Abstract
The Mediator complex controls RNA polymerase II (pol II) activity by coordinating the assembly of pol II regulatory factors at transcription start sites, and by mediating interactions between enhancer-bound transcription factors (TFs) and the pol II enzyme. Mediator structure and function is completely altered upon binding the Mediator kinase module, a multi-subunit complex that contains CDK8 or its vertebrate-specific paralog CDK19. Here, we review the mechanisms by which the Mediator kinase module controls pol II transcription, emphasizing its impact on TF activity, pol II elongation, enhancer function, and chromatin architecture. We also highlight how the Mediator kinase module integrates signaling pathways with transcription to enable rapid, stimulus-specific responses, as well as its links to human disease.
Keywords: MED12, MED13, eRNA, enhancer-promoter loops, pol II pausing, metabolism
Kinase module structure and Mediator interaction
The Mediator complex (see Glossary) is a genome-wide regulator of RNA polymerase II (pol II) transcription; consequently, Mediator itself is targeted by an array of factors that regulate its function. For example, sequence-specific, DNA-binding transcription factors (TFs) bind Mediator and control its recruitment to specific genomic loci. Also, the Mediator kinase module reversibly associates with Mediator (forming what we here call CDK-Mediator) and regulates Mediator function in several ways. Conserved from yeast to humans, the Mediator kinase module consists of four subunits: the CDK8 kinase, CCNC, MED12, and MED13. However, vertebrates evolved subunit paralogs, called CDK19, MED12L, and MED13L (Box 1), which expand the functional diversity of the kinase module in ways that remain poorly defined. Not surprisingly, Mediator kinase module subunits are required for mammalian embryogenesis [1-4] and are linked to myriad diseases (Box 2).
Box 1: Vertebrate-specific paralogs of Mediator kinase module subunits.
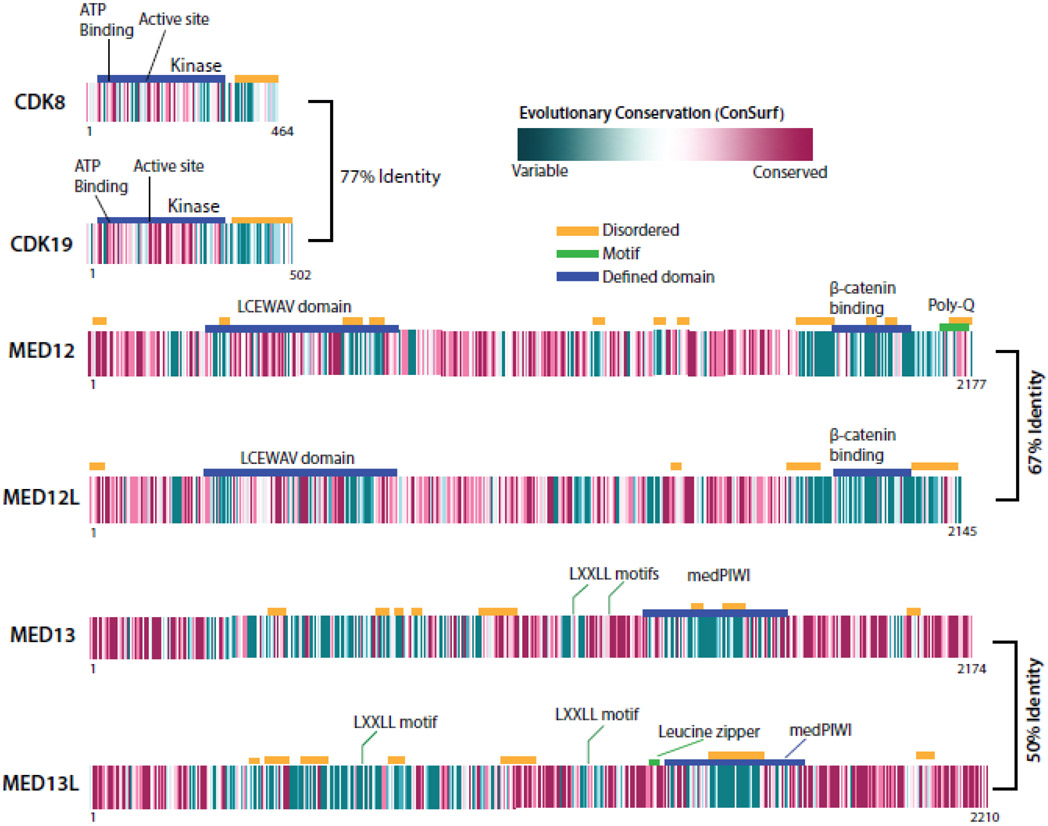
The Mediator kinase module is conserved among eukaryotes, but paralogs for CDK8, MED12, and MED13 emerged in vertebrates (see Figure I). Each paralog is expressed on different chromosomes and appears to be mutually exclusive within the kinase module [52]. Comparatively little is known about how these paralogs function, but connections to human disease have been discovered (Supplemental Table 1).
CDK8 and CDK19 have the highest sequence identity among the kinase module paralogs, and inhibitors invariably block both proteins. CDK8 and CDK19 show evidence of redundant [109] and non-redundant [52] functions, and each has been shown to regulate transcription in both kinase-dependent and kinase-independent ways [25, 87, 109, 117].
The MED12 protein is implicated in numerous X-linked diseases (Supplemental Table 1) and is expressed on the X chromosome whereas the MED12L gene resides on chromosome 3. Interestingly, CDK8 function has been linked to Xist repression (CDK19 no effect) in mice [118]. MED12L shows more restricted expression across tissues compared with MED12. Whereas MED12 is necessary for activation of Mediator kinase activity [8, 81], it is unknown whether MED12L activates CDK8/19 function. The sequence similarity between MED12 and MED12L in the N-terminal activation helix (Figure I) suggests a similar role in kinase activation.
One basic function for MED13 is to link the kinase module to the Mediator complex [10, 11]. Notably, proteomics data suggest that kinase modules containing MED13L (instead of MED13) maintain association with Mediator [39, 119]. Moreover, both MED13 and MED13L are ubiquitylated by FBW7, which initiates subunit dissociation and degradation [110]. Clinical data suggest similar, but not identical, biological roles for MED13 and MED13L (see Supplemental Table 1).
BOX 1 Figure. Sequence conservation and domain structure of human Mediator kinase module subunits.
Regions marked “disordered” are predicted to be intrinsically disordered, and defined domains are individually labeled. Evolutionary conservation was calculated from 150 homologs of at least 35% identity, using ConSurf [120]. Sequence identity between paralogs is noted.
Box 2: Mediator kinase module and disease.
Mutations in Mediator kinase module subunits cause disability and disease (see Supplementary Table 1), which can be broadly grouped into two categories: cancer or neurological/developmental disorders (reviewed in [7]). In addition, a wide range of cancers are linked to altered expression of Mediator kinase module subunits, which are summarized in recent reviews [121, 122].
Three medically related intellectual/developmental syndromes have been linked to mutations in MED12: Opitz-Kaveggia, Lujan, and Ohdo syndrome. Furthermore, mutations in the CDK8 or CDK19 kinase domains are associated with intellectual disorders that exhibit comparable phenotypes to individuals with MED12 mutations (Supplemental Table 1). Notably, whereas introduction of CDK19 could compensate for CDK8 deletion in Drosophila, mutant CDK19 associated with human neurological disorders could not, resulting in seizures and reduced fitness in surviving flies [123]. Likewise, induced expression of pathogenic human MED12 mutants in mice resulted in developmental defects [124]. These results suggest conserved biological functions for disease-associated mutations in MED12 and CDK19.
Mutations in MED12 associated with non-malignant uterine leiomyoma are among the most well-studied (Supplemental Table 1), and cause changes in enhancer-promoter looping and chromatin architecture [79] and negatively impact CDK8 or CDK19 activity [8, 9]. Such functional defects are consistent with the role of the Mediator kinase module in TF regulation and super-enhancer function [67, 74, 75].
Targeting Mediator kinase function for therapeutic benefit remains a work in progress, but novel strategies continue to emerge. For example, the Firestein lab showed that bromodomain and extraterminal domain (BET) inhibitors (e.g. JQ1) may complement CDK8 + CDK19 kinase inhibition in certain cancers [109]. Moreover, compensatory increases in enhancer occupancy of MED12 and the BET protein BRD4 were observed in CDK8 + CDK19 double knockout cells (HCT116 or DLD1), suggesting functional cooperativity between the Mediator kinase module and BRD4 [109], in agreement with other studies [51, 125].
Although current structural data for the human Mediator kinase module consists only of the CDK8-CCNC dimer [5], a cryoEM structure of the yeast (Saccharomyces cerevisiae) kinase module was recently determined by the Tsai lab [6]. This structure provided the first high-resolution data for the large Med12 and Med13 subunits. Notably, Med12 was identified as a key structural component in the yeast Mediator kinase module. The Med12 N-terminus interacts with the Cdk8–Ccnc dimer, and the Med12 C-terminus interacts with Med13 (Figure 1). The N-terminus of human MED12 is mutated in a variety of cancers [7], with mutations clustering around residues 36-44 (Supplementary Table 1). Structural and functional data from Tsai and co-workers show that these residues occupy a conserved activation helix in the yeast complex. This Med12 activation helix interacts with Cdk8 and stabilizes an otherwise disordered Cdk8 kinase activation loop, which allows substrate access to the active site [6]. Additionally, prior work from the Boyer lab showed reduced CDK8 or CDK19 kinase activity with MED12 oncogenic mutations in its N-terminus [8, 9], suggesting that the structural model for Med12-dependent activation of yeast Cdk8 is likely conserved in humans.
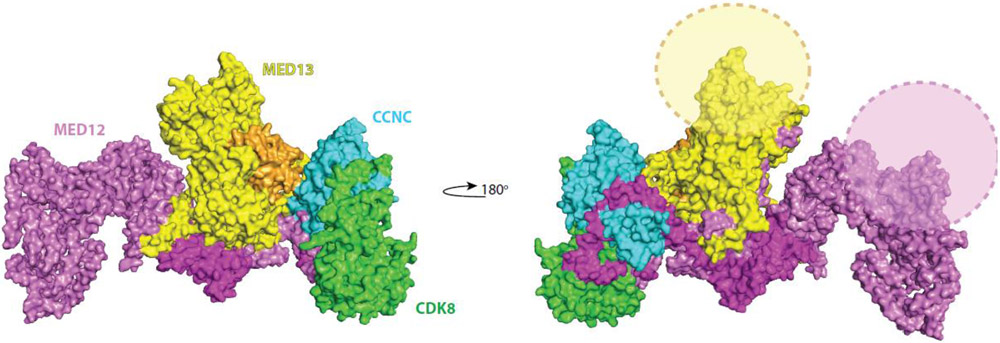
Figure 1. Structural model of the yeast (S. cerevisiae) Mediator kinase module [6].
Note the MED12 N-terminus (dark pink; at right) interacts with CDK8 and CCNC whereas the MED12 C-terminus (light pink) interacts with MED13. High-resolution data for the human complex are lacking but can be expected to show similar features. The subunit sequence similarity is 38%, 40%, 24%, and 22% for CDK8, CCNC, MED12, and MED13, respectively; also, human MED12 and MED13 each contain about 750 additional amino acids compared with yeast subunits. These additional amino acids are designated with semi-transparent ovals (right panel). PDB ID: 7KPX
While high resolution structural details of the kinase module interaction with Mediator are not available, the interaction requires MED13 [10, 11]. The Cramer lab recently completed a crosslinking-mass spectrometry analysis with yeast (S. cerevisiae) CDK-Mediator [12]. The data suggest extensive interactions between the kinase module and Mediator, including evidence for Med12 and/or Med13 interactions with Med19 and Med10, among other subunits. Med19 and Med10 reside in the hook domain of Mediator [13], which also represents a structural interface between Mediator and the TFIIH-associated kinase module (see below).
In this review, we highlight some of the regulatory functions of the Mediator kinase module, considering past data in the context of current results. We start by emphasizing how the Mediator kinase module enables transcriptional responses to cell signaling cascades to help "reprogram" gene expression patterns to changing conditions. We then discuss Mediator kinase module-dependent regulation of pol II function at different transcriptional stages (initiation, pausing, elongation), and highlight new structural data that have clarified and expanded upon results from biochemical and cell-based experiments. Finally, enhancers represent powerful genomic regulatory elements that coordinate cell type-specific and stimulus-specific gene expression programs, and we outline how the Mediator kinase module may contribute to enhancer function, through control of TF activity and enhancer-promoter looping. Throughout, we highlight areas in which understanding remains limited, and we conclude with open questions for future research.
Mediator kinase module connects cell signaling with transcription
TFs control pol II function, genome-wide, through recruitment of Mediator, chromatin remodelers, and other factors to specific genomic loci. In this way, TFs serve as "master regulators" of pre-initiation complex (PIC) assembly and function. Activation of signaling pathways causes changes in TF phosphorylation that impact TF nuclear localization and/or TF activity on chromatin (Figure 2). As examples, i) interferon-induced phosphorylation of STAT TFs triggers their nuclear localization to allow target gene activation, and ii) the ELK1 TF is phosphorylated on chromatin during MAPK pathway activation, which enhances ELK1-dependent recruitment of Mediator [14]. In each of these representative examples, TFs are the endpoints of signaling cascades. Importantly, TFs are common targets of Mediator kinases (see below), yielding a direct link to cell signaling.
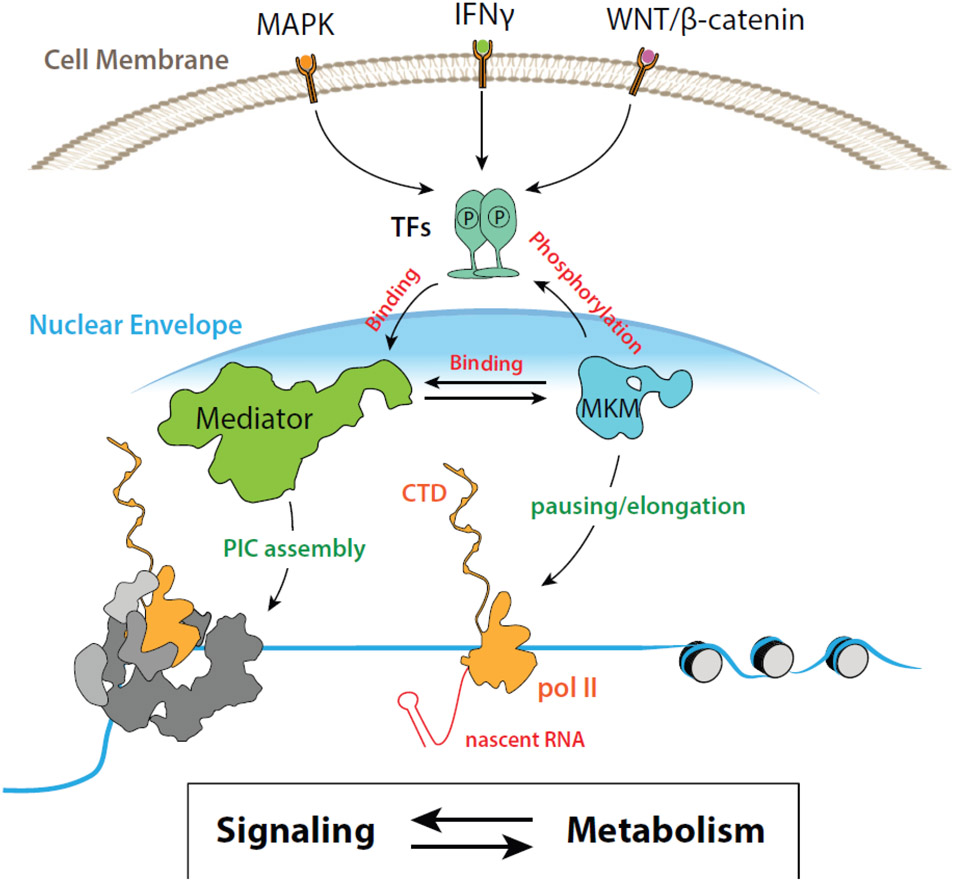
Figure 2. The Mediator kinase module coordinates signaling inputs and transcriptional outputs.
Signaling pathways such as MAPK, IFN, and WNT/β-catenin, direct transcriptional responses through transcription factors (TFs); TFs then recruit Mediator to enhancers and promoters to enable pre-initiation complex (PIC) assembly and activation of pol II transcription. The Mediator kinase module (MKM) controls TF function through phosphorylation; upon binding Mediator, the MKM blocks Mediator-pol II interaction and therefore regulates PIC assembly. The MKM also appears to influence pol II pausing and elongation via its kinase activity and through physical association with the super elongation complex (SEC). However, the molecular mechanisms remain unclear.
Coordination between the Mediator kinase module and cell signaling is evident from ancient and conserved links to metabolism. Signaling and metabolic pathways are integrated and interdependent, such that altered signaling will trigger metabolic adaptation, and vice versa (Figure 2) [15]. Whereas signaling cascades can impact metabolic flux by directly changing the activity of metabolic enzymes (e.g. through phosphorylation), they modulate metabolic pathways through transcriptional changes as well. For example, in model organisms such as S. cerevisiae, Cdk8 coordinates metabolic changes in response to nutrients [16-18] via phosphorylation of TFs that control expression of metabolic genes [19, 20]. Similarly, CDK8 indirectly controls metabolism in metazoans through modification of TFs. In mouse and human cells, CDK8 phosphorylates TFs that are major regulators of cell metabolism, such as SREBP [21], Notch ICD [1, 22], SMAD1/3 [23], and STAT1/3/5a [24]. These TFs are endpoints for the insulin, WNT/β-catenin, TGFβ, and interferon signaling cascades, respectively. Importantly, CDK8-dependent phosphorylation altered the stability [21-23] or the activity [25] of these TFs. In this way, Mediator kinases directly regulate downstream transcriptional responses to signaling cascades.
The Espinosa lab has shown that CDK8 kinase activity controls expression of glycolysis genes [26], and numerous signaling pathways converge on glycolysis because of its central role in cell metabolism. For example, glycolytic intermediates can serve as building blocks for nucleic acids (i.e. DNA and RNA), amino acids (proteins), and fatty acids (membranes), which are essential for cellular homeostasis or proliferation. In addition to CDK8, numerous labs have shown that MED12 and MED13 impact metabolism. A common theme among these studies, completed mainly in flies and mice using overexpression or knockdown/knockout methods, was that MED12 or MED13 affect lipid biosynthesis and homeostasis [21, 27-29]. Signaling cascades that influence glycolysis and/or lipid biosynthesis include MAPK, interferon, and WNT/β-catenin; experiments in model organisms have linked Mediator kinase module subunits to WNT/β-catenin [4, 30-32], interferon [24], and Ras/MAPK [30, 33] pathways. Based upon these and other results [34], the kinase module serves as an intermediary between cell signaling and transcription, at least in part, by acting through TFs and/or Mediator (Figure 2).
CDK-Mediator and pol II initiation, pausing, and elongation
Transcription initiation
The pol II enzyme initiates transcription as part of the PIC (Figure 3), which assembles at transcription start sites throughout the genome [35, 36]. As a PIC factor, the Mediator complex is required for pol II transcription, and Mediator serves as a conduit between promoter- and enhancer-bound TFs and the pol II enzyme. Mediator also controls the assembly and activity of other PIC factors, including TFIIH and pol II itself. For instance, biochemical and structural data have shown that Mediator stimulates TFIIH-dependent phosphorylation of the pol II C-terminal domain (CTD), and Mediator is required for TF-dependent activation of transcription [37, 38]. These functions are disrupted upon Mediator binding to the kinase module, as it blocks pol II assembly into the PIC, as shown by biochemical [10, 39] and structural data [6, 11, 12]. That is, kinase module binding to Mediator is mutually exclusive with pol II. The interaction between Mediator and the kinase module is dependent on MED13 [10, 11], based upon biochemical and structural data; however, the Robert lab has shown that, in yeast (S. cerevisiae), Cdk8 kinase activity may also regulate the interaction [40]. Recent cryoEM data of the human PIC also suggest that kinase module-Mediator binding is mutually exclusive with TFIIH [41-43], as depicted schematically in Figure 3. Consequently, CDK-Mediator can block PIC assembly to prevent transcription initiation.
Figure 3. Schematic of the RNA polymerase II Pre-initiation complex (PIC).
PIC structure is based upon cryoEM data from human complexes [41-43]. Each red X denotes Mediator kinase phosphorylation sites (MED14, MED26, TAF10, POLR2M) and their approximate location in the PIC. Because PIC structure is dynamic, specific sites may be phosphorylated only during select transcriptional stages (e.g. pol II pause release). The TFIIH-associated CDK-activating kinase module (CAK) binds Mediator at a similar site compared with the Mediator kinase module (MKM). This site would position the MKM downstream of the transcription start site, which may be important for MKM-dependent regulation of pol II pause release or elongation. IIA: TFIIA, IIB: TFIIB, IID, TFIID, IIE, TFIIE, IIF: TFIIF, IIH: TFIIH, TBP: TATA binding protein, CTD: C-terminal domain, CAK: CDK activating kinase module (CDK7, CCNH, MNAT1), part of TFIIH.
Pol II pausing
The Mediator kinase module can also positively impact pol II transcription. How does this occur? Whereas the mechanisms remain incompletely understood, it is evident that the Mediator kinase module can regulate post-initiation events, which we define here as occurring after pol II initiates transcription, breaks contacts with the PIC, and "escapes" the promoter. Following promoter escape, pol II typically pauses after generating a 20-80 nucleotide transcript [44]. This paused intermediate represents a key regulatory step in which pol II can either continue to transcribe (pause release) or terminate transcription (promoter-proximal termination). Pausing is controlled by many factors, including NELF and TFIID [44, 45]; whereas premature termination is less well-studied, it appears to be regulated in part by SETX and XRN2 [46, 47] (Figure 4). Mediator kinases phosphorylate NELF, TFIID, SETX, and XRN2 in human cells [48], suggesting mechanisms by which CDK8 and/or CDK19 may regulate promoter-proximal pausing or termination. In support, Mediator kinase inhibition increased pol II pausing in the context of IFNγ stimulation, and this activity was linked exclusively to CDK8, not CDK19 [25].
Figure 4. Speculative models for Mediator kinase module regulation of enhancer function and pol II transcription.
(A) CDK-Mediator may prime enhancers for future activation [98] by regulating the activity of enhancer-bound TFs and nearby chromatin modifying or remodeling complexes, through phosphorylation [48]. (B) CDK8/CDK19 phosphorylation of Mediator may promote Mediator kinase module (MKM) dissociation [12], to promote Mediator-dependent enhancer-promoter looping, PIC assembly, and pol II transcription initiation. Mediator kinase activity activates eRNA transcription at enhancers [25] and the Mediator kinase module (MKM) may remain associated with the enhancer through interactions with TFs [94] or eRNA transcripts [95]. (C) This could position the MKM for downstream phosphorylation events (e.g. to regulate pause release or elongation) without requiring association with PIC-bound Mediator. Transcriptional bursting [115] may be favored through formation of transcriptional condensates [116] (green shading) that may compartmentalize initiation factors including multiple pol II enzymes for rapid re-initiation following pause release. Premature termination can also occur at promoter-proximal sites, regulated in part by XRN2 and SETX [46, 47]. The SEC, NELF, XRN2, SETX, are each phosphorylated by the MKM [48]. (D) Re-binding of the MKM to Mediator will block PIC assembly and inhibit further transcription initiation [10], which may serve as a means to shut off transcription and disassemble the PIC. Re-assembly of the PIC could occur over time, to initiate transcription from the promoter once again.
Pol II elongation
Release of pol II pausing is controlled in part by the CDK9 kinase [49], which functions within a two-subunit P-TEFb complex or as part of the larger super elongation complex (SEC) [50]. Proteomics experiments revealed that the CDK-Mediator complex, but not Mediator itself, physically interacts with P-TEFb/SEC [39], and functional cooperativity among the Mediator kinase module and P-TEFb has been observed in response to a variety of stimuli [51, 52]. CDK9 and the Mediator kinases target some of the same proteins (e.g. NELFA, XRN2) [48, 53, 54], and structural data for the human PIC suggest that the Mediator kinase module would orient downstream of the transcription start site [41-43], toward CDK9/SEC, at promoter-proximal pause sites upon Mediator binding (Figures 3 and 4). Given the evidence for functional cooperativity, we speculate that CDK8/CDK19 and CDK9 may co-regulate each other, similar to CDK7-dependent regulation of CDK9 activity [55]. In this way, Mediator kinases could also indirectly regulate pol II pause release and elongation through CDK9/SEC.
The Mediator kinase module and enhancer function
Enhancers are one of the most fundamentally important regulators of pol II transcription in mammalian cells [56], but precisely how enhancers function remains incompletely understood. What is understood, however, is that enhancer function is dependent on TF binding and TFs mediate their biological functions in large part through enhancers. Enhancers are sequences of genomic DNA that contain clustered TF binding sites and can control pol II transcription at promoter regions that may be separated by tens of kilobases (or more) of genomic DNA. Active enhancers will have bound TFs and are typically bidirectionally transcribed by pol II. These bidirectional transcripts, called enhancer RNAs (eRNAs), are capped but unstable, and their regulatory roles remain unclear (but see below). Enhancer-bound TFs mediate their function through recruitment of chromatin remodelers, co-activators such as CBP/p300, and through interaction with promoter-bound PICs (Figure 4). The enhancer-PIC interaction may occur primarily through TF-Mediator-pol II binding, but this remains controversial (see below).
Enhancer sequences are commonly mutated in human diseases [57, 58], and enhancers drive cell differentiation programs to establish lineage-specific transcription [59-61]. Thus, enhancers are cell-type specific [60, 62, 63]. Within the past decade, different types of enhancers have been identified: so-called “typical” enhancers activate sets of genes at moderate levels, whereas “super-enhancers” drive high-level gene expression [64]. Super-enhancers are also distinguished by their large size (encompassing a few to perhaps several dozen kilobases) and their high-level occupancy of TFs, Mediator, pol II, and other factors [63, 65]. Several labs have linked Mediator kinase module components to super-enhancer function [66, 67] and, like Mediator, genomic occupancy of kinase module subunits serves as a marker for super-enhancers in human cells [68]. The Mediator kinase module regulates enhancer function in several ways, which are summarized below.
Control of TF activity
Mediator kinases directly impact enhancer function through phosphorylation of TFs, which can alter TF function or stability. TFs represent a major category of proteins that are modified by Mediator kinases in human cells [48], suggesting that CDK8/CDK19 evolved to modulate TF activity. In support of this idea, the CDK8 ortholog in yeast appears to function primarily through TF phosphorylation [19, 20, 69]. Whereas the functional consequences of most CDK8/CDK19-dependent TF phosphorylation events remain unknown in mammalian cells [48, 70], those that have been studied include Notch ICD [22, 34], SMAD1/3 [23], and STAT1 [24]. In these cases, TF phosphorylation altered its activity (i.e. target gene expression patterns) and/or stability. Additional direct evidence for Mediator kinases regulating of TF function was obtained through the use of CDK8/CDK19 inhibitors [25, 71, 72] and through chemical genetics, with an engineered CDK8 analog-sensitive cell line [73].
The development and discovery of Mediator kinase inhibitors has yielded additional insight about the biological roles of CDK8 and CDK19. The Shair lab showed that inhibition of Mediator kinase activity further activated genes regulated by super-enhancers in AML cells [74]. This caused apoptosis, and the cytotoxic effect was linked to Mediator kinase activity specifically, as inhibitor-resistant mutations in CDK8 or CDK19 rescued cell death. A more recent study by the Serrano lab also implicated Mediator kinase inhibition as a means to further activate super-enhancer target genes; in this case, mouse and human embryonic stem (ES) cells were examined [75]. Given that super-enhancers represent high-occupancy sites for TF binding [64], these data are consistent with Mediator kinase-dependent control of TF function.
Cell type and context specificity of Mediator kinase function has been widely reported [52, 74, 76] and likely derives from cell type and context-specific TF requirements. For example, select TFs are required to maintain pluripotency, and if a subset of these TFs require Mediator kinases for normal function, cell state will be disrupted upon kinase inhibition. In AML cells, Mediator kinase inhibition triggered apoptosis through induction of differentiation programs, in part through disruption of STAT1 TF function [72]. Similarly, Moraga et al. observed that Mediator kinase inhibition induced differentiation in IL-6 stimulated Th-17 cells, and this occurred via STAT3 [71], whereas Sakaguchi et al. reported that differentiation of regulatory T cells was induced by Mediator kinase inhibition, acting in part through TFs FOXP3 and STAT5 [77]. Consistent with these findings, earlier results from Firestein and co-workers, using mouse xenografts derived from human tumors, showed that CDK8 helped maintain human tumor cells in a pluripotent state [78], and CDK8 knockdown (via short hairpin (sh)RNA) caused differentiation. Interestingly, this CDK8-dependent effect was observed in tumors with amplification or overexpression of CDK8. By contrast, a groundbreaking study by Serrano and co-workers showed that Mediator kinase inhibition prevented differentiation in mouse and human ES cells; moreover, similar results were obtained upon depletion of CDK8 or CDK19 via shRNA methods [75]. These contradictory findings probably reflect different cell types and contexts (e.g. AML versus ES cells; tumor xenografts versus cultured cells), which will maintain different expression levels of TFs and other transcriptional regulators. Indeed, additional xenograft models tested by Firestein and co-workers showed that those derived from cells with lower CDK8 expression maintained hallmarks of pluripotency even after CDK8 depletion [78], showing that CDK8 dependence was cell type-specific.
MED12 also appears to control enhancer activation through TFs. A subset of uterine leiomyomas is caused by mutations in MED12; in these cells, expression and enhancer occupancy of AP-1 TFs was markedly reduced [79]. Similarly, the Egly group observed that MED12 mutations correlated with reduced expression of AP-1 subunits in neurons [80]. Because MED12 activates CDK8/CDK19 kinase function [8, 81], these results could manifest through CDK8 and/or CDK19. Indeed, the Espinosa lab has shown that CDK8 drives expression of AP-1 TFs in HCT116 cells [51]. If the link between expression of AP-1 subunits and Mediator kinase module function is shared across cell types, it could help explain many phenotypes associated with Mediator kinase module inhibition or mutation. For example, proliferation [26, 31, 51], drug resistance [33, 82], and cell migration [83, 84] are each linked to the Mediator kinase module, and each is impacted by AP-1 TFs [85, 86].
Enabling rapid stimulus-specific transcriptional responses
Also consistent with a role in enhancer function, Mediator kinase activity is important for directing rapid changes in gene expression patterns (e.g. in response to a stimulus). For example, disruption of CDK8 and/or CDK19 function negatively affects transcriptional responses to p53 activation [87, 88], serum [51], hypoxia [52, 73], or inflammatory cytokines [25, 89, 90]. An extracellular stimulus will immediately activate signaling cascades (e.g. MAPK activation upon serum induction), which will ultimately coordinate transcriptional responses in the nucleus.
Stimulus-specific changes in gene expression patterns are first observed at enhancers [91], typically within minutes of a stimulus. Newly activated enhancers can be detected through pol II-dependent induction of eRNA transcription, and eRNA transcripts can then be mapped to consensus TF sequences at the active enhancers [92]. Analysis of eRNA transcription immediately following IFNγ stimulation revealed that TFs responsive to IFNγ (e.g. STAT1, IRF1) failed to induce eRNA transcription in cells treated with the CDK8/19 inhibitor cortistatin A. Accordingly, induction of interferon responsive genes was defective, which resulted in an increased sensitivity to viral infection [25]. Because STAT1 is phosphorylated by CDK8 [24, 48], these and other results [73] establish a direct role for Mediator kinase-dependent regulation of enhancer function, and suggest that kinase-dependent transcriptional effects may be broadly mediated through enhancer-bound TFs. Because enhancer-bound, signal-responsive TFs recruit Mediator to activate stimulus-specific target genes, this ensures coincident changes in CDK8 or CDK19 genomic occupancy, through kinase module-Mediator interactions. In this way, CDK8 or CDK19 substrate specificity can rapidly shift in coordination with cell signaling cascades.
Enhancer-promoter looping
The Mediator complex is recruited to enhancers genome-wide [63] through direct protein-protein interactions with TFs. The kinase module forms a stable complex with Mediator [10, 11], through interactions dependent on the MED13 subunit. Through its interaction with Mediator, the kinase module can likewise occupy enhancer regions genome-wide [74, 93]; however, direct binding to TFs [94] or eRNA [95] may also help retain the kinase module at enhancer sequences (see below).
Mediator is a large complex (human Mediator: 26 subunits, 1.4 MDa) that can simultaneously interact with TFs and the pol II enzyme [41-43]. In mammalian cells, Mediator also interacts with Cohesin [39], which can form cell-type specific chromatin loops that may help juxtapose enhancers and promoters [93, 96, 97]. By studying mouse ES cells, the Klose lab has shown that the CpG DNA-binding protein FBXL19 acts to link enhancer-bound CDK-Mediator to CpG-rich promoters [98]. Because CDK-Mediator blocks Mediator-pol II interactions [10], this link does not immediately activate transcription. Rather, enhancer-promoter loops formed through FBXL19 and CDK-Mediator appear to "bookmark" genes for future activation during mouse ES cell differentiation. Incidentally, conserved signaling pathways govern cell differentiation, and therefore this bookmarking function may represent a distinct mechanism by which the Mediator kinase module controls transcriptional responses to signaling cascades.
The results from the Klose lab are related to findings from the Brickner group, which studied stimulus-specific transcriptional responses as a function of human CDK8 or its yeast ortholog [99]. Upon re-introduction of a stimulus, human or yeast cells depleted of CDK8 had diminished transcriptional responses, suggesting a similar "bookmarking" function. The precise mechanisms remain unclear but may involve formation of enhancer-promoter loops, as suggested by the FBXL19 results [98]. Other results from yeast support a role for the Mediator kinase module in the formation of looped architectures at promoter DNA. However, yeast have compact genomes compared with mammals, and lack enhancers; instead, yeast have upstream activator sequences (UAS) that reside 200 - 400 bp upstream of the transcription start site. The Struhl and Robert labs showed that the yeast (S. cerevisiae) Mediator kinase module associated with the UAS but not promoters [100, 101]. By contrast, Mediator itself was observed to interact with both the UAS and promoter regions, suggesting that the kinase module helps bridge UAS-promoter interactions through the Mediator complex [100, 101]. Although it remains unclear how the kinase module-Mediator interaction is controlled in yeast, data from the Robert [40] and Cramer labs [12] suggest that phosphorylation by the Cdk8 kinase may promote dissociation.
In human cells, the Shiekhattar lab showed that the CDK-Mediator complex, and MED12 in particular, was important for the formation of enhancer-promoter loops and that looped interactions may be mediated in part through eRNAs [95]. Subsequently, others have linked MED12 to control of enhancer function, with evidence of eRNA binding. By studying estrogen-responsive enhancers in MCF7 cells, Luo and colleagues observed that ERα-induced eRNA transcription was sensitive to JMJD6, which mediated interaction between the CARM1 methyltransferase and MED12 at active enhancers [102]. Subsequent studies revealed a CARM1-dependent RNA binding function for MED12 [103]. Others have reported MED12-dependent maintenance of p300 and H3K27 acetylation at enhancers in hematopoietic stem cells [66] or MED12-dependent looping at the IgH locus during class switch recombination [104]. Estrogen receptor activation and class switch recombination occur in response to MAPK and inflammatory signaling pathways, respectively; these studies suggest that MED12 may help establish enhancer-promoter loops to ensure robust transcriptional responses to these signaling cascades.
We emphasize, however, that the role of Mediator or CDK-Mediator in formation of enhancer-promoter loops remains unclear and controversial [105]. Because enhancer-promoter looping is transient and varies from cell-to-cell [106], population-based methods such as 3C or Hi-C are limited in their ability to resolve stimulus-specific or Mediator-dependent architectural changes. Live cell imaging experiments currently lack the spatial resolution required to confirm direct enhancer-promoter loops [107]. Based upon structural data of the human PIC [41-43], direct enhancer-promoter-PIC interaction may separate these sequences by 20-40nm, which is beyond current resolution limits for fluorescence microscopy [108]. Moreover, live cell imaging requires signal averaging over time, which may preclude detection of transient enhancer-promoter interactions [107]. Nevertheless, chromosome conformation methods and live cell imaging experiments have markedly advanced our understanding of pol II transcription in cells, and we anticipate that methodological improvements will yield new discoveries about enhancer-promoter dynamics and potential CDK-Mediator-dependent regulation.
Concluding remarks
Given its role in connecting cell signaling with transcription, the Mediator kinase module serves as a regulatory node through which changes in gene expression patterns can be initiated. Through kinase-dependent mechanisms, CDK8 and/or CDK19 regulate TF function to help "reprogram" gene expression patterns in response to a stimulus or developmental cues. The Mediator kinase module also functions in kinase-independent ways, through Mediator binding, which blocks Mediator-pol II interaction yet appears to promote post-initiation events, such as pol II pause release or elongation. The complexity of the pol II transcription machinery and cell signaling networks presents many opportunities for new discoveries, but also many challenges. Cell type and cell context (e.g. oxidative stress or growth factor induction) will remain important considerations in future work, as the set of active TFs will change in each case. Because disruption of the Mediator kinase module will broadly impact transcriptional programs [109], rapid methods to manipulate activity, such as degrons or chemical inhibitors, will be essential for delineating direct versus indirect effects. Biochemical and structural data will also continue to inform about molecular mechanisms, which are otherwise difficult to reliably assess with only cell-based or in vivo assays. Among the many questions that remain to be addressed regarding the biological functions and mechanisms of the Mediator kinase module, we highlight several in the outstanding questions section.
Outstanding questions.
Little is known about the biological or functional roles of MED12L or MED13L, and no study has revealed CDK19-specific kinase targets in any biological context. Among the high-confidence Mediator kinase substrates, few have been pursued in follow-up studies to determine whether phosphorylation alters biological function.
The kinase module interaction with Mediator controls Mediator-PIC interactions and therefore has major effects on gene expression; how is this interaction is controlled? In human cells, FBW7-dependent ubiquitination of MED13 has been identified [110], and data from yeast (S. cerevisiae) suggest that CDK8 kinase activity may regulate the interaction [12], but additional mechanisms are likely.
How do subunits function apart from the Mediator kinase module? Together with CCNC (but not MED12 or MED13), CDK8 or CDK19 help regulate DNA replication through interaction with MTBP, as part of a replication initiation complex [111]. In addition, the Strich lab has shown that CCNC translocates to mitochondria, which is dependent on MED13 degradation, during cell stress [16]. Mitochondrial localization of CCNC induces mitochondrial fission [112], providing a means by which CCNC could directly impact metabolism. Separately, the Olson lab observed that elevated expression of MED13 in mice increased mitochondrial content, coupled with expected effects on oxygen consumption [27]. Data from Strich and co-workers suggest that these MED13-specific effects could result from reduced trafficking of CCNC to mitochondria [16].
Live cell imaging experiments have revealed that active PIC assemblies and enhancer-promoter loops are rare and transient [113, 114], causing infrequent bursts of transcription [115]. These bursts result from multiple rounds of pol II initiation from the same promoter; how bursting is shut off remains unclear. Could it involve the Mediator kinase module, given its ability to block Mediator-pol II interactions?
Finally, what about molecular condensates? Liquid-liquid phase separation is a common biophysical property of TFs and Mediator [116], and formation of condensates may help control pol II transcription by compartmentalization of distinct transcriptional stages (e.g. initiation vs. elongation). It is currently unknown whether the Mediator kinase module influences gene expression via condensate-dependent mechanisms.
Supplementary Material
Highlights.
The Mediator kinase module transforms Mediator function through physical interaction and its kinase activity.
The Mediator kinase module regulates transcription by altering Mediator and transcription factor function at enhancers and promoters.
Rapid, stimulus-specific transcriptional responses are enabled by the kinase module.
By controlling stimulus-specific TF function and pol II activity, the Mediator kinase module helps convert signaling inputs to transcriptional outputs.
Acknowledgments:
Due to space and citation limitations, we were unable to cover all relevant topic areas or cite all relevant studies. Work in the Taatjes lab is supported in part by the NIH (R35 GM139550; R01 AI156739) and the NSF (MCB-1818147). OL is supported in part by the NCI (F31 CA254478).
Glossary
- CDK-activating kinase (CAK) module
module of the general transcription factor TFIIH. Contains CDK7, MNAT1, and CCNH
- CDK-Mediator
Mediator bound to the Mediator kinase module (MKM); may contain CDK8 or CDK19. Because the Mediator subunit MED26 dissociates upon MKM-Mediator binding, the human CDK-Mediator complex contains 29 subunits
- Cyclin-dependent kinase (CDK)
member of the serine-threonine kinase family
- Cytokine
a small signaling peptide, produced during inflammatory responses (for example)
- Degron
a peptide sequence that marks a protein for rapid degradation
- Enhancer-promoter loops
Enhancer elements are brought into spatial proximity with promoters through looping of intervening genomic DNA
- Enhancer RNA (eRNA)
results from pol II transcription of enhancer DNA
- Mediator complex
A 26-subunit assembly (in humans) that lacks the Mediator Kinase Module (MKM)
- Mediator Kinase Module (MKM)
composed of the CDK8/19, MED12/L, MED13/L, and CCNC subunits in vertebrates
- Pol II C-terminal domain (CTD)
The intrinsically disordered domain of the largest subunit of pol II, RPB1/POLR2A
- Transcription pre-initiation complex (PIC)
consists of TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Mediator, and pol II
- Upstream activator sequence (UAS)
a cis-acting regulatory sequence that enhances transcription from a nearby promoter in yeast
Footnotes
Competing interests: DJT is a member of the SAB at Dewpoint Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li N et al. (2014) Cyclin C is a haploinsufficient tumour suppressor. Nat Cell Biol 16 (11), 1080–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao YL et al. (2018) Mediator complex component MED13 regulates zygotic genome activation and is required for postimplantation development in the mouse. Biol Reprod 98 (4), 449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westerling T et al. (2007) Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 27 (17), 6177–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha PP et al. (2010) Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 137 (16), 2723–31. [DOI] [PubMed] [Google Scholar]
- 5.Schneider EV et al. (2011) The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J Mol Biol 412 (2), 251–66. [DOI] [PubMed] [Google Scholar]
- 6.Li YC et al. (2021) Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Sci Adv 7 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava S and Kulshreshtha R (2021) Insights into the regulatory role and clinical relevance of mediator subunit, MED12, in human diseases. J Cell Physiol 236 (5), 3163–3177. [DOI] [PubMed] [Google Scholar]
- 8.Park MJ et al. (2018) Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J Biol Chem 293 (13), 4870–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turunen M et al. (2014) Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep 7 (3), 654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knuesel MT et al. (2009) The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes Dev 23, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai KL et al. (2013) A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20 (5), 611–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman S et al. (2021) The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II. J Biol Chem 296, 100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H et al. (2021) Structure of mammalian Mediator complex reveals Tail module architecture and interaction with a conserved core. Nat Commun 12 (1), 1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balamotis MA et al. (2009) Complexity in transcription control at the activation domain-Mediator interface. Sci Signal 2 (69), ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J and Thompson CB (2019) Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol 20 (7), 436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khakhina S et al. (2014) Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol Biol Cell 25 (18), 2807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsay AK et al. (2014) Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS Genet 10 (10), e1004567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mousley CJ et al. (2012) A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell 148 (4), 702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirst M et al. (1999) GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell 3 (5), 673–8. [DOI] [PubMed] [Google Scholar]
- 20.Nelson C et al. (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421 (6919), 187–190. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X et al. (2012) Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Invest 122 (7), 2417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fryer CJ et al. (2004) Mastermind recruits CycC:Cdk8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol Cell 16, 509–520. [DOI] [PubMed] [Google Scholar]
- 23.Alarcon C et al. (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-b pathways. Cell 139, 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bancerek J et al. (2013) CDK8 Kinase Phosphorylates Transcription Factor STAT1 to Selectively Regulate the Interferon Response. Immunity 38 (2), 250–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinparzer I et al. (2019) Transcriptional Responses to IFN-gamma Require Mediator Kinase-Dependent Pause Release and Mechanistically Distinct CDK8 and CDK19 Functions. Mol Cell 76 (3), 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galbraith MD et al. (2017) CDK8 Kinase Activity Promotes Glycolysis. Cell Rep 21 (6), 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grueter CE et al. (2012) A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 149 (3), 671–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH et al. (2014) Heart- and muscle-derived signaling system dependent on MED13 and Wingless controls obesity in Drosophila. Proc Natl Acad Sci U S A 111 (26), 9491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie XJ et al. (2015) CDK8-Cyclin C Mediates Nutritional Regulation of Developmental Transitions through the Ecdysone Receptor in Drosophila. PLoS Biol 13 (7), e1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angeles-Albores D and Sternberg PW (2018) Using Transcriptomes as Mutant Phenotypes Reveals Functional Regions of a Mediator Subunit in Caenorhabditis elegans. Genetics 210 (1), 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firestein R et al. (2008) CDK8 is a colorectal cancer oncogene that regulates b-catenin activity. Nature 455, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H and Emmons SW (2000) A C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes & Development 14 (17), 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S et al. (2012) MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell 151 (5), 937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang Y et al. (2020) Enhancer architecture sensitizes cell specific responses to Notch gene dose via a bind and discard mechanism. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cramer P (2019) Organization and regulation of gene transcription. Nature 573 (7772), 45–54. [DOI] [PubMed] [Google Scholar]
- 36.Schier AC and Taatjes DJ (2020) Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34 (7-8), 465–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fondell JD et al. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y et al. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608. [DOI] [PubMed] [Google Scholar]
- 39.Ebmeier CC and Taatjes DJ (2010) Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A 107 (25), 11283–11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeronimo C et al. (2016) The RNA Polymerase II CTD: The Increasing Complexity of a Low-Complexity Protein Domain. J Mol Biol 428 (12), 2607–22. [DOI] [PubMed] [Google Scholar]
- 41.Abdella R et al. (2021) Structure of the human Mediator-bound transcription preinitiation complex. Science 372 (6537), 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X et al. (2021) Structures of the human Mediator and Mediator-bound preinitiation complex. Science 372 (6546). [DOI] [PubMed] [Google Scholar]
- 43.Rengachari S et al. (2021) Structure of human Mediator-RNA polymerase II pre-initiation complex. Nature. [DOI] [PubMed] [Google Scholar]
- 44.Core L and Adelman K (2019) Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33 (15-16), 960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fant CB et al. (2020) TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Mol Cell 78 (4), 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brannan K et al. (2012) mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell 46 (3), 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Z et al. (2020) Termination of non-coding transcription in yeast relies on both an RNA Pol II CTD interaction domain and a CTD-mimicking region in Sen1. EMBO J 39 (7), e101548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poss ZC et al. (2016) Identification of Mediator Kinase Substrates in Human Cells using Cortistatin A and Quantitative Phosphoproteomics. Cell Rep 15 (2), 436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gressel S et al. (2017) CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 6, e29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Z et al. (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13 (9), 543–7. [DOI] [PubMed] [Google Scholar]
- 51.Donner AJ et al. (2010) CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17 (2), 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galbraith MD et al. (2013) HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell 153 (6), 1327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanso M et al. (2016) P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev 30 (1), 117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vos SM et al. (2018) Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560 (7720), 607–612. [DOI] [PubMed] [Google Scholar]
- 55.Larochelle S et al. (2012) Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 19 (11), 1108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine M et al. (2014) Looping back to leap forward: transcription enters a new era. Cell 157 (1), 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sur I and Taipale J (2016) The role of enhancers in cancer. Nat Rev Cancer 16 (8), 483–93. [DOI] [PubMed] [Google Scholar]
- 58.Hnisz D et al. (2013) Super-enhancers in the control of cell identity and disease. Cell 155 (4), 934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S et al. (2015) The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16 (3), 144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parker SC et al. (2013) Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A 110 (44), 17921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siersbaek R et al. (2011) Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J 30 (8), 1459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siersbaek R et al. (2014) Molecular architecture of transcription factor hotspots in early adipogenesis. Cell Rep 7 (5), 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whyte WA et al. (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153 (2), 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blobel GA et al. (2021) Testing the super-enhancer concept. Nat Rev Genet. [DOI] [PubMed] [Google Scholar]
- 65.Kwiatkowski N et al. (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511 (7511), 616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aranda-Orgilles B et al. (2016) MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis. Cell Stem Cell 19 (6), 784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuuluvainen E et al. (2018) Depletion of Mediator Kinase Module Subunits Represses Superenhancer-Associated Genes in Colon Cancer Cells. Mol Cell Biol 38 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan A and Zhang X (2019) Integrative modeling reveals key chromatin and sequence signatures predicting super-enhancers. Sci Rep 9 (1), 2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chi Y et al. (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15 (9), 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez-Val A et al. (2021) Dissection of two routes to naive pluripotency using different kinase inhibitors. Nat Commun 12 (1), 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez-Fabregas J et al. (2020) CDK8 Fine-Tunes IL-6 Transcriptional Activities by Limiting STAT3 Resident Time at the Gene Loci. Cell Rep 33 (12), 108545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nitulescu II et al. (2017) Mediator Kinase Phosphorylation of STAT1 S727 Promotes Growth of Neoplasms With JAK-STAT Activation. EBioMedicine 26, 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andrysik Z et al. (2021) Multi-omics analysis reveals contextual tumor suppressive and oncogenic gene modules within the acute hypoxic response. Nat Commun 12 (1), 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pelish HE et al. (2015) Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526 (7572), 273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lynch CJ et al. (2020) Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 Mediator kinases. Nat Cell Biol 22 (10), 1223–1238. [DOI] [PubMed] [Google Scholar]
- 76.McCleland ML et al. (2015) Cdk8 deletion in the Apc(Min) murine tumour model represses EZH2 activity and accelerates tumourigenesis. J Pathol 237 (4), 508–19. [DOI] [PubMed] [Google Scholar]
- 77.Akamatsu M et al. (2019) Conversion of antigen-specific effector/memory T cells into Foxp3-expressing Treg cells by inhibition of CDK8/19. Sci Immunol 4 (40). [DOI] [PubMed] [Google Scholar]
- 78.Adler AS et al. (2012) CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res. 72 (8), 2129–39. [DOI] [PubMed] [Google Scholar]
- 79.Moyo MB et al. (2020) Altered chromatin landscape and enhancer engagement underlie transcriptional dysregulation in MED12 mutant uterine leiomyomas. Nat Commun 11 (1), 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donnio LM et al. (2017) MED12-related XLID disorders are dose-dependent of immediate early genes (IEGs) expression. Hum Mol Genet 26 (11), 2062–2075. [DOI] [PubMed] [Google Scholar]
- 81.Knuesel MT et al. (2009) The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of Mediator. Mol Cell Biol 29 (3), 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharko AC et al. (2021) The Inhibition of cDk8/19 Mediator Kinases Prevents the Development of Resistance to EGFR-Targeting Drugs. Cells 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serrao A et al. (2018) Mediator kinase CDK8/CDK19 drives YAP1-dependent BMP4-induced EMT in cancer. Oncogene 37 (35), 4792–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou J et al. (2018) Zyxin promotes colon cancer tumorigenesis in a mitotic phosphorylation-dependent manner and through CDK8-mediated YAP activation. Proc Natl Acad Sci U S A 115 (29), E6760–E6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bejjani F et al. (2019) The AP-1 transcriptional complex: Local switch or remote command? Biochim Biophys Acta Rev Cancer 1872 (1), 11–23. [DOI] [PubMed] [Google Scholar]
- 86.Fallahi-Sichani M et al. (2017) Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol 13 (1), 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Audetat KA et al. (2017) A Kinase-Independent Role for Cyclin-Dependent Kinase 19 in p53 Response. Mol Cell Biol 37 (13), e00626–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donner AJ et al. (2007) CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell 27 (1), 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen M et al. (2017) CDK8/19 Mediator kinases potentiate induction of transcription by NFkappaB. Proc Natl Acad Sci U S A 114 (38), 10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johannessen L et al. (2017) Small-molecule studies identify CDK8 as a regulator of IL-10 in myeloid cells. Nat Chem Biol 13 (10), 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arner E et al. (2015) Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347 (6225), 1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Azofeifa JG et al. (2018) Enhancer RNA profiling predicts transcription factor activity. Genome Res 28, 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kagey M et al. (2010) Mediator and Cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou H et al. (2006) Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol Cell Biol 26 (23), 8667–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lai F et al. (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dowen JM et al. (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159 (2), 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Phillips-Cremins JE et al. (2013) Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 153 (6), 1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dimitrova E et al. (2018) FBXL19 recruits cDK-Mediator to CpG islands of developmental genes priming them for activation during lineage commitment. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.D'Urso A et al. (2016) Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeronimo C et al. (2016) Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Mol Cell 64 (3), 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petrenko N et al. (2016) Mediator Undergoes a Compositional Change during Transcriptional Activation. Mol Cell 64 (3), 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao WW et al. (2018) JMJD6 Licenses ERalpha-Dependent Enhancer and Coding Gene Activation by Modulating the Recruitment of the CARM1/MED12 Co-activator Complex. Mol Cell 70 (2), 340–357 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng D et al. (2018) CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance 1 (5), e201800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomas-Claudepierre AS et al. (2016) Mediator facilitates transcriptional activation and dynamic long-range contacts at the IgH locus during class switch recombination. J Exp Med 213 (3), 303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.El Khattabi L et al. (2019) A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 178 (5), 1145–1158 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Finn EH and Misteli T (2019) Molecular basis and biological function of variability in spatial genome organization. Science 365 (6457). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patange S et al. (2021) Towards a ‘spot on’ understanding of transcription in the nucleus. J Mol Biol, 167016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Valli J et al. (2021) Seeing beyond the limit: A guide to choosing the right super-resolution microscopy technique. J Biol Chem 297 (1), 100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sooraj D et al. (2022) MED12 and BRD4 cooperate to sustain cancer growth upon loss of mediator kinase. Mol Cell 82 (1), 123–139 e7. [DOI] [PubMed] [Google Scholar]
- 110.Davis MA et al. (2013) The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev 27 (2), 151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kohler K et al. (2019) The Cdk8/19-cyclin C transcription regulator functions in genome replication through metazoan Sld7. PLoS Biol 17 (1), e2006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ganesan V et al. (2019) Cyclin C directly stimulates Drp1 GTP affinity to mediate stress-induced mitochondrial hyperfission. Mol Biol Cell 30 (3), 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fukaya T et al. (2016) Enhancer Control of Transcriptional Bursting. Cell 166 (2), 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nguyen VQ et al. (2021) Spatiotemporal coordination of transcription preinitiation complex assembly in live cells. Mol Cell 81 (17), 3560–3575 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rodriguez J and Larson DR (2020) Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu Rev Biochem 89, 189–212. [DOI] [PubMed] [Google Scholar]
- 116.Sabari BR et al. (2020) Biomolecular Condensates in the Nucleus. Trends Biochem Sci 45 (11), 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Menzl I et al. (2019) A kinase-independent role for CDK8 in BCR-ABL1(+) leukemia. Nat Commun 10 (1), 4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Postlmayr A et al. (2020) Cdk8 is required for establishment of H3K27me3 and gene repression by Xist and mouse development. Development 147 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sato S et al. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 14, 685–691. [DOI] [PubMed] [Google Scholar]
- 120.Ashkenazy H et al. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44 (W1), W344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dannappel MV et al. (2018) Molecular and in vivo Functions of the CDK8 and CDK19 Kinase Modules. Front Cell Dev Biol 6, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roninson IB et al. (2019) Identifying Cancers Impacted by CDK8/19. Cells 8 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chung HL et al. (2020) De Novo Variants in CDK19 Are Associated with a Syndrome Involving Intellectual Disability and Epileptic Encephalopathy. Am J Hum Genet 106 (5), 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tian T et al. (2021) Somatic and de novo Germline Variants of MEDs in Human Neural Tube Defects. Front Cell Dev Biol 9, 641831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhagwat AS et al. (2016) BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep 15 (3), 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.