FIGURE 2.
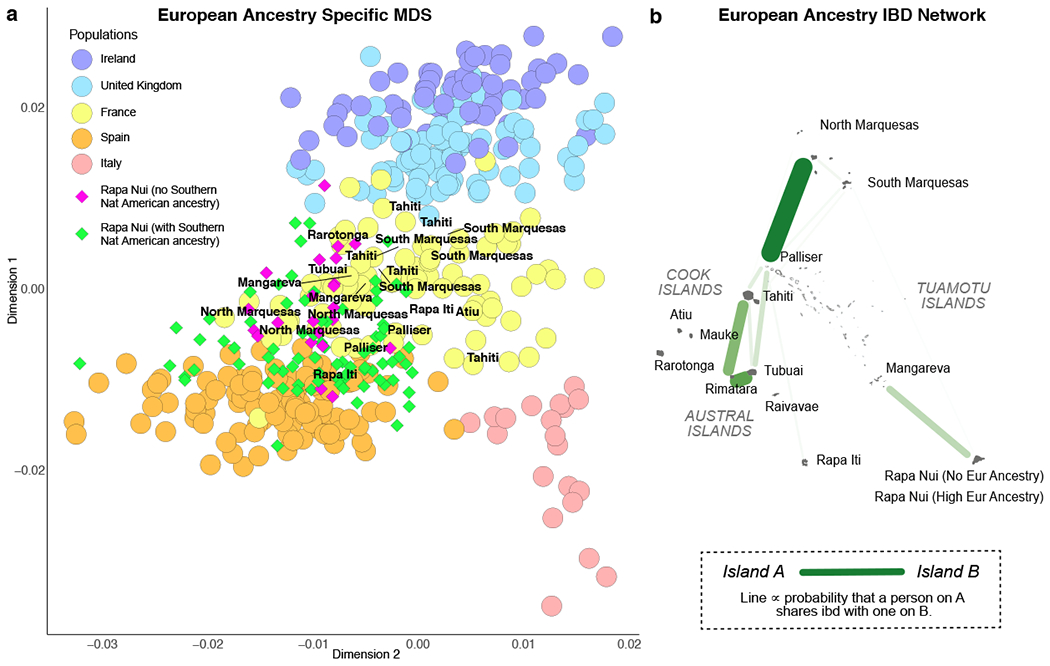
Relationship between Polynesian, Native American, and European ancestries. (a) Random-forest based local ancestry inference of a Rapanui individual showing small (old) Native American ancestry tracts embedded in Polynesian ancestry tracts. The ancestry of each haploid genome is coloured (top and bottom for each homologous chromosome pair); the autosome pairs are numbered along the vertical axis. (b) Ternary plot of ADMIXTURE ancestry fractions in Rapanui individuals having Polynesian, European, and central Native American, but no other, ancestries (each point corresponds to an individual). The first principal component in the centered log-ratio transform space27 is projected onto the figure as a dashed curve. The ancestries’ log-ratio variances are discussed in Supplementary Data Tables 7–10. (c-d) Length distribution analyses for ancestry tracts in the six Rapanui individuals having no European ancestry (c) and in North Marquesan individuals (d). Plotted points show the aggregate tract length counts, lines show the maximum likelihood best fit tract length distributions, and shading shows the one standard deviation confidence intervals assuming Gaussian noise. The best fit admixture chronology is plotted above the timeline as a line-history with each colour representing an ancestry as indicated in the key (see a).
