PURPOSE
Lehmann et al have identified four molecular subtypes of triple-negative breast cancer (TNBC)—basal-like (BL) 1, BL2, mesenchymal (M), and luminal androgen receptor—and an immunomodulatory (IM) gene expression signature modifier. Our group previously showed that the response of TNBC to neoadjuvant systemic chemotherapy (NST) differs by molecular subtype, but whether NST affects the subtype was unknown. Here, we tested the hypothesis that in patients without pathologic complete response, TNBC subtypes can change after NST. Moreover, in cases with the changed subtype, we determined whether epithelial-to-mesenchymal transition (EMT) had occurred.
MATERIALS AND METHODS
From the Pan-Pacific TNBC Consortium data set containing TNBC patient samples from four countries, we examined 64 formalin-fixed, paraffin-embedded pairs of matched pre- and post-NST tumor samples. The TNBC subtype was determined using the TNBCtype-IM assay. We analyzed a partial EMT gene expression scoring metric using mRNA data.
RESULTS
Of the 64 matched pairs, 36 (56%) showed a change in the TNBC subtype after NST. The most frequent change was from BL1 to M subtypes (38%). No tumors changed from M to BL1. The IM signature was positive in 14 (22%) patients before NST and eight (12.5%) patients after NST. The EMT score increased after NST in 28 (78%) of the 36 patients with the changed subtype (v 39% of the 28 patients without change; P = .002254).
CONCLUSION
We report, to our knowledge, for the first time that the TNBC molecular subtype and IM signature frequently change after NST. Our results also suggest that EMT is promoted by NST. Our findings may lead to innovative adjuvant therapy strategies in TNBC cases with residual tumor after NST.
INTRODUCTION
Many studies have elucidated triple-negative breast cancer (TNBC) heterogeneity.1 For example, Lehmann et al used mRNA gene expression profiling to identify TNBC molecular subtypes. Initially, they identified six subtypes: basal-like (BL) 1, BL2, mesenchymal (M), luminal androgen receptor (LAR), mesenchymal stem-like, and immunomodulatory (IM).2 The group's subsequent study,3 using laser capture microdissection and histopathologic quantification, reduced these subtypes to four (BL1, BL2, M, and LAR). IM status was found to be a modifier of the other TNBC molecular subtypes4; it was shown to be primarily driven by tumor-infiltrating lymphocytes and thus can be used to evaluate a tumor's immune status. The four TNBC subtypes can be identified in the Clinical Laboratory Improvement Amendments environment by Oncocyte Corporation (formerly Insight Genetics; Nashville, TN) using a highly modified lean version of the TNBCtype algorithm consisting of 101 genes (TNBCtype-IM).
CONTEXT
Key Objective
Triple-negative breast cancer (TNBC) can be classified into various molecular subtypes on the basis of the unique gene signatures. The impact of chemotherapy on molecular subtypes is unknown. We hypothesized that neoadjuvant systemic chemotherapy (NST) can change the TNBC molecular subtypes. This study evaluated TNBC patient samples using the TNBCtype-immunomodulatory assay on the basis of the TNBC molecular subtypes identified by Lehmann et al.
Knowledge Generated
We found that the TNBC molecular subtype and immunomodulatory signatures frequently change after NST. Furthermore, the post-NST residual samples showed evidence of epithelial-to-mesenchymal transition.
Relevance
To our knowledge, this is the first report of TNBC molecular subtypes changing after NST. Thus, the residual tumor's TNBC molecular subtype may assist in innovative targeted therapy strategies for patients with nonpathologic complete response after NST.
In TNBC, about 30%-40% of patients have been shown to have a pathologic complete response (pCR) to current standard neoadjuvant systemic chemotherapy (NST). Previous studies have shown strong associations of pCR with longer overall survival and event-free survival durations5,6; by contrast, patients with breast cancer who did not have a pCR had significantly shorter survival durations because of higher relapse rates, especially in the TNBC subpopulation.6,7 Thus, the approach to non-pCR patients is very important. Postoperative adjuvant therapy for non-pCR TNBC has proven to be effective in several clinical trials.8 However, to establish the optimal adjuvant treatment, we propose that it is important to know the TNBC molecular subtype of the residual tumor.
In this study, we investigated TNBC molecular subtypes before and after NST in patients without pCR to test the hypothesis that TNBC subtypes can change after NST. Moreover, in cases with the changed subtype, we determined whether epithelial-to-mesenchymal transition (EMT) had occurred because EMT is a malignant phenotype constituting the first step in the potential metastatic process of residual tumors.
MATERIALS AND METHODS
Patients and Samples: Pan-Pacific TNBC Consortium Data Set
Four institutions participated in the study: Department of Breast Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX; Department of Medical Oncology, Chulalongkorn University, Bangkok, Thailand; Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea; and Department of Breast Surgical Oncology, Showa University, Tokyo, Japan. We retrospectively collected patients' samples and clinical data using the following criteria: (1) Patients had pathologically diagnosed stage I to III TNBC and received NST and subsequent surgery between January 2009 and December 2014. (2) Tumors had triple-negative status as determined by immunohistochemistry (IHC) or fluorescence in situ hybridization. Estrogen receptor and progesterone receptor status was considered negative if < 1% of cells stained positively IHC. Human epidermal growth factor receptor 2 status was considered negative if (a) the IHC result was 0 to +1 or (b) the IHC result was +2 and fluorescence in situ hybridization results were negative. (3) Patients did not have pCR after NST, and both baseline (pretreatment) formalin-fixed, paraffin-embedded (FFPE) core-needle biopsy specimens and FFPE resection specimens (residual disease) were available. (4) Clinical data (clinical and pathologic stage and NST regimens) were available. All samples were subjected to centralized review, and the presence of tumor in the samples was confirmed by pathologists from MD Anderson and Showa University. We collected the following data from the medical records: patient age, clinical stage, treatment regimen, nuclear grade, and pathologic information for the residual tumor. The median follow-up from diagnosis was 41 (range 7-133) months.
The study was approved by the ethics committees at Showa University (number: 2125) and The University of Texas MD Anderson Cancer Center (number: PA14-0544). A waiver of informed consent was granted on the basis of the study's retrospective nature.
TNBC Subtype Classification (TNBCtype-IM Assay)
The pre- and post-NST pairs of TNBC samples that met our selection criteria were classified by the TNBC molecular subtype by Oncocyte Corporation Transcriptome libraries, which were constructed using a TruSeq RNA Exome Library Prep Kit (Illumina, San Diego, CA) using 100 ng of total RNA extracted from FFPE tissue sections according to the manufacturer's recommendations. Libraries were sequenced on an Illumina NextSeq 500 with 150 paired-end cycles and a mean of 25 million reads per sample. Transcripts were aligned to the human reference assembly GRCh37 (Ensembl) using the STAR application (v. 020201). Assembly and expression quantification were performed using Cufflinks tools (v. 2.2.1). The resulting FPKM data for each sample were compiled and analyzed with the TNBCtype-IM algorithm.4 Samples that were unclassified were labeled UNS, indicating that the patient's TNBC expression pattern did not correlate with a specific subtype or IM modifier contained within the TNBCtype-IM assay; these samples could express a unique and unknown signature. Additional information about TNBCtype-IM assay can be found in the Data Supplement.
EMT Score
We next evaluated whether EMT was accelerated in residual tumor after primary systemic chemotherapy. We used the previously described EMT scoring metric developed by George et al.9 This metric quantifies the EMT spectrum; it was developed via an iterative method that ranks candidate gene products on the basis of their ability to resolve NCI-60 cell line samples with regard to their respective EMT status. The EMT scoring metric was applied to transcriptomic data to quantify the extent of EMT-ness on a scale of 0 (fully epithelial [E]) to 2 (fully M). This scoring method, on the basis of a set of EMT-relevant predictor transcripts and a set of normalizers for cross-platform application, categorizes samples into E, M, or hybrid E/M phenotypes on the basis of an ordered triple Si = (P_E, P_E/M, P_M), which represents the probability of group membership for each of the phenotypes. These probabilities are then projected on a scale of 0-2: E samples are assigned values close to 0; M samples, close to 2; and maximally hybrid E/M samples, close to 1.
Apocrine Status
Apocrine status was assessed using hematoxylin and eosin–stained slides (pre-NST samples only). We defined apocrine differentiation as the presence of abundant eosinophilic cytoplasm and large nuclei with prominent nucleoli. Three pathologists each independently reviewed the case slides and classified them as apocrine and nonapocrine accordingly.
E-cadherin and Vimentin Status
We assessed E-cadherin and vimentin status by IHC to evaluate intratumor heterogeneity in regard to EMT features. The analysis is described in the Data Supplement, and the results are shown in the Data Supplement.
Residual Cancer Burden Index
The residual cancer burden (RCB) index was assessed for all cases. The RCB index was developed by Symmans et al to evaluate the RCB after NST.10,11 The index score is derived from the largest area and cellularity of residual invasive primary cancer, the number of involved lymph nodes, and the size of largest nodal metastasis. The RCB index was scored as 0 for pathologic complete response (stage yp-T0/is, ypN0), and residual disease was categorized into three RCB index classes—RCB-I (minimal), RCB-II (moderate), and RCB-III (extensive)—on the basis of predefined cut points of 1.36 and 3.28 index scores.
Statistical Analysis
Statistical analyses were performed using R software (v 3.5.1). The associations between features were analyzed using the Fisher's exact test.
RESULTS
Patients and Samples
We collected 78 paired archived pre- and post-NST samples from patients in the Pan-Pacific TNBC Consortium data set who had residual tumor after NST. Of those, eight pre-NST samples and six post-NST samples did not pass quality control (because of inadequate sample quality or insufficient sequencing coverage depth) for TNBCtype-IM classification. The remaining 64 matched pairs were analyzed.
The median patient age was 53 years, and 52% of patients were premenopausal at diagnosis (Table 1). Among the 64 patients, 20% had clinical T4 disease and 53% had lymph node metastasis. Sixty-nine percent of patients received anthracycline and taxane regimens, and 16% of patients received anthracycline alone.
TABLE 1.
Patient Characteristics at Diagnosis
Chemotherapy Impact on TNBC Subtype Classification
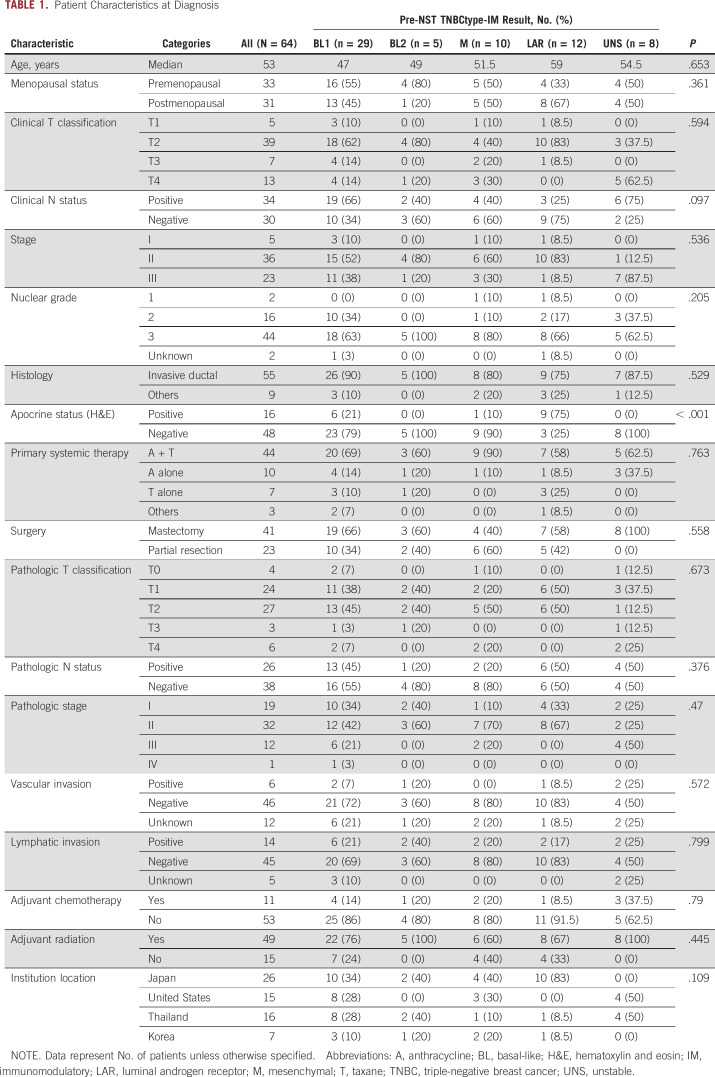
For the 64 matched pairs of samples, the distributions of TNBC subtypes in pre- and post-NST samples are shown in Figures 1 and 2. Compared with previous reports in TNBC (pCR and non-pCR),12-14 there were fewer patients with the BL1 subtype and fewer IM-positive patients; this result is expected in this non-pCR population because the BL1 subtype and IM positivity are known to be predictive markers of pCR. However, the most common pre-NST subtype was still BL1, 45% (29 of 64) of patients, followed by LAR, 19% (12 of 64) of patients (Fig 1).
FIG 1.
Distribution of triple-negative breast cancer subtypes before NST (core needle samples) for patients who did not have pathologic complete response (n = 64). BL, basal-like; LAR, luminal androgen receptor; M, mesenchymal; UNS, unstable.
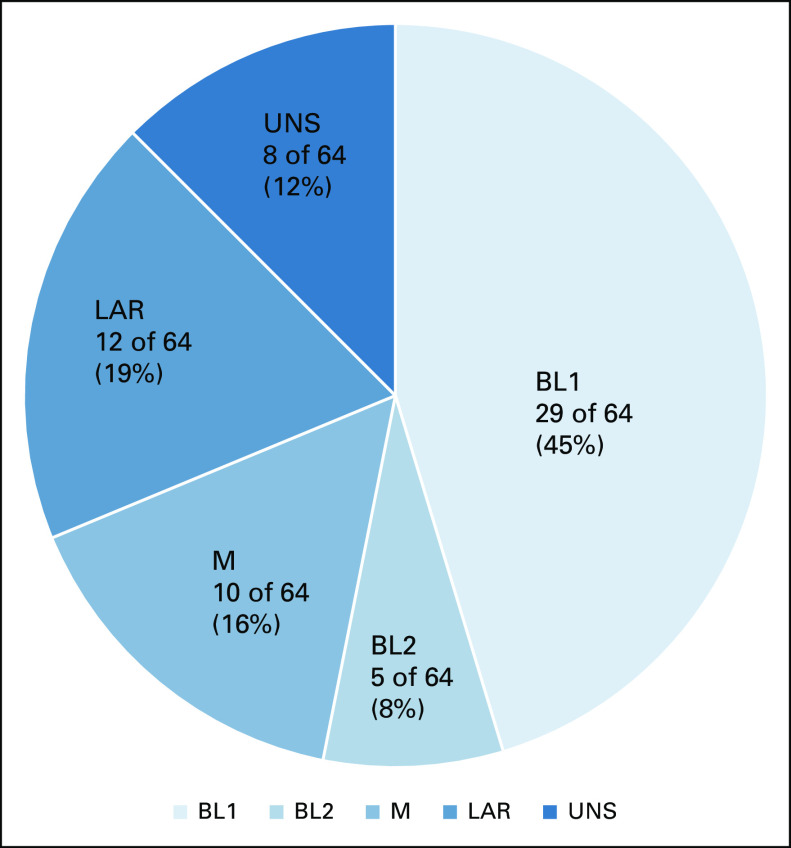
FIG 2.
Distribution of triple-negative breast cancer subtypes after NST (surgical samples) for patients who did not have pathologic complete response (n = 64). BL, basal-like; LAR, luminal androgen receptor; M, mesenchymal; UNS, unstable.
Of the 64 patients, 36 (56%) showed a change in the TNBC subtype after NST, and the distribution of TNBC subtypes changed (Fig 3). The most frequent change was from BL1, which was the dominant subtype before NST, to M, which was the dominant subtype after NST. By contrast, no tumors changed from M to BL1 subtypes. After NST, 14% (9 of 64) of patients were classified as the BL1 subtype (v 45% before NST) and 31% (20 of 64) were classified as M (v 16% before NST).
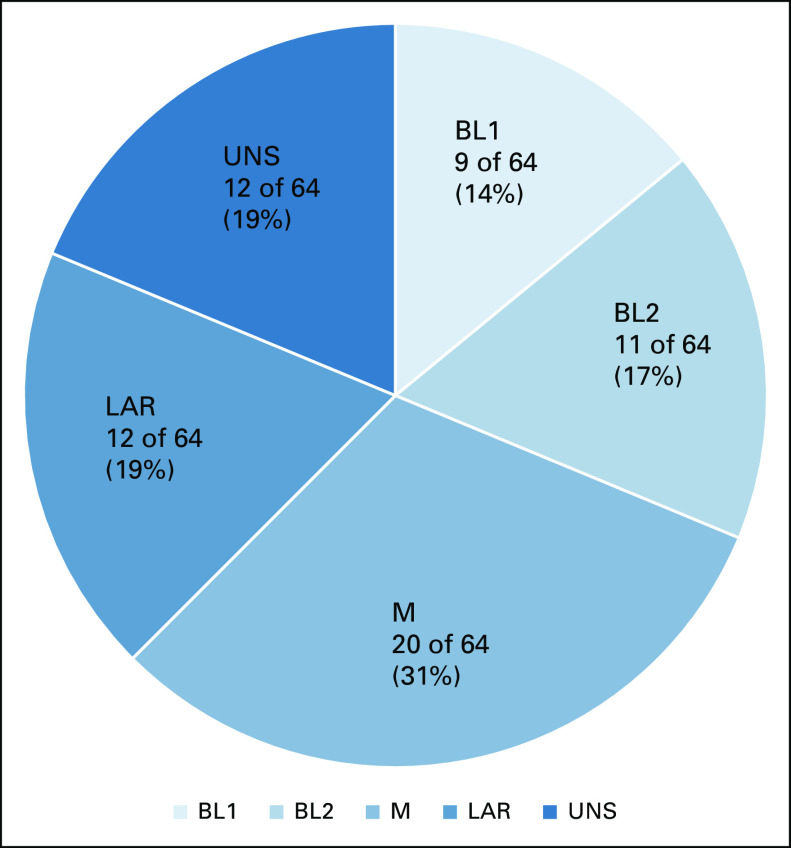
FIG 3.
Post-NST triple-negative breast cancer subtypes for patients in each pre-NST subtype group. BL, basal-like; LAR, luminal androgen receptor; M, mesenchymal; UNS, unstable.
Figure 3 shows the distribution of post-NST subtypes for each pre-NST subtype. Among the 29 tumors that were of BL1 subtype before NST, 31% (9 of 29) did not change subtype after NST and 38% (11 of 29) converted to M subtype. Among the five tumors that were of BL2 subtype before NST, two tumors did not change subtype after NST, two tumors converted to LAR subtype, and one tumor converted to M subtype. Among the 10 tumors that were of M subtype before NST, six tumors did not change subtype after NST, one tumor converted to BL2, one tumor converted to LAR, and the other two tumors were classified as UNS. Among the 12 tumors that were of LAR subtype before NST, 59% (7 of 12) did not change subtype after NST and 33% (4 of 12) converted to BL2.
Pre-NST IM signature positivity strongly correlated with the BL1 subtype; 86% of IM signature–positive samples belonged to the BL1 group (Data Supplement). Fourteen (22%) patients had a positive IM signature in their pre-NST samples, and eight (12.5%) had a positive IM signature in their post-NST samples. Nine patients converted to IM-negative status, and three patients acquired IM-positive status after NST (Data Supplement).
To assess how morphological findings correlate with molecular subtypes, we examined hematoxylin and eosin–stained pre-NST tumor samples. Apocrine differentiation correlated with the LAR subtype; 75% (9 of 12) of LAR patients showed apocrine differentiation. In addition, 21% (6 of 29) of BL1 patients showed apocrine differentiation (Table 1).
Chemotherapy Impact on EMT Score
An increased EMT score after NST was seen in 78% (28 of 36) of patients whose subtype changed, compared with only 39% (11 of 28) of patients whose subtype did not change (P = .00225; Table 2). Among the patients with a subtype change from BL1 to M, 82% (9 of 11) of patients had increased EMT scores and one patient had a decreased EMT score (Data Supplement).
TABLE 2.
Correlation Between the Subtype Change and the EMT Score
Correlation Between the Subtype Change and the RCB Score
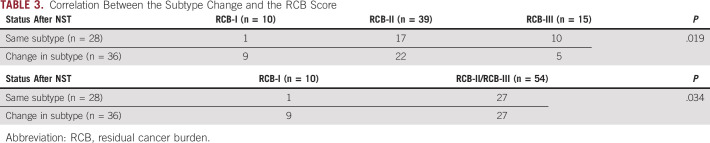
RCB was evaluable for all samples. Subtype changes were observed regardless of the residual tumor volume, but were more pronounced in tumors with a low residual tumor volume (Table 3).
TABLE 3.
Correlation Between the Subtype Change and the RCB Score
Correlation Between the EMT Score and the RCB Score
We also determined the correlation between the EMT score and the RCB score to eliminate the possibility that in tumors with a low residual tumor volume, more stromal components were included in the analysis, which may increase EMT. There was no statistically significant difference between these scores (Data Supplement).
DISCUSSION
To our knowledge, this is the first report that NST can change the molecular subtype of the residual tumor in TNBC. We found that the TNBC molecular subtype and IM signature frequently changed after NST. In addition, we showed that EMT was promoted by chemotherapy as measured by a partial EMT gene expression scoring metric. Our findings may lead to innovative adjuvant therapy strategies in TNBCs that do not achieve pCR after NST. Moreover, although the number of samples was small, we demonstrated that there are occasions in which chemotherapy may induce an IM signature, which could expand treatment options with immunotherapies for non-pCR patients.
Evidence has shown that cancer treatments affect the tumor biology and can lead to acquired epigenetic changes and mutations, some of which cause resistance.15-17
The risk of relapse is higher in patients with pathologic residual invasive disease after NST than in patients with pCR.6 Although it has been unclear whether there is a survival benefit from postoperative systemic chemotherapy after NST, several studies recently reported that adjuvant therapy for the non-pCR population increased disease-free and overall survival.8,18 According to these results, adjuvant therapies are promising treatment options for patients with breast cancer who have residual tumor after NST. To seek optimal adjuvant treatments, we must evaluate the biology of the residual tumor and elucidate the effect of NST. Our study is a valuable first step in revealing the effect of NST in inducing molecular subtype changes in more than 50% of patients; mRNA profiling analysis after NST may suggest the optimal adjuvant treatment for the individual patient. Moreover, this study showed that unlike intrinsic subtypes,19 TNBCtype-IM dynamically captured changes in molecular biology through temporal mRNA profiling.
On the other hand, intratumor heterogeneity is attracting attention because of the development of the technique of genome-wide profiling, which distinguishes single tumor cells and circulating tumor cells.20-22 Intratumor heterogeneity in breast cancers has been confirmed genetically and epigenetically in recent genome profiling reports.22 Thus, the possibility exists that pre-NST samples may not represent the characteristics of the tumor as a whole because of the small region of tumor tissue that is obtained through a core needle biopsy.
RCB was evaluable for all samples. As we expected, a low RCB-I was more common in patients with subtype change (9 of 36 [25%]) than in patients without subtype change (1 of 28 [3.5%]; Table 3). However, focusing on RCB-II and RCB-III, which reflect a moderate or extensive amount of residual tumor, 50% (27 of 54) changed subtype and the most frequent change was from BL1 to M subtypes (37%). Thus, the subtype changed even in cases in which the effect of chemotherapy was poor and high RCB remained.
Confirming our hypothesis, certain TNBC subtypes were found to have changed after NST. In fact, more patients had the M and BL2 subtypes after NST. Consistent with our previous knowledge of populations with pCR, in which 80% of patients had the BL1 subtype,23 the non-pCR population was under-represented in the BL1 subtype (45%) and had fewer IM-positive samples because the BL1 subtype and IM positivity are predictive markers for achieving pCR.12,13,23,24
The most frequent subtype change was from the BL1 to the M subtype (38% after NST), and no tumors did the opposite. This subtype is characterized by genes involved in motility, the extracellular matrix, cell differentiation pathways, and EMT.23 This subtype is also enriched in gene expression related to cell motility (the Rho pathway), cellular differentiation, and growth pathways (the anaplastic lymphoma kinase pathway, transforming growth factor beta signaling, and the Wnt/β-catenin pathway). This finding suggests that either chemotherapy accelerates the development of M features and EMT or there was heterogeneity of cell types within tumors and selective resistance and outgrowth of M cells during NST.
To clarify the validity of this hypothesis, we evaluated EMT using a gene expression scoring metric.9 EMT is a cellular process involving a multitude of phenotypic and morphologic changes that drive increased migratory and invasive potential.9 It has been implicated in acceleration of metastasis, acquisition of tumor initiation potential, resistance to anoikis, refractory response to chemotherapy, and the ability to evade the immune system.25-27 Recent studies have shown that cells need not undergo complete EMT for dissemination and that cells in one or more hybrid E/M phenotypes may be more metastatic than those that have undergone complete EMT.28 Thus, quantification of EMT status in a given sample can indicate metastatic aggressiveness. We evaluated the EMT score to infer whether the change in subtype was acquired because of the effects of chemotherapy. Although the small number of patients prevented determining the precise relationship with the EMT score for each subtype, the increased EMT score after NST in the patients with subtype changes (P = .00225) suggests that EMT is promoted by chemotherapy in at least some of the population. This result provides support that subtype changes occurred not only because of intratumor heterogeneity.
The limitations of this study include the small number of patients; although we created a pan-Pacific data set, the number of patients with each molecular subtype was limited and the patients had varying characteristics because of the retrospective nature of the study. For example, there were several NST regimens although the majority of patients received anthracycline and taxane regimens. Because of these diverse characteristics, it was difficult to measure clinically relevant outcomes, such as overall survival and disease-free survival. A further limitation was that the institutions had different specimen storage conditions.
Our findings suggest that when considering adjuvant treatment for non-pCR patients, the biology of the residual tumor should be considered and residual tumor might have more M properties than the pretreatment TNBC. Thus, targeting M features and EMT has potential as optimal therapy for non-pCR patients. A number of EMT targets have potential. These include Ras-mitogen–activated protein kinase activation, which cooperates to promote EMT and metastasis29 and Hedgehog signaling, which regulates EMT.30 Phosphatidylinositol 3-kinase has also been suggested to trigger EMT.31-33 EMT is regulated by transforming growth factor beta signaling, which promotes tumor growth, invasion, and evasion of immune surveillance.34,35 Similarly, FGF-2 induces EMT and is another druggable target.36-38 Inhibitors of these growth factors may have potential as targeted therapies for non-pCR patients.
In summary, we found that the TNBC molecular subtype and IM signature frequently changed in patients with residual disease after NST. In addition, we provide evidence suggesting that EMT is promoted by chemotherapy in some patients. Further investigation is necessary to determine whether the cause of the subtype change is intratumor heterogeneity or acquired biologic changes. Our findings may lead to innovative adjuvant chemotherapy strategies in TNBCs that do not show pCR after NST. Our findings support re-evaluation of residual tumor in future clinical trials for non-PCR patients. Furthermore, since the most frequent subtype change was from the BL1 to the M subtype and there were no cases of the reverse, targeting M features and EMT might have potential for the optimal treatment of non-pCR patients. Finally, re-evaluating immune status (ie, the IM signature) may expand the opportunity to use immunotherapies. These findings warrant independent confirmation in future studies.
ACKNOWLEDGMENT
We thank the study participants. Editorial assistance was provided by Sunita Patterson, Senior Scientific Editor, Research Medical Library, The University of Texas MD Anderson Cancer Center.
Hiroko Masuda
Speakers' Bureau: AstraZeneca, Chugai Pharma, Lilly, Novartis, Eisai
Research Funding: Eisai, Lilly (Inst)
Kenichi Harano
Honoraria: AstraZeneca, Chugai Pharma, Eisai, MSD K.K, Takeda
Consulting or Advisory Role: Takeda, Lilly Japan, AstraZeneca, Chugai Pharma
Research Funding: Merck, Daiichi-Sankyo
Oi Harada
Consulting or Advisory Role: Philips Japan
Bora Lim
Honoraria: Department of Defense, San Antonio Breast Cancer symposium
Consulting or Advisory Role: AstraZeneca, Novartis, Natera, Lilly
Research Funding: Genentech, Merck, Puma Biotechnology, Calithera Biosciences, Takeda
Napa Parinyanitikul
Consulting or Advisory Role: Roche, MSD Oncology
Speakers' Bureau: Novartis, Roche, Pfizer, AstraZeneca
Hee Jin Lee
Stock and Other Ownership Interests: NeogenTC Corp
Gyungyub Gong
Consulting or Advisory Role: Roche
Jangsoon Lee
Research Funding: AnHeart Therapeutics, CytoDyn, ChemDiv
Anthony Lucci
Speakers' Bureau: Exact Sciences
Arvind Rao
Honoraria: Cambridge Healthtech Institute
Consulting or Advisory Role: Genophyll LLC, Voxel Analytics LLC
Research Funding: Agilent (Inst)
Patents, Royalties, Other Intellectual Property: Patents pending
Travel, Accommodations, Expenses: Cambridge Healthtech Institute
Brock L. Schweitzer
Employment: OncoCyte
Research Funding: OncoCyte (Inst)
Patents, Royalties, Other Intellectual Property: I am included as an inventor on pending patents for the 27-gene algorithm used for classifying tumors
Robert S. Seitz
Employment: OncoCyte
Leadership: Oncocy
Stock and Other Ownership Interests: Oncocy
Consulting or Advisory Role: Thermo Fisher Scientific
Stephan W. Morris
Employment: Greenfire Bio
David R. Hout
Employment: Insight Genetics, OncoCyte
Leadership: Insight Genetics
Stock and Other Ownership Interests: Oncocyte
Patents, Royalties, Other Intellectual Property: I hold patents with Insight Genetics, but no royalties
Travel, Accommodations, Expenses: Insight Genetics
Seigo Nakamura
Honoraria: Chugai Pharma, Daiichi Sankyo Co Ltd
Naoto T. Ueno
Honoraria: Kyowa Hakko Kirin, Amgen, Chugai/Roche, Henry Stewart Talks, Taiho Pharmaceutical, Eisai, Rakuten Medical, Daiichi Sankyo, Pfizer, Novartis, Gilead Sciences, Puma Biotechnology
Consulting or Advisory Role: Daiichi Sankyo, Immunomedics, KeChow Pharma, Phoenix Design, Takeda, OncoCyte
Research Funding: Medivation, Bayer, Amgen, Puma Biotechnology, Merck, Daiichi Sankyo, Celgene, GlaxoSmithKline, Kyowa Hakko Kirin, Bio-Path Holdings, Novartis, Sysmex, Preferred Medicine
No other potential conflicts of interest were reported.
DISCLAIMER
The funders did not participate in the design of the study, the analysis or interpretation of data, or the writing or approval of the manuscript. The TNBC molecular subtype sample analysis was performed by Oncocyte Corporation.
PRIOR PRESENTATION
Presented in part as an abstract at the American Society of Clinical Oncology Annual Meeting, Chicago, IL, June 1-5, 2018.
SUPPORT
Supported by a Grant-in-Aid for Scientific Research 16K21374 (to H.M.); the National Institutes of Health through award Nos 1R01CA205043-01A1 (to N.T.U.), R01CA123318 (to N.T.U.), and P30CA016672 (MD Anderson's Cancer Center Support Grant; used the Clinical Trials Office and Bioinformatics Shared Resource); Breast Cancer Research Foundation grants BCRF-18-164 and APR-18-006 (to N.T.U.); the Morgan Welch Inflammatory Breast Cancer Research Program and Clinic; and a State of Texas Rare and Aggressive Breast Cancer Research Program grant (to N.T.U.).
S.K. and N.T.U. contributed equally to this work.
DATA SHARING STATEMENT
The RNA-seq data generated and/or analyzed during this study are publicly available.
AUTHOR CONTRIBUTIONS
Conception and design: Hiroko Masuda, Kenichi Harano, Sakiko Miura, Yuko Hirota, Oi Harada, Napa Parinyanitikul, Jangsoon Lee, Xiaoping Wang, Robert S. Seitz, David R. Hout, Savitri Krishnamurthy, Naoto T. Ueno
Financial support: Hiroko Masuda, Robert S. Seitz, David R. Hout, Naoto T. Ueno
Administrative support: Hiroko Masuda, David R. Hout, Naoto T. Ueno
Provision of study materials or patients: Hiroko Masuda, Yuki Matsunaga, Anita L. Wood, Napa Parinyanitikul, Hee Jin Lee, Gyungyub Gong, Anthony Lucci, Brock L. Schweitzer, Robert S. Seitz, Naoto T. Ueno
Collection and assembly of data: Hiroko Masuda, Kenichi Harano, Sakiko Miura, Yuko Hirota, Oi Harada, Yuki Matsunaga, Bora Lim, Anita L. Wood, Napa Parinyanitikul, Hee Jin Lee, Gyungyub Gong, Anthony Lucci, Brock L. Schweitzer, O. Rayne Lawrence, Robert S. Seitz, Seigo Nakamura, Naoto T. Ueno
Data analysis and interpretation: Hiroko Masuda, Kenichi Harano, Sakiko Miura, Ying Wang, Yuko Hirota, Oi Harada, Mohit Kumar Jolly, Bora Lim, Jason T. George, Herbert Levine, Anthony Lucci, Arvind Rao, O. Rayne Lawrence, Robert S. Seitz, Stephan W. Morris, Naoto T. Ueno
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hiroko Masuda
Speakers' Bureau: AstraZeneca, Chugai Pharma, Lilly, Novartis, Eisai
Research Funding: Eisai, Lilly (Inst)
Kenichi Harano
Honoraria: AstraZeneca, Chugai Pharma, Eisai, MSD K.K, Takeda
Consulting or Advisory Role: Takeda, Lilly Japan, AstraZeneca, Chugai Pharma
Research Funding: Merck, Daiichi-Sankyo
Oi Harada
Consulting or Advisory Role: Philips Japan
Bora Lim
Honoraria: Department of Defense, San Antonio Breast Cancer symposium
Consulting or Advisory Role: AstraZeneca, Novartis, Natera, Lilly
Research Funding: Genentech, Merck, Puma Biotechnology, Calithera Biosciences, Takeda
Napa Parinyanitikul
Consulting or Advisory Role: Roche, MSD Oncology
Speakers' Bureau: Novartis, Roche, Pfizer, AstraZeneca
Hee Jin Lee
Stock and Other Ownership Interests: NeogenTC Corp
Gyungyub Gong
Consulting or Advisory Role: Roche
Jangsoon Lee
Research Funding: AnHeart Therapeutics, CytoDyn, ChemDiv
Anthony Lucci
Speakers' Bureau: Exact Sciences
Arvind Rao
Honoraria: Cambridge Healthtech Institute
Consulting or Advisory Role: Genophyll LLC, Voxel Analytics LLC
Research Funding: Agilent (Inst)
Patents, Royalties, Other Intellectual Property: Patents pending
Travel, Accommodations, Expenses: Cambridge Healthtech Institute
Brock L. Schweitzer
Employment: OncoCyte
Research Funding: OncoCyte (Inst)
Patents, Royalties, Other Intellectual Property: I am included as an inventor on pending patents for the 27-gene algorithm used for classifying tumors
Robert S. Seitz
Employment: OncoCyte
Leadership: Oncocy
Stock and Other Ownership Interests: Oncocy
Consulting or Advisory Role: Thermo Fisher Scientific
Stephan W. Morris
Employment: Greenfire Bio
David R. Hout
Employment: Insight Genetics, OncoCyte
Leadership: Insight Genetics
Stock and Other Ownership Interests: Oncocyte
Patents, Royalties, Other Intellectual Property: I hold patents with Insight Genetics, but no royalties
Travel, Accommodations, Expenses: Insight Genetics
Seigo Nakamura
Honoraria: Chugai Pharma, Daiichi Sankyo Co Ltd
Naoto T. Ueno
Honoraria: Kyowa Hakko Kirin, Amgen, Chugai/Roche, Henry Stewart Talks, Taiho Pharmaceutical, Eisai, Rakuten Medical, Daiichi Sankyo, Pfizer, Novartis, Gilead Sciences, Puma Biotechnology
Consulting or Advisory Role: Daiichi Sankyo, Immunomedics, KeChow Pharma, Phoenix Design, Takeda, OncoCyte
Research Funding: Medivation, Bayer, Amgen, Puma Biotechnology, Merck, Daiichi Sankyo, Celgene, GlaxoSmithKline, Kyowa Hakko Kirin, Bio-Path Holdings, Novartis, Sysmex, Preferred Medicine
No other potential conflicts of interest were reported.
REFERENCES
- 1. Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 2. Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lehmann BD, Jovanovic B, Chen X, et al. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ring BZ, Hout DR, Morris SW, et al. Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients. BMC Cancer. 2016;16:143. doi: 10.1186/s12885-016-2198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 6. Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 7. von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 8. Toi M, Masuda N, Ohashi Y. Adjuvant capecitabine for breast cancer. N Engl J Med. 2017;377:791–792. doi: 10.1056/NEJMc1708487. [DOI] [PubMed] [Google Scholar]
- 9. George JT, Jolly MK, Xu S, et al. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 11. Symmans WF, Wei C, Gould R, et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol. 2017;35:1049–1060. doi: 10.1200/JCO.2015.63.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masuda H, Baggerly KA, Wang Y, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harano K, Wang Y, Lim B, et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS One. 2018;13:e0204513. doi: 10.1371/journal.pone.0204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carey LA. Directed therapy of subtypes of triple-negative breast cancer. Oncologist. 2011;16(suppl 1):71–78. doi: 10.1634/theoncologist.2011-S1-71. [DOI] [PubMed] [Google Scholar]
- 15. Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van de Ven S, Smit VT, Dekker TJ, et al. Discordances in ER, PR and HER2 receptors after neoadjuvant chemotherapy in breast cancer. Cancer Treat Rev. 2011;37:422–430. doi: 10.1016/j.ctrv.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 17. Amir E, Miller N, Geddie W, et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J Clin Oncol. 2012;30:587–592. doi: 10.1200/JCO.2010.33.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Minckwitz G, Huang CS, Mano MS, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 19. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 20. Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 21. Pestrin M, Bessi S, Galardi F, et al. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res Treat. 2009;118:523–530. doi: 10.1007/s10549-009-0461-7. [DOI] [PubMed] [Google Scholar]
- 22. Martelotto LG, Ng CK, Piscuoglio S, et al. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014;16:210. doi: 10.1186/bcr3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232:142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prat A, Adamo B, Cheang MC, et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jolly MK, Boareto M, Huang B, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tripathi SC, Peters HL, Taguchi A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci USA. 2016;113:E1555–E1564. doi: 10.1073/pnas.1521812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang RY, Wong MK, Tan TZ, et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530) Cell Death Dis. 2013;4:e915. doi: 10.1038/cddis.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jolly MK, Mani SA, Levine H. Hybrid epithelial/mesenchymal phenotype(s): The “fittest” for metastasis? Biochim Biophys Acta Rev Cancer. 2018;1870:151–157. doi: 10.1016/j.bbcan.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 29. Tse JC, Kalluri R. Mechanisms of metastasis: Epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem. 2007;101:816–829. doi: 10.1002/jcb.21215. [DOI] [PubMed] [Google Scholar]
- 30. Yoo YA, Kang MH, Lee HJ, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
- 31. Vlahopoulos SA, Logotheti S, Mikas D, et al. The role of ATF-2 in oncogenesis. Bioessays. 2008;30:314–327. doi: 10.1002/bies.20734. [DOI] [PubMed] [Google Scholar]
- 32. Huber MA, Beug H, Wirth T. Epithelial-mesenchymal transition: NF-kappaB takes center stage. Cell Cycle. 2004;3:1477–1480. doi: 10.4161/cc.3.12.1280. [DOI] [PubMed] [Google Scholar]
- 33. Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review) Int J Mol Med. 2008;22:271–275. [PubMed] [Google Scholar]
- 34. Fuxe J, Karlsson MC. TGF-beta-induced epithelial-mesenchymal transition: A link between cancer and inflammation. Semin Cancer Biol. 2012;22:455–461. doi: 10.1016/j.semcancer.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 35. Zhang J, Tian XJ, Zhang H, et al. TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 36. Lee JG, Kay EP. FGF-2-mediated signal transduction during endothelial mesenchymal transformation in corneal endothelial cells. Exp Eye Res. 2006;83:1309–1316. doi: 10.1016/j.exer.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 37. Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 38. Chakrabarti R, Hwang J, Andres Blanco M, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data generated and/or analyzed during this study are publicly available.