multi-Marker Analysis for Genomic Annotation (MAGMA) tool to conduct gene-based tests as well as a gene set analysis.26 Briefly, gene-based test with summary statistics involves computation of a gene-level test statistics by combining that of all variants within a gene, after which gene-based P was derived from an approximate sampling distribution estimated from a reference dataset. The gene-based statistics were then aggregated to predefined sets for association tests with cIMT traits, with LD between genes corrected by computing gene correlation matrix using the reference dataset. MAGMA analysis was done via the platform Functional Mapping and Annotation of GWAS v1.3.6a with MAGMA v1.08a, using default settings. Gene-based analyses were done with an SNP-wide mean model and the gene set analysis was performed using 15 496 gene sets (5500 curated gene sets and 9996 Gene Ontology terms respectively) from MsigDB v7.0.27,28 We applied Bonferroni corrections to assess the statistical significance of results from all gene-based and gene set analyses.
Heritability Estimation of cIMT and Genetic Correlation With Cardiovascular Diseases and Risk Factors
The heritability of cIMT measurements was estimated using the restricted maximum likelihood algorithm from the BOLT-LMM software. The proportion of additive variance explained by the lead variants was estimated by fitting a multivariable linear regression model on the cIMT measurements, assuming an additive genetic model. To investigate the shared genetic component between cIMT and vascular diseases namely, abdominal aortic aneurysm (AAA), CAD, stroke, and PAD as well as cardiovascular diseases (CVD) risk factors, we estimated genetic correlations between cIMT phenotypes and the diseases/risk factors both within our data and with the addition of external cohorts using linkage disequilibrium score regression (LDSC) v1.0.1.29 The CVD risk factors investigated in the current study include diastolic blood pressure (DBP), systolic blood pressure (SBP), HDL-C (high-density lipoprotein cholesterol), LDL-C (low-density lipoprotein cholesterol), total cholesterol, and triglyceride levels. Ascertainment of AAA, CAD, ischemic stroke, and PAD in the UK Biobank was based on a combination of hospital inpatient and self-reported records during an interview with a trained nurse (Table S2 in the Supplemental Material). Measurements of CVD risk factors in the UK Biobank are described in the supplement (Supplementary Method). Genetic associations of the cardiovascular disease were done within the subset of unrelated UK Biobank participants that was not included in the cIMT GWAS. With regards to the analysis using external cohorts, we used summary statistics data from the CARDIoGRAMplusC4D (Coronary Artery Disease Genome-Wide Replication and Meta-Analysis Plus the Coronary Artery Disease Genetics) consortium, a meta-analysis with 60 801 CAD cases and 123 504 controls to estimate the genetic correlation between cIMT and CAD.30 Summary statistics of the transethnic meta-analysis MEGASTROKE collaboration (60 341 cases; 454 450 controls) were used for the genetic correlation between cIMT and stroke including its subtypes.31 The summary statistics from transethnic analysis on the GERA (Genetic Epidemiology Research on Adult Health and Aging) cohort consisting of 99 785 individuals were used for the genetic correlation with blood pressure traits.32 For the analysis with lipid traits, we used the summary statistics from the meta-analysis (joint analysis of Metabochip and GWAS with a total of 188 577 individuals) from the GLGC (Global Lipids Genetics Consortium).33 We additionally compared the cIMT loci with the loci identified for these cardiovascular diseases using Functional Mapping and Annotation of GWAS, which searched the GWAS Catalog (version e96_r2019-09-24) for variants in high LD (R2>0.6) with lead variants in our GWAS.34
Results
Study Population Characteristics
Out of 502 493 UK Biobank participants, we included 45 185 individuals with available cIMT measurements that passed the quality control and had available genetic data (Figure S1 in the Supplemental Material). Baseline characteristics of the study population are shown in Table S1 in the Supplemental Material. This population consisted of a majority of White participants (96.6%) along with participants of Asian descent (1.4%), African descent (0.6%), of a mixed descent (0.5%), and of other/unknown ethnic background (0.6%). Overall, the mean (SD) cIMTmin was 0.58 (0.11) mm, the mean cIMTmean was 0.69 (0.13) mm, and the mean cIMTmax was 0.80 (0.15) mm. Smaller cIMT measurements were observed in women compared with men with cIMTmin 0.57 (0.10) versus 0.60 (0.12), cIMTmean 0.67 (0.11) versus 0.71 (0.14), and cIMTmax 0.77 (0.13) versus 0.83 (0.16). The cIMT measurements were strongly correlated (r2 0.86-0.96; Figure S2 in the Supplemental Material).
Novel Loci Associated With cIMT
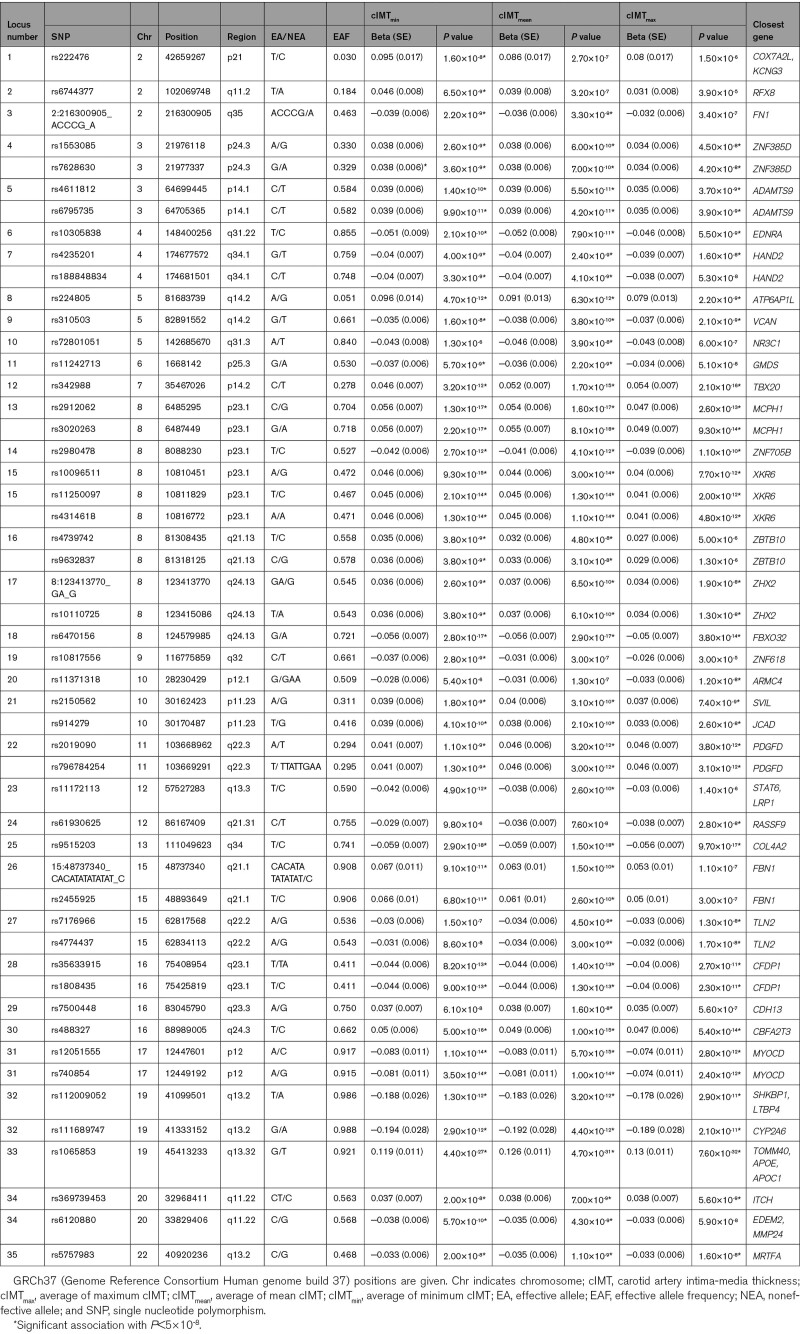
A total of 11 247 984 variants were tested in the current analyses. A total of 2837 variants in 30 loci were associated with cIMTmin, 2781 variants in 30 loci with cIMTmean, and 1789 variants in 25 loci with cIMTmax. A summary of all loci that were identified can be found in Table 1. Among these, a total of 20 loci have not been reported in previous studies. Moreover, all loci except 5q31.3 (NR3C1) and 12q21.31 (RASSF9) remained significantly associated with the cIMT phenotypes at a stringent Bonferroni adjusted P<1.67×10-8 (5×10-8/3). Replication of loci identified in the current study in a previous large meta-analysis of the CHARGE consortium12 can be found in Table S3 in the Supplemental Material. Of the 20 novel loci identified in the current study, 15 lead variants had P<0.05 in the external cohort with consistent direction of effect. Conversely, we looked up previously reported associations in the current result. Association results of the five previously reported loci that were not replicated in the current analysis are provided in Table S4 in the Supplemental Material.
Table 1.
Independent Loci Identified for All cIMT Measurements
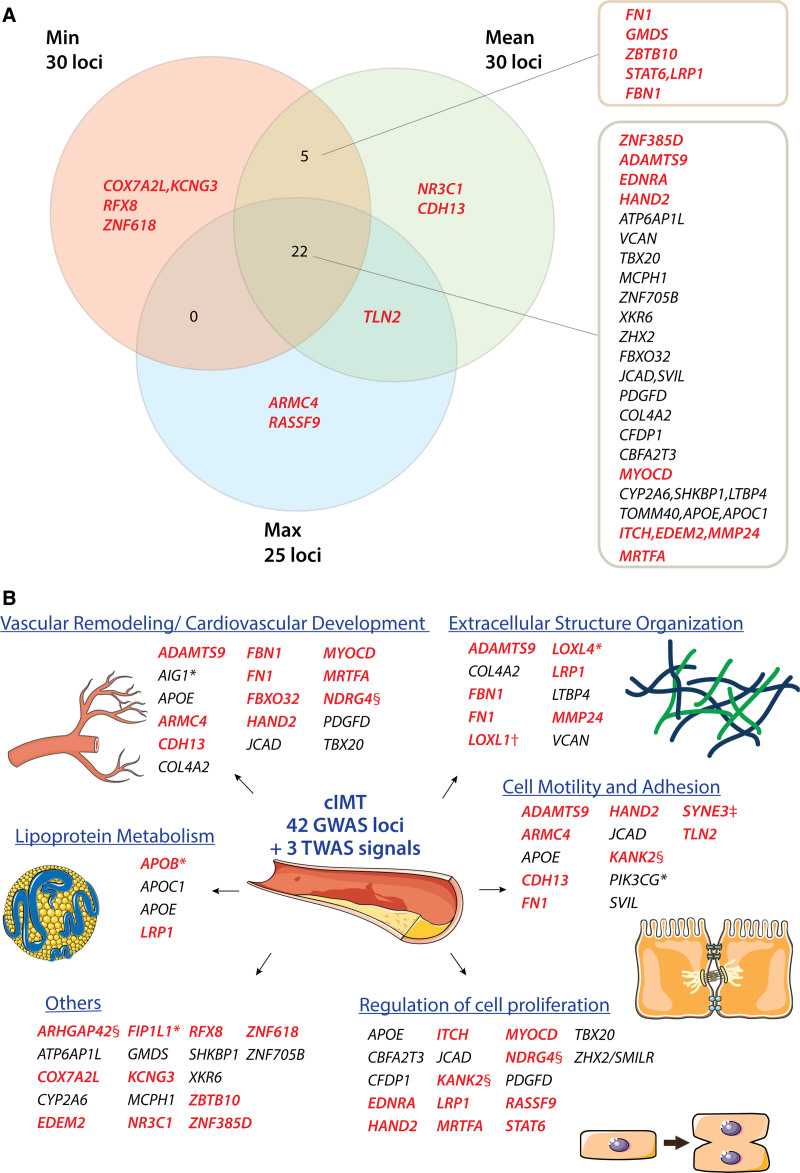
Comparing the 3 GWAS that were performed in the current study, there was a high degree of overlap in the loci identified at P<5×10-8 (Figure 1). Notably, cIMTmin and cIMTmean shared 27 out of 30 loci, whereas 22 out of 25 loci identified for cIMTmax were also detected for cIMTmean. A total of 22 loci overlapped between the 3 GWAS, 7 of which are novel: 3p24.3 (ZNF385D), 3p14.1 (ADAMTS9), 4q31.22 (EDNRA), 4q34.1 (HAND2), 17p12 (MYOCD), 22q11.22 (ITCH, EDEM2, MMP24), and 22q13.2 (MRTFA). Association results by each cIMT measurement are provided in Table S5 through S7 in the Supplemental Material. Regional association plots for each locus are provided in Figure S4 in the Supplemental Material.
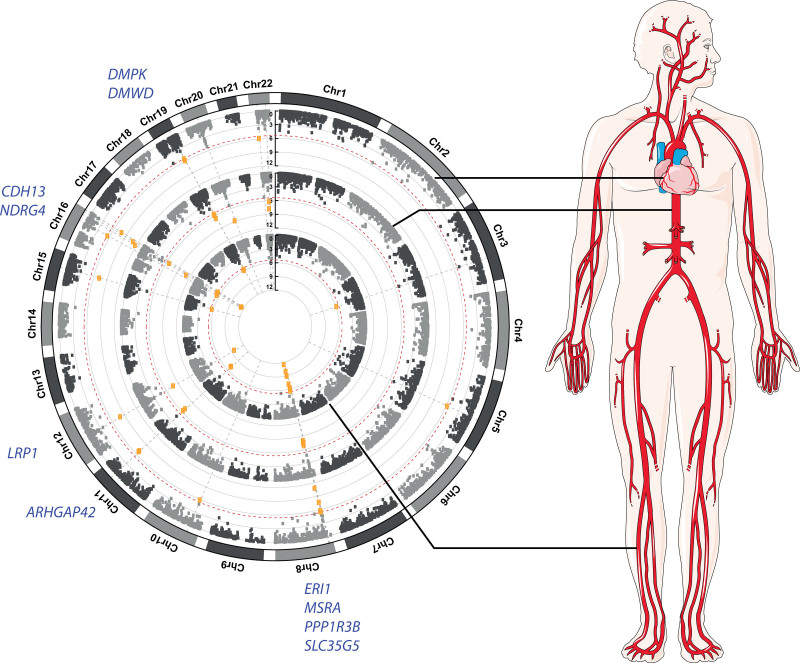
Figure 1.
Overview of independent loci identified for carotid intima-media thickness (cIMT). A, Overlaps of independent loci identified between genome-wide association study (GWAS) for three cIMT phenotypes. Each locus is marked with the nearest protein-coding gene to the lead variant. Genes marked in bold red font were the closest genes from the respective lead variants of novel loci. B, Candidate genes from main GWAS, meta-analyses, and sex interaction analysis were manually grouped into functional categories relevant to atherosclerosis. Genes marked in bold red font were from novel loci; *genes discovered in the meta-analysis of UKB (UK Biobank) and CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology)/UCLEB (UCL-LSHTM-Edinburgh-Bristol) consortia; †discovered in meta-analysis of three cIMT measurements; ‡sex-specific locus; §genes discovered in transcriptome-wide association study (TWAS) not belonging to any GWAS loci. Parts of the figure were modified from materials provided by Servier Medical art, licensed under a Creative Common Attribution 3.0 Generic License (http://servier.com/Powerpoint-image-bank). max indicates average of maximum cIMT; mean, average of mean cIMT; and min, average of minimum cIMT.
To refine the signals identified at these loci, we performed statistical fine-mapping using FINEMAP and then looked up the coding variants in high LD (R2>0.8) with the fine-mapped variants in the dbNSFP database. The closest gene(s) of the fine-mapped variants were the same as the lead variants for the majority of loci except for the locus at 2p21—the fine-mapped variant rs6736913 is close to EML4 (Table S8 in the Supplemental Material). Additional signals were identified by FINEMAP at 5q31.3 (rs258811, intronic variant of ARHGAP26), at 12q13.3 (rs7484541, intronic variant of R3HDM2), and at 16q23.1 (rs4888367, intronic variant of BCAR1). A total of nine coding variants were found in dbNSFP for 8 loci (Table S8 in the Supplemental Material).
MTAG Analysis
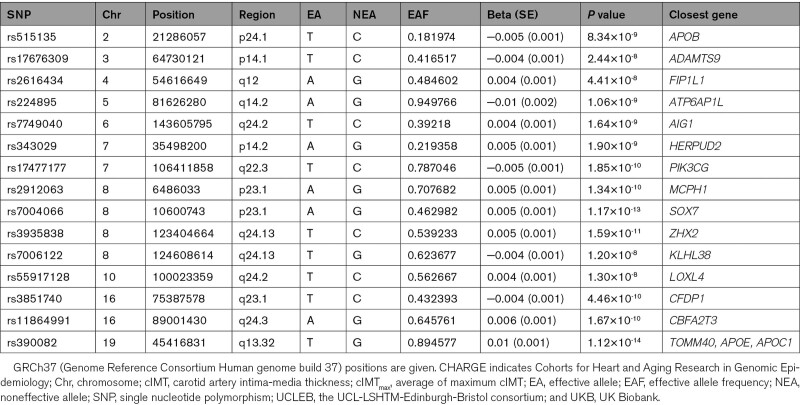
A total of 6 725 452 variants were analyzed in the meta-analysis of UK Biobank and the CHARGE/UCLEB consortia. The GWAS-equivalent sample sizes estimated by MTAG under the assumptions of LD score regression were 31 887 for UK Biobank cohort and 68 366 for the CHARGE/UCLEB consortia data. Table 2 presents the loci identified in this meta-analysis. Of the 15 loci identified, 10 were also detected in the main individual GWAS within the UK Biobank cohort (Figure 1); loci at 6q24.2 (AIG1) and 7q22.3 (PIK3CG) were reported in the CHARGE/UCLEB consortia meta-analysis. Three novel loci are 2p24.1 (APOB), 4q12 (FIP1L1), and 10q24.2 (LOXL4), although suggestive evidence for association (P<5×10-7) was already reported in the CHARGE/UCLEB consortia meta-analysis for APOB and LOXL4. The regional plots of these loci are presented in Figure S5 in the Supplemental Material.
Table 2.
Independent Loci for cIMTmax in the Meta-Analysis of UKB and CHARGE/UCLEB Consortia
Meta-analysis of 7 320 435 variants within the UK Biobank across all cIMT measurements revealed 23 loci, 22 of which were the overlapping loci detected in the individual GWAS (Figure 1). The signal at 15q24.1 (LOXL1), which did not pass the genome-wide significance threshold in the individual GWAS, was amplified in the meta-analysis (tagged by rs12441130, Beta=−0.033, SE=0.006, PMTAG=7.87×10-9). The regional plot of this new locus is presented in Figure S6 in the Supplemental Material. The stop-lost variant rs12441130 was also prioritized by FINEMAP as the most probable causal variant in this region.
Sex-Specific Loci
Sex-stratified GWAS identified 12, 14, and 8 loci for cIMTmin, cIMTmean, and cIMTmax, respectively, among female participants; among male participants, there were 9, 7, and 7 loci for cIMTmin, cIMTmean, and cIMTmax respectively. All of these loci were reported in the main GWAS except for one male-specific locus at 14q32.13 tagged by rs1209079 (SYNE3). These sex-stratified associations were then submitted to the interaction analysis. Out of 42 loci examined, 7 showed significant differences (Psexdiff <0.05/42=1.19×10-3) between female and male participants (Table S9 and Figure S7 in the Supplemental Material). Two loci 5q14.2 (VCAN, HAPLN1) and 8p23.1 (MCPH1) have been reported in previous sex-stratified analysis on cIMTmean.13 Among the 5 new loci, 17p12 (MAP2K4/MYOCD) were significantly associated with cIMTmean and cIMTmax in females only, whereas associations at 14q32.13 (SYNE3) and the 3 loci at 8p23.1 (SOX7, GATA4/C8orf49, ZNF705B) were only significant in males.
Gene Expression Analyses
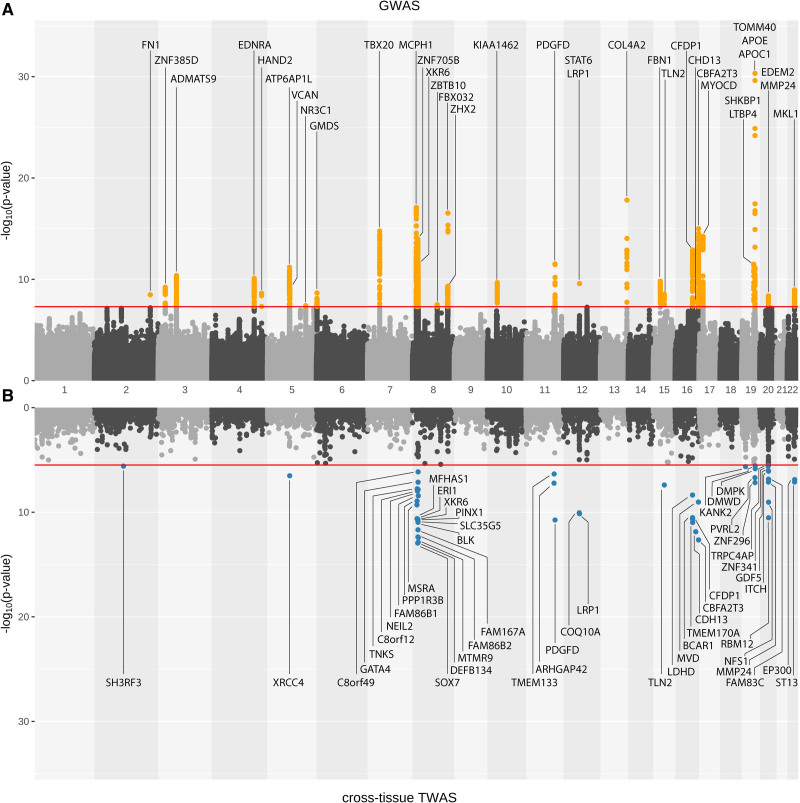
Test statistics were available for 15 004 out of 17 290 genes in the cross-tissue analysis on all 44 tissues. From the cross-tissue analysis, we identified 42, 49, and 42 genes with P<3.33×10-6 (0.05/15 004) for cIMTmin, cIMTmean, and cIMTmax, respectively (Table S10 in the Supplemental Material). Among these prioritized genes, 35 genes were identified for all three cIMT measurements and were located primarily on chromosome 8, chromosome 16, and 20 nearby the loci identified in the GWAS (Figure 2). Numerous prioritized genes, namely PINX1, BCAR1, CDFP1, and CBFA2T3, have been previously associated with cIMT. Although some of the genes prioritized in the TWAS are also the nearest protein-coding gene to the lead variant in the GWAS (XKR6 at 8p23.1, PDGFD at 11q23, LRP1 at 12q13.3, TLN2 at 15q22.2, CDH13 at 16q23.3, ITCH, and MMP24 at 20q11.22), TWAS put forward additional candidate genes such as XRCC4 at 5q14.2, and ST13 at 22q13.2 among the GWAS loci. A new association represented by KANK2 at 19p13.2, which was not found at the GWAS stage, was identified through the cross-tissue TWAS (Figure 2, Figures S8 and S9 in the Supplemental Material).
Figure 2.
Miami plot of average of mean carotid intima-media thickness. A shows genome-wide association study (GWAS) results, whereas B shows results from cross-tissue transcriptome-wide association study (TWAS). Orange dots: variants passing genome-wide significance threshold (P<5×10–8) in GWAS; Blue dots: genes passing significance threshold after Bonferroni correction (P<3.3×10–6) in TWAS.
Figure 3 depicts the results of the single-tissue tests for cIMTmean with 3 arterial tissues (aorta, coronary, and tibial). A total of 30 genes were prioritized, with 17 from the aorta, 16 from the coronary artery, and 20 from the tibial artery that passed the Bonferroni-corrected P value thresholds (4.18×10-6, 4.12×10-6, and 4.14×10-6 respectively; Table S11 in the Supplemental Material). Genes that were consistently prioritized in arterial tissues include ERI1, MSRA, and SLC35G5 on chromosome 8; ARHGAP42 on chromosome 11; LRP1 on chromosome 12; CDH13 and NDRG4 on chromosome 16; and DMPK as well as DMWD on chromosome 19. Of note, LRP1 and CDH13 were also identified in the GWAS; ARHGAP42 and NDRG4, uncovered in arterial-specific TWAS, did not belong to any GWAS loci.
Figure 3.
Circular Manhattan plots of transcriptome-wide association study (TWAS) results in arterial tissues for average of mean carotid intima-media thickness. Single-tissue TWAS was performed using Genotype-Tissue Expression data on aorta, coronary artery, and tibial artery tissue. Red dotted line: Bonferroni-corrected significance thresholds. Highlighted are the genes that were significant in all arterial tissues. Part of the figure was modified from materials provided by Servier Medical art, licensed under a Creative Common Attribution 3.0 Generic License. Chr indicates chromosome.
We also performed colocalization analysis with the summary data-based Mendelian randomization (Supplementary Methods). Among the 9 significant genes in summary data-based Mendelian randomization analysis (Table S12 in the Supplemental Material), the majority was also prioritized in either cross-TWAS (TLN2 and BCAR1), arterial TWAS (ARHGAP42 and LRP1), or both (CDH13). Summary data-based Mendelian randomization analysis also identified SVIL which was first reported in a previous GWAS.13
Finally, we examined the expression of the genes prioritized on a single-cell level in human carotid atherosclerotic plaques samples. UMAP (Uniform Manifold Approximation and Projection) clustering of 6191 cells from 38 patients yielded 18 distinct cell populations including endothelial cells, smooth muscle cells, and various immune cells (Figure S10 in the Supplemental Material). Of the 76 genes with expression data available, 14 genes were highly expressed in smooth muscle cells (AIG1, EDNRA, FBN1, FBXO32, FDFT1, KANK2, LRP1, LTBP4, PDGFD, PPP1R3B, STAT6, SVIL, TLN2, and VCAN) and 9 were highly expressed in endothelial cells (ADAMTS9, BCAR1, CDH13, COL4A1, COL4A2, JCAD, NDRG4, SOX7, and SVIL).
Gene-Based and Gene Set Analysis
Gene-based analyses with MAGMA yielded 44, 54, and 35 genes with P<2.63×10-6 (0.05/18 991) for cIMTmin, cIMTmean, and cIMTmax, respectively (Table S13 in the Supplemental Material). We additionally performed gene prioritization using DEPICT (Data-Driven Expression Prioritized Integration for Complex Traits) and found only GATA4 had a false discovery rate of <5% (Table S14 in the Supplemental Material). An overview of gene prioritization results for GWAS loci identified in the UK Biobank is presented in Table S15 in the Supplemental Material.
Three gene sets passed the Bonferroni-corrected significance threshold in the gene set analyses for cIMT measurements. These included gene sets related to formation and functions of structural protein collagen, platelet-derived growth factor signaling pathway, as well as a gene set within the 20q11 amplicon identified in breast tumor sample. The top 10 gene sets for each cIMT measurements are provided in Table S16 in the Supplemental Material.
Heritability of cIMT
Using the BOLT-REML algorithm, we estimated the SNP-heritability () to be 0.263 (SE=0.013), 0.284 (SE=0.013), and 0.248 (SE=0.013) for cIMTmin, cIMTmean, and cIMTmax, respectively. Consistent with phenotypic correlation, the cIMT measurements showed very high genetic correlations: cIMTmin and cIMTmean: rg =0.989 (SE=0.003); cIMTmin and cIMTmax: rg =0.975 (SE=0.009); cIMTmean and cIMTmax: rg =0.996 (SE=0.002).
A weighted genetic risk score was constructed for each cIMT phenotype, consisting of a summation of the number of effect alleles multiplied by their effect estimate for the respective sentinel SNPs. We found the fractions of cIMT variance explained by the weighted genetic scores of the genome-wide significant SNPs were 2.3%, 2.2%, and 1.8% for cIMTmin, cIMTmean, and cIMTmax, respectively.
Shared Genetic Components With CVD and Risk Factors
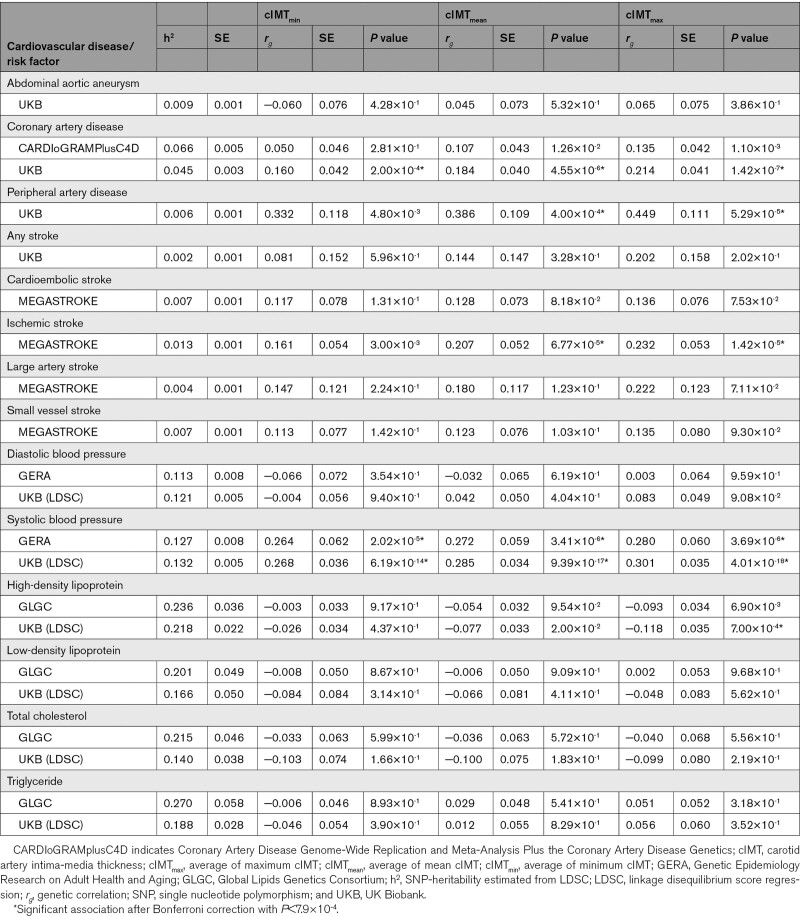
When testing the genetic correlation with the cardiovascular diseases, the strongest correlations were found for cIMTmax and the weakest for cIMTmin (Table 3). Among the 3 diseases, the strongest genetic correlations with cIMT were observed for PAD: rg =0.33 (P=0.005), 0.39 (P=0.0004), 0.45 (P=5.3×10-5) for cIMTmin, cIMTmean, and cIMTmax, respectively. Positive genetic correlation with CAD was observed for all cIMT measurement in UK Biobank with the highest correlation for cIMTmax: rg=0.21 (P=1.4×10-7); a similar trend but lower correlations were observed using data from the CARDIoGRAMplusC4D cohort. Positive genetic correlations between ischemic stroke and cIMT measurements were observed using the summary statistics from the MEGASTROKE consortium with the highest correlation for cIMTmax: rg=0.23 (P=1.4×10-5), whereas no significant correlation was observed with other stroke subtypes within the MEGASTROKE data nor with stroke within the UK Biobank cohort. Likewise, we found no significant genetic correlations with AAA within the UK Biobank cohort. Among the correlation with CVD risk factors, we found significant positive correlations between all cIMT measurements and SBP in both GERA and UK Biobank cohort with the highest correlation for cIMTmax: rg=0.30 (P=4.0×10-18) observed in the UK Biobank. A negative correlation was observed for cIMTmax with HDL-C: rg=−0.12 (P=7.0×10-4).
Table 3.
Genetic Correlation Analyses With Cardiovascular Diseases and Risk Factors
The shared genetic background was evident from overlaps of loci between the current GWAS and previous GWAS on CVD/risk factors as listed in GWAS Catalog.34 Studies reported association of APOB with CAD,35 pulse pressure,36 LDL-C,37–43 and total cholesterol.42,44 ADAMTS9 has been implicated in previous GWAS of CAD,35 DBP, SBP32,45 as well as total cholesterol level.43 Association with CAD,35,46,47 ischemic large artery stroke,31,46 intracranial aneurysm,48 PAD,49 pulse pressure,32,50,51 and SBP32,52 were previously reported for endothelin receptor gene EDNRA. AIG1 was found to be associated with pulse pressure.51 TBX20 was found associated with DBP in a large meta-analysis on blood pressure traits.36,50 PIK3CG was found to be associated with CAD,35 DBP,51 SBP,32,45,52,53 and pulse pressure.32,51,54–57 The region at 8p23.1, marked by 2 loci in the current study (ZNF705 and XKR6), was associated with CVD risk factors namely, pressure traits including DBP,32,45 SBP32,45,51–53 and pulse pressure,32,51 and lipid traits, including LDL-C43 and triglycerides.58,59 Numerous studies also reported this region may be associated with the interplay between CVD risk factors.60,61 ARMC4 was found to be associated with SBP.36,52 Several studies reported PDGFD to be associated with CAD.35,53,62,63 LRP1 has been reported to be associated with AAA64 as well as cervical artery dissection65 and CAD.35 In addition to the association with CAD,35,63 the COL4A1/COL4A2 region has also been found to be associated with ischemic stroke (small vessel).31 The FBN1 gene, whose detrimental mutation leads to genetic connective disorder Marfan syndrome, perhaps expectedly, was found to associate with pressure traits as well as aneurysms and dissections.66,67 The signal identified at TLN2 was also a locus for DBP and pulse pressure.51 Association with CAD35,53 and blood pressure traits32,50–53 were reported for the locus at 16q23.1 (CFDP1) as well as 16q23.3 (CDH13). The locus 19q13.32 (TOMM40/APOE/APOC1) is a widely replicated locus for lipid traits including apolipoproteins levels,68 LDL-C,33,37–40,42,43,69–77 HDL-C,33,42–44,56,72,74,76,78 total cholesterol,33,42,43,56,72,74–76,78 triglycerides42 as well as for CAD.35,47,63 Table S17 shows the 14 cIMT variants which were also significantly associated with CAD from the meta-analysis of UK Biobank and CARDIoGRAMplusC4D.35
Discussion
We investigated the genetic architecture of cIMT, a widely used marker of preclinical atherosclerosis, by performing GWAS in over 45 000 individuals of the UK Biobank. We performed secondary analyses including sex-stratified analysis and the meta-analysis of the three cIMT measurements which revealed one additional novel locus, respectively. We also conducted a meta-analysis combining the UK Biobank data and the CHARGE/UCLEB consortia data (GWAS-equivalent sample size=100 253) which resulted in 3 additional new loci. In total, we identified 25 novel loci associated with cIMT. We followed up with our findings by a series of bioinformatic analyses including statistical fine-mapping, expression analyses, gene-based analyses to refine the signals and prioritize candidate genes in the respective loci. Gene set analysis revealed enrichment of candidate genes in biological pathways related to structural components and formation of extracellular matrix, regulation of angiogenesis as well as lipid metabolism. Our results provide novel leads to dissect the complex process of atherosclerosis and targets of therapy.
Comparison With Previous Studies
The results confirm previous reports on genetics of cIMT traits. We replicated all SNPs associated with cIMTmean, and all except one (rs11025608) for cIMTmax in a previous GWAS that used a proportion of the current UK Biobank samples.13 We also confirmed 7 of 11 loci that were identified in the CHARGE/UCLEB consortia meta-analysis, including 31 studies with various study designs.12 These include variants at ATP6AP1L, MCPH1, SGK223 (moderate LD of 0.267 between the 2 lead variants rs11785239 and rs2980478 near ZNF705B identified in the current analysis), ZHX2, PINX1, CBFA2T3, and APOE. In addition to replicating these 15 previously reported loci, we report an additional 25 novel GWAS loci for cIMT, thereby doubling the number of loci associated with cIMT. It is of note that ADAMTS9 was previously found in colocalization analysis and EDNRA was associated with carotid plaque in the mentioned meta-analysis.12 Moreover, our TWAS analyses suggested a link between cIMT and KANK2 from cross-tissue analysis, as well as ARHGAP42 and NDRG4 from arterial-specific analysis, which were signals that had not been identified by GWAS.
Biological Insights of Selected Candidate Genes
Based on the multiple findings of the current study, either by proximity to the lead variants, identification of nonsynonymous coding variants in high LD or various gene-based prioritization analyses (Table S15 in the Supplemental Material) as well as existing literature, we discuss the candidate genes more relevant to vascular biology below. Various candidate genes identified in the present GWAS have known effects on atherosclerosis progression, for instance through their relation with vascular remodeling, extracellular matrix organization, and abnormal lipid metabolism (Figure 1). Other than the Nikolsky breast cancer 20q11 amplicon, whose region coincides with the locus at 20q11.22 in our GWAS, gene sets prioritized in the enrichment analysis with MAGMA revolve mainly around remodeling of connective tissues. Both collagen IV with COL4A1/COL4A2 (encoding 2 of the 6 alpha chains) and peptidyl lysine oxidases (LOXL1 and LOXL4) are involved in the organization of extracellular matrix (crosslinking of collagen fibrils). Current analysis also identified fellow extracellular matrix components fibrillin-1 (FBN1, the gene responsible for the congenital connective disorder Marfan syndrome) and fibronectin-1 (FN1). It is of note that both ADAMTS9 and MMP24 encode metalloproteinases also involved in maintenance of extracellular matrix. ADAMTS9 belongs to the same subgroup as ADAMTS1 as this group cleaves the proteoglycan versican, a structural component of the extracellular matrix encoded by VCAN.79 Antiangiogenic activity of ADAMTS9 has been reported with a gene knockdown study,80 whereas in human studies, ADAMTS9 has been associated with an interaction effect with smoking in coronary artery calcification81 and higher serum ADAMTS9 levels have been found in CAD patients.82 MMP24 encodes a metalloproteinase which activates gelatinase A, another matrix metalloproteinase expressed in vascular cells and encoded by MMP2.83 Extensive studies on both animal and human arteries suggest an important role of MMP2 in vascular remodeling by degrading collagen IV at basement membrane which facilitates vascular smooth muscle cell (VSMC) migration.84,85 It is worth noting that increased MMP activities were observed which contributes to aortic aneurysm formation in human86 and mouse87,88 with defective FBN1 via interaction with the large latent complex (of which fibrillin binding protein LTBP4 [latent transforming growth factor beta-binding protein 4] is a member). This further suggests involvement of metalloproteinases in arteriopathy. Altered metalloproteinases activities at atherosclerotic lesions may be attributed to endothelial to mesenchymal transition promoted by FBXO32.89,90 In the current study, we also identified another endothelial to mesenchymal transition associated gene: heart and neural crest derivatives-expressed 2 (HAND2) encodes a transcription factor involved in embryonic cardiogenesis and postnatal ventricular structural remodeling91 but has also been implicated in the regulation of angiogenesis.92 Loss-of-function mutations of HAND2 were previously associated with familial dilated cardiomyopathy93 and congenital heart defects.94
In addition to degradation of extracellular matrix, atherosclerosis also involves accumulation of VSMC at the lesion site. The PDGF signaling pathway, identified in the gene set analysis, is important for migration and proliferation of VSMC. Notably, STAT6, a gene close to the new locus identified on chromosome 12, encodes for one of the members of the PDGF pathway, whereas LRP1 at the same locus encodes for a signaling receptor, a known modulator of the PDGF signaling pathway and has been found to be an important protein in maintaining vascular wall integrity.95,96 Interestingly, a recent study reported SMILR, a long noncoding RNA located in the consistently replicated locus at 8q24.14 (ZHX2), as important mediator for VSMC proliferation in atherosclerosis upon activation by the PDGF signaling pathway.97 Of note, the PDGF receptor alpha gene (PGDFRA) is located in the novel locus detected in the meta-analysis of UK Biobank and CHARGE/UCLEB consortia in the current study.
Other novel genes identified in the current study may also be involved in regulation of VSMC proliferation, including CDH13,98,99 TLN2,100 KANK2, NDRG4, MYOCD, MRTFA, and EDNRA. KN motif and ankyrin repeat domains 2 (KANK2), which was found through the cross-tissue TWAS, encodes a protein with roles in cytoskeletal formation. KANK2 was found to be more expressed in smooth muscle cells and endothelial cells in atherosclerotic plaques and has been reported to interact with talins.101,102 It has been proposed as a candidate causal gene for CAD in a recent study.103 Animal studies reported the role of NDRG4 in cell proliferations in brain104 and the heart105 as well as in VSMCs via PDGF signaling pathway.106 Myocardin (MYOCD) encodes a nuclear protein that is expressed in cardiomyocytes and smooth muscle cells–containing tissues and has been shown to regulate inflammatory and lipid metabolism, both important to the pathogenesis of atherosclerosis, in VSMCs.107,108 Myocardin-related transcription factor A (MRTFA) encodes a protein that interacts with myocardin. A previous study in a Japanese population reported an association between MRTFA and a higher risk of CAD as well as with the severity of CAD.109 In mouse models, MRTFA was reported to be associated with the accumulation of proatherogenic macrophages in atherosclerotic plaques.110 Endothelin receptor type A (EDNRA), expressed in smooth muscle cells, encodes a receptor for endothelin-1, the most potent endogenous vasoconstrictive peptide. Numerous studies have found binding of endothelin-1 to the receptor promotes cell proliferation contributing to atherosclerosis.111 eQTL (expression quantitative trait loci) analyses in the current study prioritized other vascular molecules related to vascular tone regulation, Rho GTPase-activating protein 42 (ARHGAP42) and ARHGAP26 (closest gene of fine-mapped variant at chromosome 5), although its link to atherosclerosis is unclear.112
The loci at 8p23.1 have been consistently reported to be associated with cIMT, although the exact genes involved and the associated molecular mechanisms remain unclear. Among genes in this region, the X Kell blood group complex subunit-related family member 6 (XKR6) gene was not only found in all three GWAS but also in TWAS and MAGMA analyses for all cIMT measurements. Although the exact function of XKR6 is currently unknown, a recent study reported associations between XKR6, CAD, and ischemic stroke.113 Interestingly, this region was also found significant in the sex interaction analysis with the male-specific effects. Among the highlighted genes are GATA4 and SOX7, which were also prioritized in MAGMA, TWAS, and DEPICT. Studies in individuals with rare 8p23.1 duplication/deletion syndrome suggested possible involvement of both genes in cardiovascular malformations among the patients.114–116
Strengths and Limitations
A major strength of this study is the use of a large sample size from a single cohort. This study is the largest single-sample study of cIMT to date. Previous studies have meta-analyzed various cohorts which were heterogenic in population and phenotyping.12 By using a single cohort with a standardized method for cIMT measurements and rigid quality control, our analyses likely suffer less from heterogeneity. There is no consensus on the optimal cIMT assessment in the literature with reporting of mean or maximum cIMT values most common, although it has been suggested measurement of maximum values may be more susceptible to sampling error.117,118 In the current study, we analyzed all cIMT values available in the UK Biobank, namely cIMTmin, cIMTmean, and cIMTmax, which may facilitate comparison with other literature. Second, we performed not only GWAS but also sought to further characterize and prioritize genes from loci identified in GWAS by performing follow-up analyses leveraging external datasets. This allowed us to gather additional evidence, also in specific arterial tissues, for multiple genetic loci and provide additional insights into other novel loci that were not identified by GWAS but maybe interesting candidate genes for cIMT. The combination of evidence from these multiple data sources warrants further study of these genes in future (experimental) studies. The current study also has limitations. First, our analyses were performed in mostly Europeans, which may limit the generalizability of our results to other ethnic groups. Second, TWAS analyses have intrinsic limitations due to their methodology. The method is based on the prediction of the genetically-regulated component of gene expression and does not consider other factors that influence the expression (environmental and technical components). Additionally, false-positive signals may have arisen due to co-regulation of gene expression and TWAS may suffer in power to detect true risk genes in tissues not related to the trait.119 We, therefore, presented TWAS results on 3 arterial tissues (aorta, coronary, and tibial) which are mechanistically most relevant to cIMT. The prediction of gene expression in TWAS is based on the external reference panel which has a limited size of available samples for several tissues. The models for coronary artery, aorta, and tibial artery are based on 200 to 600 samples, which limits the definitive power of expression prediction. To tackle this issue, we additionally applied a TWAS approach that utilizes a joint imputation of gene expression in multiple tissues.24
Conclusions
In conclusion, the present study characterized the genetic architecture underlying cIMT and identified 25 novel loci. We found sex differences in 7 cIMT loci. We prioritized genes that are potentially more relevant for cIMT through extensive-expression analyses using external datasets (Genotype-Tissue Expression and AtheroExpress). Gene set analyses provided additional evidence for candidate genes and highlighted biological pathways. Finally, we found shared genetic components between cIMT and various cardiovascular diseases and risk factors, namely CAD, PAD, SBP, and HDL-C. These findings warrant further investigation for the role of these genetic loci in the development and treatment options of atherosclerosis.
Article Information
Acknowledgments
We thank the CARDIoGRAMplusC4D (Coronary Artery Disease Genome-Wide Replication and Meta-Analysis Plus the Coronary Artery Disease Genetics), the GERA study (Genetic Epidemiology Research on Adult Health and Aging), GLGC (Global Lipids Genetics Consortium) and MEGASTROKE investigators for making their data publicly available. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html. We would like to thank the Centre for Information Technology of the University of Groningen for their support and for providing access to the Peregrine high-performance computing cluster. In addition, we thank Ruben N. Eppinga, MD, PhD, Tom Hendriks, MD, PhD, M. Yldau van der Ende, MD, PhD, Hilde E. Groot, MD, Jan Walter Benjamins, BEng, and Yanick Hagemeijer, MSc, University of Groningen, University Medical Center Groningen, Department of Cardiology, for their contributions to the extraction and processing of data in the UK Biobank. None of the mentioned contributors received compensation, except for their employment at the University Medical Center Groningen.
Sources of Funding
None.
Disclosures
N. Verweij is a paid consultant for Regeneron Pharmaceuticals. The other authors report no conflicts.
Supplemental Materials
Supplementary Methods
Tables S1–S17
Figures S1–S10
Major Resources Table
Supplementary Note
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AAA
- abdominal aortic aneurysm
- CAD
- coronary artery disease
- CARDIoGRAMplusC4D
- Coronary Artery Disease Genome-Wide Replication and Meta-Analysis Plus the Coronary Artery Disease Genetics
- CHARGE
- Cohorts for Heart and Aging Research in Genomic Epidemiology
- cIMT
- carotid artery intima-media thickness
- cIMTmax
- average of maximum cIMT
- cIMTmean
- average of mean cIMT
- cIMTmin
- average of minimum cIMT
- DBP
- diastolic blood pressure
- GERA
- Genetic Epidemiology Research on Adult Health and Aging
- GLGC
- Global Lipids Genetics Consortium
- GWAS
- genome-wide association study
- HDL-C
- high-density lipoprotein cholesterol
- LDL-C
- low-density lipoprotein cholesterol
- MAGMA
- Multi-Marker Analysis for Genomic Annotation
- MTAG
- Multi-Trait Analysis of GWAS
- PAD
- peripheral artery disease
- SBP
- systolic blood pressure
- SNP
- single nucleotide polymorphism
- TWAS
- transcriptome-wide association study
- VSMC
- vascular smooth muscle cell
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.121.317007.
For Sources of Funding and Disclosures, see page 497.
Contributor Information
Ming Wai Yeung, Email: m.w.yeung@umcg.nl.
Siqi Wang, Email: s.wang01@umcg.nl.
Yordi J. van de Vegte, Email: y.j.van.de.vegte@umcg.nl.
Oleg Borisov, Email: olegbor77@gmail.com.
Jessica van Setten, Email: j.vansetten@umcutrecht.nl.
Harold Snieder, Email: h.snieder@umcg.nl.
Niek Verweij, Email: n.verweij@umcg.nl.
M. Abdullah Said, Email: m.a.said@umcg.nl.
References
- 1.Gisterå A, Hansson GK. The immunology of atherosclerosis. Nat Rev Nephrol. 2017; 13:368–380. doi: 10.1038/nrneph.2017.51 [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Xian X, Wang Z, Bi Y, Chen Q, Han X, Tang D, Chen R. Research Progress on the Relationship between Atherosclerosis and Inflammation. Biomolecules. 2018; 8:E80. doi: 10.3390/biom8030080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014; 114:1852–1866. doi: 10.1161/CIRCRESAHA.114.302721 [DOI] [PubMed] [Google Scholar]
- 4.Palasubramaniam J, Wang X, Peter K. Myocardial infarction-from atherosclerosis to thrombosis. Arterioscler Thromb Vasc Biol. 2019; 39:e176–e185. doi: 10.1161/ATVBAHA.119.312578 [DOI] [PubMed] [Google Scholar]
- 5.Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, Tokgözoğlu L, Lewis EF. Atherosclerosis. Nat Rev Dis Primers. 2019; 5:56. doi: 10.1038/s41572-019-0106-z [DOI] [PubMed] [Google Scholar]
- 6.James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 Diseases and Injuries for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017 Lancet. 2018; 392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein JH, Korcarz CE, Hurst RT, Lonn E, Kendall CB, Mohler ER, Najjar SS, Rembold CM, Post WS; American Society of Echocardiography Carotid Intima-Media Thickness Task Force. Use of carotid ultrasound to identify subclinical vascular disease and evaluate cardiovascular disease risk: a consensus statement from the American Society of Echocardiography Carotid Intima-Media Thickness Task Force. Endorsed by the Society for Vascular Medicine. J Am Soc Echocardiogr. 2008; 21:93–111. quiz 189. doi: 10.1016/j.echo.2007.11.011 [DOI] [PubMed] [Google Scholar]
- 8.Bots ML, Hoes AW, Koudstaal PJ, Hofman A, Grobbee DE. Common carotid intima-media thickness and risk of stroke and myocardial infarction: the Rotterdam Study. Circulation. 1997; 96:1432–1437. doi: 10.1161/01.cir.96.5.1432 [DOI] [PubMed] [Google Scholar]
- 9.O’Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK, Jr. Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. Cardiovascular Health Study Collaborative Research Group. N Engl J Med. 1999; 340:14–22. doi: 10.1056/NEJM199901073400103 [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Cheema FA, Bremner JD, Goldberg J, Su S, Snieder H, Maisano C, Jones L, Javed F, Murrah N, et al. Heritability of carotid intima-media thickness: a twin study. Atherosclerosis. 2008; 197:814–820. doi: 10.1016/j.atherosclerosis.2007.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gertow K, Sennblad B, Strawbridge RJ, Ohrvik J, Zabaneh D, Shah S, Veglia F, Fava C, Kavousi M, McLachlan S, et al. Identification of the BCAR1-CFDP1-TMEM170A locus as a determinant of carotid intima-media thickness and coronary artery disease risk. Circ Cardiovasc Genet. 2012; 5:656–665. doi: 10.1161/CIRCGENETICS.112.963660 [DOI] [PubMed] [Google Scholar]
- 12.Franceschini N, Giambartolomei C, de Vries PS, Finan C, Bis JC, Huntley RP, Lovering RC, Tajuddin SM, Winkler TW, Graff M, et al. ; MEGASTROKE Consortium. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun. 2018; 9:1–14. doi: 10.1038/s41467-018-07340-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strawbridge RJ, Ward J, Bailey MES, Cullen B, Ferguson A, Graham N, Johnston KJA, Lyall LM, Pearsall R, Pell J, et al. Carotid Intima-Media Thickness: Novel Loci, Sex-Specific Effects, and Genetic Correlations With Obesity and Glucometabolic Traits in UK Biobank. Arterioscler Thromb Vasc Biol. 2020; 40:446–461. doi: 10.1161/ATVBAHA.119.313226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12:e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UK Biobank Imaging Modality. Carotid Ultrasound. 2015. Accessed December 23, 2020. https://biobank.ndph.ox.ac.uk/showcase/ukb/docs/carult_explan_doc.pdf
- 16.Coffey S, Lewandowski AJ, Garratt S, Meijer R, Lynum S, Bedi R, Paterson J, Yaqub M, Noble JA, Neubauer S, et al. Protocol and quality assurance for carotid imaging in 100,000 participants of UK Biobank: development and assessment. Eur J Prev Cardiol. 2017; 24:1799–1806. doi: 10.1177/2047487317732273 [DOI] [PubMed] [Google Scholar]
- 17.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018; 562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018; 50:906–908. doi: 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015; 4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benner C, Spencer CC, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016; 32:1493–1501. doi: 10.1093/bioinformatics/btw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Li C, Mou C, Dong Y, Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020; 12:1–8. doi: 10.1186/S13073-020-00803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Wedow R, Zacher M, Furlotte NA, et al. ; 23andMe Research Team; Social Science Genetic Association Consortium. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018; 50:229–237. doi: 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, Czajkowski J, Esko T, Fall T, Kilpeläinen TO, et al. ; CHARGE Consortium; DIAGRAM Consortium; GLGC Consortium; Global-BPGen Consortium; ICBP Consortium; MAGIC Consortium. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015; 11:e1005378. doi: 10.1371/journal.pgen.1005378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Li M, Lu Q, Weng H, Wang J, Zekavat SM, Yu Z, Li B, Gu J, Muchnik S, et al. ; Alzheimer’s Disease Genetics Consortium. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat Genet. 2019; 51:568–576. doi: 10.1038/s41588-019-0345-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, Mohammadi P, Park YS, Parsana P, Segrè AV, et al. Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015; 11:e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017; 8:1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015; 47:1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015; 47:1121–1130. doi: 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, et al. ; AFGen Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; INVENT Consortium; STARNET; BioBank Japan Cooperative Hospital Group; COMPASS Consortium; EPIC-CVD Consortium; EPIC-InterAct Consortium; International Stroke Genetics Consortium (ISGC); METASTROKE Consortium; Neurology Working Group of the CHARGE Consortium; NINDS Stroke Genetics Network (SiGN); UK Young Lacunar DNA Study; MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018; 50:524–537. doi: 10.1038/s41588-018-0058-329531354 [Google Scholar]
- 32.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017; 49:54–64. doi: 10.1038/ng.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. ; Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013; 45:1274–1283. doi: 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47(D1):D1005–D1012. doi: 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Harst P, Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018; 122:433–443. doi: 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. ; Million Veteran Program. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018; 50:1412–1425. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008; 40:161–169. doi: 10.1038/ng.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, et al. ; Wellcome Trust Case Control Consortium. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008; 371:483–491. doi: 10.1016/S0140-6736(08)60208-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009; 41:56–65. doi: 10.1038/ng.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, et al. ; Wellcome Trust Case Control Consortium. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010; 30:2264–2276. doi: 10.1161/ATVBAHA.109.201020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011; 7:e1001300. doi: 10.1371/journal.pgen.1001300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann TJ, Theusch E, Haldar T, Ranatunga DK, Jorgenson E, Medina MW, Kvale MN, Kwok PY, Schaefer C, Krauss RM, et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat Genet. 2018; 50:401–413. doi: 10.1038/s41588-018-0064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, et al. ; Global Lipids Genetics Consortium; Myocardial Infarction Genetics (MIGen) Consortium; Geisinger-Regeneron DiscovEHR Collaboration; VA Million Veteran Program. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018; 50:1514–1523. doi: 10.1038/s41588-018-0222-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy R, Boutin TS, Marten J, Huffman JE, Kerr SM, Campbell A, Evenden L, Gibson J, Amador C, Howard DM, et al. Exploration of haplotype research consortium imputation for genome-wide association studies in 20,032 Generation Scotland participants. Genome Med. 2017; 9:1. doi: 10.1186/s13073-017-0414-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA, Kanoni S, et al. ; CHARGE-EchoGen consortium; CHARGE-HF consortium; Wellcome Trust Case Control Consortium. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016; 48:1171–1184. doi: 10.1038/ng.3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dichgans M, Malik R, König IR, Rosand J, Clarke R, Gretarsdottir S, Thorleifsson G, Mitchell BD, Assimes TL, Levi C, et al. ; METASTROKE Consortium; CARDIoGRAM Consortium; C4D Consortium; International Stroke Genetics Consortium. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014; 45:24–36. doi: 10.1161/STROKEAHA.113.002707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. ; EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017; 49:1385–1391. doi: 10.1038/ng.3913 [DOI] [PubMed] [Google Scholar]
- 48.Low SK, Takahashi A, Cha PC, Zembutsu H, Kamatani N, Kubo M, Nakamura Y. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum Mol Genet. 2012; 21:2102–2110. doi: 10.1093/hmg/dds020 [DOI] [PubMed] [Google Scholar]
- 49.Matsukura M, Ozaki K, Takahashi A, Onouchi Y, Morizono T, Komai H, Shigematsu H, Kudo T, Inoue Y, Kimura H, et al. Genome-wide association study of peripheral arterial disease in a Japanese Population. PLoS One. 2015; 10:e0139262. doi: 10.1371/journal.pone.0139262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. ; International Consortium of Blood Pressure (ICBP) 1000G Analyses; BIOS Consortium; Lifelines Cohort Study; Understanding Society Scientific group; CHD Exome+ Consortium; ExomeBP Consortium; T2D-GENES Consortium; GoT2DGenes Consortium; Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; UK Biobank CardioMetabolic Consortium BP working group. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017; 49:403–415. doi: 10.1038/ng.376828135244 [Google Scholar]
- 51.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, et al. ; Understanding Society Scientific Group; International Consortium for Blood Pressure; Blood Pressure-International Consortium of Exome Chip Studies; Million Veteran Program. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019; 51:51–62. doi: 10.1038/s41588-018-0303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019; 104:65–75. doi: 10.1016/j.ajhg.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O’Reilly PF, Cabrera CP, Warren HR, Rose LM, et al. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 2017; 70:e4–e19. doi: 10.1161/HYPERTENSIONAHA.117.09438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. ; LifeLines Cohort Study; EchoGen consortium; AortaGen Consortium; CHARGE Consortium Heart Failure Working Group; KidneyGen consortium; CKDGen consortium; Cardiogenics consortium; CardioGram. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011; 43:1005–1012. doi: 10.1038/ng.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Leach IM, et al. ; BIOS-consortium; CARDIo GRAMplusCD; LifeLines Cohort Study; InterAct Consortium. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015; 47:1282–1293. doi: 10.1038/ng.3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018; 50:390–400. doi: 10.1038/s41588-018-0047-6 [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi F, Akiyama M, Matoba N, Katsuya T, Nakatochi M, Tabara Y, Narita A, Saw WY, Moon S, Spracklen CN, et al. ; International Genomics of Blood Pressure (iGEN-BP) Consortium. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat Commun. 2018; 9:1–16. doi: 10.1038/s41467-018-07345-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008; 358:1240–1249. doi: 10.1056/NEJMoa0706728 [DOI] [PubMed] [Google Scholar]
- 59.Ligthart S, Vaez A, Hsu YH, Stolk R, Uitterlinden AG, Hofman A, Alizadeh BZ, Franco OH, Dehghan A; Inflammation Working Group of the CHARGE Consortium; PMI-WG-XCP; LifeLines Cohort Study. Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genomics. 2016; 17:443. doi: 10.1186/s12864-016-2712-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung YJ, Winkler TW, de Las Fuentes L, Bentley AR, Brown MR, Kraja AT, Schwander K, Ntalla I, Guo X, Franceschini N, et al. ; CHARGE Neurology Working Group; COGENT-Kidney Consortium; GIANT Consortium; Lifelines Cohort Study. A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. Am J Hum Genet. 2018; 102:375–400. doi: 10.1016/j.ajhg.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kilpeläinen TO, Bentley AR, Noordam R, Sung YJ, Schwander K, Winkler TW, Jakupović H, Chasman DI, Manning A, Ntalla I, et al. ; Lifelines Cohort Study. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat Commun. 2019; 10:376. doi: 10.1038/s41467-018-08008-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease Nat Genet. 2011; 43:339–344. doi: 10.1038/ng.782 [DOI] [PubMed] [Google Scholar]
- 63.The CARDIoGRAMplusC4D Consortium. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015; 47:1121–1130. doi: 10.1038/NG.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bown MJ, Jones GT, Harrison SC, Wright BJ, Bumpstead S, Baas AF, Gretarsdottir S, Badger SA, Bradley DT, Burnand K, et al. ; CARDIoGRAM Consortium; Global BPgen Consortium; DIAGRAM Consortium; VRCNZ Consortium. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011; 89:619–627. doi: 10.1016/j.ajhg.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, Pezzini A, Thijs V, Markus HS, Dichgans M, et al. ; International Stroke Genetics Consortium; CADISP Group. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015; 47:78–83. doi: 10.1038/ng.3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van ’t Hof FNG, Ruigrok YM, Lee CH, Ripke S, Anderson G, de Andrade M, Baas AF, Blankensteijn JD, Böttinger EP, Bown MJ, et al. Shared genetic risk factors of intracranial, abdominal, and thoracic aneurysms. J Am Heart Assoc. 2016; 5:e002603. doi: 10.1161/JAHA.115.002603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LeMaire SA, McDonald ML, Guo DC, Russell L, Miller CC, 3rd, Johnson RJ, Bekheirnia MR, Franco LM, Nguyen M, Pyeritz RE, et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat Genet. 2011; 43:996–1000. doi: 10.1038/ng.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Surakka I, Whitfield JB, Perola M, Visscher PM, Montgomery GW, Falchi M, Willemsen G, de Geus EJ, Magnusson PK, Christensen K, et al. ; GenomEUtwin Project. A genome-wide association study of monozygotic twin-pairs suggests a locus related to variability of serum high-density lipoprotein cholesterol. Twin Res Hum Genet. 2012; 15:691–699. doi: 10.1017/thg.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saxena R, Voight BF, Lyssenko V, Burtt NPNP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for Type 2 diabetes and triglyceride levels. Science. 2007; 316:1331–1336. doi: 10.1126/SCIENCE.1142358 [DOI] [PubMed] [Google Scholar]
- 70.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008; 40:189–197. doi: 10.1038/ng.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burkhardt R, Kenny EE, Lowe JK, Birkeland A, Josowitz R, Noel M, Salit J, Maller JB, Pe’er I, Daly MJ, et al. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler Thromb Vasc Biol. 2008; 28:2078–2084. doi: 10.1161/ATVBAHA.108.172288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010; 466:707–713. doi: 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, Thompson WK, Ritchie MD, Kho AN, Muthalagu A, Hayes MG, Armstrong LL, Scheftner DA, et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin Transl Sci. 2012; 5:394–399. doi: 10.1111/j.1752-8062.2012.00446.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Surakka I, Horikoshi M, Mägi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, et al. ; ENGAGE Consortium. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015; 47:589–597. doi: 10.1038/ng.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Southam L, Gilly A, Süveges D, Farmaki AE, Schwartzentruber J, Tachmazidou I, Matchan A, Rayner NW, Tsafantakis E, Karaleftheri M, et al. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat Commun. 2017; 8:15606. doi: 10.1038/ncomms15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spracklen CN, Chen P, Kim YJ, Wang X, Cai H, Li S, Long J, Wu Y, Wang YX, Takeuchi F, et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum Mol Genet. 2017; 26:1770–1784. doi: 10.1093/hmg/ddx062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu Y, Zhang D, Zhou D, Li Z, Li Z, Fang L, Yang M, Shan Z, Li H, Chen J, et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: a multi-stage genome-wide association study. J Cell Mol Med. 2017; 21:1106–1116. doi: 10.1111/jcmm.13042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, Lu Y, Yoon K, Jang HM, Kim YK, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019; 9:1–11. doi: 10.1038/s41598-018-37832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015; 16:113. doi: 10.1186/s13059-015-0676-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koo BH, Coe DM, Dixon LJ, Somerville RP, Nelson CM, Wang LW, Young ME, Lindner DJ, Apte SS. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol. 2010; 176:1494–1504. doi: 10.2353/ajpath.2010.090655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Polfus LM, Smith JA, Shimmin LC, Bielak LF, Morrison AC, Kardia SL, Peyser PA, Hixson JE. Genome-wide association study of gene by smoking interactions in coronary artery calcification. PLoS One. 2013; 8:e74642. doi: 10.1371/journal.pone.0074642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei M, Pan H, Guo K. Association between plasma ADAMTS-9 levels and severity of coronary artery disease. Angiology. 2021; 72:371–380. doi: 10.1177/0003319720979238 [DOI] [PubMed] [Google Scholar]
- 83.Llano E, Pendás AM, Freije JP, Nakano A, Knäuper V, Murphy G, López-Otin C. Identification and characterization of human MT5-MMP, a new membrane-bound activator of progelatinase a overexpressed in brain tumors. Cancer Res. 1999; 59:2570–2576 [PubMed] [Google Scholar]
- 84.Raines EW. The extracellular matrix can regulate vascular cell migration, proliferation, and survival: relationships to vascular disease. Int J Exp Pathol. 2000; 81:173–182. doi: 10.1046/j.1365-2613.2000.00155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galis ZS, Khatri JJ. Matrix metalloproteinases in vascular remodeling and atherogenesis: the good, the bad, and the ugly. Circ Res. 2002; 90:251–262. doi:10.1161/res.90.3.251 [PubMed] [Google Scholar]
- 86.Segura AM, Luna RE, Horiba K, Stetler-Stevenson WG, McAllister HA, Jr, Willerson JT, Ferrans VJ. Immunohistochemistry of matrix metalloproteinases and their inhibitors in thoracic aortic aneurysms and aortic valves of patients with Marfan’s syndrome. Circulation. 1998; 98(19 suppl):II331–7. discussion II337 [PubMed] [Google Scholar]
- 87.Chung AW, Au Yeung K, Sandor GG, Judge DP, Dietz HC, van Breemen C. Loss of elastic fiber integrity and reduction of vascular smooth muscle contraction resulting from the upregulated activities of matrix metalloproteinase-2 and -9 in the thoracic aortic aneurysm in Marfan syndrome. Circ Res. 2007; 101:512–522. doi: 10.1161/CIRCRESAHA.107.157776 [DOI] [PubMed] [Google Scholar]
- 88.Xiong W, Meisinger T, Knispel R, Worth JM, Baxter BT. MMP-2 regulates Erk1/2 phosphorylation and aortic dilatation in Marfan syndrome. Circ Res. 2012; 110:e92–e101. doi: 10.1161/CIRCRESAHA.112.268268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evrard SM, Lecce L, Michelis KC, Nomura-Kitabayashi A, Pandey G, Purushothaman KR, d’Escamard V, Li JR, Hadri L, Fujitani K, et al. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat Commun. 2016; 7:11853. doi: 10.1038/ncomms11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sahu SK, Tiwari N, Pataskar A, Zhuang Y, Borisova M, Diken M, Strand S, Beli P, Tiwari VK. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat Commun. 2017; 8:1523. doi: 10.1038/s41467-017-01366-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Šućurović S, Nikolić T, Brosens JJ, Mulac-Jeričević B. Analysis of heart and neural crest derivatives-expressed protein 2 (HAND2)-progesterone interactions in peri-implantation endometrium†. Biol Reprod. 2020; 102:1111–1121. doi: 10.1093/biolre/ioaa013 [DOI] [PubMed] [Google Scholar]
- 92.Yamagishi H, Olson EN, Srivastava D. The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J Clin Invest. 2000; 105:261–270. doi: 10.1172/JCI8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu H, Xu YJ, Li RG, Wang ZS, Zhang M, Qu XK, Qiao Q, Li XM, Di RM, Qiu XB, et al. HAND2 loss-of-function mutation causes familial dilated cardiomyopathy. Eur J Med Genet. 2019; 62:103540. doi: 10.1016/j.ejmg.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 94.Sun YM, Wang J, Qiu XB, Yuan F, Li RG, Xu YJ, Qu XK, Shi HY, Hou XM, Huang RT, et al. A HAND2 loss-of-function mutation causes familial ventricular septal defect and pulmonary stenosis. G3 (Bethesda). 2016; 6:987–992. doi: 10.1534/g3.115.026518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boucher P, Li WP, Matz RL, Takayama Y, Auwerx J, Anderson RG, Herz J. LRP1 functions as an atheroprotective integrator of TGFbeta and PDFG signals in the vascular wall: implications for Marfan syndrome. PLoS One. 2007; 2:e448. doi: 10.1371/journal.pone.0000448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strickland DK, Au DT, Cunfer P, Muratoglu SC. Low-density lipoprotein receptor-related protein-1: role in the regulation of vascular integrity. Arterioscler Thromb Vasc Biol. 2014; 34:487–498. doi: 10.1161/ATVBAHA.113.301924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mahmoud AD, Ballantyne MD, Miscianinov V, Pinel K, Hung J, Scanlon JP, Iyinikkel J, Kaczynski J, Tavares AS, Bradshaw AC, et al. The human-specific and smooth muscle cell-enriched LncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circ Res. 2019; 125:535–551. doi: 10.1161/CIRCRESAHA.119.314876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ivanov D, Philippova M, Allenspach R, Erne P, Resink T. T-cadherin upregulation correlates with cell-cycle progression and promotes proliferation of vascular cells. Cardiovasc Res. 2004; 64:132–143. doi: 10.1016/j.cardiores.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 99.Frismantiene A, Dasen B, Pfaff D, Erne P, Resink TJ, Philippova M. T-cadherin promotes vascular smooth muscle cell dedifferentiation via a GSK3β-inactivation dependent mechanism. Cell Signal. 2016; 28:516–530. doi: 10.1016/j.cellsig.2016.02.014 [DOI] [PubMed] [Google Scholar]
- 100.Essen M von, Rahikainen R, Oksala N, Raitoharju E, Seppälä I, Mennander A, Sioris T, Kholová I, Klopp N, Illig T, et al. Talin and vinculin are downregulated in atherosclerotic plaque; Tampere Vascular Study. Atherosclerosis. 2016; 255:43–53. doi: 10.1016/J.ATHEROSCLEROSIS.2016.10.031 [DOI] [PubMed] [Google Scholar]
- 101.Depuydt MAC, Prange KHM, Slenders L, Örd T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res. 2020; 127:1437–1455. doi: 10.1161/CIRCRESAHA.120.316770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun Z, Tseng HY, Tan S, Senger F, Kurzawa L, Dedden D, Mizuno N, Wasik AA, Thery M, Dunn AR, et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat Cell Biol. 2016; 18:941–953. doi: 10.1038/ncb3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shadrina AS, Shashkova TI, Torgasheva AA, Sharapov SZ, Klarić L, Pakhomov ED, Alexeev DG, Wilson JF, Tsepilov YA, Joshi PK, et al. Prioritization of causal genes for coronary artery disease based on cumulative evidence from experimental and in silico studies. Sci Rep. 2020; 10:10486. doi: 10.1038/s41598-020-67001-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamamoto H, Kokame K, Okuda T, Nakajo Y, Yanamoto H, Miyata T. NDRG4 protein-deficient mice exhibit spatial learning deficits and vulnerabilities to cerebral ischemia. J Biol Chem. 2011; 286:26158–26165. doi: 10.1074/jbc.M111.256446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qu X, Jia H, Garrity DM, Tompkins K, Batts L, Appel B, Zhong TP, Baldwin HS. Ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev Biol. 2008; 317:486–496. doi: 10.1016/j.ydbio.2008.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nishimoto S, Tawara J, Toyoda H, Kitamura K, Komurasaki T. A novel homocysteine-responsive gene, smap8, modulates mitogenesis in rat vascular smooth muscle cells. Eur J Biochem. 2003; 270:2521–2531. doi: 10.1046/j.1432-1033.2003.03626.x [DOI] [PubMed] [Google Scholar]
- 107.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001; 105:851–862. doi: 10.1016/s0092-8674(01)00404-4 [DOI] [PubMed] [Google Scholar]
- 108.Ackers-Johnson M, Talasila A, Sage AP, Long X, Bot I, Morrell NW, Bennett MR, Miano JM, Sinha S. Myocardin regulates vascular smooth muscle cell inflammatory activation and disease. Arterioscler Thromb Vasc Biol. 2015; 35:817–828. doi: 10.1161/ATVBAHA.114.305218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hinohara K, Nakajima T, Yasunami M, Houda S, Sasaoka T, Yamamoto K, Lee BS, Shibata H, Tanaka-Takahashi Y, Takahashi M, et al. Megakaryoblastic leukemia factor-1 gene in the susceptibility to coronary artery disease. Hum Genet. 2009; 126:539–547. doi: 10.1007/s00439-009-0698-6 [DOI] [PubMed] [Google Scholar]
- 110.An J, Naruse TK, Hinohara K, Soejima Y, Sawabe M, Nakagawa Y, Kuwahara K, Kimura A. MRTF-A regulates proliferation and survival properties of pro-atherogenic macrophages. J Mol Cell Cardiol. 2019; 133:26–35. doi: 10.1016/j.yjmcc.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 111.Sandoo A, van Zanten JJ, Metsios GS, Carroll D, Kitas GD. The endothelium and its role in regulating vascular tone. Open Cardiovasc Med J. 2010; 4:302–312. doi: 10.2174/1874192401004010302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bai X, Lenhart KC, Bird KE, Suen AA, Rojas M, Kakoki M, Li F, Smithies O, Mack CP, Taylor JM. The smooth muscle-selective RhoGAP GRAF3 is a critical regulator of vascular tone and hypertension. Nat Commun. 2013; 4:2910. doi: 10.1038/ncomms3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng PF, Yin RX, Deng GX, Guan YZ, Wei BL, Liu CX. Association between the XKR6 rs7819412 SNP and serum lipid levels and the risk of coronary artery disease and ischemic stroke. BMC Cardiovasc Disord. 2019; 19:202. doi: 10.1186/s12872-019-1179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Páez MT, Yamamoto T, Hayashi K, Yasuda T, Harada N, Matsumoto N, Kurosawa K, Furutani Y, Asakawa S, Shimizu N, et al. Two patients with atypical interstitial deletions of 8p23.1: mapping of phenotypical traits. Am J Med Genet A. 2008; 146A:1158–1165. doi: 10.1002/ajmg.a.32205 [DOI] [PubMed] [Google Scholar]
- 115.Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009; 149A:1661–1677. doi: 10.1002/ajmg.a.32896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barber JC, Rosenfeld JA, Graham JM, Kramer N, Lachlan KL, Bateman MS, Collinson MN, Stadheim BF, Turner CL, Gauthier JN, et al. Inside the 8p23.1 duplication syndrome; eight microduplications of likely or uncertain clinical significance. Am J Med Genet A. 2015; 167A:2052–2064. doi: 10.1002/ajmg.a.37120 [DOI] [PubMed] [Google Scholar]
- 117.Touboul PJ, Hennerici MG, Meairs S, Adams H, Amarenco P, Bornstein N, Csiba L, Desvarieux M, Ebrahim S, Hernandez Hernandez R, et al. Mannheim carotid intima-media thickness and plaque consensus (2004-2006-2011). An update on behalf of the advisory board of the 3rd, 4th and 5th watching the risk symposia, at the 13th, 15th and 20th European Stroke Conferences, Mannheim, Germany, 2004, Brussels, Belgium, 2006, and Hamburg, Germany, 2011. Cerebrovasc Dis. 2012; 34:290–296. doi: 10.1159/000343145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simova I. Intima-media thickness: appropriate evaluation and proper measurement. E-Journal of Cardiology Practice. Published May 5, 2015. Accessed June 3, 2021. https://www.escardio.org/Journals/E-Journal-of-Cardiology-Practice/Volume-13/Intima-media-thickness-Appropriate-evaluation-and-proper-measurement-described
- 119.Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019; 51:592–599. doi: 10.1038/s41588-019-0385-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fry D, Almond R, Moffat S, Gordon M, Singh P. UK Biobank Biomarker Project. Companion Document to Accompany Serum Biomarker Data. Accessed August 5, 2021. http://www.ukbiobank.ac.uk/uk-biobankbiomarker-panel/
- 121.UK Biobank Blood Pressure. Accessed July 31, 2021. https://biobank.ndph.ox.ac.uk/showcase/ukb/docs/Bloodpressure.pdf.
- 122.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016; 48:481–487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 123.Ma WF, Hodonsky CJ, Turner AW, Wong D, Song Y, Mosquera JV, Ligay AV, Slenders L, Gancayco C, Pan H, et al. Enhanced single-cell RNA-seq workflow reveals coronary artery disease cellular cross-talk and candidate drug targets. Atherosclerosis. 2022; 340:12–22. doi:10.1016/J.ATHEROSCLEROSIS.2021.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, et al. Biological interpretation of genomewide association studies using predicted gene functions. Nat Commun. 2015; 6:5890. doi: 10.1038/ncomms6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.