Abstract
Anopheles stephensi is an important vector of malaria in the South Asia, the Middle East, and Eastern Africa. The olfactory system of An. stephensi plays an important role in host-seeking, oviposition, and feeding. Odorant binding proteins (OBPs) are globular proteins that play a pivotal role in insect olfaction by transporting semiochemicals through the sensillum lymph to odorant receptors (ORs). Custom motifs designed from annotated OBPs of Aedes aegypti, Drosophila melanogaster, and Anopheles gambiae were used for the identification of putative OBPs from protein sequences of the An. stephensi Indian strain. Further, BLASTp was also performed to identify missing OBPs and ORs. Subsequently, the presence of domains common to OBPs was confirmed. Identified OBPs were further classified into three sub-classes. Phylogenetic and syntenic analyses were carried out to find homology, and thus the evolutionary relationship between An. stephensi OBPs and ORs with those of An. gambiae, Ae. aegypti and D. melanogaster. Gene structure and physicochemical properties of the OBPs and ORs were also predicted. A total of 44 OBPs and 45 ORs were predicted from the protein sequences of An. stephensi. OBPs were further classified into the classic (27), atypical (10) and plus-C (7) OBP subclasses. The phylogeny revealed close relationship of An. stephensi OBPs and ORs with An. gambiae homologs whereas only five OBPs and two ORs of An. stephensi were related to Ae. aegypti OBPs and ORs, respectively. However, D. melanogaster OBPs and ORs were distantly rooted. Synteny analyses showed the presence of collinear block between the OBPs and ORs of An. stephensi and An. gambiae as well as Ae. aegypti’s. No homology was found with D. melanogaster OBPs and ORs. As an important component of the olfactory system, correctly identifying a species’ OBPs and ORs provide a valuable resource for downstream translational research that will ultimately aim to better control the malaria vector An. stephensi.
Introduction
Anopheles stephensi, is vector of several Plasmodium species causing malaria [1,2]. Despite advances in prophylactic treatments, malaria continues to impose heavy burdens on global healthcare systems. An. stephensi has a wide geographic distribution throughout the South Asia, the Middle East and recently East Africa [3–7]. An. stephensi predominately transmits malaria in urban areas. Thus, control of An. stephensi can significantly reduce the malaria transmission. Olfaction mechanisms are central to insects including An. stephensi for host-seeking, oviposition and feeding [8,9]. Olfactory sensing mechanisms are located within the antennal sensillum of the An. stephensi [10,11]. Excitatory behaviour modifying compounds or semiochemicals enter sensilla through pores and cross the sensillum lymph with the aid of odorant binding proteins (OBPs) to induce olfactory sensing in insects [12].
The odorant binding proteins (OBPs) are globular in nature and specifically transport semiochemicals through sensillum lymph to odorant receptors (ORs) that ultimately send electrical signals to the insect brain [13,14]. OBPs also have an additional role of protecting the odorants from odorant degrading enzymes [15]. In Antheraea polyphemus (polyphemus moth), first OBP was identified 40 years ago [16]. Binding of semiochemicals to OBPs is pH dependent wherein binding takes place at pH6.5 whereas bound chemicals are released from OBPs near ORs where the pH measures around 4.5 [17–20].
The number of OBPs varies between insect species, with OBPs being highly expressed in sensillum lymph [21–23]. The number of OBPs identified in Drosophila melanogaster, Aedes aegypti and Anopheles gambiae are 51, 66 and 57, respectively [24–27]. There are several classes of OBPs: classic, atypical, plus-C, and minus-C OBPs [21]. Classification of OBPs is based on the presence of conserved cysteine residues that are considered as motifs. Six conserved cysteines are present in the classic OBPs. In each class, cysteine 2 (C2) and cysteine 3 (C3), being three amino acid residues apart, are conserved. Similarly, cysteine 5 (C5) and cysteine (C6) are conserved having eight amino acid residues between them. However, number of amino acid residues varies between C1-C2, C3-C4 and C4-C5. Classic and atypical OBPs contain same number of conserved cysteines but the number of amino acids between conserved cysteine residues vary. However, plus-C OBPs contain the two extra cysteines 4a and 6a along with a conserved proline immediately after the C6. Most of insect OBPs belong to classic group of OBPs [28]. In An. stephensi, two OBPs and one OR that is OR8 have been identified [29–31].
Odorant receptors (ORs) are transmembrane proteins containing seven helices like G-protein coupled receptors (GPCRs). ORs have been previously identified based on in situ hybridization using RNA probes and transcriptomic data from different organisms [32]. Similarly, ORs have also been identified by analysing the genomic data as in An. gambiae [33]. The number of ORs varies with the organisms. ORs are coupled with the Odorant Receptor co-receptors (Orco). Orco are highly conserved across all the insect species having been originally named as OR83b in D. melanogaster [34]. Insect olfaction relies on the interplay of OBPs that carry odorant molecules to the ORs and Orco, which results in electrical signals being sent to the insect brain.
In this study, genome wide analysis of the An. stephensi OBPs was performed to identify and classify the odorant binding proteins (OBPs) using the custom motifs and odorant receptors (ORs). Phylogenetic analyses were conducted to establish the evolutionary relationship of the OBPs and ORs with the closely related organisms Ae. aegypti, An. gambiae, and D. melanogaster. Further, we investigated synteny between the OBPs and ORs to identify the syntenic regions between these insects. This study has analyzed the gene structure and predicted the physicochemical properties and subcellular localization of the OBPs and ORs as well for confirmation of the OBPs and ORs. Being a vital component of insect olfaction, OBPs and ORs of An. stephensi play crucial roles in the mosquito life cycle and therefore, are likely to have a major influence on the potential for the mosquito to transmit malarial diseases. This study serves as the basis for the structural and functional characterization of the OBPs and ORs of An. stephensi. Identified OBPs and ORs will help in understanding the pathways sensitive to attractants and repellents in the olfactory mechanism and vector control strategies. It provides the foundation for comparative studies based on olfactory mechanisms in other insect species as well.
Methodology
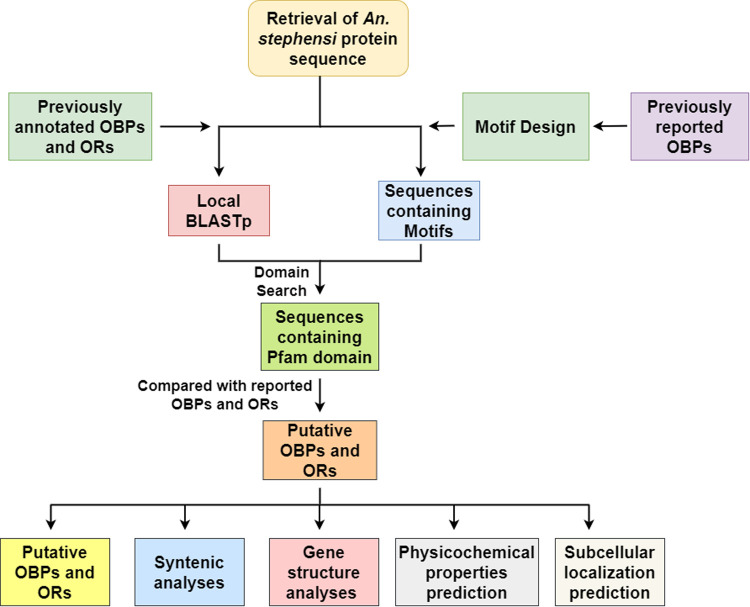
The methodology is summarized as a flow chart shown in Fig 1.
Fig 1. Pipeline of the methodology.
Pipeline of the methodology used for this study is represented.
Identification of the OBPs and ORs
Protein FASTA file of the Anopheles stephensi Indian strain, sequenced by UC Irvine, was downloaded from the NCBI database (Refseq: GCF_013141755.1). Retrieved protein FASTA file was used as a protein database in BLAST+ software for local BLASTp using the OBP sequences of Ae. aegypti, An. gambiae and D. melanogaster as query sequences. Custom motifs were designed on earlier studies according to the ScanProsite input format. These motifs are based on the conserved cysteine residues present in three classes of OBPs: classic [Cx(15,39)Cx(3)Cx(21,44)Cx(7,12)Cx(8)C], atypical [Cx(26,27)Cx(3)Cx(36,38)Cx(11,15)Cx(8)C], and plus-C [Cx(8,41)Cx(3)Cx(39,47)Cx(17,29)Cx(9)Cx(8)CPx(9,11)C]. BLASTp output was used to detect the presence of these motifs using ScanProsite (https://prosite.expasy.org/scanprosite/) to extract OBPs sequences based on their classification [35]. The retrieved sequences were further analyzed to detect the presence of the PBP/GOBP domain (Pfam: PF01395) using Pfam web server (http://pfam.xfam.org/). Pfam is a large collection of the protein families and domains [36]. Retrieved OBPs sequences were used to perform a BLASTp search against protein database of An. stephensi to search for the missing OBPs sequences in. OBPs were named according to their position on the chromosomes as AsteOBP. Positions of the OBP encoding genes on An. stephensi chromosomes were visualized using phenogram tool (http://visualization.ritchielab.org/phenograms/plot) [37]. Whereas open reading frame (ORF) were predicted using the ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/).
Like OBPs, OR sequences of the Ae. aegypti, An. gambiae and D. melanogaster were used as the query sequences for the local BLASTp [38]. The protein FASTA file of An. stephensi was used as the database. BLASTp output sequences were further checked for the presence of the 7tm_6 domain (Pfam ID: PF02949) using the Pfam web server. Further, redundant sequences having 100% sequence identity were removed. Similarly, positions of OR encoding genes on An. stephensi chromosomes were visualized using phenogram.
Multiple sequence alignment and phylogenetic tree analyses
Multiple sequence alignment for the OBPs was done using Clustal-W in Mega X-V10.2 with gap opening penalty 10. The gap extension penalty was set to 0.1 for pairwise alignment and 0.2 for the multiple sequence alignment [39]. Multiple sequence alignment was performed separately for the classic, atypical and plus-C OBPs to visualize the motif regions in each OBP subtype. Multiple sequence alignments were visualized using Jalview [40].
Multiple sequence alignments of the OBPs of the An. stephensi, An. gambiae, Ae. aegypti and D. melanogaster were performed using the Clustal-W on the Galaxy server (https://usegalaxy.org/). This alignment was used for the generation of the Maximum-Likelihood tree using the FastTree 2 on the galaxy webserver [41]. Jones-Taylor-Thornton 1992 model (JTT-Model) was used as evolutionary model that uses protein sequences for the faster generation of mutation metrices of proteins. The tree was further modified using the iTOL web server [42]. Similarly, Clustal W and FastTree 2 were used for the construction of phylogenetic tree of the Odorant Receptors (ORs).
Synteny prediction of the OBPs and ORs
Synteny analysis was performed using TBTools [43]. TBTools uses MCScanX to find the syntenic regions between the chromosomes of two organisms [44]. MCScanX is an algorithm that is used to scan the multiple genomes and identify putative homologous chromosomal regions by aligning those using genes as anchors. OBPs of the An. stephensi were analyzed against the genome of An. gambiae (Genbank: GCA_000005575.1), Ae. aegypti (Genbank: GCA_002204515.1) and D. melanogaster (Genbank: GCA_000001215.4) to identify the collinear blocks between their genomes. Likewise, syntenic regions were also predicted in the ORs of An. stephensi with the ORs of An. gambiae, Ae. aegypti and D. melanogaster.
Gene structure analysis of OBPs and ORs
Gene structures of the OBPs and ORs were visualized using the TBTools [43]. A Genome Feature File (GFF) file of the genomic sequences of An. stephensi was used for the identification of the Untranslated Regions (UTRs) and Coding DNA Sequences (CDS) in OBPs and ORs genes. MEME web server (https://meme-suite.org/meme/tools/meme) was used for the presence of conserved motifs in the peptides sequences of the OBPs and ORs [45]. Conserved Domain Database (CDD) was used to visualize the conserved Pfam domains in peptide sequences of OBPs and ORs [46]. Phylogenetic tree of OBPs and ORs of An. stephensi was constructed using the Neighbor Joining Method in Mega X-V10.2 for the use of cladogram in the gene structure representation.
Physiochemical properties and sub-cellular localization prediction of OBPs and ORs
Physiochemical properties of the OBPs and ORs including molecular weight and isoelectric points were predicted using ProtParam in the Expasy webserver (https://web.expasy.org/protparam/). Sub-cellular localization of the OBPs and ORs was predicted using the WoLF PSORT (https://wolfpsort.hgc.jp/) and CELLO web server (http://cello.life.nctu.edu.tw/cgi/main.cgi).
Results
Identification of OBPs and ORs
A total 44 OBPs were identified in the An. stephensi having complete conserved motifs. There were 27 classic, 10 atypical, and 7 plus-C OBPs as their NCBI peptide accessions and gene IDs are represented in the Table 1. Genomic and protein sequences of putative OBPs are given in S1 and S2 Data, respectively. Similarly, ORF length and number of amino acids in the OBPs have been provided in the S1 Table.
Table 1. Classification and NCBI accessions of identified OBPs.
| OBP | Protein Accession | Gene ID | Classes of OBPs | Chromosome | Molecular weight | Isoelectric Point |
|---|---|---|---|---|---|---|
| AsteOBP1 | XP_035917841.1 | LOC118504557 | Classic | X | 18669 | 4.7 |
| AsteOBP2 | XP_035891203.1 | LOC118504570 | Classic | X | 16593 | 6.5 |
| AsteOBP3 | XP_035891204.1 | LOC118510310 | Classic | X | 16564 | 5.6 |
| AsteOBP4 | XP_035891207.1 | LOC118510318 | Classic | X | 16813 | 5.6 |
| AsteOBP5 | XP_035891740.1 | LOC118510325 | Classic | X | 15898 | 4.2 |
| AsteOBP6 | XP_035891741.1 | LOC118510331 | Classic | X | 14940 | 6.2 |
| AsteOBP7 | XP_035892015.1 | LOC118502745 | Classic | 2 | 15105 | 8.4 |
| AsteOBP8 | XP_035892546.1 | LOC118502746 | Atypical | 2 | 50208 | 8.2 |
| AsteOBP9 | XP_035892547.1 | LOC118502748 | Atypical | 2 | 39006 | 5.1 |
| AsteOBP10 | XP_035893080.1 | LOC118503023 | Classic | 2 | 17788 | 4.9 |
| AsteOBP11 | XP_035894183.1 | LOC118503024 | Plus-C | 2 | 22832 | 4.7 |
| AsteOBP12 | XP_035894335.1 | LOC118503161 | Classic | 2 | 15761 | 4.3 |
| AsteOBP13 | XP_035894381.1 | LOC118503418 | Classic | 2 | 16437 | 6.5 |
| AsteOBP14 | XP_035895118.1 | LOC118503419 | Atypical | 2 | 31279 | 8.4 |
| AsteOBP15 | XP_035895684.1 | LOC118503662 | Classic | 2 | 15844 | 6.3 |
| AsteOBP16 | XP_035895818.1 | LOC118504157 | Classic | 2 | 17738 | 8.9 |
| AsteOBP17 | XP_035895821.1 | LOC118504227 | Classic | 2 | 15028 | 4.8 |
| AsteOBP18 | XP_035897326.1 | LOC118504259 | Atypical | 2 | 24233 | 6.6 |
| AsteOBP19 | XP_035897725.1 | LOC118504578 | Classic | 2 | 16412 | 5.2 |
| AsteOBP20 | XP_035898485.1 | LOC118504803 | Classic | 2 | 16227 | 8.1 |
| AsteOBP21 | XP_035898834.1 | LOC118504874 | Classic | 2 | 19676 | 8.9 |
| AsteOBP22 | XP_035898836.1 | LOC118504876 | Classic | 2 | 16709 | 8.7 |
| AsteOBP23 | XP_035899548.1 | LOC118505522 | Classic | 2 | 17444 | 5.8 |
| AsteOBP24 | XP_035900970.1 | LOC118505693 | Classic | 2 | 13982 | 8.4 |
| AsteOBP25 | XP_035902367.1 | LOC118506007 | Classic | 2 | 16094 | 4.8 |
| AsteOBP26 | XP_035903187.1 | LOC118506171 | Classic | 2 | 14920 | 4.7 |
| AsteOBP27 | XP_035903789.1 | LOC118506172 | Classic | 2 | 18610 | 4.6 |
| AsteOBP28 | XP_035905778.1 | LOC118506477 | Classic | 2 | 17744 | 9.3 |
| AsteOBP29 | XP_035907589.1 | LOC118507089 | Plus-C | 2 | 21790 | 8.5 |
| AsteOBP30 | XP_035907591.1 | LOC118507680 | Plus-C | 2 | 21161 | 5.9 |
| AsteOBP31 | XP_035907600.1 | LOC118507965 | Plus-C | 2 | 22467 | 8.8 |
| AsteOBP32 | XP_035908250.1 | LOC118508265 | Plus-C | 2 | 23618 | 7.5 |
| AsteOBP33 | XP_035908252.1 | LOC118509362 | Plus-C | 3 | 19688 | 4.9 |
| AsteOBP34 | XP_035908253.1 | LOC118510176 | Plus-C | 3 | 19627 | 4.9 |
| AsteOBP35 | XP_035909003.1 | LOC118510178 | Atypical | 3 | 36926 | 6 |
| AsteOBP36 | XP_035910841.1 | LOC118510185 | Classic | 3 | 17618 | 8.4 |
| AsteOBP37 | XP_035914753.1 | LOC118510478 | Atypical | 3 | 31775 | 5.3 |
| AsteOBP38 | XP_035917598.1 | LOC118510479 | Classic | 3 | 15299 | 4.8 |
| AsteOBP39 | XP_035895071.1 | LOC118510480 | Classic | 3 | 20736 | 5.2 |
| AsteOBP40 | XP_035895083.1 | LOC118510798 | Classic | 3 | 15612 | 5.3 |
| AsteOBP41 | XP_035907843.1 | LOC118511635 | Atypical | 3 | 35623 | 5.8 |
| AsteOBP42 | XP_035907857.1 | LOC118513288 | Atypical | 3 | 33435 | 5.2 |
| AsteOBP43 | XP_035907868.1 | LOC118514663 | Atypical | 3 | 33188 | 5.2 |
| AsteOBP44 | XP_035907882.1 | LOC118515263 | Atypical | Unknown | 31914 | 5.4 |
Classification of the identified odorant binding proteins (OBPs) along with their NCBI peptide accession and gene IDs are presented in the table. Molecular weight and isoelectric point of OBPs are also given.
Further, putative ORs identified in the genome of the An. stephensi were 45. These genes were renamed according to their respective position on the chromosomes of An. stephensi. The name, NCBI protein accessions and gene IDs of the sequences are presented in the Table 2. Genomic and peptide sequences of putative ORs are given in S3 and S4 Data, respectively. Similarly, ORF length and number of amino acids in the ORs have been provided in the S2 Table.
Table 2. Renaming of odorant receptor (ORs) and chromosome.
| ORs | NCBI Protein Accession | NCBI Gene ID | Chromosome | Molecular weight | Isoelectric Point |
|---|---|---|---|---|---|
| AsteOR1 | XP_035892088.1 | LOC118502993 | 1 | 47.484 | 9.49 |
| AsteOR2 | XP_035917717.1 | LOC118514681 | 1 | 47.46 | 9.51 |
| AsteOR3 | XP_035900263.1 | LOC118506771 | 1 | 11.761 | 9.51 |
| AsteOR4 | XP_035918152.1 | LOC118516006 | 1 | 29.498 | 8.69 |
| AsteOR5 | XP_035893454.1 | LOC118503850 | 2 | 50.216 | 9.17 |
| AsteOR6 | XP_035893486.1 | LOC118503863 | 2 | 30.798 | 7 |
| AsteOR7 | XP_035901553.1 | LOC118507296 | 2 | 45.165 | 8.73 |
| AsteOR8 | XP_035899159.1 | LOC118506317 | 2 | 97.083 | 9.08 |
| AsteOR9 | XP_035898901.1 | LOC118506200 | 2 | 49.355 | 8.23 |
| AsteOR10 | XP_035898518.1 | LOC118506022 | 2 | 45.771 | 8.7 |
| AsteOR11 | XP_035901800.1 | LOC118507433 | 2 | 46.713 | 6.22 |
| AsteOR12 | XP_035903051.1 | LOC118507903 | 2 | 46.521 | 6.55 |
| AsteOR13 | XP_035891168.1 | LOC118502720 | 2 | 46.096 | 5.9 |
| AsteOR14 | XP_035899865.1 | LOC118506605 | 2 | 52.647 | 8.83 |
| AsteOR15 | XP_035893072.1 | LOC118503656 | 2 | 39.869 | 9.08 |
| AsteOR16 | XP_035893066.1 | LOC118503653 | 2 | 47.581 | 9.26 |
| AsteOR17 | XP_035895309.1 | LOC118504650 | 2 | 14.085 | 6.7 |
| AsteOR18 | XP_035890596.1 | LOC118502471 | 2 | 43.074 | 7.17 |
| AsteOR19 | XP_035890597.1 | LOC118502472 | 2 | 48.388 | 8.52 |
| AsteOR20 | XP_035897380.1 | LOC118505546 | 2 | 43.686 | 7.6 |
| AsteOR21 | XP_035890600.1 | LOC118502475 | 2 | 51.037 | 9.41 |
| AsteOR22 | XP_035903785.1 | LOC118508262 | 2 | 53.873 | 8.11 |
| AsteOR23 | XP_035903780.1 | LOC118508261 | 2 | 55.875 | 8.31 |
| AsteOR24 | XP_035890608.1 | LOC118502484 | 2 | 46.469 | 8.91 |
| AsteOR25 | XP_035896260.1 | LOC118505078 | 2 | 46.133 | 7.55 |
| AsteOR26 | XP_035914390.1 | LOC118513096 | 3 | 46.464 | 9.36 |
| AsteOR27 | XP_035913964.1 | LOC118512920 | 3 | 48.938 | 6.37 |
| AsteOR28 | XP_035910189.1 | LOC118511343 | 3 | 53.323 | 6.59 |
| AsteOR29 | XP_035904929.1 | LOC118509018 | 3 | 42.07 | 8.72 |
| AsteOR30 | XP_035904961.1 | LOC118509031 | 3 | 43.592 | 9.29 |
| AsteOR31 | XP_035904965.1 | LOC118509034 | 3 | 43.903 | 5.75 |
| AsteOR32 | XP_035907269.1 | LOC118509993 | 3 | 44.707 | 8.63 |
| AsteOR33 | XP_035907274.1 | LOC118509997 | 3 | 17.289 | 4.97 |
| AsteOR34 | XP_035907276.1 | LOC118509999 | 3 | 43.583 | 5.65 |
| AsteOR35 | XP_035904780.1 | LOC118508959 | 3 | 10.478 | 8.73 |
| AsteOR36 | XP_035907277.1 | LOC118510000 | 3 | 32.545 | 6.1 |
| AsteOR37 | XP_035910820.1 | LOC118511624 | 3 | 48.887 | 8.47 |
| AsteOR38 | XP_035908757.1 | LOC118510691 | 3 | 43.658 | 8.99 |
| AsteOR39 | XP_035908756.1 | LOC118510690 | 3 | 43.438 | 8.56 |
| AsteOR40 | XP_035908759.1 | LOC118510692 | 3 | 43.242 | 9.03 |
| AsteOR41 | XP_035908990.1 | LOC118510790 | 3 | 15.858 | 5.27 |
| AsteOR42 | XP_035907332.1 | LOC118510050 | 3 | 32.201 | 8.45 |
| AsteOR43 | XP_035907338.1 | LOC118510056 | 3 | 45.4 | 9.52 |
| AsteOR44 | XP_035916306.1 | LOC118513983 | 3 | 33.276 | 5.81 |
| AsteOR45 | XP_035919246.1 | LOC118517353 | Unknown | 31.278 | 6.55 |
Identified odorant receptors (ORs) along with their NCBI peptide accession, chromosome and gene IDs are presented in the table. Molecular weight and isoelectric points of ORs are also given.
Chromosomal location of the OBPs and ORs
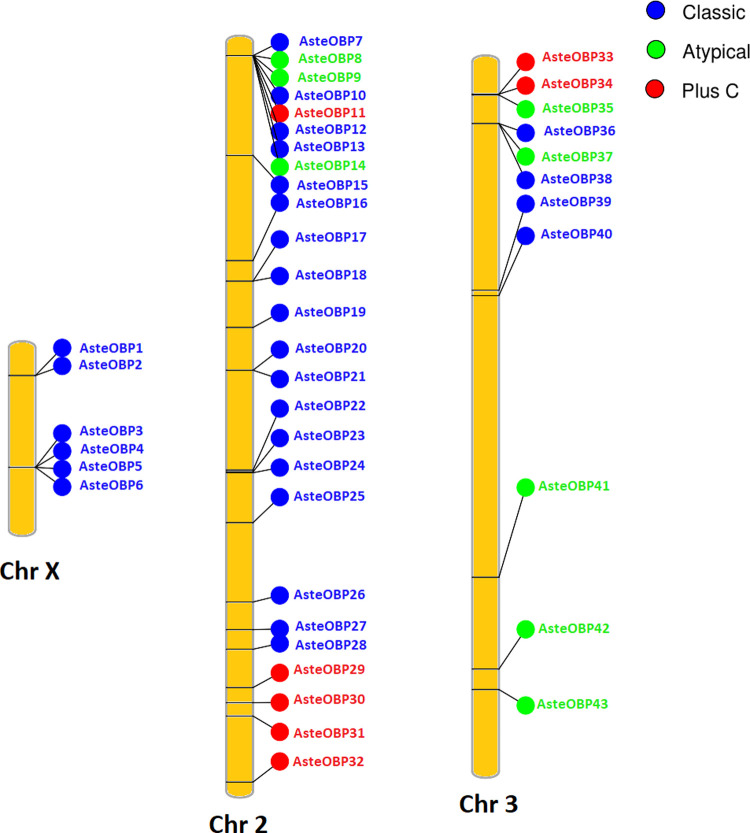
Chromosomal location of the OBPs was depicted using the Phenogram web server as shown in the Fig 2. The highest numbers of the OBPs were present on chromosome 2 containing 26 OBPs, whereas chromosome X and chromosome 3 contained 6 and 11 OBPs, respectively. Eight OBPs, starting from AsteOBP7 to AsteOBP14, were clustered on chromosome 2. Whilst on chromosome X, AsteOBP3, AsteOBP4, AsteOBP5 and AsteOBP6 were clustered together. Similarly, AsteOBP33-AsteOBP35 and AsteOBP36-AsteOBP38 were clustered on the chromosome 3. Chromosome X only contained the classic OBPs whereas chromosome 2 and 3 contained all three types. AsteOBP44 gene had not been placed on the chromosomes to date due to limitations of genomic assembly.
Fig 2. Chromosomal location of Odorant Binding Proteins (OBPs).
AsteOBPs are shown based on their position on chromosomes of the An. stephensi. Chromosome 2 contained highest number of OBP genes. Some OBPs are clustered on the chromosomes.
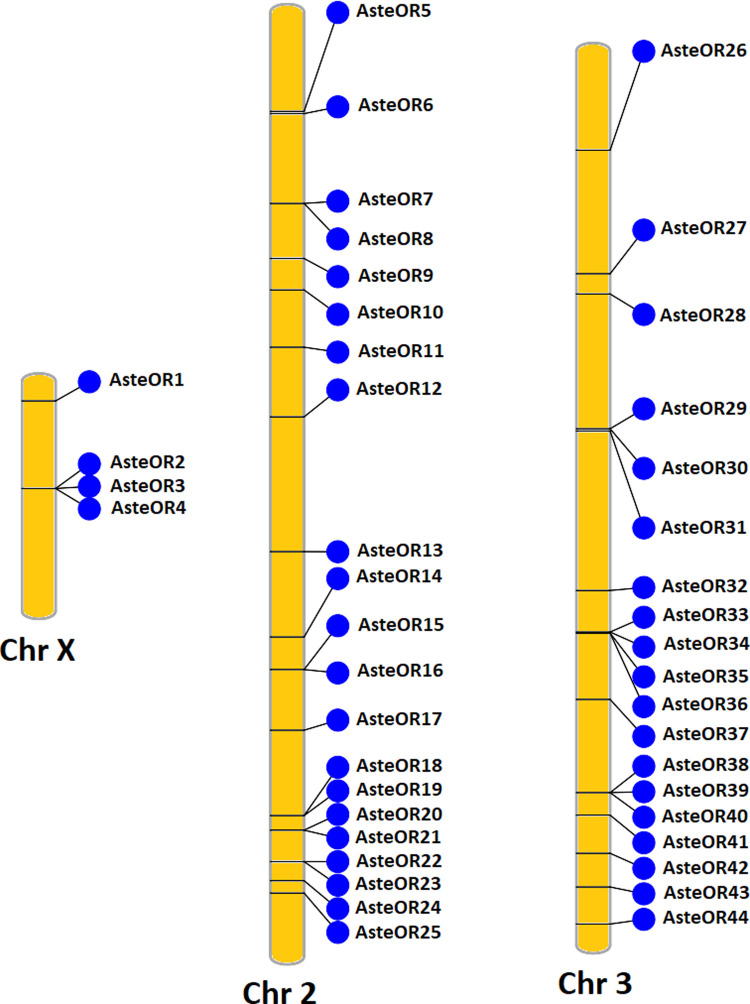
ORs were clustered on chromosome 2 of An. stephensi as there were 21 ORs’ gene located on it as represented in Fig 3. Chromosome X contained the least OR gene as it contained only 4 genes. Chromosome 3 contained 19 OR genes. Most of the OR genes were scattered except a few that were clustered together. AsteOR2-AsteOR4 were clustered together on the chromosome X. Similarly, some genes were clustered in pairs at the chromosome 2. But on chromosome 3, only three clusters of the OR genes were present where each cluster had three OR genes. AsteOR45 was present on a scaffold that was not placed on the chromosomes the genome assembly.
Fig 3. Chromosomal location of Odorant Receptors (ORs).
Position of the OR genes is shown on the chromosomes of the An. stephensi. Chromosome 2 contained highest number of OR genes. Some ORs are clustered on the chromosomes.
Multiple sequence alignment and phylogenetic analysis of the OBPs and ORs
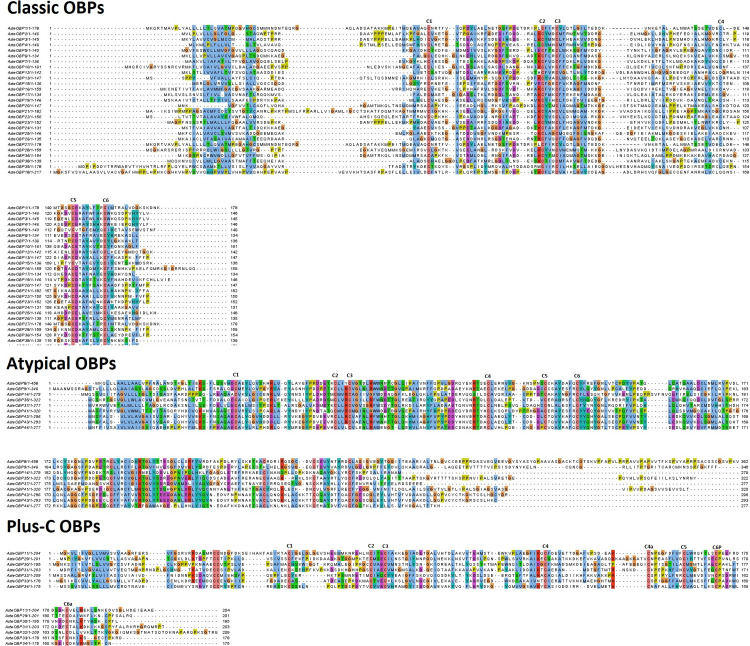
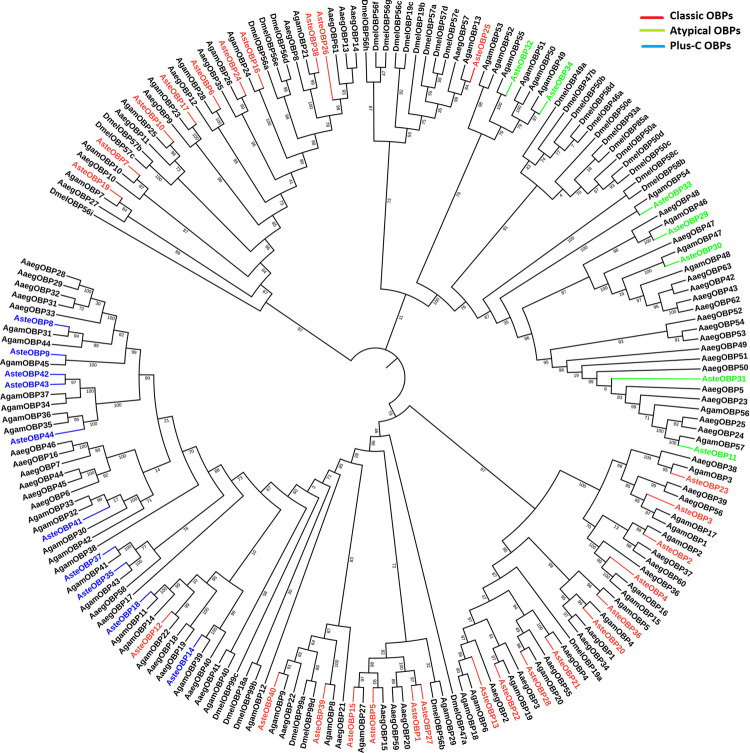
Alignments are presented in Fig 4. Clear motifs patterns depicting the conserved cysteines were visible in all three types of OBPs. Phylogenetic tree was constructed using the maximum-likelihood method. OBP sequences of the An. stephensi, An. gambiae, D. melanogaster, and Ae. aegypti were used for the construction of phylogenetic tree in Mega-X as shown in Fig 5. Most of the OBPs showed close relationship with OBPs of An. gambiae but, none of the OBPs showed close relationship with the OBPs of D. melanogaster. Similarly, only AsteOBP1, AsteOBP5, AsteOBP21, AsteOBP27, and AsteOBP31 were closely related to the AaegOBP20, AaegOBP15, AaegOBP4, AaegOBP59 and AaegOBP5 of the Ae. aegypti, respectively. It confirms the close relationship of the An. stephensi with the An. gambiae whereas D. melanogaster was distantly rooted in the phylogenetic tree among the compared organisms.
Fig 4. Multiple sequence alignment of the OBPs.
Multiple Sequence Alignment of the OBPs is shown in figure. Conserved motifs have been highlighted in all three classes of the OBPs: Classic, atypical and plus-C OBPs.
Fig 5. Phylogenetic analysis of OBPs.
Phylogenetic analysis of the OBPs of Anopheles stephensi, Anopheles gambiae, Aedes aegypti and Drosophila melanogaster were carried out using FastTree 2. An. stephensi OBPs showed closer relationship to An. gambiae OBPs. The bootstrap values have been represented in the figure.
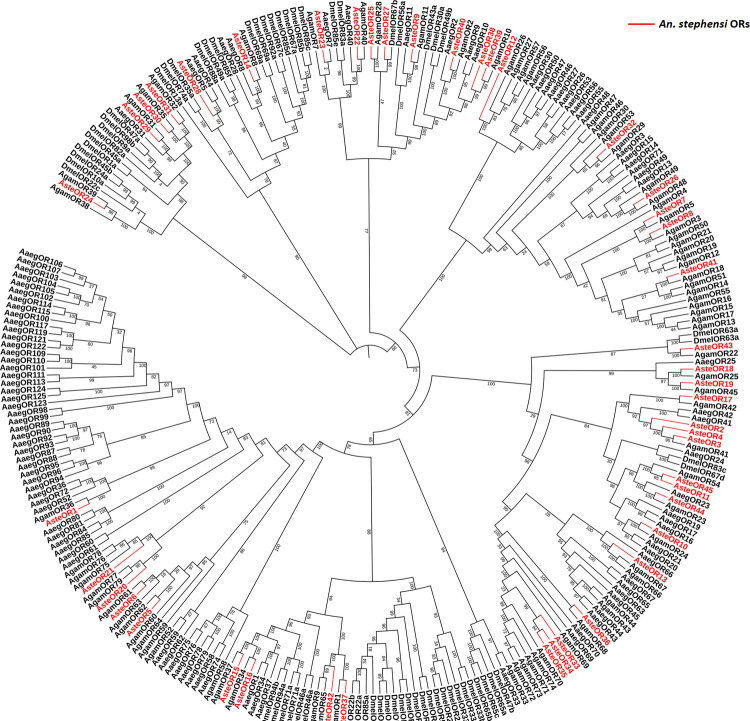
ORs showed close phylogenetic relationship with the ORs of An. gambiae as represented in Fig 6. However, only two AsteORs: AaegOR28 and AaegOR23 were closely related to AaegORs: AaegOR5 and AaegOR7, respectively. None of the An. stephensi ORs were closely rooted to the D. melanogaster ORs.
Fig 6. Phylogenetic analysis of ORs.
Phylogenetic analysis of Anopheles stephensi, Anopheles gambiae, Aedes aegypti and Drosophila melanogaster ORs were carried out using FastTree 2 on galaxy web server. An. stephensi ORs showed closer relationship to An. gambiae ORs. The bootstrap values have been represented in the figure.
Synteny analysis of OBPs and ORs
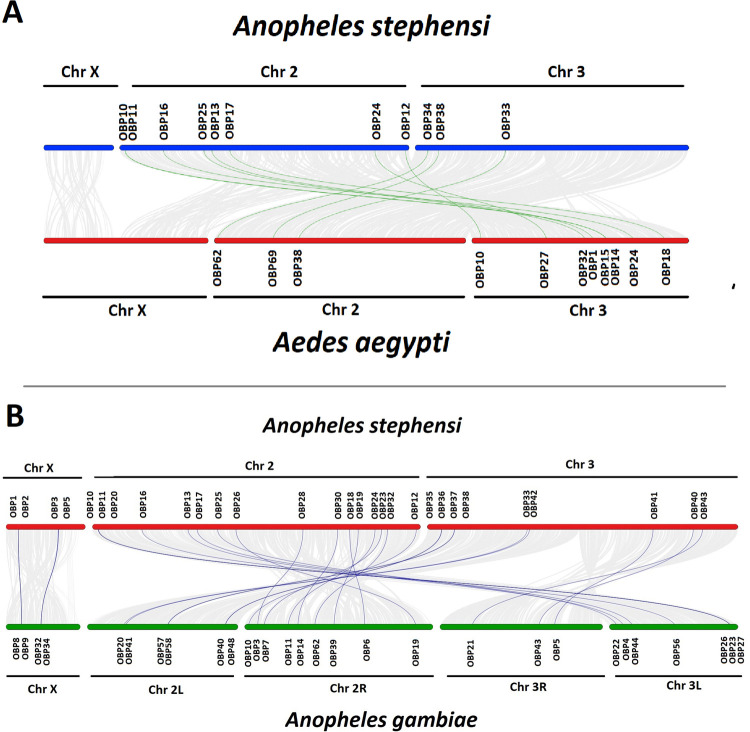
Synteny analysis was performed between the An. stephensi and Ae. aegypti. Total 11 OBPs of An. stephensi had the syntenic region with the OBPs of the Ae. aegypti as shown in the Fig 7A. OBPs present on the chromosome X of the An. stephensi and Ae. aegypti did not share any orthologues. Syntenic regions were present between the OBPs of the chromosome 2 of An. stephensi and the OBPs present on the chromosome 3 of the Ae. aegypti whereas chromosome 3 OBPs of An. stephensi contained collinear with the OBPs of chromosome 2 of Ae. aegypti.
Fig 7. Synteny analysis of OBPs.
Synteny analysis were carried out to find the collinear blocks between the OBPs of An. stephensi and OBPs of (A) Ae. aegypti and (B) Ae. gambiae. Chromosomes and chromosomal position of the OBPs have been shown. Green lines (A) show the syntenic relationship between the OBPs of An. stephensi and Ae. aegypti whereas blue lines (B) represent the syntenic relationship between the OBPs of An. stephensi and An. gambiae.
Synteny analysis were also performed between the An. stephensi and An. gambiae genome as represented in Fig 7B. A total of 29 OBPs of the An. stephensi contained the collinear blocks with the OBPs of An. gambiae. Four syntenic regions were present between the chromosome X of both organisms. Whereas others were distributed between chromosome 2 and chromosome 3 of the An. stephensi and chromosome 2L, 2R, 3L and 3R of the An. gambiae. In comparison to An. gambiae OBPs, OBPs of Ae. aegypti had less synteny with the OBPs of An. stephensi.
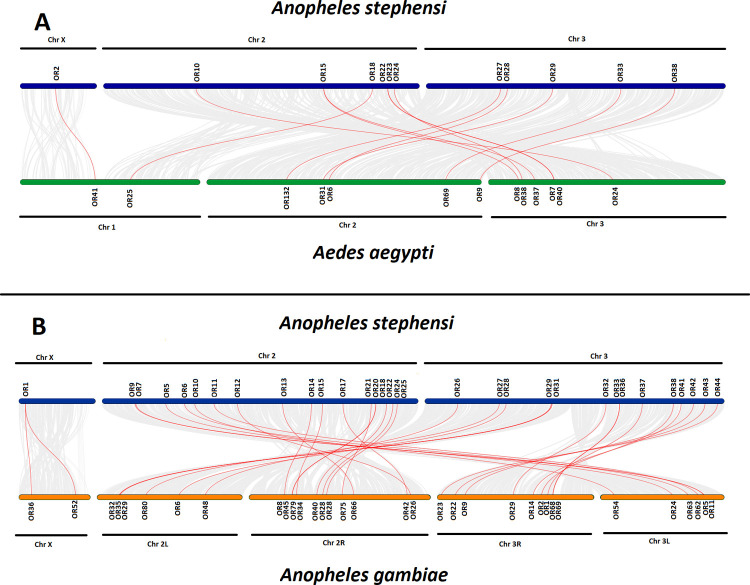
Syntenic analyses of the ORs of An. stephensi and Ae. aegypti are shown in Fig 8A. A total of 12 ORs of An. stephensi shared syntenic regions with 13 OR genes of the Ae. aegypti. Only one OR2 gene present on the chromosome X of An. stephensi showed homologous region with the OR41 of the Ae. aegypti. OR genes present on the chromosome 2 of An. stephensi showed close homology with the ORs present on the Chromosome X and 3 of Ae. aegypti. However, OR15 on chromosome 2 shared homology with the two ORs: OR8 and OR37 of Ae. aegypti. Whereas OR genes present on the chromosome 3 of An. stephensi were homologous to ORs of chromosome 2 of Ae. aegypti.
Fig 8. Synteny analysis of ORs.
Synteny analysis were carried out to find the collinear blocks between the ORs of An. stephensi and ORs of (A) Ae. aegypti and (B) Ae. gambiae. Chromosomes and chromosomal position of the ORs have been shown along with the lines showing synteny between the ORs. Red lines represent the syntenic relationship between the ORs of An. stephensi with Ae. aegypti (A) and An. gambiae (B).
Synteny analysis of An. stephensi with An. gambiae ORs is shown in Fig 8B. A total of 32 ORs of An. stephensi shared homology with 34 ORs of An. gambiae. OR1 gene present on the chromosome X of An. stephensi showed homology with the OR36 and OR52 genes present on the chromosome X of An. gambiae. However, ORs present on the chromosome 2 of the An. stephensi were homologous to the ORs present on chromosome 2R and 3L of An. gambiae. But ORs present on the chromosome 3 of the An. stephensi shared syntenic region with the ORs of chromosome 2L and 3R in An. gambiae. None of the An. stephensi OBPs and ORs shared homology with the OBPs and ORs of D. melanogaster.
Gene structure analysis of OBPs and ORs
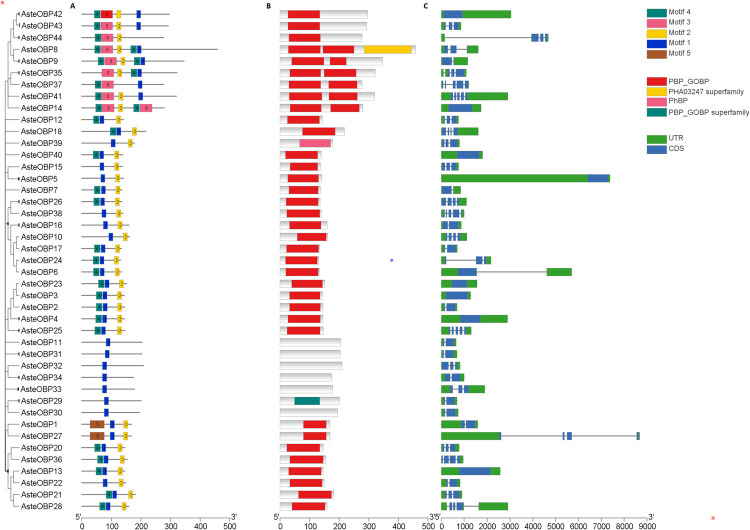
Gene structure analysis of the OBPs was carried out using the TBTool as depicted in the Fig 9. Conserved motifs present in the OBPs were identified using the MEME web server. Motif 1 was present in all the OBPs. Whereas motifs 2, 3 and 4 were present in the most of the OBPs. Conserved domains were identified using the Conserved Domain Database (CDD). CDD results indicated OBPs 11, 30, 31, 32, 33 and 34 had insignificant similarity with the Pfam PBP/GOBP domain but these were Plus-C OBPs having complete motifs. Whereas all other OBPs contained the PBP/GOBP domains.
Fig 9. Gene structure of the OBPs.
Conserved MEME motifs (A), Pfam domains (B) and UTR/CDS (C) are shown. Conserved motifs are present in all the OBPs along with the gene structure. Presence of UTRs and CDS is shown in genes.
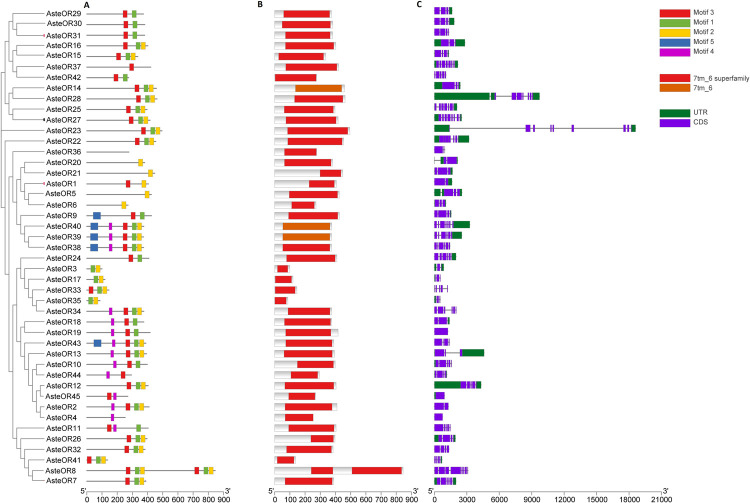
Gene structure of the ORs of An. stephensi has been represented in the Fig 10. 7tm_6 domain was present in all the sequences. CDS were present in the sequences whereas some sequences lacked UTRs. Motif 2 and Motif 3, predicted by MEME, were present in most of the sequences except OR36. Whereas motif 1, 4, and 5 were present in few sequences.
Fig 10. Gene structure of the ORs.
Conserved MEME motifs (A), Pfam domains (B) and UTR/CDS (C) are represented in the figure. Structural domain 7tm_6 of the odorant receptors is present in all the ORs. Whereas some sequences lack the conserved motifs due to partial sequence. UTRs/CDS along with intronic regions are also represented.
Prediction of physicochemical properties and subcellular localization of the OBPs and ORs
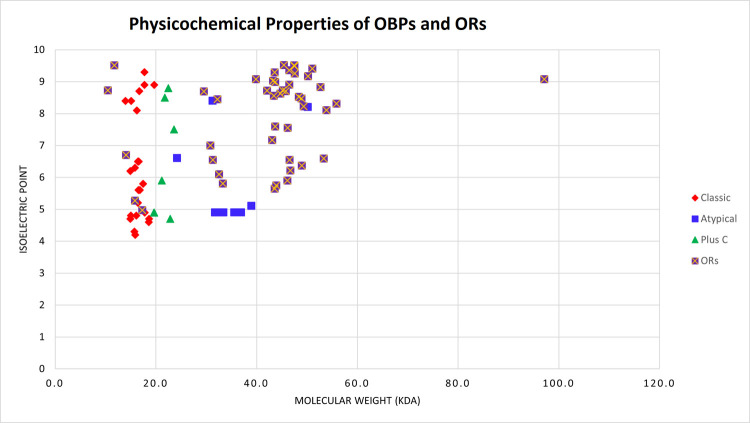
The molecular weight of the atypical OBPs varied from the 24.2 kDa to 39.0 kDa except for the OBP8 that had 50.2 kDa as presented in Table 1. In classic OBPs, molecular weight ranged between the 14 kDa to 20.7 kDa. Plus-C OBPs had the molecular weight in the range of 19.7–23.6 kDa. Isoelectric point, a pH at which molecule is neutral, ranged from 4.2 to 8.9 for all the OBPs as given in Table 1.
The molecular weight of the ORs varied from 30.798 kDa to 97.083 kDa for OR6 and OR8 respectively as shown in Table 2. However, some sequences that had partial 7tm_6 (Odorant Receptor Domain) had less molecular weight. Isoelectric point ranged between the 4.97 and 9.52 for OR33 and OR43, respectively. Isoelectric points of the OBPs and ORs in relation to the molecular weight are shown in the Fig 11.
Fig 11. Physicochemical properties of OBPs and ORs.
Molecular weight and isoelectric point of the OBPs and ORs are shown as these are important indicators of the OBPs and ORs functions. Atypical OBPs had highest molecular weight among other classes whereas ORs have higher molecular weight than OBPs.
CELLO and WoLF PSORT predicted extra-cellular localization of the OBPs indicating that OBPs are secretory proteins whereas ORs were predicted to be membrane embedded that proved their transmembrane propensity.
Discussion
In An. stephensi, 44 OBPs are identified. There are 27 classic, 10 atypical and 7 plus-C OBPs. Similarly, a total of 66 OBP encoding genes were identified in the Ae. aegypti, 66 in An. gambiae, and 49 in D. melanogaster [47–49]. Like An. stephensi, classic OBPs are abundant than atypical and plus-C OBPs in other insect species including Ae. aegypti and An. gambiae as well [14,50,51]. There were total 41, 29, and 30 classic OBPs identified in Ae. aegypti, An. gambiae, and D. melanogaster respectively [25,26,52]. Atypical OBPs are more abundant than the plus-C OBPs in insects. In Ae. aegypti, there were more plus-C OBPs than the atypical contradicting with other insects. There were 10, 16 and 12 atypical OBPs identified in Ae. aegypti, An. gambiae, and D. melanogaster, respectively. However, plus-C OBPs were 16, 12 and 12 in Ae. aegypti, An. gambiae, and D. melanogaster, respectively. Odorant Receptors (ORs) also vary in their number between insect species. A total of 45 ORs have been identified in the An. stephensi. Like An. stephensi, the numbers of identified ORs were 75 in An. gambiae, 61 in D. melanogaster, and 131 in Ae. aegypti [33,53,54]. However, a total of 226 ORs were identified in Aenasius bambawalei belonging to order Hymenoptera [55]. Some species can contain less ORs, for instance, only 8 ORs have been identified in Tomicus yunnanensis that belongs to order Coleoptera [56].
Most of the OBP genes are localized on chromosome 2 of An. stephensi. Whereas Chromosome X contains six OBP genes. Similarly, the least number of OBP genes were found on chromosome X whereas most were present on chromosome 3L in An. gambiae [52]. Clusters of OBP encoding genes are present on all chromosomes whereas some of these genes are scattered all over the chromosomes in An. stephensi. A similar pattern of clustering has been reported for D. melanogaster that have four clusters of OBPs genes [25]. Like OBPs, ORs are also abundant on chromosome 2 whereas chromosome X contains only 4 OR genes in An. stephensi. This indicates a pattern that odorant genes are abundant on the chromosome 2 whereas chromosome X contains only few odorant genes in the insects.
Phylogenetic analysis showed a close relationship of the OBPs and ORs of An. stephensi with An. gambiae as both are closely related organisms. Whereas few OBPs and ORs of An. stephensi showed relationship with the OBPs and ORs of Ae. aegypti. However, OBPs and ORs were distantly related to the OBPs and ORs of the D. melanogaster among compared organisms. Because An. stephensi shares same genus with An. gambiae whereas Ae. aegypti belong to different genus. However, D. melanogaster belonged to different family as compared to An. stephensi. Like Anopheles stephensi, OBPs and ORs of the Ae. aegypti also showed close relationship with An. gambiae OBPs and ORs because of the closer relationship between both organism than the distantly related D. melanogaster [26]. Likewise, Culex quinquefasciatus OBPs and ORs showed the closer relationship with OBPs and ORs of Ae. aegypti as compared to D. melanogaster [57]. Syntenic analysis showed more homologous OBPs and ORs between An. stephensi and An. gambiae genomes. Whereas less syntenic regions of An. stephensi OBPs and ORs are predicted with the OBPs and ORs of Ae. aegypti. None of the OBPs and ORs of An. stephensi shared homology with the D. melanogaster OBPs and ORs, syntenic analysis revealed. Similarly, in Ae. aegypti, OBPs were more related to An. gambiae than D. melanogaster. It would have been interesting to compare An. stephensi, a hematophagous species, with a non-hematophagous mosquito species but genome assemblies of the later, such as like Toxorhynchites splendens are currently yet to be sequenced.
PBP/GOBP domain (Pfam ID: PF01395) is conserved in all the OBPs despite of their subclasses. Like OBPs of other insects, classic and atypical OBPs of An. stephensi had significant similarity with PBP/GOBP domain and conserved motifs as well. Whereas plus-C OBPs had insignificant similarity to PBP/GOBP domain even though conserved plus-C OBP motif is present in these OBPs as checked with multiple sequence alignment. Similarly, ORs contain 7tm_6 domain (Pfam ID: PF02949) that is a transmembrane protein domain having 7 helices [54,58].
Physicochemical analysis of the OBPs predicted that atypical OBPs have the highest molecular weight in An. stephensi that is followed by the plus-C and classic OBPs. Similar results have been found in the OBPs of Ae. aegypti and An. gambiae as classic OBPs had molecular weight less than 15.5 kDa [47,49]. Whereas atypical OBPs of Ae. aegypti and An. stephensi had molecular weight between 27 and 38 kDa along with plus-C OBPs having molecular weight between 25–35 kDa [47,49]. However, ORs are high molecular weight proteins as their molecular weight varies from 30 kDa to 97 kDa in An. stephensi. The isoelectric point of the OBPs of dipteran species is found between 4 and 10 as is the case with Ae. aegypti OBPs [26]. Similarly, pI was between 4 and 10 for the An. stephensi OBPs. ORs also had pI ranged between 4 and 10 but it has not reported previously in any insect species.
With advancements in computation biology, new techniques are being devised for the functional role of the OBPs. New attractants and repellents are being discovered on the basis of molecular docking, quantitative structure-activity-relationship (QSAR), and molecular dynamic simulations [59,60]. QSAR has been used extensively for the identification of new repellents and for the discovery of novel attractants [61,62]. Similarly, molecular docking is a fast and reliable method to screen multiple ligands for the identification of OBP-semiochemicals interactions [63]. Molecular simulations helps in the retention of ligand-receptor interactions for an extended period to induce behavioral response, thus helping to predict novel attractants [64,65]. Identification of OBPs is the first step towards the computational biology-based discovery of the novel attractants and repellents. It will not only be cost efficient but also time efficient for the in-silico screening of large chemical libraries. So, the identification of the OBPs and ORs of An. stephensi, given their vital role in olfaction process, will help in further understand mosquito olfactory system. It will help in the identification of novel attractants and repellents to control human malaria vector, An. stephensi.
Conclusion
Odorant Binding Proteins (OBPs) are the first responder in the insect olfactory mechanism delivering the semiochemicals to the Odorant Receptors (ORs) in insects including An. stephensi. Total of the 44 OBPs and 45 ORs have been identified in the An. stephensi by this study. OBPs were further classified into the classic (27), atypical (10), and plus-C OBPs (7) based on the presence of the conserved motifs. Phylogenetic analysis revealed close relationship of OBPs and ORs of An. stephensi with An. gambiae. However, very few OBPs and ORs were related to Ae. aegypti OBPs and ORs but no relationship was established with D. melanogaster OBPs and ORs. Syntenic analysis showed close homology with An. gambiae OBPs. Physicochemical properties predicted the molecular weight and isoelectric point of the OBPs and ORs within previously reported range of OBPs and ORs. This study revealed the OBPs and ORs in the Anopheles stephensi which can be further characterized by NMR and crystallographic studies and help in the identification of novel compounds to control the spread of An. stephensi.
Supporting information
(TXT)
(TXT)
(TXT)
(TXT)
ORFs were predicted by ORF Finder. Similarly, OBP’s names and accessions have also been given.
(DOCX)
ORFs were predicted by ORF Finder. Similarly, OR’s names and accessions have also been given.
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Rickman LS, Jones TR, Long GW, Paparello S, Schneider I, Paul CF, et al. Plasmodium falciparum-infected Anopheles stephensi inconsistently transmit malaria to humans. NAVAL MEDICAL RESEARCH INST BETHESDA MD; 1990. [DOI] [PubMed] [Google Scholar]
- 2.Tadesse FG, Ashine T, Teka H, Esayas E, Messenger LA, Chali W, et al. Anopheles stephensi Mosquitoes as Vectors of Plasmodium vivax and falciparum, Horn of Africa, 2019. 2021;27(2):603. doi: 10.3201/eid2702.200019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh SK, Tiwari S, Raghavendra K, Sathyanarayan TS, Dash APJJob. Observations on sporozoite detection in naturally infected sibling species of the Anopheles culicifacies complex and variant of Anopheles stephensi in India. 2008;33(3):333–6. doi: 10.1007/s12038-008-0052-5 [DOI] [PubMed] [Google Scholar]
- 4.Djadid ND, Gholizadeh S, Aghajari M, Zehi AH, Raeisi A, Zakeri SJAt. Genetic analysis of rDNA-ITS2 and RAPD loci in field populations of the malaria vector, Anopheles stephensi (Diptera: Culicidae): implications for the control program in Iran. 2006;97(1):65–74. doi: 10.1016/j.actatropica.2005.08.003 [DOI] [PubMed] [Google Scholar]
- 5.Seyfarth M, Khaireh BA, Abdi AA, Bouh SM, Faulde MKJPr. Five years following first detection of Anopheles stephensi (Diptera: Culicidae) in Djibouti, Horn of Africa: populations established—malaria emerging. 2019;118(3):725–32. doi: 10.1007/s00436-019-06213-0 [DOI] [PubMed] [Google Scholar]
- 6.Faulde MK, Rueda LM, Khaireh BAJAt. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti, Horn of Africa. 2014;139:39–43. doi: 10.1016/j.actatropica.2014.06.016 [DOI] [PubMed] [Google Scholar]
- 7.Takken W, Lindsay SJEid. Increased threat of urban malaria from Anopheles stephensi mosquitoes, Africa. 2019;25(7):1431. doi: 10.3201/eid2507.190301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gakhar S, Sharma R, Sharma A. Population genetic structure of malaria vector Anopheles stephensi Liston (Diptera: Culicidae). 2013. [PubMed] [Google Scholar]
- 9.Gholizadeh S, Djadid ND, Nouroozi B, Bekmohammadi MJAt. Molecular phylogenetic analysis of Anopheles and Cellia subgenus anophelines (Diptera: Culicidae) in temperate and tropical regions of Iran. 2013;126(1):63–74. doi: 10.1016/j.actatropica.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 10.Healy T, Copland M, Cork A, Przyborowska A, Halket JJM, entomology v. Landing responses of Anopheles gambiae elicited by oxocarboxylic acids. 2002;16(2):126–32. doi: 10.1046/j.1365-2915.2002.00353.x [DOI] [PubMed] [Google Scholar]
- 11.Dekker T, Steib B, Cardé R, Geier MJM, entomology v. L‐lactic acid: a human‐signifying host cue for the anthropophilic mosquito Anopheles gambiae. 2002;16(1):91–8. doi: 10.1046/j.0269-283x.2002.00345.x [DOI] [PubMed] [Google Scholar]
- 12.Hoffman SA, Aravind L, Velmurugan SJP, vectors. Female Anopheles gambiae antennae: increased transcript accumulation of the mosquito-specific odorant-binding-protein OBP2. 2012;5(1):1–6. doi: 10.1186/1756-3305-5-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun JS, Xiao S, Carlson JRJRSOB. The diverse small proteins called odorant-binding proteins. 2018;8(12):180208. doi: 10.1098/rsob.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venthur H, Zhou J-JJFip. Odorant receptors and odorant-binding proteins as insect pest control targets: a comparative analysis. 2018;9:1163. doi: 10.3389/fphys.2018.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chertemps T, Maïbèche M. Odor degrading enzymes and signal termination. Insect Pheromone Biochemistry and Molecular Biology: Elsevier; 2021. p. 619–44. [Google Scholar]
- 16.Vogt RG, Riddiford LMJN. Pheromone binding and inactivation by moth antennae. 1981;293(5828):161–3. doi: 10.1038/293161a0 [DOI] [PubMed] [Google Scholar]
- 17.Mazumder S, Chaudhary BP, Dahal SR, Al-Danoon O, Mohanty SJB. Pheromone Perception: Mechanism of the Reversible Coil–Helix Transition in Antheraea polyphemus Pheromone-Binding Protein 1. 2019;58(45):4530–42. doi: 10.1021/acs.biochem.9b00737 [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Murphy EJ, Nix JC, Jones DNJSr. Aedes aegypti Odorant Binding Protein 22 selectively binds fatty acids through a conformational change in its C-terminal tail. 2020;10(1):1–15. doi: 10.1038/s41598-020-60242-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G-Q, Li D-Z, Lu Y-L, Wang Y-Q, Kong D-X, Wang M-QJSr. Deciphering the Odorant Binding, Activation, and Discrimination Mechanism of Dhelobp21 from Dastarus Helophoroides. 2018;8(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damberger FF, Michel E, Ishida Y, Leal WS, Wüthrich KJPotNAoS. Pheromone discrimination by a pH-tuned polymorphism of the Bombyx mori pheromone-binding protein. 2013;110(46):18680–5. doi: 10.1073/pnas.1317706110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J-JJV, Hormones. Odorant-binding proteins in insects. 2010;83:241–72. [DOI] [PubMed] [Google Scholar]
- 22.He P, Zhang J, Liu N-Y, Zhang Y-N, Yang K, Dong S-LJPO. Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stål. 2011;6(12):e28921. doi: 10.1371/journal.pone.0028921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brito NF, Moreira MF, Melo ACJJoIP. A look inside odorant-binding proteins in insect chemoreception. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 24.Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MAJGr. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. 2002;12(9):1357–69. doi: 10.1101/gr.239402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham LA, Davies PLJG. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. 2002;292(1–2):43–55. doi: 10.1016/s0378-1119(02)00672-8 [DOI] [PubMed] [Google Scholar]
- 26.Zhou JJ, He XL, Pickett J, Field LJImb. Identification of odorant‐binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. 2008;17(2):147–63. doi: 10.1111/j.1365-2583.2007.00789.x [DOI] [PubMed] [Google Scholar]
- 27.Vogt RGJJoce. Odorant binding protein homologues of the malaria mosquito Anopheles gambiae; possible orthologues of the OS-E and OS-F OBPs of Drosophila melanogaster. 2002;28(11):2371–6. doi: 10.1023/a:1021009311977 [DOI] [PubMed] [Google Scholar]
- 28.Pelletier J, Leal WSJPo. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. 2009;4(7):e6237. doi: 10.1371/journal.pone.0006237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengul M, Tu ZJImb. Characterization and expression of the odorant‐binding protein 7 gene in Anopheles stephensi and comparative analysis among five mosquito species. 2008;17(6):631–45. doi: 10.1111/j.1365-2583.2008.00837.x [DOI] [PubMed] [Google Scholar]
- 30.Sengul MS, Tu ZJImb. Identification and characterization of odorant‐binding protein 1 gene from the Asian malaria mosquito, Anopheles stephensi. 2010;19(1):49–60. doi: 10.1111/j.1365-2583.2009.00929.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speth Z, Kaur G, Mazolewski D, Sisomphou R, Siao DDC, Pooraiiouby R, et al. Characterization of Anopheles stephensi Odorant Receptor 8, an Abundant Component of the Mouthpart Chemosensory Transcriptome. Insects. 2021;12(7):593. doi: 10.3390/insects12070593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Q, Chess AJG. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. 1999;60(1):31–9. doi: 10.1006/geno.1999.5894 [DOI] [PubMed] [Google Scholar]
- 33.Rinker DC, Zhou X, Pitts RJ, Rokas A, Zwiebel LJ. Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles gambiae. BMC genomics. 2013;14(1):1–15. doi: 10.1186/1471-2164-14-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vosshall LB, Hansson BSJCs. A unified nomenclature system for the insect olfactory coreceptor. 2011;36(6):497–8. doi: 10.1093/chemse/bjr022 [DOI] [PubMed] [Google Scholar]
- 35.De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. 2006;34(suppl_2):W362–W5. doi: 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths‐Jones S, et al. The Pfam protein families database. 2004;32(suppl_1):D138–D41. doi: 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe D, Dudek S, Ritchie MD, Pendergrass SAJBm. Visualizing genomic information across chromosomes with PhenoGram. 2013;6(1):1–12. doi: 10.1186/1756-0381-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall T, Biosciences I, Carlsbad CJGBB. BioEdit: an important software for molecular biology. 2011;2(1):60–1. [Google Scholar]
- 39.Kumar S, Stecher G, Li M, Knyaz C, Tamura KJMb, evolution. MEGA X: molecular evolutionary genetics analysis across computing platforms. 2018;35(6):1547. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJJB. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. 2009;25(9):1189–91. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin APJPo. FastTree 2–approximately maximum-likelihood trees for large alignments. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork PJNar. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. 2019;47(W1):W256–W9. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Chen H, He Y, Xia RJB. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. 2018:289660. [Google Scholar]
- 44.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. 2012;40(7):e49–e. doi: 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey TL, Johnson J, Grant CE, Noble WSJNar. The MEME suite. 2015;43(W1):W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. 2010;39(suppl_1):D225–D9. doi: 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu P, Zwiebel L, Smith D. Identification of a distinct family of genes encoding atypical odorant‐binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect molecular biology. 2003;12(6):549–60. doi: 10.1046/j.1365-2583.2003.00440.x [DOI] [PubMed] [Google Scholar]
- 48.Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome research. 2002;12(9):1357–69. doi: 10.1101/gr.239402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou JJ, He XL, Pickett J, Field L. Identification of odorant‐binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect molecular biology. 2008;17(2):147–63. doi: 10.1111/j.1365-2583.2007.00789.x [DOI] [PubMed] [Google Scholar]
- 50.Pelosi P, Maida RJCB, Biochemistry PPB, Biology M. Odorant-binding proteins in insects. 1995;111(3):503–14. doi: 10.1016/0305-0491(95)00019-5 [DOI] [PubMed] [Google Scholar]
- 51.Pelosi P, Zhu J, Knoll WJS. Odorant-binding proteins as sensing elements for odour monitoring. 2018;18(10):3248. doi: 10.3390/s18103248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li ZX, Pickett JA, Field LM, Zhou JJJAoIB, America PPiCwtESo. Identification and expression of odorant‐binding proteins of the malaria‐carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. 2005;58(3):175–89. doi: 10.1002/arch.20047 [DOI] [PubMed] [Google Scholar]
- 53.Shiao M-S, Fan W-L, Fang S, Lu M-YJ, Kondo R, Li W-H. Transcriptional profiling of adult Drosophila antennae by high-throughput sequencing. Zoological Studies. 2013;52(1):1–10. [Google Scholar]
- 54.Bohbot J, Pitts R, Kwon HW, Rützler M, Robertson H, Zwiebel LJImb. Molecular characterization of the Aedes aegypti odorant receptor gene family. 2007;16(5):525–37. doi: 10.1111/j.1365-2583.2007.00748.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nie X, Li Q, Xu C, Li D, Zhang Z, Wang MQ, et al. Antennal transcriptome and odorant binding protein expression profiles of an invasive mealybug and its parasitoid. 2018;142(1–2):149–61. [Google Scholar]
- 56.Liu N-Y, Li Z-B, Zhao N, Song Q-S, Zhu J-Y, Yang BJCB, et al. Identification and characterization of chemosensory gene families in the bark beetle, Tomicus yunnanensis. 2018;25:73–85. doi: 10.1016/j.cbd.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 57.Manoharan M, Ng Fuk Chong M, Vaïtinadapoulé A, Frumence E, Sowdhamini R, Offmann BJGb, et al. Comparative genomics of odorant binding proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. 2013;5(1):163–80. doi: 10.1093/gbe/evs131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ray A, Van Naters WVDG, Carlson JRJJob. Molecular determinants of odorant receptor function in insects. 2014;39(4):555–63. doi: 10.1007/s12038-014-9447-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basak SC, Bhattacharjee AKJCmc. Computational approaches for the design of mosquito repellent chemicals. 2020;27(1):32–41. doi: 10.2174/0929867325666181029165413 [DOI] [PubMed] [Google Scholar]
- 60.Oliferenko PV, Oliferenko AA, Poda G, Pillai GG, Bernier UR, Agramonte NM, et al. Insect olfactory system as target for computer-aided design of mosquito repellents. Computational Design of Chemicals for the Control of Mosquitoes and their Diseases: CRC Press; 2017. p. 39–64. [Google Scholar]
- 61.Devillers JJS, Research QiE. 2d and 3d structure–activity modelling of mosquito repellents: A review. 2018;29(9):693–723. doi: 10.1080/1062936X.2018.1513218 [DOI] [PubMed] [Google Scholar]
- 62.Bhadoriya KS, Sharma MC, Jain SV, Raut GS, Rananaware JRJMCR. Three-dimensional quantitative structure–activity relationship (3D-QSAR) analysis and molecular docking-based combined in silico rational approach to design potent and novel TRPV1 antagonists. 2013;22(5):2312–27. [Google Scholar]
- 63.Gopal JV, Kannabiran KJISCLS. Studies on interaction of insect repellent compounds with odorant binding receptor proteins by in silico molecular docking approach. 2013;5(4):280–5. doi: 10.1007/s12539-013-0152-2 [DOI] [PubMed] [Google Scholar]
- 64.Bezerra-Silva PC, Dutra KA, Santos GK, Silva RC, Iulek J, Milet-Pinheiro P, et al. Evaluation of the activity of the essential oil from an ornamental flower against Aedes aegypti: electrophysiology, molecular dynamics and behavioral assays. 2016;11(2):e0150008. doi: 10.1371/journal.pone.0150008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta KK, Sethi G, Jayaraman MJJovbd. Molecular docking and simulation studies of gustatory receptor of Aedes aegypti: A potent drug target to distract host-seeking behaviour in mosquitoes. 2016;53(2):179. [PubMed] [Google Scholar]