Abstract
Allogeneic hematopoietic cell transplantation (allo-HCT) induces profound shifts in the intestinal bacterial microbiota. The dynamics of intestinal fungi and their impact on clinical outcomes during allo-HCT are not fully understood. Here, we combined parallel high-throughput fungal ITS1 amplicon sequencing, bacterial 16S amplicon sequencing, and fungal cultures of 1279 fecal samples from a cohort of 156 allo-HCT patients to reveal potential trans-kingdom dynamics and their association with patient outcomes. We saw that the overall density and the biodiversity of intestinal fungi were stable during allo-HCT, but the species composition changed drastically from day to day. We identified a subset of patients with fungal dysbiosis defined by culture positivity (n=53) and stable expansion of Candida parapsilosis complex species (n=19). They presented with distinct trans-kingdom microbiota profiles, characterized by a decreased intestinal bacterial biomass. These patients had worse overall survival and higher transplant-related mortality independent of candidemia. This expands our understanding of the clinical significance of the mycobiota and suggests that targeting fungal dysbiosis may help to improve long-term patient survival.
Introduction
Allogeneic hematopoietic cell transplantation (allo-HCT) is a curative treatment for certain malignant and non-malignant hematological disorders. Patients that undergo allo-HCT receive cytotoxic conditioning chemotherapy prior to infusion of donor immune cells. To prevent opportunistic infections during this period of intense immunosuppression, patients typically receive routine antibacterial and antifungal prophylaxis 1–6. The impact of allo-HCT on the bacterial microbiota has been characterized in prior studies: intestinal bacterial burden and α-diversity start to decline in the week prior to donor hematopoietic cell infusion, concurrent with the initiation of conditioning chemotherapy and antimicrobial therapy 7–10. Damage to the intestinal microbiota primarily affects anaerobic bacterial taxa that mediate colonization resistance against pathobionts, e.g., Enterococcus and Enterobacteriaceae 10,11. Low bacterial diversity and enterococcal domination have been associated with lower overall survival, higher transplant-related mortality, and higher graft-versus-host-related mortality in allo-HCT patients 7,12. Domination by bacterial pathobionts in the gut precedes bacterial translocation and the development of bloodstream infections 10,13,14.
In a recent case-control study of 15 patients using culture-dependent and -independent analyses of paired fecal samples and bloodstream fungal isolates, we reported that intestinal expansion of Candida species preceded bloodstream invasion, linking a perturbation in the fungal mycobiota to invasive fungal disease 15. In addition, exposure to antifungal prophylaxis and the presence of Candida colonization by culture-based surveys has been linked to acute intestinal graft versus host disease 16,17. In studies of inflammatory bowel disease and intestinal wound healing a number of fungal species, including Malassezia, Candida albicans, and Debaryomyces, have been associated with colitis exacerbations, responsiveness to fecal microbiota transplantation, or defective wound healing 18–20, though it is unknown whether shifts in intestinal fungi predict long-term survival in inflammatory bowel disease or HCT.
Here, we characterize the composition and dynamic changes of intestinal fungal communities by both longitudinal amplicon-based and culture-dependent analyses in fecal samples collected from 156 allo-HCT patients. Allo-HCT was associated with a substantial increase in viable fungi that were recovered in fecal samples, in conjunction with an expansion of Candida parapsilosis complex species in a subset of patient. These patients had distinct trans-kingdom microbiota profiles. Patients with intestinal C. parapsilosis complex species expansion had a reduced overall survival that was explained by increased transplant-related mortality.
Results
Study characteristics
To analyze intestinal fungi during allo-HCT, we performed a cohort study that included 156 patients with 1279 prospectively collected fecal samples (median: 10 per patient). Collection started 10 days prior to and continued until 30 days after allo-HCT. These subjects were randomly drawn from a large-scale, prospective stool biobanking effort at our institution, which has been described previously 7,10. Patients underwent transplantation between January 1st, 2017, and December 31st, 2018, and had a median clinical follow-up of 32 months for this study (see cohort characteristics in Extended Data Table 1). The most common indication for transplantation was acute leukemia and most patients received unmodified grafts (40% received unmodified PBSC, and 20% unmodified bone marrow grafts). In this cohort, the median hematopoietic-cell transplantation comorbidity index (HCT-CI) risk score 21 was 2 (interquartile range: 1 to 4), corresponding to an intermediate mortality risk. Patients received antifungal prophylaxis with intravenous micafungin (150 mg/day), an echinocandin drug starting around day −7 days prior to HCT for a median of 17 days (IQR 15 to 22 days), prior to advancing to azole-based fungal prophylaxis, consistent with current institutional practices.
The fungal mycobiota and bacterial microbiota exhibit distinct dynamics during allo-HCT.
To assess the temporal dynamics of the intestinal mycobiota, we quantified fungal burden and diversity in longitudinally obtained samples. Fungal density was quantified by qPCR with the fungal 18S ribosomal DNA as a target 22. While the fecal fungal DNA content exhibited a large variation at all time points examined, there was no overall time-dependent change and no difference in fungal DNA abundance pre- and post-transplantation (Fig. 1a, Extended Data Fig. 1a). Similarly, the fungal α-diversity at the species-level varied only marginally over the time course of allo-HCT (Fig. 1b, Extended Data Fig. 1b). These results contrasted with a significant loss in bacterial density (Fig. 1c, Extended Data Fig. 1c) and diversity (Fig. 1d, Extended Data Fig. 1d) around the time of HCT.
Figure 1: Dynamics of the fungal and bacterial microbiota during allo-HCT.
(a) Fungal density measured by 18S qPCR per gram of stool (n = 1224 samples) and (b) fungal diversity at species level measured by the Inverse Simpson Index (n = 1279 samples) over time. (c) Bacterial density measured by 16S qPCR per gram of stool (n = 1217 samples) and (d) bacterial diversity at species level measured by the Inverse Simpson Index (n = 1184 samples) over time. (a-d) The dashed line represents day 0, the day of donor cell infusion. Each dot represents a single sample. (e) Fungal (n = 5916 sample pairs) and (f) bacterial (n = 5171 sample pairs) similarity during allo-HCT. The Chao-Jaccard index at species level. was used to calculate similarity between two samples from one individual. A value of 1 (1-Chao Jaccard) corresponds to two samples with identical composition, while a value of corresponds to two samples not sharing any species. (e-f) Each dot represents a sample pair from a single patient. Statistics: The solid black line corresponds to the smoothed average using Loess smoothing. See also Extended Data Fig. 1.
The distinct temporal dynamics of the fungal and bacterial microbiota included a lower day-to-day similarity, as measured by the Chao-Jaccard similarity index 23 of the fungal mycobiota composition compared to that of the bacterial microbiota (Fig. 1e and 1f). In the ensuing analysis, a value of 1 corresponds to an identical composition of 2 samples, while a value of 0 corresponds to no taxa being present in both samples. For sample pairs collected one day apart, the fungal mycobiota had a median similarity (1-Chao-Jaccard) of 0.79 and the bacterial microbiota a median similarity of 0.98. For samples collected 30 days apart, the fungal mycobiota had a median similarity of 0.73, while the bacterial microbiota had a median similarity of 0.19. This observation was consistent with prior reports of a decline in the similarity of the bacterial microbiota over time, best fitting to a power law 24. Thus, fecal samples collected on consecutive days differed substantially in their mycobiota composition and were on average not more similar than two samples from the same patient collected 30 days apart. In contrast, two samples collected on consecutive days had a much more similar bacterial composition than two samples from the same patient collected 30 days apart.
Fungal culture-positivity and expansion of single taxa define intestinal fungal dysbiosis.
Dysbiosis of the gut mycobiota is not necessarily reflected in summary measures such as density and diversity. Thus, we further characterized the mycobiota composition by (1) analyzing culturable fungi recovered from fecal samples on Sabouraud agar supplemented with vancomycin and gentamicin, and (2) performing taxonomic profiling at the species level via analysis of internal transcribed spacer 1 (ITS1) amplicons that were derived from a segment of spacer DNA located between the fungal 18S and 5.8S rRNA genes 25. Culturable fungi were recovered from 195 out of 1279 samples (15%). The proportion of fecal samples from which fungi could be cultured was substantially higher in the post-transplant period (day 0 to +30) compared to the pre-transplant period (day −10 to −1; Fig. 2a). C. parapsilosis complex species (C. parapsilosis, C. orthopsilosis, and C. metapsilosis) were the most common cultured fungi, followed by S. cerevisiae, and other Candida species (Fig. 2b). The presence of viable fungi was associated with an increase in intestinal fungal biomass (Fig. 2c), and with a reduction in fungal α-diversity (Fig. 2d).
Figure 2: Distinct characteristics of fungal culture-positive and -negative fecal samples.
(a) Percentage of fecal samples with a positive fungal culture per day (n = 251 pre-transplant and n = 1028 post-transplant). Each circle represents the proportion for a single day. The pre-transplant period is defined by days −10 to −1, the post-transplant period by days 0 to 30. P-value was calculated by a two-sided Wilcoxon rank sum test. (b) Number of samples from which the specified fungal species could be cultured. The column labeled “other” groups fungal species found in five samples or less. Viable fungi were cultured from 195 samples and include 219 species. (c) Fungal density by 18S copies per gram of stool (n = 179 fecal culture positive samples and n= 1045 fecal culture negative samples) and (d) diversity by Inverse Simpson (n = 194 fecal culture positive samples and n=1085 fecal culture negative samples) in fecal samples with positive and negative fungal cultures. (c-d): Two-sided P-values were calculated from generalized estimating equations with patient ID as cluster variable. (a,c,d): The upper and lower limit of the boxes represent the 75th and 25th percentile with the central line showing the median of the values. The upper whisker represents the maximum value or the 75th percentile value plus 1.5 times the interquartile range, whichever is larger. The lower whisker represents the minimum value or the 25th percentile value minus 1.5 times the interquartile range, whichever is larger.
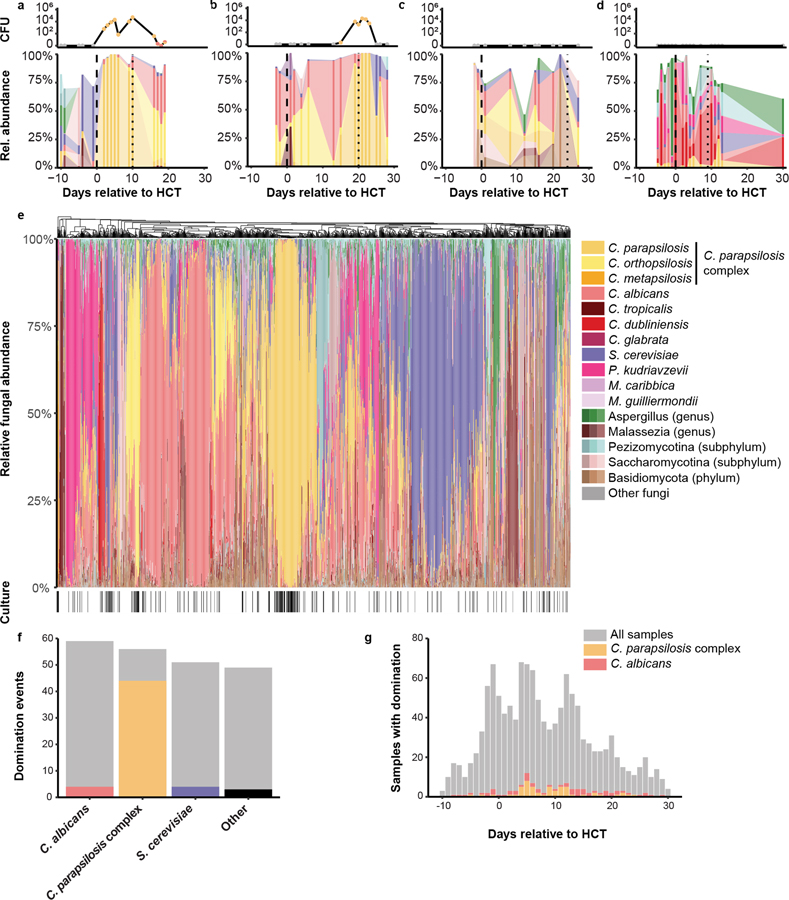
To assess the relationship between culture-dependent and independent methods of sample analysis in our dataset, we compared the culture results to results obtained by ITS1 amplicon sequencing. In 177 of the 194 (91%) culture-positive samples, the cultured fungus could be detected in ITS1 sequences (Extended Data Fig 2). Figures 3A–3D show both the longitudinal culture results and the ITS1 amplicon composition of fecal samples from representative patients during allo-HCT. Data from representative subjects in Fig. 3a and 3b suggest that culture positivity and the relative expansion of corresponding taxa in the ITS1 sequences coincide temporally. To explore the association between culture-positivity and sequence expansion by ITS1 amplicon sequencing, we defined fungal domination by the presence of a single species at 90% relative abundance or higher. We found that if a sample was dominated by a fungal species on one day, the mycobiota composition of the next day was more similar than a pair of samples collected on consecutive days in which the first sample was not dominated by a single taxon (Extended Data Fig 2). This similarity in β-diversity was highest for initial samples dominated by C. parapsilosis complex, followed by samples dominated by C. albicans (Extended Data Fig. 2). This is consistent with our finding that domination by C. parapsilosis complex species and subsequent culture-positivity could persist over several days (Fig 3a and b).
Figure 3: Fungal composition in fecal samples during allo-HCT.
(a-d) Representative plots for individual patients (a, b) with viable fungi and (c, d) without viable fungi in fecal samples. The top rows depict the number of colony-forming units of fungi cultured from individual samples, color-coded by the cultured fungal species. The bottom rows depict the taxonomic composition of each sample. Fungal species present in a single sample or present below a peak abundance of 10% have been blanked in the graphs. The dashed lines represent day 0 and the day of engraftment, respectively. (e) Hierarchical clustering at genus level of all samples and culture-positivity. A black line in the lower ribbon represents a positive culture from the respective sample (n = 1279 samples). When three shades of a color appear in the legend next to a taxon this indicates that multiple variegated shades of color were applied to individual ASVs within indicated taxa. (f) Domination events and relation to culture-positivity for major taxa. The grey bars represent the number of samples with domination (≥90% relative abundance) of the specific taxon. The colored bars represent the number of samples with domination and a positive culture by the specific taxon. (g) Histogram of the temporal relation of domination by C. parapsilosis complex and C. albicans. See also Extended Data Fig. 2
Overall, 215 (17%) samples that belonged to 96 patients (62% of cohort) showed fungal domination with the 90% threshold in relative abundance (Fig 3e). C. albicans (59 samples, 5% of all samples), C. parapsilosis complex species samples (56 samples, 5% of all samples), and S. cerevisiae (51 samples, 4% of all samples) were the most common taxa that dominated fecal samples. Collectively, all other fungal taxa dominated less than 10 samples. Comparison of patients with culture-positive and culture-negative fecal samples revealed striking differences (Fig. 3e and 3f). Samples with C. parapsilosis complex species domination yielded positives cultures in 43/56 (77%) cases. In contrast, culture positivity was rare for samples with C. albicans 10/59 (17%) or with S. cerevisiae 10/51 (20%) domination (Fig. 3f). Domination events with C. parapsilosis complex species clustered in the immediate post-transplant period. In contrast, samples dominated by C. albicans were more evenly distributed across the study period (Fig. 3g). Time to engraftment did not differ between patients with C. parapsilosis domination and those without domination (median of 12 days for both groups). In sum, C. parapsilosis complex species expanded in the intestine in a subset of patients after HCT, and this domination state was frequently associated with the recovery of viable fungi from fecal samples.
Cross-kingdom interactions between intestinal fungi and bacteria in allo-HCT patients
To examine microbiota-intrinsic factors that led to fungal domination, we assessed intestinal trans-kingdom interactions between bacteria and fungi. Samples with C. parapsilosis complex domination had a substantial reduction in bacterial density than samples without any fungal domination. S. cerevisiae domination states showed a modest, yet significant reduction in bacterial density, while C. albicans domination states were not associated with a lower bacterial density (Fig. 4a). Bacterial α-diversity did not vary by large amounts between different fungal domination states (Fig. 4b). To assess whether the lack of association between bacterial diversity and fungal composition might be due to a different composition of the bacterial microbiota, we first assessed co-domination patterns between bacteria and fungi (Fig 4c). Most fungal domination states were not associated with concurrent bacterial domination states – commonly defined as presence of a species at 30% relative abundance or higher 7,10,13. Intriguingly, co-domination between C. parapsilosis complex and Enterococcus was extremely rare, although Enterococcus domination is one of the most common bacterial domination states in this cohort 7,10–12. In contrast, samples with C. albicans domination were commonly concurrently dominated by Enterococcus. In addition, co-domination events of Staphylococcus and Streptococcus species were more commonly observed with C. parapsilosis complex than with C. albicans or S. cerevisiae. To explore these observations further, we correlated changes in the bacterial and fungal mycobiota. Variations in the relative abundance of C. parapsilosis complex, C. albicans, and S. cerevisiae correlated with distinct bacterial signatures (Fig. 4d). Increases in C. parapsilosis complex abundance correlated with an increase in taxa belonging to the family Staphylococcaceae, and negatively correlated with Enterococcaceae (Fig. 4d), as expected from the co-domination analysis (Fig. 4c). Mirroring the findings in the co-domination analysis, the relative abundance of C. albicans in fecal samples also correlated with the relative abundance of Enterococcus. The relative abundance of C. parapsilosis complex species correlated positively with Aerococcaceae, Flavobacteriaceae, Mycobacteriaceae, Enterobacteriaceae, Mycoplasmataceae, and Verrucomicrobiaceae. Importantly, C. albicans and S. cerevisiae were associated with distinct bacterial family correlation profiles compared to C. parapsilosis complex species (Fig 4d).
Figure 4: Trans-kingdom bacterial and fungal interactions during allo-HCT.
(a) Comparison of bacterial density per gram of stool (n = 1175 samples: 1018 with no domination, 51 with C. parapsilosis complex domination, 58 with C. albicans domination, and 48 with S. cerevisiae domination) and (b) diversity (n = 1104 samples: 967 with no domination, 43 with C. parapsilosis complex domination, 50 with C. albicans domination, and 44 with S. cerevisiae domination) in fecal samples with different dominating fungal taxa (≥ 90% relative abundance). Two-sided p-values for between-group differences were evaluated by generalized estimating equations with patient ID as cluster variable without accounting for multiple comparisons. The upper whisker represents the maximum value or the 75th percentile value plus 1.5 times the interquartile range, whichever is larger. (c) Co-domination matrix between bacterial taxa (defined as relative abundance of a bacterial species ≥ 30%) and fungal domination (n = 140). (d) Spearman correlation matrix of the relative abundances of bacterial families and fungal taxa of interest. q-values corrected by false-discovery (FDR) algorithm. (n = 1554 samples)
Impact of intestinal fungal dysbiosis on fungal bloodstream infections
C. parapsilosis domination was observed in 19/156 patients (12% incidence). Three of these patients (16%) subsequently developed candidemia, two during the fecal sampling period and one at day +47. All three patients with C. parapsilosis bloodstream infection survived the infectious episode. One patient developed C. glabrata bloodstream infection on day +84, after relapse of the primary disease and one day prior to death. In this patient, C. glabrata was present at a maximum of 50% relative abundance in two consecutive stool samples on days +18 and +19 with no intervening stool samples. No other episodes of fungal bloodstream infections occurred in the follow-up period. Thus, all Candida bloodstream infection in this cohort were preceded by expansion of the same Candida species as detected in blood cultures, as expected based on prior findings26.
Impact of intestinal fungal dysbiosis on allo-HCT survival.
Finally, we examined whether intestinal fungal dysbiosis in the pre-engraftment period (day 0 to engraftment) could predict other adverse outcomes in allo-HCT patients. We defined fungal dysbiosis as the presence of culturable fecal fungi or the expansion and domination of a single fungal species. Patients with at least one fungal culture-positive fecal sample in the pre-engraftment period had worse overall survival compared to patients without viable fungi in any specimen (Fig 5a). The lower overall survival in patients with viable fungi in fecal specimens was due to a higher transplant-associated mortality (Fig. 5b).
Figure 5: The intestinal fungal mycobiota and clinical outcomes.
(a) Overall survival and (b) transplant-related mortality in relation to viable fungi recovered from samples within the pre-engraftment period (days 0 to the first day of an absolute neutrophil count > 500/mm3). (c) Overall survival and (d) transplant-related mortality in relation to C. parapsilosis complex domination (≥90% relative abundance) within the pre-engraftment period (days 0 to the first day of an absolute neutrophil count > 500/mm3). (a, c) Kaplan-Meier curves tested by two-sided log-rank test. (b, d) Cumulative incidence curves with relapse and progression of primary disease as competing interest. Differences tested by two-sided Gray’s test. See also Extended Data Fig. 3. (a-d) Numbers below the graphs correspond to the number of patients (still) at risk at the specific time point.
To assess the impact of a different threshold of relative abundance to define a fungal domination event, we calculated the median relative abundance of the most common taxon in all samples during the pre-engraftment period (82%) and re-examined clinical outcomes using this alternate threshold. We found that incorporating the alternate threshold in our analyses did not alter the association between fungal dysbiosis and overall survival or between fungal dysbiosis and transplant-related mortality (Extended Data Fig. 3).
Since C. parapsilosis complex was most commonly detectable as viable cultures, we tested whether domination with a species belonging to the C. parapsilosis complex was similarly predictive of adverse clinical outcomes. Patients with pre-engraftment domination by C. parapsilosis complex species had a worse overall survival and higher transplant-related mortality than patients with no pre-engraftment domination by C. parapsilosis complex species (Fig. 5c and 5d). In contrast, patients with C. albicans domination did not have a lower probability of overall survival or a higher probability of transplant-related mortality compared to patients without a fungal domination event (Extended Data Fig. 3).
C. parapsilosis complex domination remained associated with worse overall survival even after adjusting for age, comorbidities, graft type, primary disease, and receipt of anti-anaerobic antibiotics in the pre-engraftment period, all of which affect outcomes in allo-HCT patients (Extended Data Table 2). For both measures of intestinal fungal dysbiosis, the study was not powered to detect differences in specific causes of transplant-related mortality (Supplementary Tables 1 and 2) according to the scheme proposed by Copelan et al. 27.
In summary, fungal dysbiosis in allo-HCT patients was characterized by domination of C. parapsilosis complex species in conjunction with expansion of culturable intestinal fungi. This fungal expansion and domination were facilitated by a reduction in the intestinal bacterial density. Patients with C. parapsilosis and patients with culturable intestinal fungi had a substantially lower overall survival and higher transplant-related mortality after allo-HCT.
Discussion
In this study, we characterized the dynamics of the fungal intestinal community during allo-HCT by high-density longitudinal sampling. The mycobiota had highly variable densities and compositions across time during HCT. This result is consistent with findings from the Human Microbiome Project, which also demonstrated higher intra- and interindividual variability of intestinal fungi compared to bacteria in healthy individuals 28,29. A large proportion of fungal DNA and fungi detected in feces from healthy adults is thought to be due to transient passage rather than direct colonization. Specifically, C. albicans in fecal samples has been linked to oral hygiene, while S. cerevisiae has been linked to food sources 29.
While fungal density did not change over time on a global scale in our cohort, more viable fungi were cultured from samples in the period after HCT. The vast majority of culturable fungi consisted of C. parapsilosis complex species, with concurrent expansion observed by ITS1 amplicon sequencing. In contrast, expansion of C. albicans ITS1 sequences did not correlate with the presence of culturable fungi. While the efficiency of retrieving culturable fungi may have been reduced due to one cycle of freeze-thawing, sample storage conditions were unlikely to differentially affect C. parapsilosis complex and other closely related Candida species. The C. albicans ITS1 amplicon sequences may have originated from oral or intestinal communities that were impacted by the routine use of micafungin as antifungal prophylaxis during the pre-engraftment allo-HCT period, prior to advancing subjects to azole-based prophylaxis. C. parapsilosis complex species are known to have an inherently higher minimal inhibitory concentration against micafungin and other echinocandin drugs compared to other Candida species 30,31. This phenotype is due to a chromosomal P660A mutation in the fks1 gene 32, though C. parapsilosis complex species are not intrinsically resistant to echinocandin drugs. Our findings indicate that C. parapsilosis complex species were able to stably colonize the gut in a subset of allo-HCT patients that were treated with micafungin-based antifungal prophylaxis. Antifungal prophylaxis is a standard allo-HCT practice since it was shown over 25 years ago that administration of prophylaxis improves patient survival 4,5. Thus, it is not feasible to conduct mycobiota studies during allo-HCT without antifungal prophylaxis. The single-center design is a limitation of the current study. It is possible that different prophylaxis strategies employed across HCT centers worldwide may lead to different Candida species domination and fecal culture positivity that in turn may be associated with adverse clinical outcomes.
Colonization by C. parapsilosis complex species in the pre-engraftment period was associated with significantly reduced overall survival and higher transplant-related mortality. This association was independent of Candida bloodstream infections, although all Candida bloodstream infections occurred in the context of intestinal Candida expansion, consistent with recent findings 15. The presence of culturable Candida in oral or fecal samples within the first 10 days after hospitalization of allo-HCT patients has previously been linked to a higher incidence of acute graft-versus-host disease (GvHD, grade II to IV) 17. Of note, in that study, subjects did not receive routine antifungal prophylaxis, which had previously been shown to independently reduce the occurrence of severe intestinal GvHD (grades III and IV) 16. Since only twelve patients (8%) in our cohort developed acute GvHD of grades III or IV, this study was not powered to detect differences in the incidence of GvHD or GvHD-related mortality. A limitation of the current study is the absence of causal links due to the non-interventional character of the study population. Future studies are required to address the mechanistic impacts of fungal dysbiosis in mouse colonization models and in interventional trials.
Recently, profound injury to the intestinal bacterial microbiota during allo-HCT has been linked to worse clinical outcomes in a large multicenter study, including patients from our center 7. The use of carbapenem drugs and piperacillin-tazobactam diminishes the bacterial intestinal density and modulates the microbiota composition 8. Correspondingly, the use of these antibiotics is associated with higher transplant-related and higher GvHD-related mortality in allo-HCT patients 33,34. In the context of reduced bacterial diversity and under antibiotic selection pressure bacterial pathobionts, such as Enterococcus and Enterobacteriaceae, can expand in the intestine of allo-HCT patients 9–14. Intestinal enterococcal domination has been linked to lower overall survival in allo-HCT patients and been causally involved in the development of acute GvHD 12.
Intriguingly, co-domination of enterococci and C. parapsilosis was infrequent in our cohort. Prior use of carbapenems or piperacillin-tazobactam, antibiotics that target anaerobic bacteria, only had a small negative confounding effect on the association of C. parapsilosis complex domination and overall survival. Thus, the use of broad-spectrum antibiotics is likely to negatively affect patient outcomes by multiple parallel effects, such as loss of bacterial diversity, expansion of bacterial pathobionts, or, as our data suggest, expansion and intestinal colonization by fungal pathobionts.
C. parapsilosis complex expansion and an increase in fungal burden occurred in the context of severely diminished bacterial densities and a relative increase of bacterial genera not commonly seen at high abundances in the core intestinal microbiota, such as Staphylococcus 35. This complements our recent findings that patients with Candida expansion prior to development of bloodstream infections have a marked loss of intestinal anaerobic bacterial communities 15. In contrast to the results from our prior case-control study on patients with candidemia, we did not see a reduction in bacterial diversity in samples with C. parapsilosis domination in the current cohort which was much larger (n = 156) than the case-control study (n = 15). It is notable that less than 20% of patients with C. parapsilosis complex intestinal domination go on to develop candidemia. Thus, it is conceivable that there may be differences in the bacterial load in C. parapsilosis complex-dominated patients that go on to develop candidemia versus those that do not. Other differences in the patient populations among both studies may be relevant: the case-control study focused on patients with candidemia, which had a higher comorbidity index, and did not exclude patients that failed to engraft or patients that received a second allo-HCT. Both factors that may influence the bacterial intestinal composition and density.
This study is consistent with the general observation that antibiotic treatment is a prerequisite for stable gut colonization with most C. albicans and non-albicans Candida strains in mice 36. Metabolites in human feces can inhibit the growth of C. albicans, C. parapsilosis, and other fungi in vitro 37. The presence of specific anaerobic bacteria in the mouse intestinal microbiota, such as clostridial Firmicutes and Bacteroides thetaiotamicron, can confer colonization resistance against murine Candida albicans colonization 38,39. Other intestinal bacteria, such as Lactobacillus rhamnosus GG and E. fecalis can inhibit C. albicans hyphal formation and decrease its virulence 40,41. However, since C. parapsilosis complex species do not form hyphae, other trans-kingdom interactions may be at play. Determination of the specific mediators of colonization resistance against pathogenic fungi, specifically non-albicans Candida species may allow the development of prophylactic probiotic strategies to counter the detrimental effects of antibacterial therapy.
Our data support a model in which the loss of commensal bacterial flora promotes C. parapsilosis complex species expansion under selective pressure of echinocandin drugs. Fungal overgrowth by these species is in turn associated with worse clinical outcomes in allo-HCT patients. Identifying and targeting fungal dysbiosis in the context of allo-HCT may improve patient outcomes and highlights the clinical importance of the mycobiota in transplantation as therapy for cancer or organ failure.
Methods
Data availability
Fungal ITS 1 and bacterial 16S amplicon sequences specific to this manuscript are deposited in NCBI SRA under Bioproject PRJNA746305. Samples, sequences, and their accession number are provided in the Supplement. Clinical and laboratory data used for the analyses in this manuscript are listed in the Supplement. Additional requests for clinical data and materials will be reviewed by Memorial Sloan Kettering Cancer Center to determine if they are subject to intellectual property or confidentiality obligations. Data and materials that can be shared will be released via a material transfer or data-sharing agreements and can be requested from the corresponding author (hohlt@mskcc.org).
Inclusion Criteria and Sample Collection
Adult patients that undergo allo-HCT at MSKCC are invited to participate in a biospecimen protocol that prospectively and repeatedly collects fecal samples over the course of allo-HCT 10. To be included in the current study, patients had to undergo allo-HCT in 2017 or 2018, have a successful cell engraftment, and have at least one fecal sample available in the repository dating between the day of HCT and the day of engraftment. A portion of the patients and samples in this study have been previously reported 7,12. Of this patient pool, we randomly selected 156 patients using the sample function in R. The sample size was based on sample size and effect sizes seen in our early studies on the bacterial microbiota and allo-HCT 10,42,43.
All available fecal samples of the selected patients collected between ten days prior to 30 days after HCT were processed. Data on age and sex of included patients can be found in Table 1.
Sample and clinical data collection were performed prior to the development of this specific research question. Patients and personnel involved in sample and data collection can therefore be considered blinded to the exposures and outcomes of the study.
All patients provided written informed consent for biospecimen collection. The fecal biospecimen repository and this study protocol were approved by the MSKCC institutional review board. The study was performed in concordance with the Declaration of Helsinki.
Analysis of fecal samples
After homogenization in 200μl of sterile PBS, 50 μl of liquid stool or 50μg of formed stool were plated on Sabouraud dextrose agar plates complemented with 0.01 mg/ml vancomycin and 0.1 mg/ml gentamicin to culture viable fungi for 48h at 37 °C 15. Single colonies were counted to calculate colony-forming units (cfu). Mold colonies were not further classified. Yeast colonies were picked and stored in glycerol stocks at −80°C. Species identification was performed via Sanger sequencing of ITS 1 and comparison of the resulting sequences to the NCBI GenBank nucleotide collection. Our culture-based approach was not optimized to recover viable Malassezia species, a major constituent of the skin mycobiota 44, due to the requirements for fatty acid supplementation in culture media 45. However, Malassezia domination by ITS1 amplicon sequencing was observed in only two instances (0.16% of fecal samples), suggesting that very rare events of Malassezia intestinal domination did not impact clinical outcomes.
For fungal DNA extraction, aliquots of 200–300μg of solid or 200–300μl of liquid stool were weighed. They were then preprocessed with 500 μl of lyticase buffer (50 mM Tris, pH 7.6, 10 mM ethylenediaminetetraacetic acid, 28 mM β-mercaptoethanol) and 100 U lyticase (Sigma). 0.2ml 0.1-mm silicone beads and 0.3ml 0.5mm beads were added prior to vortexing and incubating the samples for 30 minutes at 32 °C. After adding 30 μl of 4 M NaCl, 47 μl of 2 M Tris-HCl, pH 8.0, 13 μl of 0.5 M ethylenediaminetetraacetic acid, 200 μl of 20% sodium dodecylsulfate and 500 μl of phenol chloroform, the samples were homogenized by bead-beating for 2 min. Detritus was spun down at 13,300g at 4 °C for 5 min. The upper aqueous layer was recovered and added to a mix of 100 μl DEB buffer (0.2M NaCl, 0.2M Tris, pH 8.0, 0.5M EDTA) and 500 μl phenol-chloroform. After inverting and spinning down, the upper aqueous layer was again recovered. This process was repeated for a total of three times. After the final round of phenol-chloroform 200 μl of the aqueous phase was mixed with 200 μl of buffer AL (QIAamp DNA Mini Kit) and 200 μl of 100% ethanol. Further processing was according to the QIAmp DNA Mini Kit instructions. DNA was eluted in 80 μl of ultrapure water.
Quantification of fungal and bacterial DNA
The amount of fungal and bacterial DNA was quantified via qPCR. 18S copy numbers were determined using the FungiQuant protocol 22: The reactions were performed with a TaqMan Fast Advanced Master Mix (Applied Biosystems 4444964) in a 96-well plate with 10 μl per well and using 1 μl of DNA eluate. Working concentrations were 20μM for the FungiQuant-F (5’-GGRAAACTCACCAGGTCCAG-3’) and FungiQuant-R (5’-GSWCTATCCCCAKCACGA-3’) primers, and 1μM for the probe (5’-GGRAAACTCACCAGGTCCAG-3’). A standard curve was obtained by serial dilutions of genomic DNA from the reference strain C. albicans SC5314. After 2 minutes at 50 °C and 10 minutes at 95 °C, cycling conditions were15 seconds at 95 °C and 1 minute at 60 °C for a total of 40 cycles.
16S copy number were determined using a protocol based on the DyNAmo SYBR Green rtPCR Kit (ThermoFisher F410L) and the broad-range bacterial 16S primers 563F and 926Rb at 0.2mM. Cycling conditions were 10 minutes at 95 °C followed by 40 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min. Standard curves were obtained by serial dilution of the PCR blunt vector (Invitrogen) containing one copy of the 16S rRNA gene derived from a member of the Porphyromonadaceae family. For both 18S and 16S copy numbers, values were normalized to the fecal weight input.
Amplicon preparation, sequencing, and data processing
ITS1 and 16S (V4-V5) amplicon preparation were performed as reported previously by our group 15,46: For both the 16S and ITS1-specific forward and reverse primers were modified with unique 12-base Golay barcodes preceding the primer for parallel sequencing of up to 60 samples per sequencing pool and subsequent sample identification. To offset primer sequencing, additional 1 to 8 nucleotides were placed in front of the barcode 46.
For ITS 1 amplicon preparation 1 μl of purified sample eluate template DNA (at 1:1, 1:5, and 1:25 dilutions) was amplified with the ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS1R (5’-GTTCAAAGAYTCGATGATTCAC-3’) primers (1.25μM) and Phusion enzyme (ThermoFisher F530L) for 35 cycles: 30 s at 98°C, 30 s at 53°C, and 30s at 72°C. The resulting three PCR amplicon mixes per sample were pooled and purified using the Axygen AxyPrep MagKit Mag PCR Clean-Up Kit (Axygen MAGPRCL50) according to the manufacturer’s protocol. For 16S amplicon preparation each sample, duplicate 50-μl PCRs were performed. 50 ng of purified DNA were amplified with 2.5 U Platinum Taq DNA polymerase, and 0.5 mM of primers 563F and 926R at 94°C for 3 min, followed by 27 cycles of 94°C for 50 s, 51°C for 30 s, and 72°C for 1 min and a final elongation step at 72°C for 5 min.
Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen 28104) after pooling of replicates. DNA content in the 16S and ITS amplicon mixes was measured by Qubit Assay (ThermoFisher Q32853). Equimolar amounts of DNA were pooled and sent to the MSKCC Integrated Genomics Operation for library preparation and sequencing on the Illumina MiSeq platform using PE300 for ITS1 and PE250 for 16S setting.
ASV Construction and taxonomic identification
Raw reads were demultiplexed on a per-sample basis according to the unique Golay barcodes. After primer and barcode removal, the DADA2 pipeline was used for quality filtering and inference of exact amplicon sequencing variants (ASVs). For 16S amplicons, the default values in DADA2 were used, for ITS1 amplicons, we used a truncation length of 257 and a maximum expected (maxEE) error of 5. To accommodate ITS1 segments exceeding PE300 settings, we concatenated forward and reverse reads only in the absence of an overlap.
Taxonomy was assigned at species level with an algorithm based on BLAST with NCBI RefSeq RNA for 16S and UNITE database for ITS1 as training sets, respectively47,48. ASV tables, taxonomy, and sample metadata were compiled in a phyloseq object in R 49. Low-quality samples with less than 200 reads were removed from further analysis, both for ITS 1 and 16S.
Statistical Analyses
All analyses were performed in R version 4.0.5 within RStudio version 1.3. Data analysis was performed unblinded. The statistical methods used are described in the figure legends. No data points were excluded from analysis. Hierarchical clustering of samples was performed by using weighted unifrac distances at the genus level (as determined by the taxonomy in the UNITE database). Fungal and bacterial diversity were estimated by the Inverse Simpson Index. Similarity between sample pairs of the same patient was estimated by calculating the Chao-Jaccard index between these two samples. For all survival and clinical outcome analyses, the exposure of interest had to occur between the day of cell infusion and the first day the absolute neutrophil count returned over 0.5 (engraftment). Outcome analyses started at engraftment. As only patients that had engrafted were included in the study, no patient died before engraftment. To compare overall survival Kaplan-Meier curves were plotted and between-group differences tested by log-rank test. Transplant-related mortality was defined as any death except deaths due to relapses or progressive primary disease. To assess transplant-related mortality, cumulative incidences were plotted with relapse and progressive disease as competing interests using the cmprsk package (version 2.2–10) in R. Between group differences were tested by Gray test. For both, overall survival and transplant-related mortality, patients were censored at 2 years after engraftment or at last contact, whichever was shorter. To estimate the hazard ratio of overall survival in patients with and without C. parapsilosis domination, a Cox proportional hazard model was used. Covariates tested included age at transplantation, sex, graft type, primary disease, HCT-CI, and receipt of piperacillin-tazobactam or a carbapenem in the pre-engraftment period. Piperacillin-tazobactam and carbapenems were chosen as they have been previously shown to have the largest detrimental impact on the anaerobic intestinal microbiota and has been associated with worse clinical outcomes in allo-HCT patients 8,34.
To correlate relative bacterial and fungal abundances, we grouped bacteria at the genus level and looked at three fungal taxonomic groups: C. parapsilosis complex, other species of the Candida genus, and S. cerevisiae. Cross-correlation was performed with the associate function in the R package microbiome (version 1.15.0) by using Spearman correlation and adjusting p-values by the false discovery rate (FDR) algorithm.
Code availability
Denoising and taxonomic annotation pipelines were customized as reported50. All analyses were performed in R using base R and publicly available packages. Analysis code can be requested from the lead author (rollingt@mskcc.org).
Extended Data
Extended Data Fig. 1. Impact of culture-positivity and TPN administration on fungal and bacterial microbiota dynamics during allo-HCT, related to Fig. 1.
(a) Comparison of fungal density per gram of stool (n = 246 pre-transplant samples and n = 978 post-transplant samples) and (b) fungal diversity (n = 251 pre-transplant samples and n = 1028 post-transplant samples) between the pre- (days −10 to −1) and the post-transplant period (days 0 to 30). (c) Comparison of bacterial density per gram of stool (n = 241 pre-transplant samples and n = 976 post-transplant samples) and (d) diversity (n = 245 pre-transplant samples and n = 939 post-transplant samples) between the pre- and post-transplant period. Statistical tests (a-d): Two-sided P-values issued from generalized estimating equations with patient ID as cluster variable are shown. The upper whisker represents the maximum value or the 75th percentile value plus 1.5 times the interquartile range, whichever is larger.
Extended Data Fig. 2. Taxonomic composition and relation to culture-positivity, related to Fig. 3.
(a) Taxonomic composition of culture-positive samples by ITS (top ribbon) and species cultured from the respective samples (bottom ribbon). (b) Comparison of fungal similarity (1-Chao-Jaccard index) according to presence of fungal domination status of the earliest sample. Each dot represents a sample pair of an individual patient collected one day apart (n= 516 sample pairs). (c) Comparison of fungal similarity (1-Chao-Jaccard index) according to the taxa responsible for fungal domination in the earliest samples. Each dot represents a sample pair of an individual patient collected one day apart (n= 88 sample pairs). (b-c) Two-sided p-values issued from generalized estimating equations with patient ID as cluster variable without accounting for multiple comparisons are shown. The upper whisker represents the maximum value or the 75th percentile value plus 1.5 times the interquartile range, whichever is larger.
Extended Data Fig. 3. Supplementary data for clinical outcomes and the fungal intestinal microbiota, related to Fig. 5.
(a) Overall survival and (b) transplant-related mortality in relation to a relative abundance of C. parapsilosis complex ≥ 82% within the pre-engraftment period (days 0 to the first day of an absolute neutrophil count > 500/mm3). (a) Kaplan-Meier curves tested by two-sided log-rank test. (b) Cumulative incidence curves with relapse and progression of primary disease as competing interest. Differences tested by two-sided Gray’s test. (c) Overall survival and (d) transplant-related mortality in relation to domination by C. albicans within the pre-engraftment period (days 0 to the first day of an absolute neutrophil count > 500/mm3). (c) Kaplan-Meier curves tested by two-sided log-rank test. (d) Cumulative incidence curves with relapse and progression of primary disease as competing interest. Differences tested by two-sided Gray’s test. (a-d) Numbers below the graphs correspond to the number of patients (still) at risk at the specific time point.
Extended Data Table 1:
Patient Characteristics.
| Patient Characteristics | N = 156 |
|---|---|
|
| |
| Age at HCT (years, median [IQR]) | 60 (50–65) |
|
| |
| Female Sex (n, %) | 63 (40%) |
|
| |
| Primary Disease (n, %) | |
| - Acute leukemia | - 80 (51%) |
| - Myelodysplastic syndrome | - 28 (18%) |
| - Lymphoma | - 25 (16%) |
| - Myeloproliferative disorder | - 10 (6%) |
| - Multiple myeloma | - 8 (5%) |
| - Other | - 5 (3%) |
|
| |
| Graft Type (n, %) | |
| - Unmodified PBSC | - 62 (40%) |
| - T-cell depleted PBSC a | - 47 (30%) |
| - Unmodified bone marrow | - 31 (20%) |
| - Umbilical cord blood | - 16 (10%) |
|
| |
| HCT-CI (median, [IQR]) | 2 (1–4) |
|
| |
| Follow-up (months, median [IQR]) | 32 (15–38) |
one patient received a graft that was selectively depleted of alpha/beta-T-cells.
Extended Data Table 2:
Cox proportional hazard model of overall survival.
| Characteristics | Univariate HR (95%CI) | Two-sided P | Adjusted HR (95%CI) | Two-sided P |
|---|---|---|---|---|
|
| ||||
| C. parapsilosis complex domination | ||||
| - No | 1.00 | 1.00 | ||
| - Yes | 2.62 (1.29–5.29) | 0.008 | 2.43 (1.11–5.32) | 0.027 |
|
| ||||
| Sex | ||||
| - Female | 1.00 | 1.00 | ||
| - Male | 0.70 (0.39–1.26) | 0.235 | 0.64 (0.34–1.19) | 0.158 |
|
| ||||
| Age (per year increase) | 1.04 (1.01–1.07) | 0.017 | 1.05 (1.02–1.09) | 0.004 |
|
| ||||
| HCT-CI (>2 vs 0–2) | ||||
| - 0–2 | 1.00 | 1.00 | ||
| - >2 | 3.64 (1.88–7.07) | <0.001 | 2.33 (1.17–4.66) | 0.017 |
|
| ||||
| Graft type | ||||
| - Unmodified a | 1.00 | 1.00 | ||
| - T-cell depleted | 0.77 (0.40–1.49) | 0.433 | 0.46 (0.22–0.96) | 0.038 |
|
| ||||
| Disease | ||||
| - Acute Leukemia | 1.00 | 1.00 | ||
| - Lymphoma | 0.48 (0.18–1.23) | 0.125 | 0.38 (0.14–1.06) | 0.065 |
| - MDS | 0.79 (0.38–1.67) | 0.545 | 0.56 (0.26–1.19) | 0.132 |
| - Other | 0.10 (0.01–0.72) | 0.023 | 0.07 (0.01–0.53) | 0.01 |
|
| ||||
| Carbapenem or Piperacillin/Tazobactam | ||||
| - No | 1.00 | 1.00 | ||
| - Yes | 2.86 (1.41–5.77) | 0.003 | 3.12 (1.45–6.65) | 0.003 |
Patients were classified as having Candida parapsilosis complex domination if they had at least one sample with 90% or higher Candida parapsilosis relative abundance between day 0 and the day of engraftment. They were classified has having received a carbapenem or piperacillin/tazobactam if they received at least one dose between days 0 and the day of engraftment.
Unmodified PBSCs, unmodified BM, and cord grafts were grouped together.
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grant RO-5328/2 (T.R.), National Institutes of Health (NIH) grants R01 AI093808 (T.M.H.), R21 AI105617 (T.M.H.), R21 AI156157 (T.M.H.), U01 AI124275 (J.B.X.), R01 AI137269 (J.B.X. and Y.T.), K08 HL143189 (J.U.P.), R01 CA228358 (M.R.M.v.d.B.), R01 CA228308 (M.R.M.v.d.B.), P01 CA023766 (M.R.M.v.d.B.), R01 HL125571 (M.R.M.v.d.B.), R01 HL123340 (M.R.M.v.d.B.), P01 AG052359 (Project 2, M.R.M.v.d.B.), a Takeda Science Foundation-Fellowship (K.Y.M.), an Investigator in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund (T.M.H.), the Ludwig Center for Cancer Immunotherapy (T.M.H.), a Tri-Institutional Stem Cell Initiative award 2016–013 (M.R.M.v.d.B.), the Lymphoma Foundation (M.R.M.v.d.B.), the Parker Institute for Cancer Immunotherapy at MSKCC (M.R.M.v.d.B., K.A.M., and J.U.P.), the Susan and Peter Solomon Divisional Genomics Program (T.M.H. and M.R.M.v.d.B.), and the DKMS (K.A.M.). All authors were supported by NIH P30 CA008748 (Cancer Center Core Grant).
Competing Interests Statement
Jonathan U. Peled reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics, and consulting fees from DaVolterra, CSL Behring, and from Maat Pharma. He has filed intellectual property applications related to the microbiome (reference numbers #62/843,849, #62/977,908, and #15/756,845). Marcel R.M. van den Brink has received research support from Seres Therapeutics; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, WindMIL Therapeutics, Rheos, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Forty Seven Inc., Priothera, Ceramedix, Lygenesis, Pluto Immunotherapeutics, Magenta Therapeutics, Merck & Co, Inc., and DKMS Medical Council (Board); has IP Licensing with Seres Therapeutics, Juno Therapeutics, and stock options from Seres and Notch Therapeutics. Tobias M. Hohl has participated in a scientific advisory board for Boehringer-Ingelheim Inc.
References
- 1.Mellinghoff SC, et al. Primary prophylaxis of invasive fungal infections in patients with haematological malignancies: 2017 update of the recommendations of the Infectious Diseases Working Party (AGIHO) of the German Society for Haematology and Medical Oncology (DGHO). Ann Hematol 97, 197–207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ullmann AJ, et al. Infectious diseases in allogeneic haematopoietic stem cell transplantation: prevention and prophylaxis strategy guidelines 2016. Ann Hematol 95, 1435–1455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomblyn M, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant 15, 1143–1238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slavin MA, et al. Efficacy and safety of fluconazole prophylaxis for fungal infections after marrow transplantation--a prospective, randomized, double-blind study. The Journal of infectious diseases 171, 1545–1552 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Goodman JL, et al. A controlled trial of fluconazole to prevent fungal infections in patients undergoing bone marrow transplantation. N Engl J Med 326, 845–851 (1992). [DOI] [PubMed] [Google Scholar]
- 6.van Burik JA, et al. Micafungin versus fluconazole for prophylaxis against invasive fungal infections during neutropenia in patients undergoing hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 39, 1407–1416 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Peled JU, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med 382, 822–834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morjaria S, et al. Antibiotic-Induced Shifts in Fecal Microbiota Density and Composition during Hematopoietic Stem Cell Transplantation. Infect Immun 87(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holler E, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 20, 640–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taur Y, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 55, 905–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120, 4332–4341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein-Thoeringer CK, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 366, 1143–1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoma I, et al. Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative bacteria. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamburini FB, et al. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med 24, 1809–1814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai B, et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marr KA, et al. Prolonged fluconazole prophylaxis is associated with persistent protection against candidiasis-related death in allogeneic marrow transplant recipients: long-term follow-up of a randomized, placebo-controlled trial. Blood 96, 2055–2061 (2000). [PubMed] [Google Scholar]
- 17.van der Velden WJ, et al. Role of the mycobiome in human acute graft-versus-host disease. Biol Blood Marrow Transplant 19, 329–332 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Jain U, et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 371, 1154–1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limon JJ, et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 25, 377–388 e376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonardi I, et al. Fungal Trans-kingdom Dynamics Linked to Responsiveness to Fecal Microbiota Transplantation (FMT) Therapy in Ulcerative Colitis. Cell Host Microbe 27, 823–829 e823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorror ML, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood 106, 2912–2919 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CM, et al. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol 12, 255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao A, Chazdon RL, Colwell RK & Shen TJ A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8, 148–159 (2005). [Google Scholar]
- 24.Faith JJ, et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson RH, et al. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 17, 95–109 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zhai B, et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med 26, 59–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copelan E, et al. A scheme for defining cause of death and its application in the T cell depletion trial. Biol Blood Marrow Transplant 13, 1469–1476 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Nash AK, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auchtung TA, et al. Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi. mSphere 3(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda F, et al. Antifungal activity of micafungin against Candida and Aspergillus spp. isolated from pediatric patients in Japan. Med Mycol 47, 145–148 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Marti-Carrizosa M, Sanchez-Reus F, March F, Canton E & Coll P Implication of Candida parapsilosis FKS1 and FKS2 mutations in reduced echinocandin susceptibility. Antimicrob Agents Chemother 59, 3570–3573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller MA, et al. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J Clin Microbiol 46, 2620–2629 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber D, et al. Microbiota Disruption Induced by Early Use of Broad-Spectrum Antibiotics Is an Independent Risk Factor of Outcome after Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant 23, 845–852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shono Y, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med 8, 339ra371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koh AY, Kohler JR, Coggshall KT, Van Rooijen N & Pier GB Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS pathogens 4, e35 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia C, et al. The Human Gut Microbial Metabolome Modulates Fungal Growth via the TOR Signaling Pathway. mSphere 2(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan D, et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 21, 808–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santus W, Devlin JR & Behnsen J Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect Immun 89(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allonsius CN, et al. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb Biotechnol 10, 1753–1763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham CE, Cruz MR, Garsin DA & Lorenz MC Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci U S A 114, 4507–4512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taur Y, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenq RR, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant 21, 1373–1383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID & Velegraki A The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25, 106–141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taur Y, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med 10(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson RH, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47, D259–D264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benson DA, et al. GenBank. Nucleic Acids Res 45, D37–D42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMurdie PJ & Holmes S Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput, 235–246 (2012). [PMC free article] [PubMed] [Google Scholar]
- 50.Rolling T, Zhai B, Frame JV, Hohl TM & Taur Y Customization of a dada2-based pipeline for fungal Internal Transcribed Spacer 1 (ITS 1) amplicon datasets. bioRxiv, 2021.2007.2019.452952 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fungal ITS 1 and bacterial 16S amplicon sequences specific to this manuscript are deposited in NCBI SRA under Bioproject PRJNA746305. Samples, sequences, and their accession number are provided in the Supplement. Clinical and laboratory data used for the analyses in this manuscript are listed in the Supplement. Additional requests for clinical data and materials will be reviewed by Memorial Sloan Kettering Cancer Center to determine if they are subject to intellectual property or confidentiality obligations. Data and materials that can be shared will be released via a material transfer or data-sharing agreements and can be requested from the corresponding author (hohlt@mskcc.org).