Abstract
In medulloblastomas (MBs), the expression and activity of RE1-silencing transcription factor (REST) is increased in tumors driven by the sonic-hedgehog (SHH) pathway, specifically SHH-α (children 3–16 years) and SHH-β (infants) sub-groups. Unexpectedly, SHH-β tumors display more neuronal maturation compared to SHH-α tumors, yet both are correlated with poor overall patient survival. We studied the contribution of REST to tumorigenesis using a novel transgenic mouse model (RESTTG). In this model, conditional NeuroD2-controlled REST transgene expression in lineage committed Patched 1 heterozygous (Ptch1+/−) cerebellar granule neuron progenitors (CGNPs) caused markedly accelerated tumorigenesis and penetrance and infiltrative disease. Mechanistic studies revealed a neuronal maturation context specific antagonistic interplay between the transcriptional repressor and activator, REST and Gli1, respectively. REST elevation and low expression of an inhibitor of Gli1, β-Arrestin1 (Arrb1), promoted Gli1 activity and Ptch1 expression in proliferating cells, in spite of increased histone H3K9 methylation at the Ptch1 promoter. However, in lineage committed RESTTG CGNPs, Arrb1 increase and decreased Gli1 activity in conjunction with increased histone H3K9 methylationat the Ptch1 locus led to premature silencing of Ptch1 expression. In human tumors, PTCH1, GLI1 and ARRB1 expression were significantly decreased in SHH-β tumors compared to SHH-α tumors. These findings as well as the synergistic decrease of MB cell proliferation in culture upon pharmacological inhibition of G9a and histone deacetylase (HDAC) activities support a role for REST in MB progression in more lineage committed cells. Lineage committed RESTTG CGNPs in comparison with proliferating progenitors, also exhibited decreased expression of the phosphatase tensin homolog (Pten), a negative regulator of Akt kinase. Consistent with this, human SHH-β tumors had significantly lower PTEN expression, although an unexpected decrease in its gene expression was seen in SHH-α tumors. Pharmacological blockade of AKT promoted apoptosis in REST-high cells in culture. Our findings linking REST to differentiation-specific chromatin remodeling, PTCH1 silencing, and AKT hyperactivation in MB tissues and models reveal potential subgroup specific therapeutic targets to explore for patients with SHH MB.
Introduction
Medulloblastoma (MB), the most common malignant brain tumor of childhood, is a molecularly diverse embryonal tumor of the posterior fossa. Genomic studies over the last few years have confirmed at least four distinct subgroups [Wingless (WNT), Sonic Hedgehog (SHH), Group 3 and Group 4] (1–4)(2–5). Retrospective studies have confirmed that long-term outcomes are subgroup specific (1). Patients with WNT-subtype MB tumors have a greater than 90% long term survival in contrast to patients with Group 3 tumors who have a 40 to 60% 5-year overall survival. Patients with SHH-driven subtype MB have an intermediate prognosis (2–4). Current treatment protocols include maximal surgical resection, cranio-spinal radiation therapy for children older than 3 years of age and aggressive multi-agent chemotherapy (1). Unfortunately, metastasis continues to be a major clinical challenge in at least 3 of the 4 subgroups (6). Additionally, survivors of MB have substantial long-term toxicities as a result of therapy (1). Therefore, in recent years, there has been an earnest push towards understanding subgroup-specific MB biology to target the molecular events that drive tumorigenesis and metastasis (1, 3, 6, 7).
SHH MBs are the best characterized subtype of the disease. The availability of multiple genetically engineered mouse models has provided valuable insights into parallels with normal cerebellar development and most importantly, has allowed identification of cerebellar granule cell progenitors (CGNP) as the cell of origin of a subset of these tumors (8–11). These studies have also implicated the developmentally important Shh signaling pathway as a driver of the early postnatal burst of proliferation of CGNPs in the cerebellar external granule layer (EGL) (12). The developmental switch to terminal neuronal differentiation coincides with a decline in Shh pathway activity. Study of this switch in Shh pathway activity and its contribution to the transition from a proliferative to a differentiation program in CGNPs will shed light on MB genesis. Shh is a secreted ligand and binds Patched1 (Ptch1), a transmembrane receptor and tumor suppressor protein, to relieve its repressive interaction with the oncogene, Smoothened (Smo). The subsequent movement of the latter to the primary cilia, allows signaling through Gli proteins to regulate the expression of cell cycle regulatory genes including cyclins (Ccnd1 and Ccnd2), the proto-oncogene N-Myc and, through a feedback loop, Ptch1, Gli1 and Gli2 (13, 14). Current evidence supports regulation of Shh signaling at multiple steps (13). Of particular interest to this report is control of Ptch1 expression during neuronal lineage commitment in CGNPs, because data from Ptch1 heterozygous mice mouse models suggests its loss of haploinsufficiency (LOH) is critical to tumor progression (8, 15, 16). A second point of focus of this study is regulation of pathway activity at the level of Gli proteins. Of the three Gli proteins expressed in vertebrates, Gli1 is a transcriptional activator, Gli2 toggles between an activator and a repressor and Gli3 functions as a transcriptional repressor. Phosphorylation and ubiquitination are known to control the activity of these proteins in a Shh ligand-dependent manner (17–19). In addition, Gli1 activity is regulated by its inhibitory acetylation by the p300 -β-Arrestin 1 (Arrb1) complex (20, 21). Clearly, knockdown and overexpression studies in CGNPs suggest that Arrb1 plays a role in the shift of CGNPs from a proliferative to a differentiation state and therefore, it is important to understand its regulation as well (21).
The RE1-silencing transcription factor (REST) is a transcriptional repressor of neuronal differentiation genes (22–29). It also maintains cell proliferation and blocks cell cycle exit by preventing stabilization of the cyclin-dependent kinase inhibitor, p27, a key event in cells undergoing terminal neuronal differentiation (29, 30). REST is expressed in neural stem cells, but its expression declines during neurogenesis (24, 31–34). It is a DNA-binding protein and serves as a scaffold for chromatin remodeling enzymes that repress gene expression including histone deacetylases 1 and 2 (HDAC1/2), the histone methyl transferase G9a, and the histone demethylase LSD1 (35). REST protein abundance is increased in over 80% of human MBs (14). In a previously reported study, we observed a tendency for poor prognosis and an increased risk for developing disseminated disease in a small cohort of patients with large increases in REST expression in their tumors compared to normal cerebella (29). This observation was particularly intriguing in a subset of patients with desmoplastic MB who had an unexpectedly poor outcome when compared to the entire SHH subgroup (29).
The overall goal of this study was to investigate the involvement of REST in the regulation of Shh signaling during neuronal differentiation of CGNPs and to determine if a perturbation of this process promotes MB progression. Human SHH-MBs are sub-divided into SHH-α, -β, -γ, -δ tumors, which differ in the age of the patients, propensity for metastasis, and overall survival (36). Through analysis of a publicly available dataset, we demonstrated that REST expression and activity, as measured by the expression of its target neuronal differentiation genes, exhibited a strong correlated with poor prognosis in specific SHH subgroups. This included a subset of SHH-α and a majority of the SHH-β subgroup of tumors (36). We also employed a novel transgenic mouse model (RESTTG) with potential for conditional elevation of human REST (hREST) transgene (Tg) in lineage committed CGNPs, to not only show poor survival of mice with REST elevation and constitutive activation of Shh signaling, but to also uncover the molecular mechanisms underlying REST-mediated tumorigenesis. Our data suggest that REST elevation increases Shh signaling in proliferating cells, but accelerates the decline in pathway activity in differentiating cells. In proliferating CGNPs, despite the increase in chromatin compaction at the Ptch1 promoter in response to elevated REST expression, Gli1 activity was maintained as a result of downregulated expression of a negative regulator, Arrb1. In lineage committed CGNPs, the increase in Arrb1 expression and consequent diminution in Gli1 activity in conjunction with an increase in chromatin compaction at the Ptch1 locus led to a decline in Shh signaling. Separately, a REST-dependent decrease in expression of Phosphatase and Tensin Homolog (Pten) gene expression, a negative regulator of Akt activity, was also seen in differentiating RESTTG CGNPs. We propose that these events, at least in part, underlie the aggressive disease course and leptomeningeal dissemination of REST-expressing Shh-MB tumors in mice. It may also explain the poor outcomes seen in patients with increased REST expression in their tumors, mainly those with SHH-α and β subgroup of tumors (36). Finally, our findings in cell culture models provide support for potential pharmacological targeting of G9a and HDACs in high-REST MBs.
RESULTS
REST expression and activity is elevated in SHH-α and -β subgroup of tumors
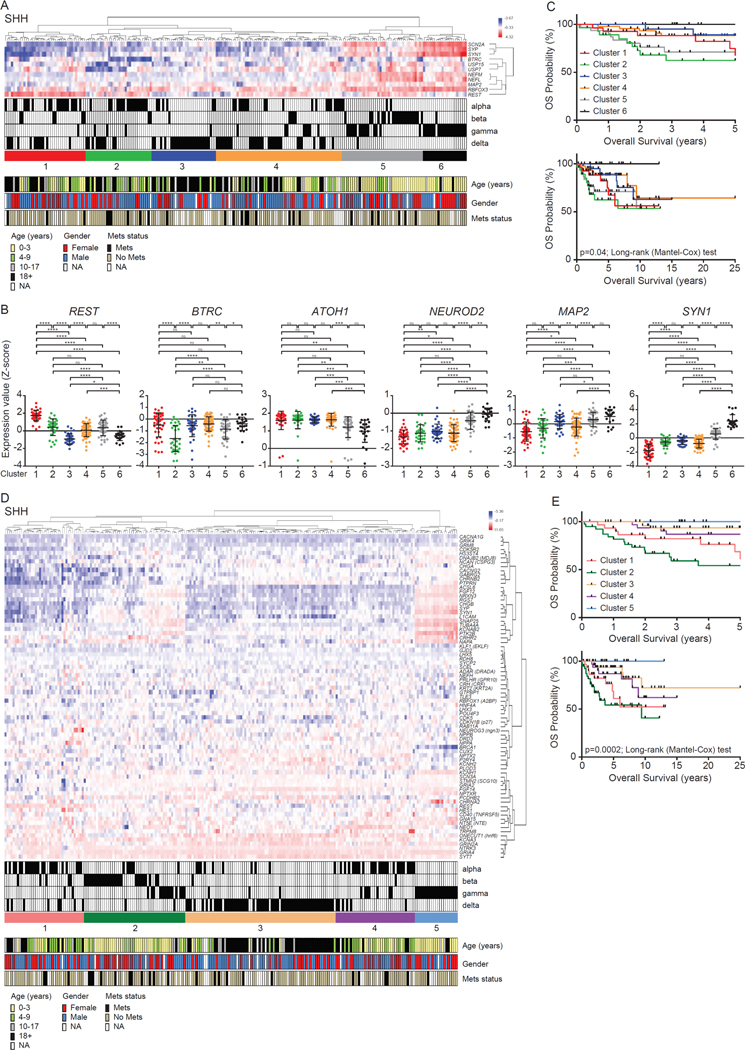
REST is an important regulator of neurogenesis and negatively controls the expression of a number genes involved in terminal neuronal differentiation (32, 37–39). In MBs, which are poorly differentiated tumors, we and others have previously demonstrated an aberrant maintenance of REST expression in a small cohort of patient samples (22, 28, 29). Additionally, knockdown of REST in MB cell lines blocks their tumorigenic potential in mouse orthotopic models, whereas its ectopic expression in v-myc immortalized neural stem cells promotes tumor progression (25, 28). These data suggest that REST contributes to the development of MB. To test this hypothesis, we studied the repressive activity of REST on neurogenesis in a larger cohort of MB samples in a publicly available database (GSE85218), by clustering samples based on transcriptome information on REST in combination with that of known regulators of its protein stability (BTRC, USP7 and USP15), and a subset of its target neuronal differentiation genes (SCN2A, SYP, SYN1, NEFM, NEFL, MAP2 and RBFOX3/NEUN) (Fig. 1A and fig. S1) (36). The WNT group of MBs (n=70 patients, 29 males and 35 females; age range 2–56 years old, median age 10.8 years old; 6 patients with known metastasis) were divided into 3 clusters (fig. S1A). Tumors in cluster 1 exhibited significantly higher REST expression compared to samples in clusters 2 and 3 (fig. S1B). However, elevation of REST mRNA was not associated with a significant difference in the 5-year or 20-year survival of patients (fig. S1C). Group 3 MBs (n=144 patients; 99 males and 38 females; age range 1–49 years old, median age 5.1 years old; 43 patients with known metastasis) were divided into 5 clusters based on their terminal differentiation (fig. S1D). Of these clusters 1 and 4 had significantly increased expression of REST mRNA, compared to the other clusters, but a correlation with 5-year or 25-year overall survival of patients was not noted (figs. S1, E and F).Group 4 MBs (n=326 patients; 216 males and 92 females; age range 1–48 years old, median age 8.0 years old; 101 patients with known metastasis) were similarly divided into 6 clusters (fig. S1G). Of these samples in clusters 2 through 6 had significantly downregulated expression of REST mRNA compared to the ones in cluster 1 (fig. S1H). Once again, REST mRNA expression did not correlate with 5-year or 25-year overall survival of patients (fig. S1I).
Figure 1: Clinical characteristics based on gene expression profiles.
(A) Hierarchical clustering analysis of SHH MB patient samples using gene expression. Hierarchical clustering assay identified six distinct clusters based on expression profiles of neuronal differentiation markers (www.ncbi.nlm.nih.gov/geo; dataset GSE85217). The blue to red color scale indicates the expression level (Z-score). The key, clinical information (subtype, age, gender and metastasis status) regarding patient samples, is provided beneath. (B) Gene expression profiles measured by microarray in six clusters. Each dot corresponds to one individual patient. Data show individual variability and means with S.D. ns; not significant, *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001. (C) Overall survival of six clusters in SHH MB patients (p-value; log rank Mantel-Cox test). (D) Hierarchical clustering analysis of SHH MB patient samples using gene expression. Hierarchical clustering assay identified five distinct clusters based on expression profiles of REST target genes. The blue to red color scale indicates the expression level based on Z-score. The clinical information (subtype, age, gender and metastasis status) regarding patient samples was shown in the bottom panel. (E) Overall survival of five clusters in SHH MB patients (p-value; log rank Mantel-Cox test).
Finally, we performed a similar analysis of SHH-MB samples (n=223 patients, 128 males and 82 females; age range 0–56 years old, median age 8.8 years old; 26 patients with known metastatic disease). Samples were divided into 6 clusters, with a significant elevation of REST mRNA seen in clusters 1, 2 and 5, and cluster 1 exhibiting the highest transcript levels (Fig. 1, A and B). Clusters 1/2 SHH-α tumors and cluster 5- SHH-β tumors exhibited significantly downregulated expression of BTRC, the gene encoding a component of the ubiquitin dependent proteasomal machinery that targets REST for degradation, suggesting a potential for REST protein stabilization and increased activity in these samples (Fig. 1B). The lowest expression of BTRC mRNA was seen in samples from cluster 2 (Fig. 1B). Expression of ubiquitin-specific protease (USP)-7 mRNA, encoding a known REST-specific deubiquitylase (DUB) was significantly reduced only in cluster 2 (fig. S1K). A second DUB that controls REST protein stability is encoded by the USP15 gene (40). USP15 expression was significantly increased in cluster 5 compared to the other clusters (fig. S1K). The expression of a panel of neuronal differentiation genes (ATOH1, NEUROD2, MAP2, NEFM, SYN1, SYP) was also investigated in the above clusters. We observed a significant reduction in ATOH1 and an upregulation of other granule neuron markers (NEUROD2, MAP2, NEFM, SYN1, SYP) in clusters 5 and 6 (Fig. 1B and fig. S1K). Together, these data suggest that REST mRNA expression is mis-regulated in samples in cluster 1, whereas a more modest increase in REST mRNA expression in conjunction with an impaired protein degradation machinery was seen in clusters 2 and 5 (Fig. 1B).
Unexpectedly, the majority of SHH-α tumors (56 out of 65) were found in clusters 1, 2 and 4, whereas cluster 5—with a notable expression of terminal neuronal differentiation genes—included 21 of a total of 35 SHH-β tumors (Fig. 1A). Most SHH-γ tumors (42 of 47) were found in clusters 4, 5 and 6, whereas SHH-δ tumors (n=76) exhibited more heterogeneity in their differentiation phenotype and were scattered between clusters 1 through 4 (Fig. 1A). Patients with SHH-α and β tumors have poor prognosis and are at a higher risk for developing metastasis compared to those with SHH-γ and -δ tumors (36). Consistent with this, patients with samples found in clusters 2 (SHH-α) and the more differentiated cluster 5 (SHH-β) had the worst 5-year overall survival (Fig. 1C). At 25 years, only a 50% overall survival rate was seen in clusters 1–5 (Fig. 1C). These data suggest that increased REST expression is associated with worse initial overall survival in a subset of patients with SHH-α (cluster 2) and a plurality of SHH-β (cluster 5) tumors.
To further evaluate the above association between increased REST activity and poor prognosis in SHH MB samples, we performed a clustering analyses of REST and a substantially expanded list of known differentiation and non-differentiation REST-target genes (n=76) on the basis of a literature search (Fig. 1D and fig. S1L) (41–48). Here, a large number of SHH-β tumors (28 out of 35) were found in cluster 2 and recapitulated our findings above (Fig. 1C) with respect to the worst 5-year survival (Fig. 1E). This cluster of tumors also exhibited increased REST expression and a significant decline in the expression of BTRC (fig. S1M). Most SHH-α tumors were found within clusters 1 and 4 (Fig. 1D). Expression of USP7 was not significantly different between the various clusters; (however, USP15 expression was decreased in clusters 4 and 5 compared to the other clusters (fig. S1M). Collectively, the above data suggest that increased REST expression can occur both in early progenitors and in more neuronal lineage-committed cells, which define a subset of SHH-α and most SHH-β MBs, respectively.
Three fourths of SHH-α tumors (in clusters 1 and 4; 49 out of 65) had notable losses in 9q (73.5%), 10q (46.9%) and 17p (40.8%), and gains in 9p (40.8%) (fig. S1J) (36). As expected, SHH-β, SHH-γ and SHH-δ patients exhibited fewer copy-number changes (arm-level aberrations) compared to SHH- α tumors (fig. S1J) (36).
Generation and characterization of a novel genetically engineered mouse model with increased REST expression in CGNPs
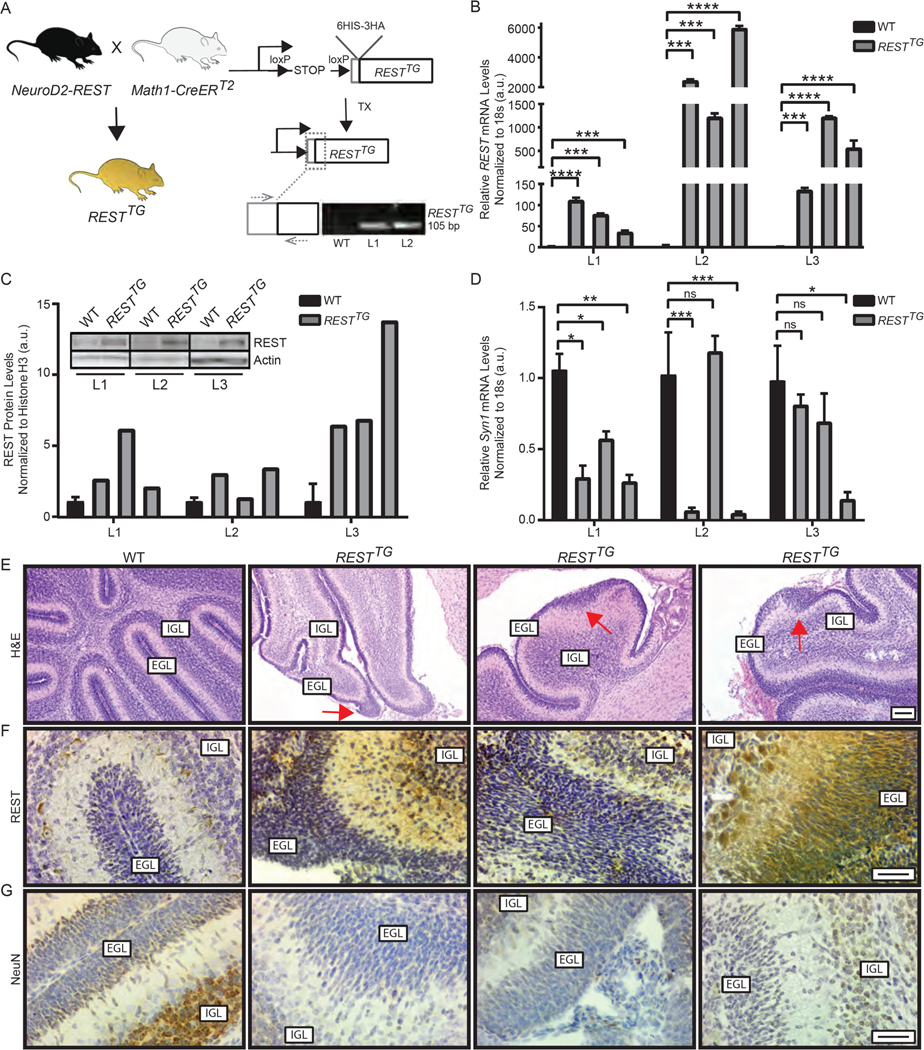
The above findings led us to hypothesize that increased REST expression in CGNPs contributes to tumor progression, and that lineage committed cells could also undergo transformation. To test this postulate and to understand the specific contribution of REST to tumorigenesis, we first generated a novel transgenic mouse model (NeuroD2-REST), where conditional expression of hREST Tg in CGNPs is driven by a 1 kb fragment of the NeuroD2 (ND2) promoter (Fig. 2A) (11). Tg presence was confirmed in NeuroD2-REST mice by qPCR, using primers against an epitope tag at the 5’end of the hREST sequence and the 5’end of the open reading frame using the same strategy as described (in Fig. 2A) for RESTTG mice. The latter were derived by crossing NeuroD2-REST mice with Math1-CreERT2 mice. Genotyping confirmed amplification of a specific 105 bp band in two RESTTG littermates (L1 and L2), but not in wild type (WT) mice (Fig. 2A).
Figure 2: Generation and characterization of a novel genetically engineered mouse model with increased REST expression in CGNPs.
(A) Schema to describe generation of a conditional RESTTG mouse model. Primers targeting the 6HIS-3HA tag and 5’end of the hREST cDNA were used for genotyping. Agarose gel of PCR product from WT, line 1 (L1) and line 2 (L2), is shown. (B and C) CGNPs harvested from postnatal day 8 (p8) pups that received TX injections on p2, p3 and p4 were cultured for up to 15 days. Cells were collected and analyzed for (B) REST Tg mRNA expression by qRT-PCR analyses and (C) REST protein abundance by Western blotting. Data are mean ± S.D. of 3 individual pups. Representative Westerns are shown. (D) Neuronal differentiation in CGNPs was evaluated by qRT-PCR measurement of Syn1 mRNA expression. Data are mean ± S.D. of 3 individual pups. (E) H&E staining of brain tissue from p8 WT (n=3) or RESTTG (n=3) pups injected with tamoxifen (TX). (F and G) Sections were analyzed by IHC for (F) REST and (G) NeuN expression using specific antibodies to assess protein expression in CGNPs in the external granule layer (EGL) and granule neurons of the internal granule layer (IGL) (n=3). For (B-C), Bars represent mean with standard deviations of fold changes relative to WT controls. p values for qRT-PCR were calculated by paired two-tailed t test of ΔCp values: significance is indicated as not significant (ns), p<0.05 (*), p<0.01 (**), p<0.001 (***), or p<0.0001 (****). Densitometry was obtained using Image Lab software (Bio-Rad). For (E), scale bar = 50 μm (10X). For (F, and G), scale bars = 20 μm (40X).
hREST Tg expression was induced in vivo by intraperitoneal (IP) injections of tamoxifen of WT and RESTTG littermates. CGNPs were harvested from brains of postnatal day 8 (p8) progeny, grown in culture for 10 days. Suspended CGNPs were collected and re-plated in fresh growth media supplemented with recombinant SHH protein for an additional 5 days. CGNPs were collected and qRT-PCR analyses was performed. A statistically significant induction of hREST expression was seen specifically in CGNPs from three lineages of RESTTG mice, but not in CGNPs from a control WT mouse (Fig. 2B), confirmed additionally by Western blot analyses of lysates from harvested CGNPs (Fig. 2C). Overall, the increase in hREST Tg expression ranged from 2-fold to 13-fold and was comparable to that previously demonstrated in human MB samples (28). As expected, a decrease in the expression of the REST-target neuronal differentiation gene, Syn1, was seen in CGNPs from RESTTG mice when compared to cells from age-matched WT mice (Fig. 2D).
We also assessed p8 RESTTG mice for changes in postnatal cerebellar development after tamoxifen administration and hREST Tg induction as compared to age-matched control WT littermates. Hematoxylin and eosin (H&E) staining identified areas of substantial EGL expansion in 90% of RESTTG animals compared to WT controls (Fig. 2E and fig. S2A). Immunohistochemistry (IHC) indicated visibly increased REST abundance in areas of EGL expansion in RESTTG mice compared to WT animals (n=3) (Fig. 2F and fig. S2B). Unlike cells in the internal granule layer (IGL)of control WT mice, a noticeable increase REST abundance was observed in IGL cells in cerebella from RESTTG animals and correlated with decreased abundance of a neuronal differentiation marker (NeuN) in CGNPs from same tissue (Fig. 2, F and G; fig. S2, B and C). Thus, increased REST expression was associated with abnormal EGL expansion and blockade of neuronal differentiation.
Increased REST expression in the context of constitutive SHH signaling activity worsens survival and promotes tumors with leptomeningeal spread
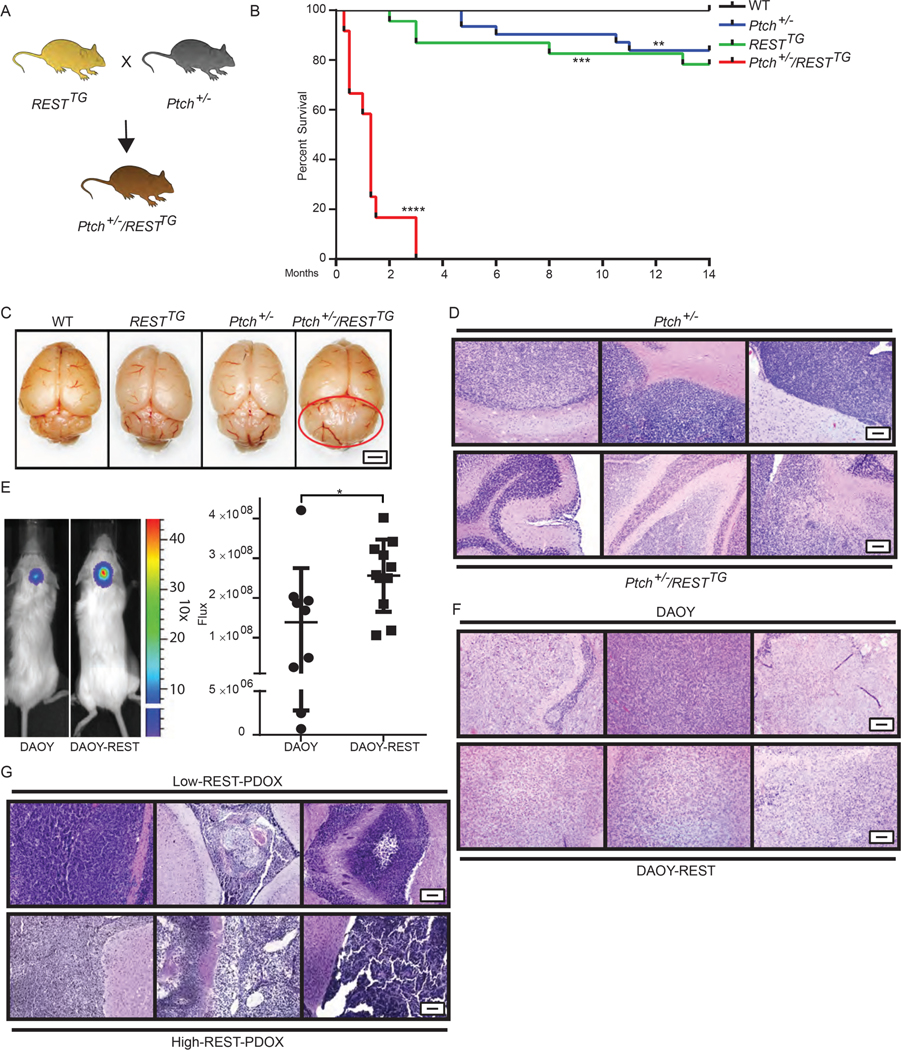
To examine the contribution of REST to progression of SHH-driven MBs, RESTTG mice were crossed with Ptch+/− mice and hREST Tg expression was induced in the resulting progeny (Ptch+/−/RESTTG) by TX injection (Fig. 3A). Kaplan Meier curves revealed a marked decrease in the survival of the 13 Ptch+/−/RESTTG mice; all died within 10 to 90 days of Tg induction (Fig. 3B). In contrast, 21.7% of RESTTG and 16.1% of Ptch+/− mice (n=23 and n=31, respectively) died by 14 months of age, whereas all 45 WT mice survived (Fig. 3B). Gross examination of brains of Ptch+/−/RESTTG mice revealed tumor burden in 100% of the mice (Fig. 3C, in which a representative brain from a 40-day-old mouse is shown). Tumor development in approximately 16% of Ptch+/− mice occurred with a considerably delayed latency between 141 and 332 days of age (Fig. 3B). The above results indicate that increased REST expression in the context of activated SHH signaling not only increased tumor penetrance but also caused a sharp decrease in tumor latency. H&E staining of the cerebella of tumor-bearing Ptch+/−/RESTTG mice revealed small blue-cell tumors with leptomeningeal dissemination (Fig. 3D and fig. S3A). In contrast, tumors in Ptch+/− mice were more localized (Fig. 3D and fig. S3A).
Figure 3: REST elevation alters kinetics and penetrance of Shh-driven MB development.
(A) Schema to describe generation of Ptch+/−/RESTTG mice. (B) Survival of WT (n=45), RESTTG (n=23), Ptch+/− (n=31), and Ptch+/−/RESTTG (n=13) mice following TX administration to induce REST Tg expression in RESTTG and Ptch+/−/RESTTG mice was assessed by Kaplan Meier analysis. (C) Representative gross images of brains from p40 WT, RESTTG, Ptch+/− and Ptch+/−/RESTTG mice are shown (n=3). Red circle indicates cerebellar tumor in p40 Ptch+/−/RESTTG mice. Scale bar = 2 mm. (D) H&E staining of brain tissue from Ptch+/− and Ptch+/−/RESTTG animals (n=3) are shown. (E) Immunodeficient mice bearing cerebellar xenografts of human DAOY cells expressing endogenous REST (n=9) or hREST (DAOY-REST (n=11) were monitored for tumor growth by BLI. Images of representative mice and relative flux for the entire cohort are shown prior to euthanasia on day 47 due to tumor burden. P values were obtained using Student t test. H&E staining of brain tissue from (F) DAOY and DAOY-REST xenografts (n=3), and (G) Low REST and High REST patient derived MB xenografts (PDOX) (n=3) are shown. For (D, F, and G), scale bars = 50 μm (10X).
Validation of these REST-dependent changes in tumor behavior was obtained using isogenic “low”-REST (DAOY) and “high”-REST (DAOY-REST) tumors obtained by implanting these cell lines in the cerebella of immunodeficient mice. DAOY-REST cells gave rise to larger tumors as assessed by bioluminescence imaging, and in a larger cohort of animals compared to animals with DAOY cell tumors (Fig. 3E). The entire cohort of mice implanted with DAOY-REST tumors also developed infiltrative tumors and extracranial tumors compared to this phenotype only in 20% of mice implanted with DAOY cells (Fig. 3F, and fig. S3B). Because the validity of DAOY as a bonafide “SHH” medulloblastoma cell line is frequently questioned, mice bearing patient-derived orthotopic xenografts (PDOXs) were also examined. These studies revealed that high-REST expressing SHH-driven MB tumors also displayed a similar propensity for leptomeningeal dissemination as DAOY-REST cells when compared to animals with low-REST SHH and DAOY-tumors (Fig. 3G, and fig. S3C).
Augmented REST expression causes Ptch1 loss-of-heterozygosity in tumors
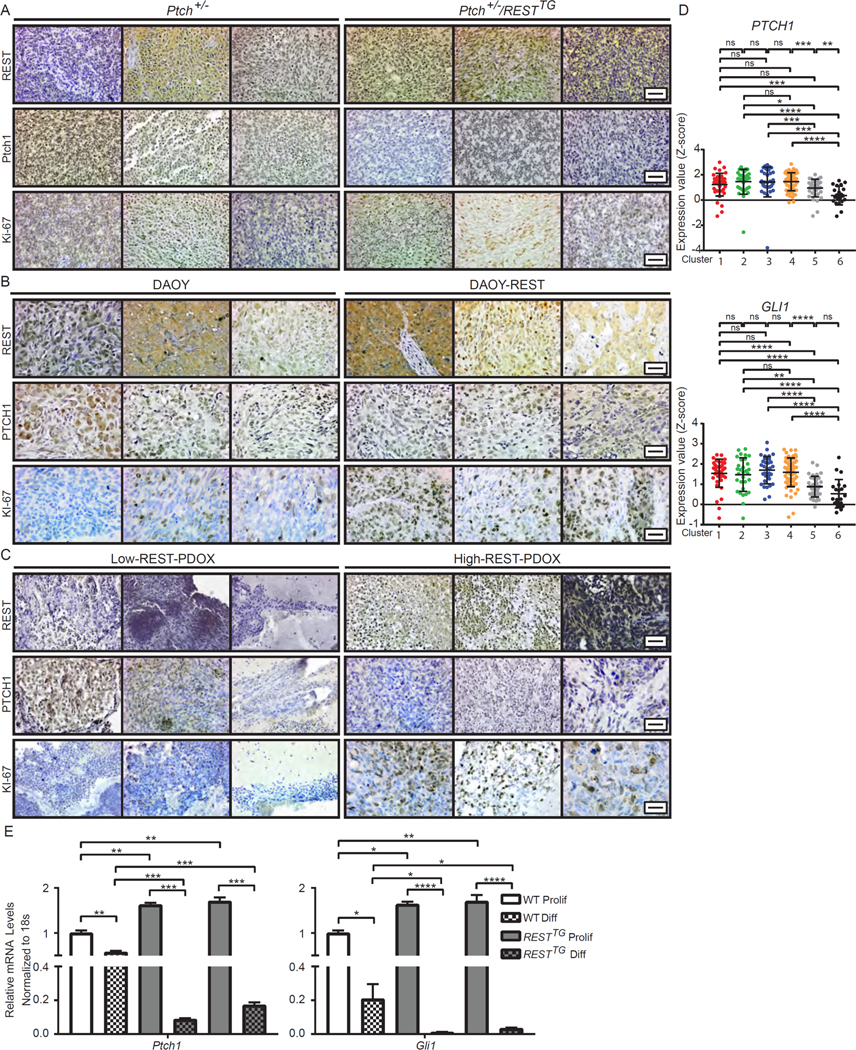
Loss of heterozygosity (LOH) in Ptch1 has been suggested as a driver of tumor development in Ptch+/− mice (8, 15, 16). Work from other groups has also shown that deletion of both alleles of Ptch1 gene in CGNPs and neural stem cells (NSCs) accelerates MB development (7, 16, 41, 49). To determine the status of Ptch1 expression in tumors from Ptch+/− and Ptch+/−/RESTTG animals, IHC analyses was performed using antibodies to PTCH1. These studies revealed strong PTCH1 staining in two of three tumor sections from Ptch+/− animals (Fig. 4A and fig. S4A). Unexpectedly, one of these tumors exhibited an increase, whereas the others had decreased REST abundance (Fig. 4A and fig. S4A). In Ptch+/−/RESTTG tumors there was a decrease in PTCH staining in two of the three brains examined and all three had high abundance of REST (Fig. 4A and fig. S4A). Sections of DAOY tumors (1 of 3) exhibited higher PTCH1 abundance compared to DAOY-REST tumors (1 of 3) (Fig. 4B and fig. S4B). REST abundance once again was increased in one of three DAOY tumors and all three DAOY-REST tumors (Fig. 4B and fig. S4B). In brain sections of mice bearing SHH-driven PDOX (n=3), two low-REST tumors had a higher intensity of PTCH1 staining compared to one of three high-REST tumors (Fig. 4C and fig. S4C). Furthermore, transcriptome analyses of SHH-group of patient tumors (described in Fig. 1A), identified samples in cluster 5 (SHH-β) as having statistically significantly decreased expression of PTCH1 and GLI1 compared to the other clusters (Fig. 4D). SHH signaling activity as measured by expression of GLI2, GLI3 and SMO, also suggested a decline in pathway-cluster 5 (SHH-β) tumors (fig. S4D). Additionally, Ki-67 staining was substantially increased in Ptch+/−/RESTTG, DAOY-REST and PDOX-high-REST tumors when compared to Ptch+/−, DAOY and PDOX-low-REST tumor sections (n=3), respectively (Fig. 4A to 4C; and fig. S4, A to C), suggesting that augmentation of REST increases proliferation in tumor cells.
Figure 4: REST downregulates PTCH1 expression.
(A to C) Cerebellar sections of (A) Tumor-bearing Ptch+/− and Ptch+/−/RESTTG mice (n=3) and (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH subgroup patient derived xenografts (n=3) were analyzed by IHC for REST, PTCH1, and KI-67 expression using specific antibodies. (D) PTCH1 and GLI1 mRNA expression profiles measured by microarray. Hierarchical clustering based on expression levels of neuronal differentiation markers divided the SHH MB patient samples into six distinct clusters. Each dot corresponds to one individual patient. (E) Ptch1 and Gli1 mRNA expression was measured in WT (white bars) and RESTTG (gray bars) CGNPs after culturing with proliferation or differentiation media. WT data represents the mean ± S.D. from triplicate samples, RESTTG data represents two individual pups. Graph represents fold change compared to WT proliferating controls. For (A, B, and C), scale bars = 20 μm (40X). For (E), p values were calculated by paired two-tailed t test of ΔCp values: significance is indicated as not significant (ns), p<0.05 (*), p<0.01 (**), p<0.001 (***), or p<0.0001 (****).
REST and Gli1 competitively control Ptch1 gene expression in a lineage commitment context-specific manner
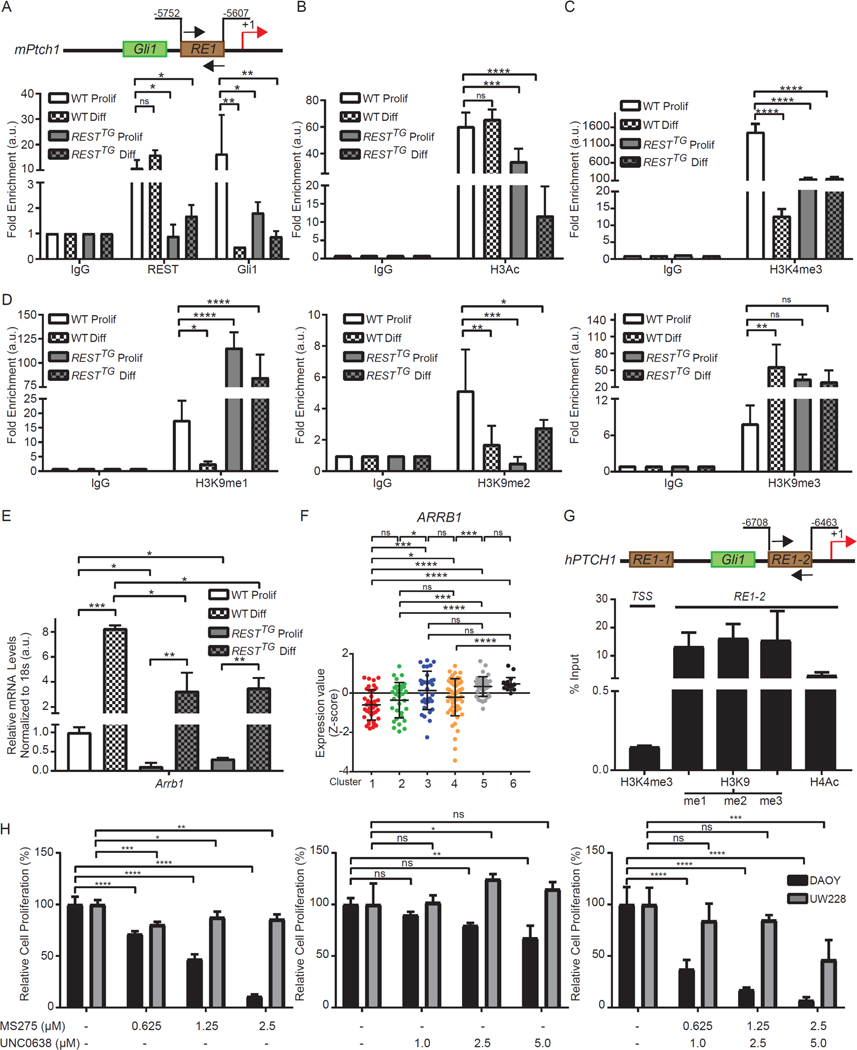
Because cells in human SHH-β tumors appeared to be more differentiated, we examined the strength of SHH signaling, as measured by Gli1 and Ptch1 mRNA expression, during differentiation of WT and RESTTG CGNPs in culture. qRT-PCR analysis was performed using mRNA extracted from WT and RESTTG CGNPs and cultivated under conditions supporting proliferation [medium supplemented with SHH, EGF, and fibroblast growth factor (FGF)] and conditions supporting differentiation [medium lacking SHH, EGF, and FGF, but containing neural growth factor (NGF)]. As expected, a 5-fold and 2-fold decline in Gli1 and Ptch1 mRNA expression, respectively, was observed under differentiation conditions relative to their expression in proliferating cells (Fig. 4E). A similar analysis performed with CGNPs from RESTTG mice showed a small but statistically significant increase in the abundance of Ptch1 and Gli1 mRNA relative to each in proliferating WT cells (Fig. 4E). However, under differentiation conditions, Gli1 and Ptch1 mRNA expression markedly less in CGNPs from RESTTG mice than in WT cells (Fig. 4E). As expected, REST was constitutively expressed in both proliferating and differentiating RESTTG CGNPs and was associated with lack of expression of the REST-target gene, Scg10 (fig. S4, E and F).
Because Ptch1 is a target of GLI1-mediated transcriptional activation, and because a search of the upstream regulatory region of Ptch1gene in mice identified a REST binding RE1 element proximal to GLI1-binding sites in the Ptch1 promoter, we explored whether Ptch1 is a direct target of REST (Fig. 5A). To this end, we examined the binding of Gli1 and REST to the mouse Ptch1 promoter in proliferating CGNPs from WT and RESTTG mice and further compared it to that under differentiation conditions (Fig. 5A). Chromatin immunoprecipitation (ChIP) assays performed using antibodies to GLI 1 and REST and control IgG revealed substantial, and comparably similar, REST binding to the RE1 site upstream of the Ptch1 promoter under proliferation and differentiation conditions in WT CGNPs (Fig. 5A). Although significantly less binding, if any compared with IgG controls, was observed in RESTTG CGNPs, REST occupancy of the RE1 site REST upstream of the Ptch1 promoter was increased 2-fold in differentiating RESTTG cells compared to its proliferating counterpart (Fig. 5A). Curiously, whereas substantial GLI1 binding to the RE1 site was observed in proliferating WT CGNPs, its binding was markedly less (by 8-fold) in proliferating CGNPs from RESTTG mice (Fig. 5A). GLI1 binding to the Ptch1 promoter was decreased in both WT and RESTTG CGNPs under differentiating relative to proliferative conditions (Fig. 5A). Thus, in REST-overexpressing GCNPs, the binding of Gli1 to the Ptch1 promoter was compromised when compared to WT cells, more so under differentiating conditions.
Figure 5: Transcription factor binding and resulting histone modification changes leads to Ptch1 loss of heterozygosity.
(A) Schematic representation of mPtch1 promoter with RE1 site and adjacent Gli1 binding site are shown. REST and Gli1 binding to RE1 site on mPtch1 promoter measured by ChIP-qPCR in WT and RESTTG proliferating and differentiating CGNPs. Data are represented as fold change over IgG (n=3 for WT and n=6 for RESTTG ). (B) Enrichment of H3Ac over IgG at mPtch1 promoter in proliferating and differentiating CGNPs. Bars represent fold change of H3Ac over IgG in the samples (n=3 for WT and n=6 for RESTTG ).(C) Enrichment of trimethylation at Histone H3 lysine 4 (H3K4me3) evaluated by ChIP-qPCR at the mPtch1 TSS site in WT and RESTTG proliferating and differentiating CGNPs (n=3). (D) Enrichment of mono, di and trimethylation at Histone H3 lysine 9 (H3K9me1, 2 and 3) evaluated by ChIP-qPCR at the mPtch1 RE1 site in WT and RESTTG proliferating and differentiating CGNPs (n=3). (E) Arrb1 mRNA expression was measured in WT and RESTTG CGNPs after culturing with proliferation or differentiation media. WT data represents the mean ± S.D. from triplicate samples, RESTTG data represents two individual pups.Graph represents fold change compared to WT proliferating controls. (F) ARRB1 mRNA expression profile measured by microarray. Hierarchical clustering based on expression levels of neuronal differentiation markers divided the SHH MB patient samples into six distinct clusters. Each dot corresponds to one individual patient. (G) Enrichment of H3K4me3 at hPTCH1 TSS and enrichment of other histone modifications at hPTCH1 RE1 site using ChIP-qPCR from a High-REST PDOX sample. (H) DAOY MB cell line treated with either HDAC inhibitor MS275 (0.625 μM to 5 μM) or G9a inhibitor UNC0638 (0.5 μM to 5 μM), or a combination of both, and MTT assay was performed at 48 hours post treatment to measure cell viability. Bars represent mean with S.D. of triplicate. For (A –D), statistical significance was obtained using two-way ANOVA in the Graphpad software and p values calculated using Dunnett’s method for multiple comparisons. For (E), p values were calculated by paired two-tailed t test of ΔCp values: significance is indicated as not significant (ns), p<0.05 (*), p<0.01 (**), p<0.001 (***), or p<0.0001 (****). For (H), statistical significance was calculated using two-way ANOVA in the Graphpad software and p values were measured by multiple comparisons using either Sidak or Tukey’s test (Graphpad 7.0).
GLI1 and REST are canonical transcriptional activator and repressor, respectively. The abundance of the activating histone H3 acetylation mark at the RE1 site was unchanged between proliferating and differentiating WT CGNPs (Fig. 5B). However, a 2-fold decline in its abundance was seen in proliferating CGNPs from RESTTG animals when compared to proliferating control WT CGNPs, with an additional 3-fold decrease in this mark noted in differentiating RESTTG CGNPs (Fig. 5B). Tri-methylation of Lys4 in histone H3 (H3K4-me3) is a mark that is found at transcriptionally active promoters (50). At the transcriptional start site (TSS) in the Ptch1 promoter, a 100-fold decrease in its abundance was noted between proliferating and differentiating WT CGNPs (Fig. 5C). A 10-fold reduction in its levels was seen in proliferating RESTTG CGNPs relative to WT cells, which was maintained under differentiation conditions (Fig. 5C). Thus, increased abundance of REST in RESTTG CGNPs, under both proliferation and differentiation conditions, led to an overall decrease in both Ptch1 promoter activity and histone acetylation around the upstream RE1 site, raising the possibility that aberrant REST expression creates a more closed chromatin architecture.
To address this possibility, we measured the activity of G9a, a histone H3K9 methyl transferase associated with the REST-CoREST complex, which canonically catalyzes the repressive monomethylation (me1) and dimethylation (me2) of histone H3K9. In rare instances, G9a also promotes histone H3K9 trimethylation (me3) (51, 52). To assess whether G9a regulates Ptch1 gene expression, we used ChIP assays to measure histone H3K9-me1, -me2 and -me3 in the cognate promoter in proliferating and differentiating CGNPs from WT and RESTTG mice. H3K9-me1 and -me2 marks decreased 6-fold and 3-fold, respectively, at the Ptch1 promoter upon induction of neurogenesis in WT CGNPs (Fig. 5D). Under these conditions, a 7-fold increase in histone H3K9-me3 was also seen in WT CGNPs (Fig. 5D). In contrast, a 5-fold increase and a 3-fold decrease in histone H3K9-me1 and -me2 abundance, respectively, relative to control WT cells was seen at the Ptch1 promoter in CGNPs from RESTTG animals (Fig. 5D). Onset of differentiation was accompanied by maintenance of H3K9-me1 and -me3 abundance and a 3-fold increase in H3K9-me2 abundance in CGNPs from RESTTG animals (Fig. 5D). These data are suggestive of a more repressive chromatin structure at the Ptch1 promoter in RESTTG CGNPs compared to WT cells, whether proliferating or differentiating. The lack of a change in abundance of the histone H3K9-me1 and -me3 marks in proliferating and differentiating RESTTG CGNPs suggests that increased REST expression leads to an overall premature compaction of the Ptch1 promoter (Fig. 5D).
The above results were unexpected in a few aspects. First, histone H3K9-me3 was seen at the Ptch1 promoter during neuronal differentiation of both WT and RESTTG CGNPs. We therefore performed co-immunoprecipitation assays to examine whether REST formed a complex with Suv39H1, a histone H3K9 trimethyl transferase; however, we did not detect a significant association between REST and Suv39H1 in the DAOY MB cell line, even though both proteins were detected by Western blotting (fig. S5). Second, despite the reduction in GLI1 binding, decreases in histone H3 acetylation and histone H3K4-me3 and an increase in histone H3K9-me1/-me3 in proliferating RESTTG CGNPs relative to WT cells, a corresponding decrease in Ptch1 transcript abundance was not seen (Fig. 4E). To explain this conundrum, we explored whether a negative regulator of GLI1 activity was downregulated in proliferating RESTTG CGNPs. Expression of Arrb1, encoding β-arrestin 1, known to facilitate inhibitory acetylation of GLI1 protein by the histone acetyl transferase (HAT) p300 was significantly (10-fold) decreased in proliferating RESTTG CGNPs compared to WT cells (Fig. 5E) (20, 21, 53). Its expression was increased in both cell types grown in differentiating conditions, but relatively and significantly less so in RESTTG CGNPs (Fig. 5E). In human MB tissue samples, ARRB1 expression was significantly less in SHH-α tumors than in SHH-β tumors (Fig. 5F). Existence of associated histone acetylation and activating and repressive histone methylation events at the RE1 site and in the promoter was also confirmed through ChIP assays in a PDOX tumor sample (Fig. 5G).
Finally, to confirm that histone H3K9 methylation and histone acetylation play a role in the viability of human SHH-MB cells, DAOY and UW228 MB cells were treated with a G9a inhibitor (UNC0638) and a HDAC inhibitor (MS275) either alone or in combination at various doses. Treating cells with UNC0638 alone had only a small effect in one cell line and a counterintuitive pro-survival effect in the other (Fig. 5H), but treating cells with MS275 or the combination had a statistically significant cytotoxic effect (Fig. 5H), including an enhanced and synergistic effect with the combination (table S1). Thus, these findings collectively indicate that Ptch1 is a REST target gene, that REST and GLI1 competitively control Ptch1 gene expression, and that pharmacologically targeting HDACs and G9a activity can synergistically decrease the proliferation of MB cells in culture
REST overexpression induces PTEN loss and AKT activation
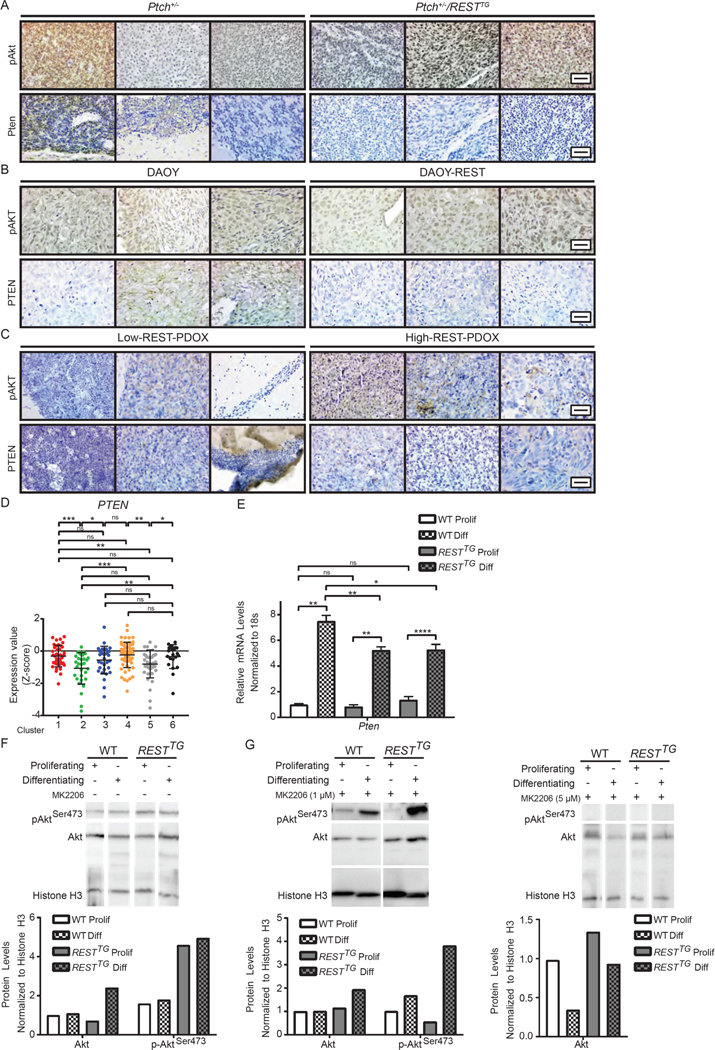
Another hallmark of our Ptch+/−/RESTTG animals was leptomeningeal dissemination and infiltrative behavior of tumor cells. Several studies have demonstrated the relevance of hyperactivation of AKT signaling in MB metastasis and dissemination (54, 55). To examine the status of AKT signaling in tumors from Ptch+/− and Ptch+/−/RESTTG animals, brain sections from these animals were stained with antibodies to Ser473-phosphorylated AKT and analyzed by IHC (n=3, each). Whereas tumors in both Ptch+/− and all Ptch+/−/RESTTG animals exhibited phosphorylation of Akt at Ser473, it was markedly stronger in the Ptch+/−/RESTTG brain sections, indicating a REST elevation-dependent hyperactivation of AKT (Fig. 6A and fig. S6A). Brain slices from mice bearing isogenic DAOY and DAOY-REST xenografts or PDOXs of low and high-REST–expressing, SHH-driven MB tumors also suggested an increase in AKT activity that was dependent on REST abundance (Fig. 6, B and C; and fig. S6, B and C).
Figure 6: REST elevation in the context of constitutive SHH signaling results in AKT hyperactivation.
(A to C) IHC was performed with p-AKTSer473 or PTEN specific antibodies in (A) Ptch+/− and Ptch+/−/RESTTG tumors (n=3) (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH- subgroup patient derived xenografts (n=3). (D) PTEN mRNA expression profile was measured by microarray. Hierarchical clustering based on expression levels of neuronal differentiation markers divided the SHH MB patient samples into six distinct clusters. Each dot corresponds to one individual patient. (E) Pten mRNA expression was measured in WT and\RESTTG CGNPs after culturing with proliferation or differentiation media. Graph represents fold change compared to WT proliferating controls. WT data represents the mean ± S.D. from triplicate samples, RESTTG data represents two individual pups. (F) Western blot analysis was used to measure p-AktSer473 and total Akt protein expression in WT (n=2) and RESTTG (n=2) CGNPs after culturing with proliferation or differentiation media. Representative Western images of p-AktSer473, Akt, and input control histone H3 are shown. (G) p-AktSer473 and total Akt protein expression were measured after 5 hours of treatment of proliferating and differentiating WT and RESTTG CGNPs with MK2206 (1 or 5μM) (n=2). Representative Western images of p-AktSer473, Akt and histone H3 (control) are shown. For (A, B, and C), scale bars = 20 μm (40X). For (E), p values for qRT-PCR were calculated by paired two-tailed t test of ΔCp values: significance is indicated as not significant (ns), p<0.05 (*), p<0.01 (**), p<0.001 (***), or p<0.0001 (****).
Copy number changes leading to loss of PTEN is a clinically relevant characteristic of human SHH-β tumors, and various studies have shown that PTEN loss contributes to MB tumor progression in mice (36, 56–58). Therefore, we investigated whether AKT hyperactivation in tumors from Ptch+/−/RESTTG animals was associated with a decline in PTEN abundance. We did not detect PTEN staining in tumors from Ptch+/−/RESTTG mice, compared with two of three tumors from Ptch+/− animals (Fig. 6A). The correlation between REST and PTEN was also evaluated in DAOY/ DAOY-REST tumors as well as high- and low-REST PDOXs. PTEN abundance was detectable in two of three DAOY and one of three DAOY-REST tumors (Fig. 6B and fig. S6B). In PDOX tumors, PTEN abundance was detectable in all three low-REST and two of three high-REST tumors (Fig. 6C, and fig. S6C). PTEN expression was decreased in cluster 2 (SHH-α) and cluster 5 (SHH-β) tumors, significantly so relative to all others except cluster 3 (Fig. 6D).
Changes in Pten expression was also assessed during proliferation and differentiation of CGNPs from WT and RESTTG mice by qRT-PCR analyses. Its expression was increased 7-fold in differentiating cells relative to that in proliferating WT CGNPs (Fig. 6E). However, a slightly less robust 5-fold upregulation was observed in differentiating RESTTG CGNPs relative to that in proliferating cells from RESTTG mice (Fig. 6E). Similar to Pten, expression of Akt1 transcript was upregulated in differentiated CGNPs from WT and RESTTG mice compared to proliferating counterparts; however, the increase was less robust (3.5-fold) in RESTTG cells than the 5- to 7-fold change in WT cells (fig. S6E), and expression of Akt2 and Akt3 mRNA was increased only in differentiating WT CGNPs (fig. S6E). In human tumors, the expression of AKT1, and AKT3 transcripts was not increased in clusters 1, 2 or 5 (fig. S6D), but that of AKT2 transcript was significantly increased in cluster 2 compared to cluster 5 (fig. S6D).
Subsequently, Western blotting in WT and RESTTG CGNPs showed similar amounts of phosphorylated (activated) AKT in proliferating compared with differentiating WT CGNPs (Fig. 6F). However, the abundance of phosphorylated AKT was increased 2.5-fold in proliferating RESTTG CGNPs relative to WT cells and was similarly high in differentiating RESTTG CGNPs (Fig. 6F). However, total AKT protein abundance was also increased 2.5-fold in differentiating RESTTG CGNPs compared to that in proliferating cells (Fig. 6F). Thus, both PTEN and AKT1 were upregulated at the transcriptional level during neurogenesis in WT and RESTTG CGNPs, albeit to a lesser extent in RESTTG CGNPs, consistent with the increase in total Akt protein levels as well as its phosphorylated form in differentiating RESTTG CGNPs relative to WT cells. AKT phosphorylation was blocked in proliferating and differentiating WT and RESTTG CGNPs by treatment with the AKT inhibitor MK2206 at a dose of 5μM (Fig. 6G, right panel), but not as efficiently at a lower dose of 1μM in differentiating RESTTG relative to WT CGNPs (Fig. 6G, left panel). Unexpectedly, at the same drug concentration, Akt phosphorylation was 2-fold lower in proliferating RESTTG CGNPs compared to WT CGNPs (Fig. 6G, left panel).
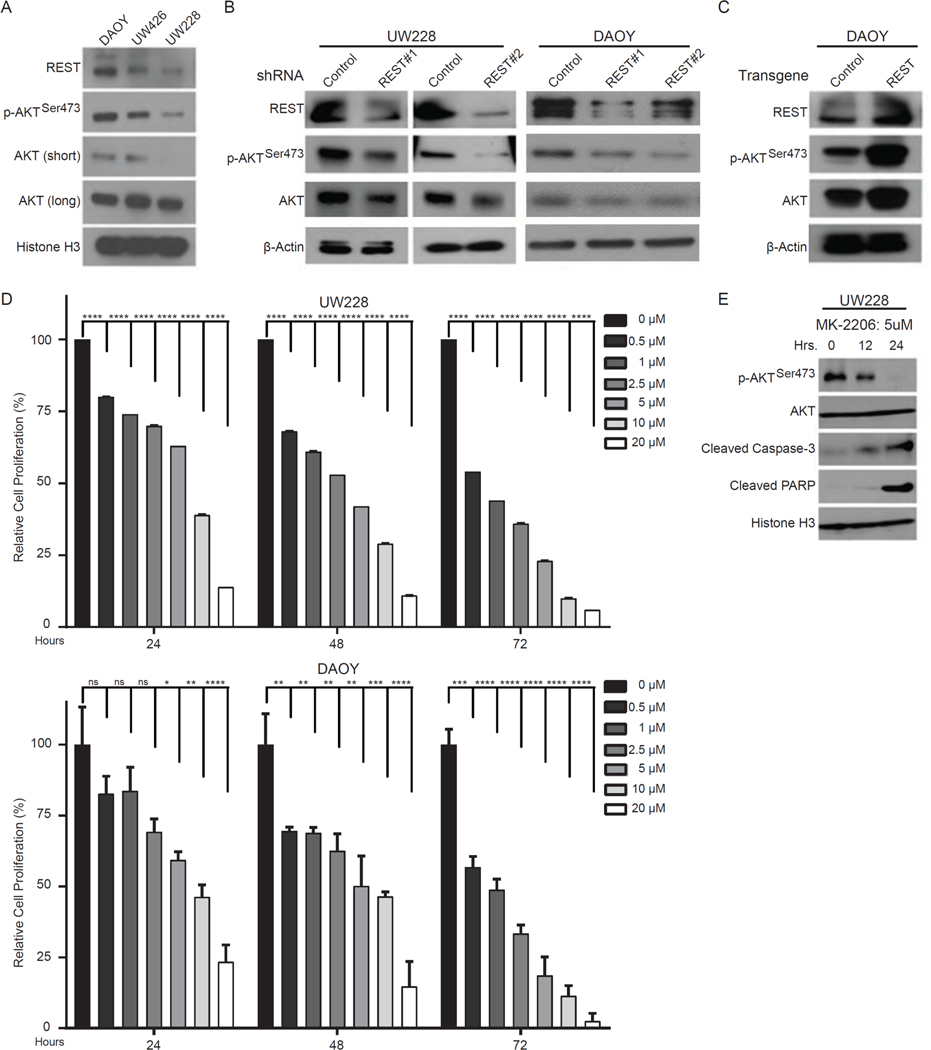
SHH-subgroup human MB cell lines (DAOY, UW426 and UW228) also exhibited a similar association between REST abundance and AKT activation as measured by Western blotting (Fig. 7A). Knockdown and overexpression approaches were then taken to establish a dependency of AKT activation on REST. REST knockdown in UW228 and DAOY cells with each of two REST-specific short hairpin RNAs (shRNAs) decreased the abundance of phosphorylated AKT at Ser473 (Fig. 7B), while total AKT abundance was also slightly, but at least with the second shRNA not proportionally, decreased (Fig. 7B, and fig. S7).
Figure 7: REST-dependent AKT phosphorylation in MB cell lines.
(A) Western blotting for basal protein abundance of REST, p-AKTSer473, total AKT, and histone H3 (control) in DAOY, UW426 and UW228 cells. Representative blots are shown; long/short indicate exposure times. (B and C) Western blotting for total and phosphorylated (p-AKTSer473) protein abundance after either (B) shRNA-mediated REST knockdown in UW228 and DAOY cells or (C) REST overexpression in DAOY cells. Blots are representative images of 3 experiments. (D) MTT assay-derived proliferation of UW228 and DAOY cells treated with various doses of MK2206 for 24, 48 or 72 hours. Data are means ± S.D. of 3 independent assays. (E) Western blotting for abundance of p-AKTSer473, total AKT, cleaved Caspase-3, cleaved PARP and histone H3 (control) to assess induction of apoptosis following treatment of UW228 cells with MK2206 (5 μM) for 12 or 24 hours. Representative blots are shown (n=3).
Conversely, REST overexpression in DAOY cells led to an increase in the abundance of both total and Ser473-phosphorylated AKT (Fig. 7C). Pharmacological inhibition of AKT activity in UW228 and DAOY cells by the AKT inhibitor MK2206 caused a dose and time dependent decrease in the viability of both cells (Fig. 7D). However, DAOY cells, with higher REST expression, were less sensitive to MK2206 compared to UW228 cells as determined by IC50 measurements (Fig. 7D, and table S2). As expected, AKT inhibition with MK2206 promoted caspase-3 activation and PARP cleavage, indicating activation of apoptosis (Fig. 7E). Thus, aberrant REST expression was associated with loss of PTEN expression and increased AKT activation in both mouse and human MBs.
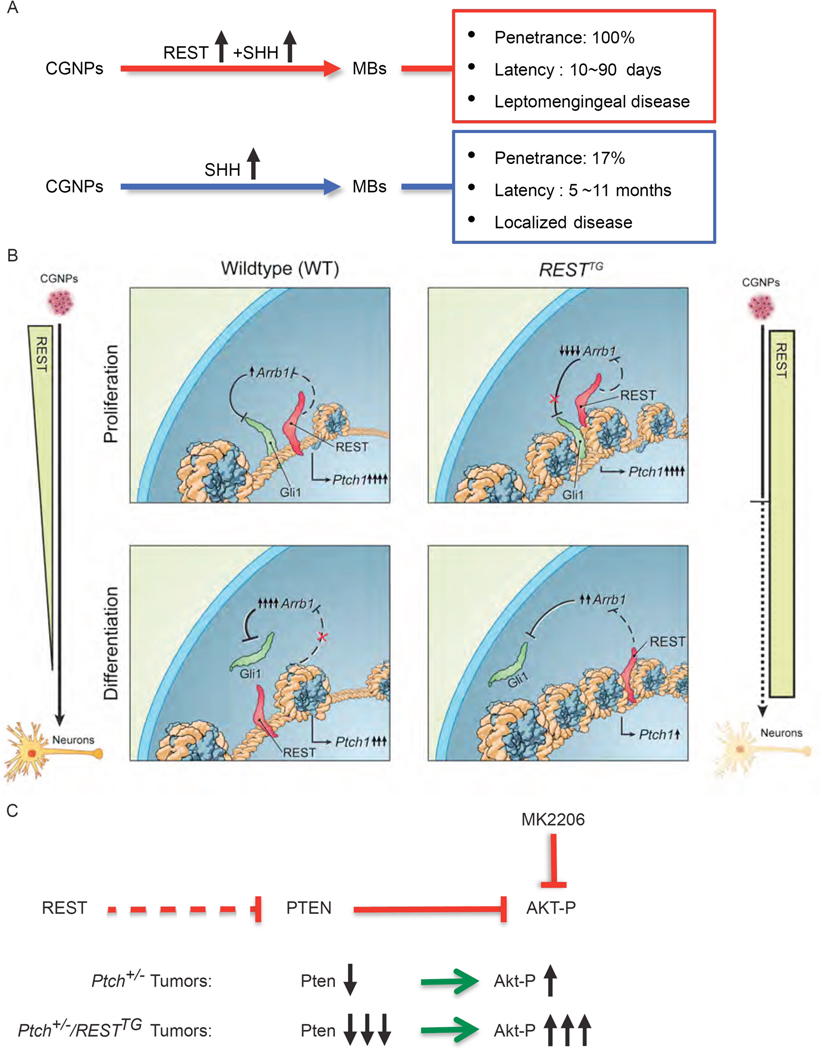
In conclusion, our findings collectively suggest that increased REST expression in SHH-MBs supports a more aggressive disease course (Fig. 8A) by promoting epigenetic repression of PTCH1 and a decrease in PTEN, thereby enhancing SHH and AKT signaling, respectively (Fig. 8, B and C). Hyperactivation of AKT and loss of PTEN have been described in human MBs and are known drive tumor progression in mouse models (56).
Figure 8: Increased REST expression drives progression of SHH-driven MBs.
(A) Schematic representation of tumor characteristics obtained from CGNPs with perturbed SHH signaling in the presence of absence of increased REST expression. (B) Graphical representation of REST-dependent chromatin remodeling of the Ptch1 gene in WT or RESTTG CGNPs during proliferation and differentiation. (C) Model depicting REST regulation of AKT signaling in SHH-driven MBs.
Discussion
Whether mis-regulation of neurogenesis contributes to MB tumorigenesis had not been evaluated until the discovery that ectopic expression of REST in neural stem cells recapitulated the poorly differentiated phenotype of human tumors in mice (23, 28). Increased REST expression in tumors was associated with a decrease in survival of a small subset of patients with desmoplastic MBs, which are SHH-driven tumors (2, 4, 29). Our analysis of human SHH tumor transcriptome data yielded a surprising result in that increased REST expression and activity correlated with two clusters of SHH-tumor samples with poor prognostic significance for patients (29). One was more immature in its lineage commitment and aligned with a SHH-α molecular profile, whereas the other displayed significant expression of neuronal differentiation markers, and aligned with a SHH-β molecular profile. These results suggest that MBs can arise from immature and more lineage committed CGNPs, which is concordant with not only our findings here, but also a previous under-appreciated observation that Smo1 oncogene expression in lineage committed cells promoted transformation (11, 58). Indeed, there is growing acceptance that cells at various stages of lineage commitment can undergo transformation (16, 59–61). Of importance, downregulated expression of BTRC, a proteasome component that degrades REST and GLI1 proteins, in human SHH-α and -β MB clusters, and its association with increased activity of these proteins, underscores the need to consider transcriptomic changes in conjunction with proteomic alterations (62–65).
Since tumor formation was not observed in RESTTG animals, the reason for their decreased survival needs investigation. However, mice with constitutive activation of Shh signaling and REST elevation formed tumors with accelerated kinetics and penetrance, highlighting its importance in driving tumor progression. The finding that REST elevation in CGNPs causes haploinsufficiency and not LOH, is consistent with the current consensus that mono-allelic loss of Ptch1 leads to pre-neoplastic lesions only (15, 16, 66, 67). However, in the context of Ptch1 haploinsufficiency, REST caused Ptch1 LOH and promoted rapid MB formation, which aligns with studies that have shown deletion of both alleles of Ptch1 in CGNPs and neural stem cells to accelerate MB kinetics in mice (16, 68). Finally, increased REST expression was also seen in Ptch+/− tumors, suggesting that deregulation of REST may be a second “hit”. However, onset of tumors in older mice suggests a greater relevance to the adult SHH-δ subgroup. The issue of potential tumor heterogeneity in our model system also remains to be verified.
Another important finding of our work is that a novel and complex interaction between REST and GLI1 controls Shh signaling. In our study, normal neurogenesis of CGNPs was accompanied by a decline in Shh pathway activity, as measured by Ptch1 and Gli1 expression. The first point of convergence between REST and Shh pathway was noted at Ptch1 gene and involved an antagonistic interaction between REST and GLI1 proteins for occupancy and chromatin remodeling at the Ptch1 promoter. The second point of convergence occurred further downstream at GLI1, and involved a REST-dependent silencing of Arrb1 gene, which encodes a negative regulator of GLI1 protein activity (53). In proliferating cells, GLI1 appeared to prevail over REST at the Ptch1 locus, possibly because of a lack of an inhibitory effect of Arrb1 on Gli1 protein. In differentiating cells, upregulation of Arrb1 and decreased Gli1 activity was associated with a decline in Shh signaling. We propose that this regulation is perturbed in tumors with increased REST expression in the context of Ptch1 haploinsufficiency. Specifically, we suggest that in tumors arising from proliferating Ptch+/−/RESTTG CGNPs, premature chromatin compaction at Ptch1 and consequent functional Ptch1 LOH in conjunction with increased GLI1 activity promoted by Arrb1 absence facilitated Shh pathway activity and drove a human SHH-α-like tumor. In contrast, in tumors arising consequent to increased REST expression in more neuronal lineage committed Ptch+/−/RESTTG CGNPs, a further compaction of the Ptch1 locus associated with diminished Gli1 activity associated with upregulation of Arrb1, results in decreased Shh signaling, and is reminiscent of human SHH-β tumors. Clearly, this apparent tumor heterogeneity in our system needs further investigation. This body of work also adds alterations in histone H3K9 methylation as an important epigenetic event in MB etiology, nevertheless, several questions remain. REST-dependent augmentation of G9a activity and a non-canonical increase in H3K9-me3 at the Ptch1 promoter is inexplicable, although enhanced chromatin retention may provide a possible explanation (69, 70). The synergistic decline of human MB cell line proliferation in vitro upon treatment with MS275 and UNC0638 may be due to a direct effect on REST-HDAC activity, or indirectly through modulation of Arrb1 activity, or both, and remains to be explored.
The REST-AKT phosphorylation link also ties chromatin remodeling to leptomeningeal MB dissemination. It is supported by data from a seminal screen that implicated REST, Pten and Akt as separate candidate drivers of metastasis in Shh-driven MB mouse models (71, 72). Increased AKT activity or genetic deletion of Pten in the context of Ptch haploinsufficiency are known drivers of murine MB metastasis (19, 33, 55, 71–75). We propose that REST-dependent loss of Pten expression may also account for increased Akt phosphorylation in MBs. In human SHH-MBs, genetic loss of PTEN is seen in SHH-β tumors (36). However, our data argues that downregulation of PTEN expression can also occur in a subset of SHH-α tumors.
In addition to the above events, other molecules, such as N-Myc, may also be deranged in our mouse model (28, 67). Although the status of N-Myc was not specifically examined here, it is a downstream target of Gli1, and we previously showed that REST knockdown downregulates N-MYC levels in MB cells (22, 76). From a therapeutic perspective, Akt signaling is known to promote MB cell survival (77). Therefore, sensitivity of REST-high SHH-driven MBs to drugs used in the clinic as standard of care, need to be carefully examined. Further pre-clinical investigation of G9a and HDAC inhibitors, either alone or in combination with AKT, SMO and GLI inhibitors, against REST-high MBs are necessary.
In conclusion, growing evidence suggests chromatin remodelers such as REST contribute to SHH-driven MBs (27, 29, 78, 79). Importantly, both immature and lineage committed CGNPs appear to contribute to SHH-driven MB formation. Their molecular dissection will provide a rationale for testing unique combinations of pharmacological agents against human SHH tumor subgroups, especially SHH-α and SHH-β tumors, where prognosis continues to be grim.
Materials and methods
Plasmids
Human REST (hREST) transgene was cloned into a modified pcDNA3.1-V5/His plasmid wherein the cytomegalovirus (CMV) promoter was replaced by a 1-kb region of the NeuroD2 (ND2) promoter from plasmid pcS2SmoA1 (54). A 6x His/3x HA epitope tag was added to the aminoterminus of hREST to generate pcDNA3.1/ND2/REST followed by insertion of a LoxP-1XStop-LoxP site from the Lox-Stop-Lox TOPO plasmid between the ND2 promoter and hREST transgene. (Addgene plasmid # 11584) (29).
Animals
Plasmid pcDNA3.1/ND2/LoxP-REST was restriction digested and the ND2-LoxP-Stop-LoxP-hREST DNA fragment was used for pronuclear injection of embryos from C57/Bl6 mice [National Cancer Institute (NCI, Bethesda, MD)] at the Institutional Genetically Engineered Mouse Facility. Three NeuroD2-REST transgenic founders were backcrossed for several generations and then crossed to mice conditionally expressing Cre recombinase under the Math1 promoter (Math1CreERT2) (Jackson Laboratories) to create RESTTG animals. hREST expression was induced by IP injections of (100 μL of 2mg/mL) tamoxifen (Sigma Aldrich) on postnatal days 2, 3, and 4. Immunocompromised NOD-scid IL2rgammanull (NSG) mice (Jackson Laboratories) were purchased for xenograft studies. The intracranial inoculation of cells into NSG mice was performed using a stereotactic device as described previously (13). All mice were housed and treated in accordance with the guidelines of The University of Texas MD Anderson Cancer Center’s Animal Care and Use Committee. NSG mice were subjected to bioluminescent imaging (BLI) weekly and were sacrificed on day 47 or at the onset of symptoms. For BLI, mice were given IP injections of 150 mg/kg D-Luciferin (Promega, Madison, WI), anesthetized with 2.5% isoflurane and imaged using the Xenogen Spectrum (IVIS-200) system.
Tumor Samples
Histopathological review of medulloblastoma samples and analyses were performed following Institutional Review Board approval to. C. Hawkins from the Hospital for Sick Children, Toronto. Sections of paraffin-embedded tissue were studied by H&E and IHC staining performed by V. Rajaram (UT Southwestern Medical Center, Dallas, TX) and C. Hawkins. All antibodies are listed in table S3.
Statistical Analysis
Statistical significance of overall survival between various mouse groups was calculated by Logrank (Mantel-Cox) test. For all qRT-PCR data, paired two-sided t tests were performed using GraphPad Prism version 7.0 for Windows (GraphPad Software Inc., San Diego, CA, USA). Data are shown as means ± SD of at least 3 independent samples. P values < 0.05 were considered to be statistically significant. Significance is indicated as, p<0.05 (*), p<0.01 (**), p<0.001 (***), or p<0.0001 (****); where necessary for clarity, lack of significance is indicated (ns). Hierarchical clustering was performed by ArrayTrack Software available at http://edkb.fda.gov/webstart/arraytrack/ using Ward’s method Samples were divided into 3–6 clusters based on the dendrogram from hierarchical clustering to keep at least 15 patients in each cluster for further analyses. p values for comparisons between every pairwise combination among clusters based on gene expression status were obtained using the unpaired t test with Welch’s correction using GraphPad Prism version 7.0.
Gene expression profile in patient samples
Microarray datasets containing the gene expression values of medulloblastoma patients were obtained from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo). We used GSE85217 dataset, which contained Affimetrix Human Gene 1.1 ST Array profiling of 763 primary MB samples to evaluate gene expression. Microarray data were normalized using the Robust Multi-array Average (RMA) method. The expression data for each gene were Z-score transformed.
Immunohistochemistry (IHC) and microscopy
Mouse brain tissues were fixed in 10% buffered formalin phosphate and embedded in paraffin. 4- μm-thick brain sections were used for IHC analysis. Sections were deparaffinized with 100% histoclear solution or xylene followed by rehydration with ethanol and water. Sections were treated with 3% H2O2 solution for 10 min to block the endogenous peroxidase. Antigen retrieval was performed using a pressure cooker (Aroma). Slides were placed in Citrate buffer (pH 6.0) and incubated for 30–60 min under steam conditions. The samples were then cooled and washed in Tris-Buffered Saline–Tween (TBST). Blocking was performed by incubating the sections in blocking buffer (1% BSA + 5% normal goat serum in 1 × TBST) for 1 hr. The sections were then incubated with primary antibodies as indicated at 4°C overnight. The primary antibody was detected using a secondary antibody conjugated to horse-radish peroxidase (HRP; Jackson Laboratories) by incubating sections for 1 hour at room temperature. All incubations were performed in humidified conditions. Finally, slides were washed with TBST and developed using 3,3′-diaminobenzidine (DAB) (Vertex) as a substrate and counterstained with Hematoxylin. After dehydration and mounting, slides were dried and visualized under microscope. All antibodies used are listed in table S3. Stained slides were viewed using a Nikon ECLIPSE E200 microscope and images were captured under 4X, 10X, and 40X magnification with an Olympus SC100 camera. Analyses were performed using Olympus CellSens Entry software. Whole mouse brains were viewed under an Olympus SZ61 microscope. Image capture and analyses were performed as described above.
Cell Culture
Human MB cell lines DAOY, UW228, and UW426 were cultured as described previously (29). Cerebellar tissues from postnatal day-8 WT and RESTTG pups were dissected and triturated using 18.5-gauge needle with 10 ml syringe to form single cell suspension. The cells were grown for 10 days in Neurobasal medium supplemented with B27 (vitamin A), Glutamax, antibiotic/antimycotic, heparin, and 20 μg/ml of EGF and FGF, triturating every 4 days to disrupt cell attachment to the plate. Neurospheres were grown for an additional 4 days in the presence of recombinant sonic hedgehog (rhSHH) at 0.1 μg/ml concentration. The proliferating cells were collected and used for further analysis. To differentiate the cells, the proliferating neurospheres were put back in culture containing 20 μg/ml of NGF without EGF and FGF. The cells were cultured in a 6-well plate for 5 days and the attached differentiated cells were collected after dissociation with TrypLE and used for further experiments. Unattached cells were discarded.
Plasmid and Cell Transfection
The pcDNA3.1/ND2/REST plasmid was stably transfected into DAOY and UW426 cells using Lipofectamine 2000 reagent in accordance with the manufacturer’s instructions (Invitrogen).
MTT Assay
DAOY, UW228, DAOY-HR vs DAOY-LR and UW426-HR vs UW426-LR cells were cultured in 96-well microplates and treated with various concentration of the AKT inhibitor MK2206 (US Biological Life Science), HDAC inhibitor MS275 (Cayman Chemicals, catalog # 13284) and G9a inhibitor UNC0638 (Cayman Chemicals, catalog # 10734), either alone or in combination for the time period indicated in the figures. After labeling with 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT/thiazolyl blue tetrazolium bromide; Sigma), cell growth was analyzed by SpectraMax Plus 384 microplate reader (Molecular Devices, LLC). Synergy between MS275 and UNC0638 was calculated using the fractional product method of Webb using the formula g1g2 = g12 following the procedure described previously (80).
Chromatin immunoprecipitation (ChIP) and qPCR from proliferating and differentiating CGNPs
CGNPs from proliferating and differentiating cells were fixed with 1% formaldehyde, cross-linked, and processed for ChIP analyses according to the manual from Millipore. Briefly, cross-linked cells were washed with 1X PBS and lysed using lysis buffer [50 mmol/L Tris-HCl (pH 8.0), 10 mmol/L EDTA (pH 8.0), 1% SDS, and protease inhibitors] and sonicated, and 1% of this material was saved as input DNA. The remainder of the samples was diluted 5-fold with ChIP dilution buffer (16.7 mmol/L Tris-HCl (pH 8.0), 167 mmol/L NaCl, 1.2 mmol/L EDTA (pH 8.0), 1.1% Triton X-100, and protease inhibitors), precleared, and incubated with various antibodies for 12 hours at 4°C. The complex was then incubated with protein-A beads (Millipore), washed, and eluted. Following reversal of the cross-link, DNA was purified with a PCR Purification Kit (Zymo Research). Bound DNA was quantified by SYBRGreen qPCR using Roche Lightcycler 96. Data were analyzed using the comparative 2−ΔΔCp method. The antibodies used for ChIP and primers used for the mouse Ptch1 are listed in tables S3 and S4, respectively.
ChIP and qPCR from PDOX samples
ChIP assays were performed by the Center for Cancer Epigenetics Core facility with following modifications to previously described high-throughput ChIP protocol (81). Briefly, 100 mg PDX tissue was dissociated in Hank’s balanced salt solution (HBSS) using MACS dissociator to obtain single cell suspension. The cell suspension was crosslinked in 1% formaldehyde for 10 min at room temperature, followed by incubation with glycine for 5 min to stop crosslinking. Cells were collected and washed with ice cold PBS and crude nuclei was isolated using cell lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5 % NP-40 supplemented with protease inhibitor) for 10 min followed by centrifugation for 5min. The nuclear pellet was lysed for 30 min on ice using lysis buffer (12 mM Tris-HCl pH 7.5, 6 mM EDTA pH 8.0, 0.5 % SDS) supplemented with protease inhibitor. Chromatin lysates were fragmented with a Bioruptor (Diagenode) to obtain DNA fragments ranging 200–600 bp. After centrifugation, the supernatant was collected and incubated with respective antibodies conjugated with Dynabeads Protein G (Invitrogen) overnight at 4°C. The immunocomplexes were collected using Dynamag, washed as described in the protocol, treated with RNase and Proteinase K, and reverse crosslinked overnight followed by DNA extraction. The DNA region of interest was detected by SYBR green real-time qPCR using primers to the human PTCH1 promoter (table S4).
Co-Immunoprecipitation (Co-IP)
Whole cell lysates (WCL) were prepared from DAOY cells. Briefly, DAOY cells were collected and washed in Phosphate Buffered Saline (PBS) (Corning Cellgro). Cells were crosslinked in 1% formaldehyde, rocked at room temperature for 10 min, and washed 2Xs in PBS. Cells were collected by centrifugation and incubated in lysis buffer (50 mM Tris HCl, pH 8.0, 50 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease/phosphatase inhibitors) for 5 min on ice with periodic mixing. The lysates were clarified by centrifugation at 13,000 g for 10 min at 4°C, and supernatants were aliquoted. WCLs were precleared at 4°C for 30 minutes with gentle rocking. Beads were collected by centrifugation at 1000 rpm for 1 min at 4°C. Supernatant was transferred to new tube and diluted 1:4 with lysis buffer. 2.5% Input was collected and stored at −80°C. Primary antibody or control IgG was added to tube and incubated for 12 hrs at 4°C with gentle rocking. The complex was then incubated with protein-A beads (Millipore) for 3 hrs at 4°C, washed 4Xs (20 mM Tris (pH 7.4), 50–300 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, 5 mM EDTA, 25% glycerol, and protease/phosphatase inhibitors) and eluted with lysis buffer plus SDS loading dye. Proteins were analyzed by Western blotting as previously described. Protein bands were developed using SuperSignalTM West Dura Extended Duration Substrate and detected using ChemiDocTM Touch Imaging System (Bio-Rad).
Quantitative Reverse Transcription Polymerase Chain Reaction
CGNPs from WT and RESTTG mice were harvested, cultured, and collected as described above. RNA was extracted using the Quick-RNA MiniPrep Kit (Zymo Research). Equal amounts of RNA up to 1 μg were reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR was performed in triplicate with a 2X SensiMix SYBR & Fluorescein Kit (Bioline) using a LightCycler 96 Real-Time PCR System (Roche Diagnostics GmbH). Relative mRNA expression normalized to 18S ribosomal RNA was determined by the comparative 2-ΔΔCp method. Relative mRNA expression was graphed as fold change compared to WT controls. Primers sequences are listed in table S4.
Lentiviral Infection
For short hairpin RNA: HEK293T cells were co-transfected with control or REST shRNA together with packaging plasmid (PAX2) and envelope plasmid (MD2). Lentiviral particles were harvested 48 hours after transfection. DAOY and UW228 cells were transduced with the collected viral supernatant in the presence of Polybrene (8 μg/ml) and incubated for 48 hours. Infected cells were cultured in medium containing 2 μg/ml puromycin for selection up to 1 week.
Western Blot Analysis
DAOY, UW228, and UW426 cells were collected and cell extracts were prepared for Western blot analysis. Briefly, cell extracts were prepared by incubation in lysis buffer [50 mM Tris HCl, pH 8.0, 50 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, plus protease and phosphatase inhibitors] for 30 min on ice. The lysates were clarified by centrifugation at 13,000 g for 10 min at 4°C, and the supernatants were collected and boiled in sodium dodecyl sulfate (SDS) loading buffer (Cell Signaling Technology). Proteins were separated by electrophoresis on 10% SDS-polyacrylamide gels (Bio-Rad Laboratories), transferred to Hybond-P PVDF membranes (GE Healthcare), and analyzed by Western blotting with the indicated primary antibodies (table S3) and HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Thermo Scientific). Protein bands were developed using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and detected using a Kodak Medical X-Ray Processor 104 (Eastman Kodak Company) and ChemiDoc™ Touch Imaging System (Bio-Rad). Images were analyzed using Image Lab Software version 5.2.1 (Bio-Rad).
Supplementary Material
fig S1: Clinical characteristics. (A-C) Clinical characteristics based on clustering analysis of WNT MB patient samples. (D-F) Clinical characteristics based on clustering analysis of Group 3 MB patient samples. (G-I) Clinical characteristics based on clustering analysis of Group 4 MB patient samples. (J) Arm-level chromosomal copy number change in SHH patients. Red and blue colored bars correspond to the gain and loss in the given chromosomal region, respectively. (K) Gene expression profiles measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. Bars represent mean with standard deviations. *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant. (L) Listed genes used for clustering analysis in the order of top-down pictured in Figure 1.
fig S2: Cerebellar architecture and protein expression in age-matched RESTTG and WT mice. (A) WT or RESTTG p8 brain sections stained with H&E. Sections were analyzed by IHC for (B) REST and (C) NeuN protein expression in CGNPs using specific antibodies. Scale bars = 200 μm (top panel −4x) and 20 μm (bottom panel - 40X).
fig S3: REST elevation contributes to infiltrative Shh-driven MB development. Comparison of H&E stained brain sections from (A) Ptch+/− and Ptch+/−/RESTTG animals with localized and infiltrative tumors, respectively (n=3), (B) DAOY and DAOY-REST xenografts (n=3), and (C) Low REST and High REST PDOXs (n=3). Scale bars = 200 μm (top panel −4x) and 20 μm (bottom panel - 40X).
fig S4: Differential regulation of PTCH1 expression by REST in MB tumors and CGNPs. Cerebellar sections of (A) tumor-bearing Ptch+/− and Ptch+/−/RESTTG mice (n=3) and (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH- subgroup PDOXs (n=3) were analyzed by IHC for REST, PTCH1, and KI-67 expression using specific antibodies. (D) GLI2, GLI3, and SMO mRNA expression profile measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. (E) hREST and (F) Scg10 mRNA expression was measured in WT and RESTTG GNPs after culturing with proliferation or differentiation media. WT data represents the mean ± SD (of fold change) of triplicate samples (n=3), RESTTG data represents 2 individual pups (n=2). For (A, B, and C) scale bars = 200 μm (top panel - 4X) and 50 μm (bottom panel −10X). For (E and F), p values were calculated by one-tailed t test of dCT values: *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant.
fig S5: REST and SUV39H1 do not co-immunoprecipitate in DAOY cells. Co-IP was performed to determine interaction between REST and SUV39H1. Whole cell extracts from DAOY were immunoprecipitated with IgG (control), REST, or no antibody, run on an SDS-PAGE and probed for REST and SUV39H1 protein. Blot shown is representative of 3 experiments.
SF6: REST elevation increases AKTSer473 phosphorylation. IHC was performed with specific antibodies targeting p-AKTS473, AKT, or PTEN protein in (A) Ptch+/− and Ptch+/−/RESTTG tumors (n=3), (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH- subgroup PDOXs (n=3). (D) AKT1, AKT2, and AKT3 mRNA expression profile measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. (E) Akt1, Akt2, and Akt3 mRNA expression was measured in WT and RESTTG GNPs after culturing with proliferation or differentiation media. WT data represents the mean ± SD (of fold change) of triplicate samples (n=3), RESTTG data represents 2 individual pups (n=2). For (A, B, and C) scale bars = 200 μm (top panel - 4X) and 50 μm (bottom panel −10X). For (E), p values were calculated by one-tailed t test of dCT values: *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant.
SF7: REST-dependent AKT phosphorylation in cell-lines. (A) Western blot analysis of p- AKTSer473 and AKT protein expression following REST knockdown in DAOY cells (Fig. 6 B, right). Densitometry was obtained using Image Lab software (Bio-Rad). Bars represent mean with standard deviations (n=3).
Table S1: Synergy between MS275 and UNC0638. Synergy was calculated DAOY and UW228 at 48H post treatment either with each drug alone or in combination.
Table S2: IC50 values for UW228 and DAOY cells treated with MK-2206. Analysis of MTT assays was used to determine IC50 values after UW228 and DAOY cells were treated with MK-2206 at various concentrations as indicated for 24, 48 or 72 hours.
Table S3: Antibodies for IHC, qChIP, and Western blotting assays.
Table S4: Primers for qChIP and qRT-PCR assays.
Acknowledgements:
We thank M.C. Hung for helpful comments, X. Shi for providing reagents, and R. DuBois for his support.
Funding: This work was supported by grants from the National Institutes of Health (NIH, 5R01-NS-079715-01 and 5R03NS077021-01 to V.G., and R01 CA185402 to X.-N.L.), American Cancer Society (RSG-09-273-01-DDC to V.G.), Cancer Prevention Research Institute of Texas (CPRIT-RP150301 to V.G.), Addis Faith Foundation and Rally Foundation for Childhood Cancers (to VG.), and The University of Texas MD Anderson Cancer Center-CCE Scholar Program (to T.D.).
Footnotes
Competing interests: S.K. is a member of the Data Safety Monitoring Board of the Nationwide Children’s Hospital. All other authors declare that they have no competing interests.
Data and materials availability: The microarray data underlying our analyses is previously published and publicly available from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; dataset GSE85217). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Coluccia D, Figuereido C, Isik S, Smith C, Rutka JT, Medulloblastoma: Tumor Biology and Relevance to Treatment and Prognosis Paradigm. Current neurology and neuroscience reports 16, 43 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM, Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123, 473–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rusert JM, Wu X, Eberhart CG, Taylor MD, Wechsler-Reya RJ, SnapShot: Medulloblastoma. Cancer cell 26, 940–940 e941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM, Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123, 465–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koestner A, Swenberg JA, Wechsler W, Transplacental production with ethylnitrosourea of neoplasms of the nervous system in Sprague-Dawley rats. Am J Pathol 63, 37–56 (1971). [PMC free article] [PubMed] [Google Scholar]
- 6.Svalina MN, Kikuchi K, Abraham J, Lal S, Davare MA, Settelmeyer TP, Young MC, Peckham JL, Cho YJ, Michalek JE, Hernandez BS, Berlow NE, Jackson M, Guillaume DJ, Selden NR, Bigner DD, Nazemi KJ, Green SC, Corless CL, Gultekin S, Mansoor A, Rubin BP, Woltjer R, Keller C, IGF1R as a Key Target in High Risk, Metastatic Medulloblastoma. Sci Rep 6, 27012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, Remke M, Cavalli FM, Ramaswamy V, Lindsay PE, Jelveh S, Donovan LK, Wang X, Luu B, Zayne K, Li Y, Mayoh C, Thiessen N, Mercier E, Mungall KL, Ma Y, Tse K, Zeng T, Shumansky K, Roth AJ, Shah S, Farooq H, Kijima N, Holgado BL, Lee JJ, Matan-Lithwick S, Liu J, Mack SC, Manno A, Michealraj KA, Nor C, Peacock J, Qin L, Reimand J, Rolider A, Thompson YY, Wu X, Pugh T, Ally A, Bilenky M, Butterfield YS, Carlsen R, Cheng Y, Chuah E, Corbett RD, Dhalla N, He A, Lee D, Li HI, Long W, Mayo M, Plettner P, Qian JQ, Schein JE, Tam A, Wong T, Birol I, Zhao Y, Faria CC, Pimentel J, Nunes S, Shalaby T, Grotzer M, Pollack IF, Hamilton RL, Li XN, Bendel AE, Fults DW, Walter AW, Kumabe T, Tominaga T, Collins VP, Cho YJ, Hoffman C, Lyden D, Wisoff JH, Garvin JH Jr., Stearns DS, Massimi L, Schuller U, Sterba J, Zitterbart K, Puget S, Ayrault O, Dunn SE, Tirapelli DP, Carlotti CG, Wheeler H, Hallahan AR, Ingram W, MacDonald TJ, Olson JJ, Van Meir EG, Lee JY, Wang KC, Kim SK, Cho BK, Pietsch T, Fleischhack G, Tippelt S, Ra YS, Bailey S, Lindsey JC, Clifford SC, Eberhart CG, Cooper MK, Packer RJ, Massimino M, Garre ML, Bartels U, Tabori U, Hawkins CE, Dirks P, Bouffet E, Rutka JT, Wechsler-Reya RJ, Weiss WA, Collier LS, Dupuy AJ, Korshunov A, Jones DT, Kool M, Northcott PA, Pfister SM, Largaespada DA, Mungall AJ, Moore RA, Jabado N, Bader GD, Jones SJ, Malkin D, Marra MA, Taylor MD, Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529, 351–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodrich LV, Milenkovic L, Higgins KM, Scott MP, Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM, The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer research 64, 7794–7800 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hatten ME, Roussel MF, Development and cancer of the cerebellum. Trends Neurosci 34, 134–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, Hansen S, Knoblaugh SE, Lee D, Eberhart CG, Hallahan AR, Olson JM, The Smo/Smo model: hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer research 68, 1768–1776 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Wechsler-Reya RJ, Scott MP, Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22, 103–114 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Choudhry Z, Rikani AA, Choudhry AM, Tariq S, Zakaria F, Asghar MW, Sarfraz MK, Haider K, Shafiq AA, Mobassarah NJ, Sonic hedgehog signalling pathway: a complex network. Ann Neurosci 21, 28–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenney AM, Cole MD, Rowitch DH, Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130, 15–28 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, Lin SM, Wechsler-Reya RJ, Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ, Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer cell 14, 135–145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R, Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell reports 6, 168–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Y, Bai CB, Joyner AL, Wang B, Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Molecular and cellular biology 26, 3365–3377 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP Jr., Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A 103, 4505–4510 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miele E, Po A, Begalli F, Antonucci L, Mastronuzzi A, Marras CE, Carai A, Cucchi D, Abballe L, Besharat ZM, Catanzaro G, Infante P, Di Marcotullio L, Canettieri G, De Smaele E, Screpanti I, Locatelli F, Ferretti E, beta-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer 17, 488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parathath SR, Mainwaring LA, Fernandez LA, Guldal CG, Nahle Z, Kenney AM, beta-Arrestin-1 links mitogenic sonic hedgehog signaling to the cell cycle exit machinery in neural precursors. Cell Cycle 9, 4013–4024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das CM, Taylor P, Gireud M, Singh A, Lee D, Fuller G, Ji L, Fangusaro J, Rajaram V, Goldman S, Eberhart C, Gopalakrishnan V, The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene 32, 1691–1701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, Majumder S, Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Molecular cancer therapeutics 4, 343–349 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Gopalakrishnan V, REST and the RESTless: in stem cells and beyond. Future neurology 4, 317–329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, Rastelli L, Marin Dias Carneiro A, Levin V, Fuller GN, Echelard Y, Majumder S, The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nature medicine 6, 826–831 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Negrini S, Prada I, D’Alessandro R, Meldolesi J, REST: an oncogene or a tumor suppressor? Trends in cell biology 23, 289–295 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Shi X, Wang Q, Gu J, Xuan Z, Wu JI, SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development. Oncogene, (2016). [DOI] [PubMed] [Google Scholar]
- 28.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S, Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Molecular and cellular biology 26, 1666–1678 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor P, Fangusaro J, Rajaram V, Goldman S, Helenowski IB, MacDonald T, Hasselblatt M, Riedemann L, Laureano A, Cooper L, Gopalakrishnan V, REST is a novel prognostic factor and therapeutic target for medulloblastoma. Molecular cancer therapeutics 11, 1713–1723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobson THW, Hatcher RJ, Swaminathan J, Das CM, Shaik S, Tao RH, Milite C, Castellano S, Taylor PH, Sbardella G, Gopalakrishnan V, Regulation of USP37 Expression by REST-Associated G9a-Dependent Histone Methylation. Mol Cancer Res 15, 1073–1084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G, REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G, REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Riobo NA, Haines GM, Emerson CP Jr., Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer research 66, 839–845 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Schoenherr CJ, Anderson DJ, Silencing is golden: negative regulation in the control of neuronal gene transcription. Current opinion in neurobiology 5, 566–571 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Ooi L, Wood IC, Chromatin crosstalk in development and disease: lessons from REST. Nature reviews. Genetics 8, 544–554 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, Garzia L, Torchia J, Nor C, Morrissy AS, Agnihotri S, Thompson YY, Kuzan-Fischer CM, Farooq H, Isaev K, Daniels C, Cho BK, Kim SK, Wang KC, Lee JY, Grajkowska WA, Perek-Polnik M, Vasiljevic A, Faure-Conter C, Jouvet A, Giannini C, Nageswara Rao AA, Li KKW, Ng HK, Eberhart CG, Pollack IF, Hamilton RL, Gillespie GY, Olson JM, Leary S, Weiss WA, Lach B, Chambless LB, Thompson RC, Cooper MK, Vibhakar R, Hauser P, van Veelen MC, Kros JM, French PJ, Ra YS, Kumabe T, Lopez-Aguilar E, Zitterbart K, Sterba J, Finocchiaro G, Massimino M, Van Meir EG, Osuka S, Shofuda T, Klekner A, Zollo M, Leonard JR, Rubin JB, Jabado N, Albrecht S, Mora J, Van Meter TE, Jung S, Moore AS, Hallahan AR, Chan JA, Tirapelli DPC, Carlotti CG, Fouladi M, Pimentel J, Faria CC, Saad AG, Massimi L, Liau LM, Wheeler H, Nakamura H, Elbabaa SK, Perezpena-Diazconti M, Chico F de Leon Ponce, Robinson S, Zapotocky M, Lassaletta A, Huang A, Hawkins CE, Tabori U, Bouffet E, Bartels U, Dirks PB, Rutka JT, Bader GD, Reimand J, Goldenberg A, Ramaswamy V, Taylor MD, Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer cell 31, 737–754 e736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J, The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci 31, 9772–9786 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HJ, Denli AM, Wright R, Baul TD, Clemenson GD, Morcos AS, Zhao C, Schafer ST, Gage FH, Kagalwala MN, REST Regulates Non-Cell-Autonomous Neuronal Differentiation and Maturation of Neural Progenitor Cells via Secretogranin II. J Neurosci 35, 14872–14884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S, REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faronato M, Patel V, Darling S, Dearden L, Clague MJ, Urbe S, Coulson JM, The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell Cycle 12, 1964–1977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ, Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A 101, 10458–10463 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruce AW, Lopez-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, Koch CM, Langford CF, Dunham I, Andrews RM, Vetrie D, Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res 19, 994–1005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, Karuturi RK, Wei CL, Buckley NJ, Stanton LW, REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol 6, e256 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saritas-Yildirim B, Childers CP, Elsik CG, Silva EM, Identification of REST targets in the Xenopus tropicalis genome. BMC Genomics 16, 380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun YM, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, Bee T, Wilde S, Buckley NJ, Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol Biol Cell 16, 5630–5638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel G, Ekici M, Rossler OG, RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res 359, 99–109 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Wagoner MP, Gunsalus KT, Schoenike B, Richardson AL, Friedl A, Roopra A, The transcription factor REST is lost in aggressive breast cancer. PLoS Genet 6, e1000979 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang D, Luo G, Ding X, Lu C, Preclinical experimental models of drug metabolism and disposition in drug discovery and development. Acta Pharmaceutica Sinica B 2, 549–561 (2012). [Google Scholar]
- 49.Wetmore C, Eberhart DE, Curran T, Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer research 61, 513–516 (2001). [PubMed] [Google Scholar]
- 50.Gu B, Lee MG, Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci 3, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S, A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37, 46–56 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T, Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25, 473–481 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, Pelloni M, De Simone G, Pedone EM, Gallinari P, Giorgi A, Steinkuhler C, Vitagliano L, Pedone C, Schinin ME, Screpanti I, Gulino A, Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol 12, 132–142 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Dimitrova V, Arcaro A, Targeting the PI3K/AKT/mTOR signaling pathway in medulloblastoma. Curr Mol Med 15, 82–93 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Hartmann W, Digon-Sontgerath B, Koch A, Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sorensen N, Wiestler OD, Pietsch T, Phosphatidylinositol 3’-kinase/AKT signaling is activated in medulloblastoma cell proliferation and is associated with reduced expression of PTEN. Clin Cancer Res 12, 3019–3027 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Castellino RC, Barwick BG, Schniederjan M, Buss MC, Becher O, Hambardzumyan D, Macdonald TJ, Brat DJ, Durden DL, Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PLoS One 5, e10849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Genovesi LA, Ng CG, Davis MJ, Remke M, Taylor MD, Adams DJ, Rust AG, Ward JM, Ban KH, Jenkins NA, Copeland NG, Wainwright BJ, Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups. Proc Natl Acad Sci U S A 110, E4325–4334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metcalfe C, Alicke B, Crow A, Lamoureux M, Dijkgraaf GJ, Peale F, Gould SE, de Sauvage FJ, PTEN loss mitigates the response of medulloblastoma to Hedgehog pathway inhibition. Cancer research 73, 7034–7042 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Brown G, Sanchez-Garcia I, Is lineage decision-making restricted during tumoral reprograming of haematopoietic stem cells? Oncotarget 6, 43326–43341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai AY, Kondo M, Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med 203, 1867–1873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL, Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer cell 14, 123–134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ, SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang R, Huang SY, Ka-Wai Li K, Li YH, Hsu WH, Zhang GJ, Chang CJ, Yang JY, Dual degradation signals destruct GLI1: AMPK inhibits GLI1 through beta-TrCP-mediated proteasome degradation. Oncotarget 8, 49869–49881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forget A, Martignetti L, Puget S, Calzone L, Brabetz S, Picard D, Montagud A, Liva S, Sta A, Dingli F, Arras G, Rivera J, Loew D, Besnard A, Lacombe J, Pages M, Varlet P, Dufour C, Yu H, Mercier AL, Indersie E, Chivet A, Leboucher S, Sieber L, Beccaria K, Gombert M, Meyer FD, Qin N, Bartl J, Chavez L, Okonechnikov K, Sharma T, Thatikonda V, Bourdeaut F, Pouponnot C, Ramaswamy V, Korshunov A, Borkhardt A, Reifenberger G, Poullet P, Taylor MD, Kool M, Pfister SM, Kawauchi D, Barillot E, Remke M, Ayrault O, Aberrant ERBB4-SRC Signaling as a Hallmark of Group 4 Medulloblastoma Revealed by Integrative Phosphoproteomic Profiling. Cancer cell 34, 379–395 e377 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Purzner T, Purzner J, Buckstaff T, Cozza G, Gholamin S, Rusert JM, Hartl TA, Sanders J, Conley N, Ge X, Langan M, Ramaswamy V, Ellis L, Litzenburger U, Bolin S, Theruvath J, Nitta R, Qi L, Li XN, Li G, Taylor MD, Wechsler-Reya RJ, Pinna LA, Cho YJ, Fuller MT, Elias JE, Scott MP, Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci Signal 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pazzaglia S, Tanori M, Mancuso M, Gessi M, Pasquali E, Leonardi S, Oliva MA, Rebessi S, Di Majo V, Covelli V, Giangaspero F, Saran A, Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene 25, 5575–5580 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Tamayo-Orrego L, Wu CL, Bouchard N, Khedher A, Swikert SM, Remke M, Skowron P, Taylor MD, Charron F, Evasion of Cell Senescence Leads to Medulloblastoma Progression. Cell reports 14, 2925–2937 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Wang J, Wechsler-Reya RJ, The role of stem cells and progenitors in the genesis of medulloblastoma. Experimental neurology 260, 69–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Esteve PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S, Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res 33, 3211–3223 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon JM, Parker JS, Liu F, Rothbart SB, Ait-Si-Ali S, Strahl BD, Jin J, Davis IJ, Mosley AL, Pattenden SG, A Role for Widely Interspaced Zinc Finger (WIZ) in Retention of the G9a Methyltransferase on Chromatin. J Biol Chem 290, 26088–26102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mumert M, Dubuc A, Wu X, Northcott PA, Chin SS, Pedone CA, Taylor MD, Fults DW, Functional genomics identifies drivers of medulloblastoma dissemination. Cancer research 72, 4944–4953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, Garzia L, Van Meter T, Zagzag D, Jabado N, Schwartzentruber J, Majewski J, Scheetz TE, Pfister SM, Korshunov A, Li XN, Scherer SW, Cho YJ, Akagi K, MacDonald TJ, Koster J, McCabe MG, Sarver AL, Collins VP, Weiss WA, Largaespada DA, Collier LS, Taylor MD, Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 482, 529–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lastowska M, Al-Afghani H, Al-Balool HH, Sheth H, Mercer E, Coxhead JM, Redfern CP, Peters H, Burt AD, Santibanez-Koref M, Bacon CM, Chesler L, Rust AG, Adams DJ, Williamson D, Clifford SC, Jackson MS, Identification of a neuronal transcription factor network involved in medulloblastoma development. Acta Neuropathol Commun 1, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, Fults DW, Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene 23, 6156–6162 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Wang JY, Del Valle L, Gordon J, Rubini M, Romano G, Croul S, Peruzzi F, Khalili K, Reiss K, Activation of the IGF-IR system contributes to malignant growth of human and mouse medulloblastomas. Oncogene 20, 3857–3868 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Thomas WD, Chen J, Gao YR, Cheung B, Koach J, Sekyere E, Norris MD, Haber M, Ellis T, Wainwright B, Marshall GM, Patched1 deletion increases N-Myc protein stability as a mechanism of medulloblastoma initiation and progression. Oncogene 28, 1605–1615 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC, PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & development 22, 436–448 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones DT, Northcott PA, Kool M, Pfister SM, The role of chromatin remodeling in medulloblastoma. Brain pathology 23, 193–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindsey JC, Kawauchi D, Schwalbe EC, Solecki DJ, Selby MP, McKinnon PJ, Olson JM, Hayden JT, Grundy RG, Ellison DW, Williamson D, Bailey S, Roussel MF, Clifford SC, Cross-species epigenetics identifies a critical role for VAV1 in SHH subgroup medulloblastoma maintenance. Oncogene 34, 4746–4757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Das CM, Aguilera D, Vasquez H, Prasad P, Zhang M, Wolff JE, Gopalakrishnan V, Valproic acid induces p21 and topoisomerase-II (alpha/beta) expression and synergistically enhances etoposide cytotoxicity in human glioblastoma cell lines. J Neurooncol 85, 159–170 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I, High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 8, 539–554 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
fig S1: Clinical characteristics. (A-C) Clinical characteristics based on clustering analysis of WNT MB patient samples. (D-F) Clinical characteristics based on clustering analysis of Group 3 MB patient samples. (G-I) Clinical characteristics based on clustering analysis of Group 4 MB patient samples. (J) Arm-level chromosomal copy number change in SHH patients. Red and blue colored bars correspond to the gain and loss in the given chromosomal region, respectively. (K) Gene expression profiles measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. Bars represent mean with standard deviations. *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant. (L) Listed genes used for clustering analysis in the order of top-down pictured in Figure 1.
fig S2: Cerebellar architecture and protein expression in age-matched RESTTG and WT mice. (A) WT or RESTTG p8 brain sections stained with H&E. Sections were analyzed by IHC for (B) REST and (C) NeuN protein expression in CGNPs using specific antibodies. Scale bars = 200 μm (top panel −4x) and 20 μm (bottom panel - 40X).
fig S3: REST elevation contributes to infiltrative Shh-driven MB development. Comparison of H&E stained brain sections from (A) Ptch+/− and Ptch+/−/RESTTG animals with localized and infiltrative tumors, respectively (n=3), (B) DAOY and DAOY-REST xenografts (n=3), and (C) Low REST and High REST PDOXs (n=3). Scale bars = 200 μm (top panel −4x) and 20 μm (bottom panel - 40X).
fig S4: Differential regulation of PTCH1 expression by REST in MB tumors and CGNPs. Cerebellar sections of (A) tumor-bearing Ptch+/− and Ptch+/−/RESTTG mice (n=3) and (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH- subgroup PDOXs (n=3) were analyzed by IHC for REST, PTCH1, and KI-67 expression using specific antibodies. (D) GLI2, GLI3, and SMO mRNA expression profile measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. (E) hREST and (F) Scg10 mRNA expression was measured in WT and RESTTG GNPs after culturing with proliferation or differentiation media. WT data represents the mean ± SD (of fold change) of triplicate samples (n=3), RESTTG data represents 2 individual pups (n=2). For (A, B, and C) scale bars = 200 μm (top panel - 4X) and 50 μm (bottom panel −10X). For (E and F), p values were calculated by one-tailed t test of dCT values: *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant.
fig S5: REST and SUV39H1 do not co-immunoprecipitate in DAOY cells. Co-IP was performed to determine interaction between REST and SUV39H1. Whole cell extracts from DAOY were immunoprecipitated with IgG (control), REST, or no antibody, run on an SDS-PAGE and probed for REST and SUV39H1 protein. Blot shown is representative of 3 experiments.
SF6: REST elevation increases AKTSer473 phosphorylation. IHC was performed with specific antibodies targeting p-AKTS473, AKT, or PTEN protein in (A) Ptch+/− and Ptch+/−/RESTTG tumors (n=3), (B) DAOY and DAOY-REST xenografts (n=3) and (C) human SHH- subgroup PDOXs (n=3). (D) AKT1, AKT2, and AKT3 mRNA expression profile measured by microarray. The patient samples were divided into six distinct clusters based on clustering assay. Each dot corresponds to one individual patient. (E) Akt1, Akt2, and Akt3 mRNA expression was measured in WT and RESTTG GNPs after culturing with proliferation or differentiation media. WT data represents the mean ± SD (of fold change) of triplicate samples (n=3), RESTTG data represents 2 individual pups (n=2). For (A, B, and C) scale bars = 200 μm (top panel - 4X) and 50 μm (bottom panel −10X). For (E), p values were calculated by one-tailed t test of dCT values: *; p<0.05, **; p<0.01, ***; p<0.001, ****; p<0.0001, ns; non-significant.
SF7: REST-dependent AKT phosphorylation in cell-lines. (A) Western blot analysis of p- AKTSer473 and AKT protein expression following REST knockdown in DAOY cells (Fig. 6 B, right). Densitometry was obtained using Image Lab software (Bio-Rad). Bars represent mean with standard deviations (n=3).
Table S1: Synergy between MS275 and UNC0638. Synergy was calculated DAOY and UW228 at 48H post treatment either with each drug alone or in combination.
Table S2: IC50 values for UW228 and DAOY cells treated with MK-2206. Analysis of MTT assays was used to determine IC50 values after UW228 and DAOY cells were treated with MK-2206 at various concentrations as indicated for 24, 48 or 72 hours.
Table S3: Antibodies for IHC, qChIP, and Western blotting assays.
Table S4: Primers for qChIP and qRT-PCR assays.