Abstract
To investigate the occurrence of multinucleoside analog resistance during therapy failure, we surveyed the drug susceptibilities and genotypes of nearly 900 human immunodeficiency virus type 1 (HIV-1) samples. For 302 of these, the 50% inhibitory concentrations of at least four of the approved nucleoside analogs had fourfold-or-greater increases. Genotypic analysis of the reverse transcriptase (RT)-coding regions from these samples revealed complex mutational patterns, including the previously recognized codon 151 multidrug resistance cluster. Surprisingly, high-level multinucleoside resistance was associated with a diverse family of amino acid insertions in addition to “conventional” point mutations. These insertions were found between RT codons 67 and 70 and were commonly 69Ser-(Ser-Ser) or 69Ser-(Ser-Gly). Treatment history information showed that a common factor for the development of these variants was AZT (3′-azido-3′-deoxythymidine, zidovudine) therapy in combination with 2′,3′-dideoxyinosine or 2′,3′-dideoxycytidine, although treatment patterns varied considerably. Site-directed mutagenesis studies confirmed that 69Ser-(Ser-Ser) in an AZT resistance mutational background conferred simultaneous resistance to multiple nucleoside analogs. The insertions are located in the “fingers” domain of RT. Modelling the 69Ser-(Ser-Ser) insertion into the RT structure demonstrated the profound direct effect that this change is likely to have in the nucleoside triphosphate binding site of the enzyme. Our data highlight the increasing problem of HIV-1 multidrug resistance and underline the importance of continued resistance surveillance with appropriate, sufficiently versatile genotyping technology and phenotypic drug susceptibility analysis.
Despite the recent success of antiretroviral combination therapy (30), the development of human immunodeficiency virus type 1 (HIV-1) drug resistance during therapy remains a major cause of antiretroviral treatment failure (4, 21). Mechanisms of resistance to HIV-1 reverse transcriptase (RT) inhibitors, both nucleoside analogs and nonnucleoside RT inhibitors, in addition to protease inhibitors have been well documented (32). Such descriptions have largely focussed on resistance to single inhibitors and the potential cross-resistance to other inhibitors of the same class. A number of genetic changes in RT are known to confer discreet resistance to specific nucleoside analogs. For example, a set of six mutations, at codons 41, 67, 70, 210, 215, and 219, result in resistance to AZT (3′-azido-3′-deoxythymidine; zidovudine) but not to other nucleoside analogs (8, 11, 16, 23). Likewise, a single mutation at codon 74 confers resistance to ddI (2′,3′-dideoxyinosine; didanosine) and ddC (2′,3′-dideoxycytidine; zalcitabine) but not to AZT (35). Interestingly, little information has been available regarding the genetic basis of d4T (2′,3′-didehydro-3′-deoxythymidine; stavudine) resistance (22, 27).
The outgrowth of HIV-1 resistant to multiple inhibitors commonly occurs in situations where frequent therapy changes have been made and a patient’s viral load has not been completely suppressed (10). This cross-resistance can be a result of the buildup of multiple, discreet resistance mutations in the same genome. For example, it has been shown that extensive cross-resistance to the current protease inhibitors occurs following the sequential acquisition of multiple mutations in the protease-coding region (3). Less information is available regarding mechanisms of broad nucleoside analog cross-resistance. Early in vitro selection studies demonstrated that AZT-ddI coresistance could occur by the combination of AZT and ddI mutations on the same genome (24). In certain circumstances, the Met184Val mutation, which confers high-level 3TC (β-l-2′,3′-dideoxy-3′-thiacytidine; lamivudine) resistance can cause resensitization of AZT resistance (2, 25, 37). Recent studies have shown that AZT-3TC coresistance can occur during extensive combination therapy (18, 28, 29). This involves the common AZT and 3TC resistance mutations in addition to other amino acid polymorphisms in RT (18, 29).
It has become apparent that multiple nucleoside resistance (MNR) can occur via a mutational pathway completely independent from the “classic” resistance mutations (13, 33). Thus, a cluster of five mutations in the RT-coding region (resulting in substitutions Ala62Val, Val75Ile, Phe77Leu, Phe116Tyr, and Gln151Met) that confer simultaneous resistance to AZT, ddI, ddC, and d4T, with minimal resistance to 3TC, have been described recently. Examination of therapy histories has indicated that the combination of AZT with ddI or ddC seems to precipitate the development of these MNR variants (13). The pivotal mutation in this cluster results in the Gln151Met substitution, as this is the first to appear and can independently confer a degree of multiple resistance (13, 33). To date, population studies have indicated that the development of the 151Met MNR complex is relatively rare, with estimates of its incidence at around 1 to 2% (13, 33).
The present study was designed to investigate the occurrence of HIV-1 MNR in a relatively large number of clinical samples and to determine the genetic basis of this resistance. In our resistance database, we surveyed 892 HIV-1 samples from patients failing therapy by using a standardized recombinant-based phenotypic assay and DNA sequence analysis. MNR was associated with complex mutational patterns in the RT-coding region. Surprisingly, this group of MNR variants included a family of 45 samples with one, two, or three amino acid insertions between codons 67 and 70 of RT. We have confirmed the significance of these insertions (with and without other mutations) by phenotypic analysis of variants constructed by site-directed mutagenesis. Furthermore, a modelling study based on a newly reported RT structure (12) showed that these insertions are likely to have direct effects on nucleoside triphosphate binding in the active site of the enzyme.
MATERIALS AND METHODS
Source of plasma samples.
Virco is a virology diagnostic company with laboratories in the United Kingdom and Belgium that test patient samples to assist practitioners in the management of viral diseases. The plasma samples described in this study came mostly from the United States with a minority (<10%) from European countries. Because of requirements for anonymity and the nature of our services, comprehensive therapy and clinical histories were not available for most of the patients. The majority (>90%) of samples in this study were from different patients. Samples were selected for the study if they had a viral load of greater than 1,000 HIV-1 RNA copies/ml; for the purposes of this study, patients with this level of plasma HIV-1 were considered to be failing therapy.
HIV-1 RNA extraction and amplification of pol gene coding regions.
Viral RNA was extracted from 200-μl samples of patient plasma with the QIAamp viral RNA extraction kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. cDNA encompassing part of the pol gene was produced by using Expand RT (Boehringer Mannheim) as described previously (9). A 2.2-kb fragment encoding the protease and RT regions was then amplified by nested PCR by using previously described primers and conditions (9). This genetic material was subsequently used in both phenotyping and genotyping experiments.
Generation of HIV-1 recombinants and inhibitor susceptibility assay.
MT-4 cells (7) were cotransfected with pol gene PCR fragments and the protease- and RT-deleted HIV-1 molecular clone pGEM3ΔPRT, as described (9). This resulted in viable recombinant viruses containing protease- and RT-coding regions from the donor PCR fragment. Phenotypic susceptibility to nucleoside analogs was determined by using an MT-4 cell viral cytopathic effect protection assay as described (9). Fold resistance values were derived by dividing the mean 50% inhibitory concentration (IC50) for a patient’s recombinant virus by the mean IC50 for wild-type control virus (strain HXB2-D).
DNA sequence analysis of HIV-1 RT from patient samples.
The PCR products obtained from patient plasma samples were genotyped by dideoxynucleotide-based sequence analysis. Samples were sequenced using the Big Dye terminator kit (Applied Biosystems) and resolved on an ABI 377 DNA sequencer as described (23, 26).
Site-directed mutagenesis of HIV-1 RT.
Mutations in the RT-coding region were created by site-directed mutagenesis of a wild-type HXB2-D EcoRI-PstI restriction enzyme fragment, encompassing the HIV-1 pol gene and cloned into pGEM3 (Promega). Single and multiple nucleotide changes were introduced into RT with the ExSite mutagenesis kit (Strategene). All mutant clones were verified by DNA sequence analysis of the entire RT. PCR fragments were prepared from the mutated clones, and the altered RT-coding regions were transferred into the HIV-1 HXB2-D genetic background by homologous recombination as described above. The susceptibility of these recombinant viruses to nucleoside analogs was subsequently determined by the MT-4 cell cytopathic effect protection assay (9).
Modelling of mutant HIV-1 RTs and the binding of template primer.
Modelling of RT mutations into the p66 subunit of the ternary RT-template primer-dTTP complex (12) was carried out by using the FRODO (14) and O (15) software programs. The Met41Leu, Thr69Ser, Leu210Trp, and Thr215Tyr substitutions were made, together with the insertion of Ser-Ser after residue 69. Following optimization of the stereochemistry, the conformation of the β2-β3 loop was selected from the library of related structures available within O.
RESULTS
Phenotypic susceptibility analysis.
Plasma samples (n = 892) were obtained from patients who had received therapy with various combinations of nucleoside analogs. These patients were defined as failing therapy if a plasma HIV-1 viral load of greater than 1,000 RNA copies/ml was detected. The Antivirogram recombinant virus assay (9) was used to simultaneously determine the susceptibilities of these samples to AZT, 3TC, d4T, ddI, and ddC. From this analysis, we identified 302 samples for which increases in IC50 (relative to a wild-type control virus) to at least four of these inhibitors were fourfold or greater. Thus, a substantial number of MNR viruses were present in the sample population.
Genotypic analysis of MNR samples.
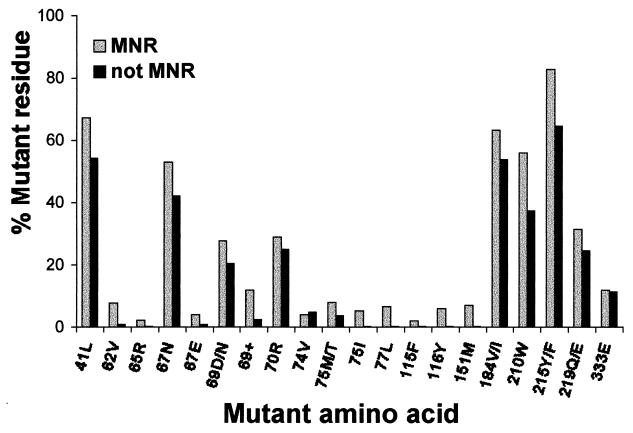
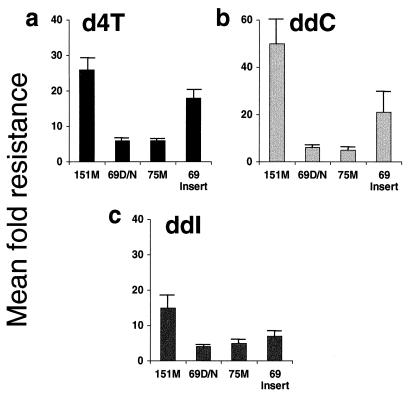
Genotypic analysis was performed on all 892 samples by dideoxynucleotide sequencing (see Materials and Methods). Complex patterns of multiple mutations were seen in the RT-coding regions of the MNR samples. These included combinations of AZT and 3TC resistance mutations (particularly 41Leu, 67Asn, 210Trp, and 215Tyr with 184Val/Ile) plus mutations at codon 69 (Thr69Asp/Asn) and/or codon 75 (Val75Met). A comparison of the incidence of specific RT mutations in MNR versus non-MNR samples in the population surveyed is shown in Fig. 1. This analysis highlighted the incidence of the codon 151 mutational cluster in the MNR group. In addition, a novel family of amino acid insertions and rearrangements between codons 67 and 70 was also prevalent in the MNR group. These two patterns of mutations were associated with high-level phenotypic MNR (Fig. 2); 27 samples had the codon 151 cluster and 45 samples had insertions and rearrangements (typically a Thr69Ser substitution followed by insertion of two amino acids). The mean fold increases in d4T, ddI, and ddC IC50s for these different groups are shown in Fig. 2. This analysis indicated that codon 69 insertion mutants had a high increase in d4T and ddC resistances (>10-fold), which was also seen with the codon 151 cluster. However, samples with AZT and 3TC resistance mutations plus Thr69Asp/Asn or Val75Met substitutions in RT showed only modest levels of resistance to these drugs (Fig. 2). Not surprisingly, all four groups shown in Fig. 2 were highly resistant to AZT and 3TC (mean fold increases of greater than 500-fold for AZT IC50 and greater than 30-fold for 3TC IC50). This was because many MNR samples contained mutations conferring AZT resistance (e.g., 41Leu, 67Asn, 210Trp, and 215Tyr) and 3TC resistance (Met184Val/Ile).
FIG. 1.
Frequency of mutations in RT from MNR and non-MNR clinical samples. The light grey bars represent the overall frequency of RT mutations found in HIV-1 samples for which the IC50s of at least four of the nucleoside analogs tested showed a fourfold increase. The dark grey bars show the mutation frequency of isolates that were not cross resistant to four or more of the nucleosides. The individual mutations analyzed in this population of 892 samples are indicated in the single-letter amino acid code. 69+ indicates amino acid insertions at codon 69. Rarely, codon 219 was found to be Asn or Arg. These variants were included in the analysis of mutant codon 219 residues.
FIG. 2.
Nucleoside analog susceptibility of MNR patient-derived recombinant HIV-1 variants. Recombinant viruses were produced from samples of patient plasma as described in Materials and Methods and were tested for susceptibility to d4T (a), ddC (b), and ddI (c). The mean fold increase in IC50 (Mean fold resistance) relative to wild-type controls are shown for groups of viruses with different genotypes, i.e., the codon 151Met multidrug resistance cluster (151M) (n = 27), viruses with 69Asp/Asn (69D/N) (n = 195) or 75Met (75M) (n = 43) in a background of AZT and 3TC resistance mutations, and codon 69 insertion mutants (69 Insert) (n = 45) in a background of AZT resistance mutations. Error bars indicate standard errors. Note that the total number of samples (n = 310) is higher than the 302 MNR samples described because a small minority were 69Asp/Asn and 75Met double mutants and are represented in both groups.
Spectrum of different insertions seen between RT codons 67 and 70.
The extensive variety of insertions between codons 67 and 70 of RT is summarized in Table 1. The largest group (n = 16) had a Thr69Ser substitution followed by an insertion of two Ser residues. The next largest group (n = 10) also had a Thr69Ser substitution but was followed by an insertion of Ser-Gly. Samples with a number of different double amino acids inserted after 69Ser were also identified. In addition, insertions of two or three amino acids between codons 68 and 69 were also seen. The positions of these insertions were based on the fact that Thr69 and Lys70 were contiguous. In some samples, there were rarely observed substitutions at codon 67 (Asp67Glu/Ser/Gly), rather than the common 67Asn AZT resistance mutation. In two samples, deletion of codon 70 was observed (after insertion of three residues between codons 68 and 69), and a single Thr69Ser substitution without an insertion was seen in four samples (Table 1). The inserted residues did not show any obvious patterns in terms of codon usage. For example, the Ser-Ser insertions were rarely direct repeats of the Ser69 codon, suggesting that simple reiterations of the Ser69 codon could not account for the appearance of these insertions in the RT (data not shown).
TABLE 1.
Spectrum of genetic heterogeneity seen between RT codons 67 and 70 in clinical samples containing amino acid insertions
| Amino acid sequencea | No. of isolates containing the sequence | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| D67 | S68 | X | X | X | T69 | X | X | K70 | |
| D | S | S | — | — | K/R | 4 | |||
| G | Y | T | D | — | R | 1 | |||
| D/E | S | S | S | S | K/R | 16 | |||
| D/E | S/N | S | S | G | K | 10 | |||
| D/E | S | S | E | E | K | 3 | |||
| D/E | S/N | S | S | A | K | 3 | |||
| D/E | S | S | V | G | K/R | 2 | |||
| D | S | S | A | G | K | 1 | |||
| D | S | A | S | G | K | 1 | |||
| D/E | S | S | V | — | T | K | 2 | ||
| D | S | S | T | — | T | K | 1 | ||
| D/E | S | S | S | — | T | K | 3 | ||
| S | S | S | S | G | T/A | — | 1 | ||
| G | S | G | G | G | T | — | 1 | ||
Wild-type amino acid residues are shown in the main headings and amino acid insertions are marked as an X (boldface). Samples having only the substitution of Thr69Ser are also shown. Alternative amino acid residues are shown at various positions for different samples, or where mixed nucleotides were detected within a single sample. —, positions where no amino acid was seen.
Patient therapy patterns in relation to codon 69 insertions.
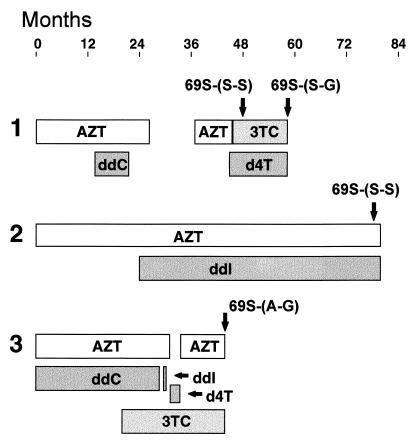
The codon 69 insertions were always present in a background of AZT resistance mutations, especially Thr215Tyr/Phe. This may not be surprising as therapy histories from many of the patients whose samples were analyzed in this study revealed a common pattern of AZT therapy, followed by combination therapy with nucleosides and protease inhibitors (data not shown). Figure 3 shows typical treatment patterns for three patients, indicating that the time samples were obtained for virological analysis. It was not possible from these histories to determine precisely the nucleoside analog(s) responsible for selecting codon 69 insertions. Sequential samples from patient 1 revealed an interesting transition of 69Ser-(Ser-Ser) to 69Ser-(Ser-Gly) during a period of 3TC-d4T combination therapy.
FIG. 3.
Therapy histories of three patients whose HIV-1 isolates developed codon 69 insertions. Nucleoside analog therapies (AZT, 3TC, ddC, ddI, or d4T) are shown as horizontal bars, indicating the time period in which each patient (1, 2, or 3) received a particular treatment. The time point at which plasma samples were obtained for genotypic and phenotypic analyses are shown by the arrows together with the specific codon 69 insertion detected. Other therapies which these patients may have been receiving are not indicated on this figure.
Susceptibility analysis of HIV-1 variants constructed by site-directed mutagenesis.
To investigate the significance of the observed mutational patterns associated with MNR virus, we constructed a series of viruses by site-directed mutagenesis with specific changes in a defined genetic background (HXB2-D). Thr69Asp or Val75Met in a background of AZT mutations conferred little or no resistance to 3TC, d4T, ddI, or ddC (data not shown). Variants were also constructed with 69Ser-(Ser-Ser), either alone or together with two AZT resistance mutations (210Trp and 215Tyr). In addition, the potential role of Ala62Val, a substitution also frequently associated with the insertions, was investigated by adding this mutation to a background of 69Ser-(Ser-Ser) plus 210Trp/215Tyr. Susceptibility data for six nucleoside analogs are summarized in Table 2. These data showed that the 69Ser-(Ser-Ser) insertion alone did not confer MNR. In fact, this virus had a significant decrease in susceptibility only to 3TC. By contrast, the variants with the insert plus AZT resistance mutations had decreased susceptibility to AZT, 3TC, d4T, ddC, and abacavir (4-[(2-amino-6-cyclopropyl-amino)-9H-purin-9-yl]-2-cyclopentene-1-metha-nol, 1592U89), confirming that the codon 69 insertions plus AZT mutations conferred the MNR phenotype.
TABLE 2.
Nucleoside analog susceptibility of HIV-1 variants constructed by site-directed mutagenesisa
| Mutation(s) | Fold increase in IC50 of:
|
|||||
|---|---|---|---|---|---|---|
| d4T | AZT | 3TC | 1592 | ddI | ddC | |
| 69S-[S-S] | 2.3 | 2.2 | 6.2 | 2.6 | 1.7 | 2.1 |
| 210W, 215Y | 1.1 | 10 | 3.8 | 1.5 | 1.2 | 1.2 |
| 210W, 215Y, 62V | 0.7 | 8 | 1.3 | 0.7 | 0.5 | 0.8 |
| 69S-[S-S], 210W, 215Y | 4.8 | 220 | 20 | 4.6 | 2.1 | 4.2 |
| 69S-[S-S], 210W, 215Y, 62V | 5.2 | >2,500 | 15 | 5.4 | 1.8 | 2.7 |
Mutations were created in HIV-1 RT by site-directed mutagenesis, and the mutant RT was transferred into the HXB2-D wild-type virus background. The specific mutations created in this background are indicated. The nucleoside susceptibility of mutant viruses (fold increase in IC50 relative to wild-type HXB2-D virus control) was assessed as described in Materials and Methods. These data represent the mean values of two independent determinations. The mean IC50 for the wild-type control virus in these experiments were as follows: AZT, 0.04 μM; d4T, 2.39 μM; 3TC, 1.67 μM; ddI, 4.6 μM; ddC, 1.41 μM; and 1592U89, 2.9 μM.
Structural implications of insertion mutants in RT.
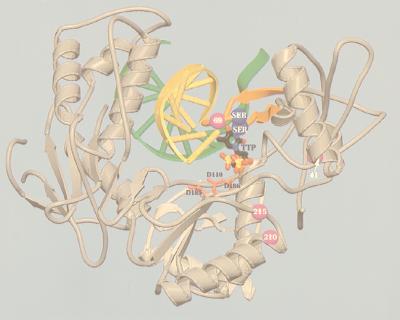
Recent insights into the location of mutant residues in the HIV-1 RT structure (12) have enabled us to examine the structural implications of amino acid insertions at codon 69. Amino acid residues 67 to 70 are located in a loop connecting antiparallel β strands 2 and 3 in the fingers domain of the enzyme (12). Unlike previous models, the recently published RT structure shows that this loop is in close proximity to the deoxynucleotide triphosphate (dNTP) binding site (12). Using this information, we have produced a structural model of HIV-1 RT containing an insertion of two Ser residues following 69Ser, together with 41Leu, 210Trp, and 215Tyr (Fig. 4). The results of this modelling show that the insertion at residue 69 is most likely to confer resistance by slightly altering the presentation of residues at the start and the end of the loop. This could be the case for residues 65 and 74, which are directly involved in the recognition of the incoming dNTP.
FIG. 4.
Schematic diagram of the HIV-1 RT heterodimer showing template-primer, dTTP, and the positions of drug resistance mutations. Mutated residues (42, 69, 210, and 215) are shown as purple spheres. The Ser-Ser insertion after position 69 is shown as blue spheres. The β2-β3 loop containing residue 69 and the dipeptide insertion is orange. The TTP is shown in normal atom color coding. The primer backbone is yellow and the template is green. Spikes between template and primer indicate the region of double-stranded DNA. The side chains of the three catalytic aspartate residues (110, 185, and 186), located within the polymerase active site, are red.
DISCUSSION
We have described the frequent occurrence of phenotypic multinucleoside analog resistance in a large collection of HIV-1 samples from treated patients who were failing antiretroviral therapy. It was apparent from genotypic analysis of these samples that a number of different molecular mechanisms exist that can lead to MNR. These include the accumulation of multiple, previously recognized nucleoside analog resistance mutations such as those conferring resistance to AZT, 3TC, and dideoxynucleosides. While such variants usually display high levels of resistance to AZT and 3TC, they tend not to be highly resistant to other nucleosides, particularly d4T. Distinct patterns of mutations specifically related to the MNR phenotype were also recognized. The first of these was the codon 151 cluster of mutations that resulted in high levels of nucleoside analog resistance. This is similar to previous reports documenting the occurrence of this genotypic pattern (13, 33). In addition, a new family of insertions and deletions between RT codons 67 and 70 plus AZT resistance mutations was also associated with highly MNR virus. Detection of the single substitution, Thr69Ser, without an insertion, suggested that Thr69Ser might develop prior to amino acid insertion. However, the precise mechanism(s) by which these insertions occur is unclear, as there was never perfect reiteration of the Ser codon at the nucleotide level.
A number of recent studies have simultaneously reported similar findings regarding the phenotypic effect of codon 69 (1, 6, 39, 40). These observations were broadly concordant, each including the fact that a codon insertion itself had minimal effect on nucleoside resistance. In our study, a virus carrying the 69Ser-(Ser-Ser) change alone only showed significant decreased 3TC susceptibility. By contrast, Winters et al. (41) reported that, alone, 69Ser-(Ser-Ala) or 69Ser-(Ser-Gly) conferred up to a 10-fold increase in IC50 to most nucleosides except d4T. These apparently discrepant observations might be related to the fact that different susceptibility assay systems were employed. Alternatively, as the specific site-directed mutant inserts that were assessed in both studies were different (Ser-Ser versus Ser-Ala or Ser-Gly), different amino acid insertions might confer subtly different MNR phenotypes. We are currently evaluating this possibility by simultaneously testing a panel of insert mutants in the same standardized recombinant phenotypic assay. A second concordant observation regarding the appearance of the inserts was that they were always present in a background of AZT resistance mutations, especially Thr215Tyr/Phe. Furthermore, analysis of site-directed mutants confirmed that the overall levels of nucleoside resistance increased when AZT resistance mutations were also present (39, 41). This was especially the case for AZT, 3TC, and d4T, demonstrating that, in the context of codon 69 insertions, AZT resistance mutations could influence susceptibility to nucleosides besides AZT. It was apparent that the magnitude of resistance to the dideoxynucleosides was not as high with the site-directed mutant viruses that we tested as with the clinical isolates containing insertions. This was probably influenced by two factors. Firstly, many of the clinical isolates contained more AZT resistance mutations than just 210W and 215Y. Secondly, there were many other insertions besides Ser-Ser in the clinical samples (see Table 1). The high degree of d4T resistance associated with the insertions was of interest since previous reports of d4T resistance have been rare in clinical samples and usually confined to HIV-1 variants with the codon 151 cluster (13, 27, 33, 34).
The presence of AZT resistance mutations together with insertions in clinical samples suggests that AZT therapy is a prerequisite for the subsequent development of the codon 69 insertion. However, since these insertions were never observed during AZT therapy alone (4, 17), combination therapy with d4T, ddI, or ddC probably played a role in their selection. This was recently confirmed by a number of groups who analyzed sequential samples from patients who developed mutant HIV-1 containing codon 69 insertions (6, 41). In particular, combination therapy with AZT and either ddI or ddC appears to be associated with the selection of insert-containing strains. Of interest in our study, sequential samples from patient 1 transitioned from 69Ser-(Ser-Ser) after a period of AZT-ddC therapy to 69Ser-(Ser-Gly) during d4T-3TC combination therapy. This implies that transition from Ser-Ser to Ser-Gly confers additional selective advantage, perhaps related to d4T resistance (discussed above). A similar transition was seen in isolates from one patient receiving ddI following AZT therapy (5). It is possible that initial development of AZT resistance may be an absolute requirement before the selection of codon 69 insertions. It will be important to establish whether these insertions also emerge in response to initial therapy with other nucleosides.
It is curious that HIV-1 RT has the inherent ability to accommodate two or three amino acid insertions in a region of the enzyme critical for polymerase function (12). However, a recent report demonstrated that the loop connecting β2 and β3 strands in the fingers region could accommodate the insertion of 15 amino acid residues without loss of RT function, although polymerase processivity was increased (19). Despite this considerable flexibility in the loop, several residues in this region appear to make important contacts with the incoming dNTP during polymerization (12). For example, residue 65 directly contacts the γ-phosphate of this dNTP. Our modelling study shows that the insertion of two amino acid residues at this position of the loop will most likely cause restrictions on the incoming dNTP. This helps to explain why such a change has a broad effect on nucleoside analog susceptibility. However, it is not obvious from our model how the AZT resistance mutations act in concert with the insertion to increase the magnitude of nucleoside resistance. It is possible that long-range conformational changes such as those observed for AZT resistance mutations Thr215Tyr and Lys219Gln (31) could play a role in this. It is likely that crystal structures of RTs containing such mutations will help resolve this issue.
The description of this novel HIV-1 drug resistance mechanism reported here and by others has significant consequences regarding the monitoring of resistance in clinical isolates. Current HIV-1 genotyping technologies that are based on hybridization (20, 36) are unlikely to detect these insertions. In fact, a recent study comparing dideoxynucleotide and solid-phase hybridization sequencing technologies demonstrated that, as expected, the hybridization-based technology failed to detect codon 69 insertions in clinical samples (38). Given the considerable sequence variation seen with this family of mutant viruses, the continual design of new probes necessary to update these assays will be extremely challenging. Therefore, surveillance of HIV-1 drug resistance should be performed using dideoxy-sequencing technologies in conjunction with phenotypic susceptibility assays. Finally, as monitoring of drug resistance in samples from treated individuals becomes more common, our study highlights that reliance solely on genotyping data could be misleading in the face of newly evolving patterns of mutations.
ACKNOWLEDGMENTS
We thank A. Brophy and M. Salim for excellent assistance with the sequencing work. R. Lanier (Glaxo Wellcome, Research Triangle Park, N.C.) is thanked for kindly supplying abacavir (1592U89).
REFERENCES
- 1.Bloor, S., K. Hertogs, R. L. Desmet, R. Pauwels, and B. A. Larder. 1998. Virological basis for HIV-1 resistance to stavudine investigated in clinical samples. International Workshop on HIV Drug Resistance and Treatment Strategies, 24–27 June 1998, Lake Maggiore, Italy. Antivir. Ther. 3(Suppl. 2) abstract 15.
- 2.Boucher C A, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High-level resistance to (-) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condra J H, Holder D J, Shleiff W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J, Rhodes A, Robbins H, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1997;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Aquila R T, Johnson V A, Welles S L, Japour A J, Kuritzkes D R, DeGruttola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, Richman D D. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. AIDS Clinical Trials Group Protocol 116B/117 Team and the Virology Committee Resistance Working Group. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 5.De Antoni A, Foli A, Lisziewiez J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 6.de Jong, J. J., S. Jurriaans, J. Goudsmit, E. Baan, R. Huismans, S. Danner, M. Hillebrand, J. H. ten Veen, and F. de Wolf. 1998. Insertion of two amino acids in reverse transcriptase (RT) during antiretroviral combination therapy: implications for resistance against nucleoside RT inhibitors. International Workshop on HIV Drug Resistance and Treatment Strategies, 24–27 June 1998, Lake Maggiore, Italy. Antivir. Ther. 3(Suppl. 2) abstract 18.
- 7.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 8.Harrigan P R, Kinghorn I, Bloor S, Kemp S D, Najera I, Kohli A, Larder B A. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertogs K, de Béthune M P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant resistant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch M S, Conway B, D’Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society—USA Panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 11.Hooker D, Tachedjian G, Soloman A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 13.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones T A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 15.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 16.Kellam P, Boucher C A B, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellam P, Boucher C A B, Tijnagel J M G H, Larder B A. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol. 1994;75:341–351. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 18.Kemp S D, Shi C, Bloor S, Harrigan P R, Mellors J W, Larder B A. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and l-2′,3′-dideoxy-3′-thiacytidine. J Virol. 1998;72:5093–5098. doi: 10.1128/jvi.72.6.5093-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kew Y, Olsen L R, Japour A J, Prasad V R. Insertions into the β3-β4 hairpin loop of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in processive polymerization. J Biol Chem. 1998;273:7529–7537. doi: 10.1074/jbc.273.13.7529. [DOI] [PubMed] [Google Scholar]
- 20.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 21.Kuritzkes D R. Clinical significance of drug resistance in HIV-1 infection. AIDS. 1996;10(Suppl. 5):S27–S31. doi: 10.1097/00002030-199612005-00005. [DOI] [PubMed] [Google Scholar]
- 22.Lacey S F, Larder B A. Novel mutation (V75T) human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-dideoxyhydro-2′,3′-dideoxythymidine in cell culture. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high level resistance to zidovudine. Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 24.Larder B A, Kellam P, Kemp S D. Convergent combination therapy can select viable multidrug resistant HIV-1 in vitro. Nature (London) 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 25.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 26.Larder B A, Kohli A, Kellam P, Kemp S D, Kronick M, Henfrey R D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature (London) 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 27.Lin P F, Samanta H, Rose R E, Patick A K, Trimble J, Bechtold C M, Revie D R, Khan N C, Federici M E, Li H, et al. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J Infect Dis. 1994;170:1157–1164. doi: 10.1093/infdis/170.5.1157. [DOI] [PubMed] [Google Scholar]
- 28.Miller V, Phillips A, Rottmann C, Staszewski S, Pauwels R, Hertogs K, de Bethune M-P, Kemp S D, Bloor S, Harrigan P R, Larder B A. Dual resistance to zidovudine (ZDV) and lamivudine (3TC) in patients treated with ZDV/3TC combination therapy: association with therapy failure. J Infect Dis. 1998;177:1521–1532. doi: 10.1086/515304. [DOI] [PubMed] [Google Scholar]
- 29.Nijhuis M, Schuurman R, de Jong D, van Leeuwen R, Lange J, Danner S, Keulen W, de Groot T, Boucher C A B. Lamivudine-resistant human immunodeficiency virus type 1 variants (184V) require multiple amino acid changes to become co-resistant to zidovudine in vivo. J Infect Dis. 1997;176:398–405. doi: 10.1086/514056. [DOI] [PubMed] [Google Scholar]
- 30.Palella F J, Jr, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 31.Ren J, Esnouf R M, Hopkins A L, Jones E Y, Kirby I, Keeling J, Ross C K, Larder B A, Stuart D I, Stammers D K. 3′-Azido-3′-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes. Proc Natl Acad Sci USA. 1998;95:9518–9523. doi: 10.1073/pnas.95.16.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:129–142. [Google Scholar]
- 33.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kaojima E, Alcaide M L, Chockekuchai S, Roy B M, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soriano V, Dietrich U, Villalba N, Immelmann A, Gil-Aguado A, Echevarria S, Clotet B, Ocana I, Santamaria J M, Bouza E, Barona V, Gatell J M, Gonzalez-Lahoz J. Lack of emergence of genotypic resistance to stavudine after two years of monotherapy. AIDS. 1997;11:696–697. [PubMed] [Google Scholar]
- 35.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 36.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhoftede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vahey M, Nau M E, Barrick S, Cooley J D, Sawyer R, Sleeker A A, Vickerman P, Bloor S, Larder B, Michael N L, Wegner S A. Limitations of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of diverse human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J Clin Microbiol. 1999;37:2533–2537. doi: 10.1128/jcm.37.8.2533-2537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitcomb, J. M., K. Limoli, T. Wrin, D. Smith, H. Tian, N. Parkin, Y. S. Lie, and C. J. Petropoulos. 1998. Phenotypic and genetic analysis of stavudine-resistant isolates of HIV-1. International Workshop on HIV Drug Resistance and Treatment Strategies, 24–27 June 1998, Lake Maggiore, Italy. Antivir. Ther. 3(Suppl. 2) abstract 17.
- 40.Winters, M. A., K. I. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, D. A. Katzenstein, R. W. Shafer, and T. C. Merigan. 1998. Phenotypic and molecular analysis of HIV-1 isolates possessing 6 bp inserts in the reverse transcriptase gene that confer resistance to nucleoside analogues. International Workshop on HIV Drug Resistance and Treatment Strategies, 24–27 June 1998, Lake Maggiore, Italy. Antivir. Ther. 3(Suppl. 2) abstract 16.
- 41.Winters M A, Coolley K I, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-base pair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside analogues. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]