Abstract
6-Anilinouracils are selective inhibitors of DNA polymerase III, the enzyme required for the replication of chromosomal DNA in gram-positive bacteria (N. C. Brown, L. W. Dudycz, and G. E. Wright, Drugs Exp. Clin. Res. 12:555–564, 1986). A new class of 6-anilinouracils based on N-3 alkyl substitution of the uracil ring was synthesized and analyzed for activity as inhibitors of the gram-positive bacterial DNA polymerase III and the growth of gram-positive bacterial pathogens. Favorable in vitro properties of N-3-alkyl derivatives prompted the synthesis of derivatives in which the R group at N-3 was replaced with more-hydrophilic methoxyalkyl and hydroxyalkyl groups. These hydroxyalkyl and methoxyalkyl derivatives displayed Ki values in the range from 0.4 to 2.8 μM against relevant gram-positive bacterial DNA polymerase IIIs and antimicrobial activity with MICs in the range from 0.5 to 15 μg/ml against a broad spectrum of gram-positive bacteria, including methicillin-resistant staphylococci and vancomycin-resistant enterococci. Two of these hydrophilic derivatives displayed protective activity in a simple mouse model of lethal staphylococcal infection.
The escalating incidence of infection with multiple-antibiotic-resistant forms of low G+C content gram-positive bacteria is a major and rapidly growing clinical problem (20). In an effort to address this problem, we have sought to identify a new gram-positive-bacterium-specific antimicrobial target and to develop selective “bullets” to hit it. The target we have selected is DNA polymerase III (pol III), the product of the polC gene (10, 13, 15, 16).
We have targeted the polC-specific pol III for three reasons. First, it is absolutely essential for the replication of the host chromosome of the low G+C content gram-positive bacteria; when its action is blocked, chromosomal DNA fails to replicate and the bacterial host dies (5, 6, 10, 15, 25). Second, this pol III is a target whose essential structure is strongly conserved in a broad group of relevant low G+C content gram-positive pathogens, including Staphylococcus, Streptococcus, Enterococcus, and Mycoplasma (2, 13). Third, the active-site domain of this enzyme incorporates a unique receptor which renders it specifically susceptible to the small molecule inhibitors of the 6-anilinouracil (AU) class (see references 6 and 7 and structure shown in Fig. 1A, below). It is this AU class of pol III-specific inhibitors which we seek to develop as gram-positive-bacterium-selective antimicrobials. The results described below summarize the progress we have made in this effort.
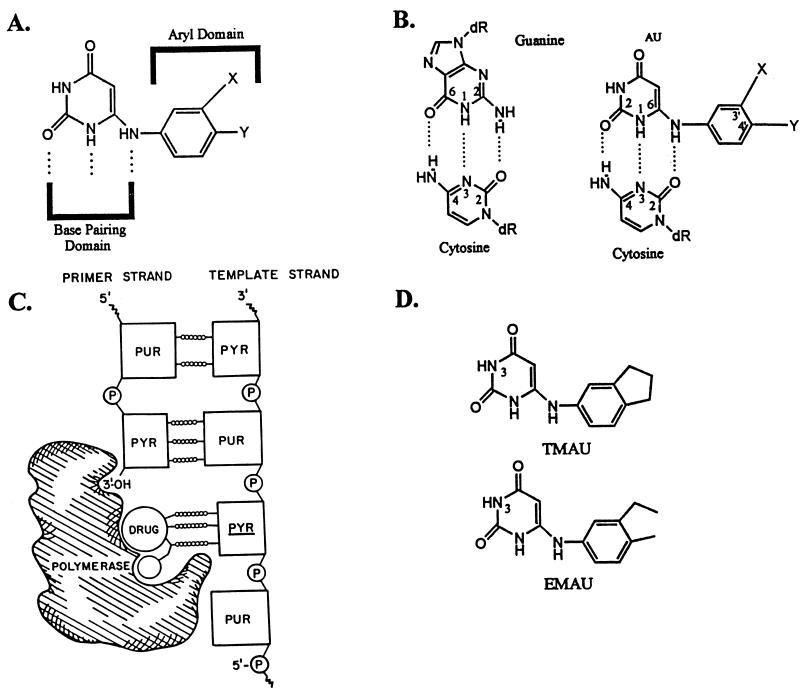
FIG. 1.
Structures and mechanisms of action of AUs which selectively inhibit the gram-positive pol III. (A) Relevant domains of the AU molecule. (B) Equivalence of the base-pairing domains of the guanine (left) and AU (right) molecules. (C) The AU molecule inhibits its pol III target by sequestering it into an inactive DNA-drug-protein complex. PUR, purine; PYR, pyrimidine. (D) Structures of two relevant AUs, EMAU and TMAU.
MATERIALS AND METHODS
Determination of water solubility.
A known mass of pure, dried compound was dissolved in dilute NaOH at pH 12.0. The solution was then neutralized to pH 7.0 with 0.1 N HCl for determination of the extinction coefficient at the absorbance maximum. To determine water solubility, an excess of each compound was stirred in deionized water (pH 7.0) at 25°C for 1 h. After removal of undissolved compound by filtration, the absorbance of the solution at the absorbance maximum was determined and used to calculate the concentration of the compound remaining in solution. Each reported value is the mean of three independent experiments.
Bacterial strains and media.
Bacillus subtilis was the standard penicillin-sensitive laboratory strain, BD54 (16). Enterococcus faecalis, Enterococcus faecium, and Staphylococcus aureus were clinically derived strains which were typed and kindly provided by Gary Doern, University of Massachusetts Medical Center. Unless noted otherwise, bacteria were grown in Luria broth medium consisting of 0.5% yeast extract (Difco), 1% tryptone (Bacto-tryptone; Difco) and 0.5% NaCl in deionized water.
Inhibitors.
N-3-substituted AUs were synthesized, purified, and analyzed as described in reference 19. 6-(p-Hydroxyphenylazo)uracil (HPUra) and other AUs were synthesized and purified as described (26).
Enzymes.
The pol IIIs of B. subtilis and S. aureus were homogeneous recombinant proteins expressed and prepared as described (12, 18). E. faecalis pol III was fraction V purified from ATCC 8043 as described by Barnes and Brown (1). The dnaE-encoded E. coli pol III was the recombinant form of the reconstituted core enzyme (alpha-epsilon-theta [14]) and was kindly provided by Charles McHenry, University of Colorado, Denver. Immunopurified calf thymus DNA pol alpha was kindly provided by Ulrich Hübscher of the University of Zürich-Irchel, Zürich, Switzerland.
Determination of MIC.
Log-phase bacterial cultures were diluted to a concentration of ∼105 CFU per ml in Luria broth, and samples of this suspension were distributed to the wells of a 48-well microassay plate. Each compound was dissolved in dimethylsulfoxide (DMSO) and added to one well at a concentration of 200 μM. The contents of this well were then serially diluted, twofold each time, through seven wells, to produce a series of eight wells containing 0.5 ml of suspension and compound at the following successive concentrations: 200, 100, 50, 25, 12.5, 6.25, 3.125, and 1.5625 μM. All wells, including controls incubated in the absence of inhibitors, contained 1% DMSO. DMSO at this concentration did not significantly affect the growth of any of the test organisms. Plates were incubated for 24 h at 37°C and read by visual inspection of the wells. MIC was defined as the lowest concentration of inhibitor at which bacterial growth was not visually apparent. Reported values are the median values for three experiments. MICs were converted from micromolar concentrations to micrograms per milliliter for tabulation.
Bactericidal activity.
Each inhibitor was dissolved in sterile DMSO and diluted 100-fold into Mueller-Hinton broth (MHB; Difco) containing log-phase methicillin-sensitive S. aureus (MSSA) (Smith strain) at a concentration of ∼106 CFU per ml. The control culture contained only 1% DMSO. The cultures were incubated at 37°C, and at intervals during a 24-h period, samples were removed, diluted extensively in sterile MHB, and plated on Luria broth-based agar plates to determine the numbers of CFU.
DNA polymerase activity.
DNA polymerases were assayed by using activated calf thymus DNA and the basic method described in reference 1. Briefly, enzyme was added to buffered solution containing Mg2+, dithiothreitol, glycerol, saturating concentrations of activated calf thymus DNA, dATP, dCTP, dTTP, and [3H]dTTP, and appropriate concentrations of inhibitor included as a dilution of a stock solution in DMSO. Assay mixtures were incubated at 30°C for 10 min, reactions were stopped with cold trichloroacetic acid, and mixtures were filtered to capture the cold acid-insoluble 3H-labeled DNA. The filters were washed in turn with cold trichloroacetic acid and ethanol and then dried. The incorporated radioactivity was measured by scintillation counting of the dried filters as described (1). Apparent inhibitor constants (Kis) of the AUs were determined directly by truncated assay in the absence of dGTP as described by Wright and Brown (23). Each reported value is the average for three independent experiments. The average standard deviation for the values presented was ±17.4%.
Assay for protection against lethal staphylococcal infection in vivo.
This assay was performed by the Pharmacology Services Laboratory of MDS Panlabs, Inc. Female Swiss-Webster mice (20 g) were infected with a single intraperitoneal injection of MSSA (ATCC Smith strain; 0.5 ml in physiological saline; 4 × 107 CFU/mouse), and 15 min thereafter, mice in each group of ten infected animals were injected intraperitoneally once with one of the following solutions: (i) 0.1 ml of physiological saline (vancomycin vehicle control); (ii) 0.1 ml of vancomycin (4 mg/ml in physiological saline; dose, 20 mg/kg of body weight); (iii) 0.1 ml of AU vehicle control (10% DMSO–90% peanut oil); or (iv) 0.1 ml of 10% DMSO–90% peanut oil containing the relevant N-3-substituted AU at a concentration sufficient to give a dose of either 5 or 10 mg/kg. Following injection, each group of ten animals was monitored for survival over a 3-day period.
Assay of DNA and RNA synthesis in B. subtilis BD54.
At time zero [3H]adenine (7 × 105 cpm/pmol; final concentration, 0.1 mM) and inhibitor or inhibitor diluent were added to a log-phase culture growing at 37°C in a minimal medium (8) with a doubling time of 35 min. After 30 min, duplicate 1-ml samples were removed to determine incorporation into alkali-soluble (RNA) and alkali-stable (DNA) fractions of cold trichloroacetic acid-insoluble material as described previously (4).
RESULTS AND DISCUSSION
Summary of the mechanism of action of AUs on the gram-positive bacterial pol III.
Although simple molecules, the AUs have two distinct domains essential for their inhibitory action. These are illustrated in Fig. 1A. One, the so-called base-pairing domain, is comprised of three uracil-specific components—the ring 1-NH, the 2-keto, and the 6-NH components. The second, enzyme-specific, domain is contributed by an appropriately substituted aryl group at N-6. Although formally pyrimidines, the AUs derive their capacity to inhibit pol III by mimicking the guanine component of the purine 2′-deoxyribonucleoside triphosphate dGTP. As shown in Fig. 1B, the AUs derive this property from the capacity of the unconventional base-pairing domain to specifically form three hydrogen bonds with the pyrimidine base cytosine. Fig. 1C summarizes how the base-pairing and enzyme-specific domains of the AU molecule cooperate to inhibit pol III as it tries to elongate the primer terminus past a cytosine residue in the DNA template. The base-pairing domain of the inhibitor forms hydrogen bonds with the cytosine, and simultaneously, the enzyme-specific aryl moiety binds to the enzyme’s unique aryl receptor. As a result, the inhibitor sequesters the enzyme with high affinity into a nonproductive ternary complex with template-primer (5, 9, 10).
Effect of alkyl substitution at N-3 on antibacterial and anti-pol III activities of AUs.
Among the AUs which we have synthesized (5, 7, 23–26), two of the most potent, 6-(3-ethyl-4-methylanilino)uracil (EMAU) and 6-([3,4-trimethylene]anilino)uracil (TMAU), contain alkyl groups in the 3 and 4 positions of the anilino ring (Fig. 1D shows structures). Although TMAU and EMAU were potent inhibitors of the gram-positive bacterial pol III (26), they showed weak antibacterial activity, yielding MICs of 9 to 18 μg/ml (Table 1). In an attempt to improve their antibacterial activities, we developed structure-activity relationships involving substituents at ring nitrogen 3 (see numbering in Fig. 1B and D), the only position in the AU molecule where nondestructive substitution is feasible (21).
TABLE 1.
In vitro antibacterial activity of N-3-substituted TMAU and EMAU derivatives
| AU | R group | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| B. subtilis BD54 | MSSA (S. aureus IP8) | MRSA6 (S. aureus B42876) | MRSA3 (S. aureus B43243) | E. faecalis ATCC 8043 | VRE | E. faecium ATCC 29212 | E. coli XL1 Blue (Stratagene) | ||
| TMAU | -H | 9 | 18 | 18 | 18 | 9 | 18 | 18 | >49 |
| -CH3 (methyl) | 1 | 10 | 13 | 19 | 10 | 5 | 13 | >51 | |
| -CH2CH3 (ethyl) | 0.6 | 3 | 7 | 5 | 3 | 3 | 3 | >54 | |
| -CH2CH⩵CH2 (allyl) | 0.7 | 5 | 7 | 5 | 7 | 4 | 5 | >57 | |
| -(CH2)2CH3 (propyl) | 4 | 21 | 21 | 21 | 21 | 11 | 21 | >57 | |
| -(CH2)3CH3 (butyl) | 1 | 22 | 22 | 22 | 15 | 7 | 22 | >60 | |
| -(CH2)5CH3 (hexyl) | >33 | >33 | >33 | >33 | >33 | >33 | >33 | >65 | |
| -CH2C6H5 (benzyl) | >33 | >33 | >33 | >33 | >33 | >33 | >33 | >67 | |
| EMAU | -H | 9 | 18 | 18 | 18 | 18 | 18 | 5 | >49 |
| -CH2CH3 (ethyl) | 0.4 | 3 | 10 | 10 | 5 | 1 | 5 | >55 | |
| -CH2CH⩵CH2 (allyl) | 0.5 | 5 | 11 | 11 | 11 | 11 | 11 | >57 | |
MICs are the medians of three independent experiments performed as described in the Materials and Methods section.
The first structure-activity relationship to exploit N-3 alkylation was performed on the TMAU platform. It had two components, one of which assessed antimicrobial potency and the other of which assessed potency for enzyme inhibition. The former utilized several strains of relevant gram-positive bacteria, while the latter utilized purified pol IIIs derived from three relevant gram-positive organisms—B. subtilis, S. aureus, and E. faecalis.
Table 1 summarizes the relative antibacterial activities (i.e., MICs) of a series of N-3-substituted TMAUs. The MIC results show that N-3 alkylation with up to three carbons generally improved potency for all organisms relative to that of the unsubstituted TMAU. The ethyl and allyl groups increased potency, but larger groups, such as the butyl group, did not. Derivatives with the largest substituents, hexyl and benzyl, had considerably weaker activities than TMAU.
Table 2 summarizes the effect of N-3 alkylation of TMAU on its potency against the three isolated pol IIIs. The results, displayed as apparent inhibitor constants (Kis [23]), indicate that potency increased with increasing size of the N-3 alkyl chain, reached a maximum (>10-fold) with addition of the butyl substituent, and diminished with addition of the larger, hexyl group. Addition of the benzyl group had a decidedly negative effect on anti-pol activity, reducing it 15- to 18-fold relative to that for the unsubstituted TMAU.
TABLE 2.
Kis of N-3-substituted TMAUs and EMAUs against the pol IIIs of B. subtilis, S. aureus, and E. faecalis
| AU | R group substituted at N-3 |
Ki (μM)a
|
||
|---|---|---|---|---|
| B. subtilis | S. aureus | E. faecalis | ||
| TMAU | -H | 1.20 | 1.59 | 1.22 |
| -CH3 (methyl) | 1.34 | 1.22 | 0.84 | |
| -CH2CH3 (ethyl) | 0.31 | 0.38 | 0.31 | |
| -CH2CH⩵CH2 (allyl) | 0.38 | 0.54 | 0.24 | |
| -(CH2)2CH3 (propyl) | 0.31 | 0.20 | 0.22 | |
| -(CH2)3CH3 (butyl) | 0.09 | 0.10 | 0.09 | |
| -(CH2)5CH3 (hexyl) | 0.24 | 0.42 | 0.14 | |
| -CH2C6H5 (benzyl) | 21.4 | 36.1 | 17.6 | |
| EMAU | -H | 1.00 | 1.16 | 0.61 |
| -CH2CH3 (ethyl) | 0.12 | 0.22 | 0.07 | |
| -CH2CH⩵CH2 (allyl) | 0.21 | 0.32 | 0.19 | |
All Kis are the averages of three independent experiments performed as described in the Materials and Methods section.
To determine whether the effects of N-3 substitution on TMAU were applicable more generally to another active AU platform, we synthesized and tested the ethyl and allyl derivatives of EMAU. The results are summarized in the bottom three rows of Tables 1 and 2.
With respect to antibacterial activity (Table 1), ethyl or allyl substitution of EMAU had essentially the same effect as it had on TMAU, i.e., it increased potency against all organisms except E. faecium. The effect of ethyl or allyl substitution was most dramatic in the case of B. subtilis, increasing potency approximately 20-fold. In all cases, the N-3 ethyl form of EMAU (Et-EMAU) displayed slightly greater potency than the allyl derivative. For the two strains of methicillin-resistant S. aureus (MRSA) both the N-3 allyl form of EMAU and Et-EMAU displayed potency less than twofold greater than that of the unsubstituted EMAU molecule. For the other strains, N-3 alkyl substitution increased potency from 2- to 18-fold.
Both N-3 substituents significantly increased the anti-pol III potency of EMAU (Table 2), and with the exception of E. faecalis pol III, the orders of relative potency were qualitatively the same as those for the ethyl and allyl derivatives of TMAU (Table 2). In the case of E. faecalis pol III, the respective potencies of the ethyl and allyl forms did not demonstrate a specific rank order. As anticipated, N-3 substitution also did not apparently change the selectivity of the molecule (5) for low G+C content gram-positive bacteria as evidenced by the lack of significant inhibitory effect of high concentrations on the growth of the gram-negative organism Escherichia coli (Table 1, right column).
In sum, the above results indicated that N-3 substitution on the AU platform can lead to increases in both the antibacterial and anti-pol III activities of these gram-positive-bacterium-selective inhibitors. This effect was apparently maximal with N-3 ethyl and allyl substitutions. As expected for compounds which putatively hit a novel target, the N-3 alkyl-substituted AUs were essentially equivalent with respect to potency against antibiotic-sensitive and antibiotic-resistant strains of relevant gram-positive organisms.
Effects of hydrophilic N-3 substitution of EMAU.
Given their strong anti-pol III actions and their favorable in vitro antimicrobial potencies, we attempted to assess Et-EMAU and Et-TMAU for their capacities to prevent gram-positive bacterial infection in mice and thus “prove principle” in vivo. However, their marginal aqueous solubilities (3 μM for Et-TMAU and 6.3 μM for Et-EMAU; determined as described in the Materials and Methods section) severely restricted their bioavailabilities following either oral or parenteral administration and therefore precluded their use for this purpose. To improve water solubility and bioavailability, we focused on the EMAU platform and proceeded to synthesize two N-3 hydroxyalkyl forms, i.e., the 2-hydroxyethyl derivative, HE-EMAU, and the 3-hydroxypropyl derivative, HP-EMAU (19). As expected, the water solubilities of both agents were increased significantly compared to that of Et-EMAU. The aqueous solubility of HE-EMAU was 32 μM, and that of HP-EMAU was 98 μM. Even the respective methoxyalkyl intermediates from which HE-EMAU and HP-EMAU were synthesized were more water soluble than Et-EMAU (the water solubility of the 2-methoxyethyl intermediate [ME-EMAU] was 23 μM, and that of the 3-methoxypropyl intermediate [MP-EMAU] was 22 μM).
Table 3 summarizes the effects of the N-3 hydroxyalkyl and methoxyalkyl substitutions on the antimicrobial activity of the EMAU platform. These substituents all enhanced antibiotic activity relative to that of EMAU; their MICs, with the exceptions of those for vancomycin-resistant E. faecalis (VRE; Table 1) and MSSA (Table 1), equalled or were better than those of Et-EMAU. Significantly, the propyl derivatives, MP-EMAU and HP-EMAU, were as active against the MRSA and VRE strains as they were against the corresponding antibiotic-sensitive strains, and like the alkyl-substituted derivatives, none of these more-hydrophilic compounds was active against E. coli at ∼60 μg/ml, the highest concentration tested (right column, Table 3).
TABLE 3.
In vitro antibacterial activity of N-3-hydroxyalkyl and methoxyalkyl EMAUs
| R group | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| B. subtilis BD54 | MSSA (S. aureus IP8) | MRSA6 (S. aureus B42876) | MRSA3 (S. aureus B43243) | E. faecalis ATCC 8043 | VRE | E. faecium ATCC 29212 | E. coli XL1 Blue (Stratagene) | |
| -(CH2)2OH (HE) | 0.7 | 7 | 14 | 14 | 5 | 7 | 7 | >58 |
| -(CH2)2OCH3 (ME) | 0.9 | 15 | 8 | 8 | 6 | 6 | 8 | >61 |
| -(CH2)3OH (HP) | 0.9 | 6 | 8 | 8 | 6 | 6 | 6 | >61 |
| -(CH2)3OCH3 (MP) | 0.5 | 6 | 6 | 6 | 3 | 3 | 6 | >63 |
MICs are the medians of three independent experiments performed as described in the Materials and Methods section.
The anti-polymerase activities of the methoxyalkyl- and hydroxyalkyl EMAUs are summarized in Table 4. These substituents did not significantly change either the potency of the EMAU derivative or its selectivity for the gram-positive bacterial pol III. All three gram-positive bacterial pol IIIs were clearly sensitive to the four compounds, with S. aureus pol III slightly less sensitive than the enzymes of B. subtilis and E. faecalis. In contrast, none of the four agents displayed significant inhibitory activity against either the dnaE-specific gram-negative bacterial (E. coli) pol III or the mammalian (calf thymus) replicative enzyme, pol alpha.
TABLE 4.
Inhibitor constants (Kis) of N-3 hydroxyalkyl and methoxyalkyl EMAU derivatives against relevant DNA polymerases
| R group |
Ki (μM)
|
||||
|---|---|---|---|---|---|
| B. subtilisb | S. aureusb | E. faecalisb | E. colib | pol alphac | |
| -(CH2)2OH (HE) | 0.59 | 2.48 | 0.48 | >160 | >160 |
| -(CH2)2OCH3 (ME) | 0.58 | 2.82 | 0.46 | >160 | >160 |
| -(CH2)3OH (HP) | 0.73 | 2.10 | 0.61 | >160 | >160 |
| -(CH2)3OCH3 (MP) | 1.00 | 1.27 | 0.42 | >160 | >160 |
All Kis are the averages of three independent experiments performed as described in the Materials and Methods section.
Denotes the DNA pol III of the indicated organism; see Materials and Methods section for source.
Derived from calf thymus; see Materials and Methods section for source.
Bactericidal properties of the methoxyalkyl and hydroxyalkyl EMAUs.
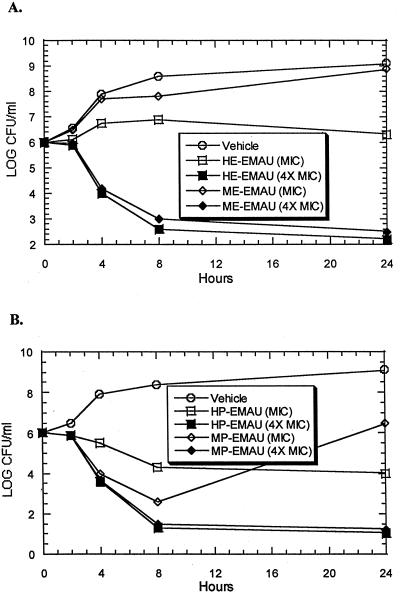
Our earlier studies of the effects of HPUra, the prototype for the AUs, indicated that it was bactericidal for E. faecalis (6). To determine if the N-3-substituted hydroxyalkyl and methoxyalkyl EMAUs also were bactericidal, time-kill studies were conducted with the MSSA Smith strain which was to be used (see below) to examine their abilities to protect against in vivo infection. The results are summarized in Fig. 2. As shown in panel A, the ethyl derivatives, HE-EMAU and ME-EMAU, were bactericidal at 4× MIC, while at MIC both of them permitted some cell replication. The two propyl compounds, HP-EMAU and MP-EMAU (Fig. 2, panel B), were clearly bactericidal at 4× MIC, while at MIC they were, at best, bacteriostatic.
FIG. 2.
Bactericidal activities of N-3 methoxyalkyl- and hydroxyalkyl-EMAUs against S. aureus (Smith strain). Each of the four agents was tested, as indicated, at its MIC and at four times (4×) its MIC. The MICs of the agents for this strain were as follows: HE-EMAU, 8 mg/ml; ME-EMAU, 16 mg/ml; HP-EMAU, 8 mg/ml; and MP-EMAU, 8 mg/ml. (A) Activities of HE-EMAU and ME-EMAU. (B) Activities of HP-EMAU and MP-EMAU.
Preservation of other key properties of HPUra.
In addition to its bactericidal effect, HPUra has three well-documented properties which characterize its antibacterial action (5, 6, 9, 10, 17, 25). The first is its spectrum. HPUra is active against only low G+C content gram-positive eubacteria and Mycoplasmatales (3) and thus has no significant effect on the growth of gram-negative eubacteria, mammalian cells, or high G+C content gram-positive eubacteria such as Streptomyces and Mycobacterium (4). The second distinct property of HPUra is its selectivity for replicative DNA synthesis. Administration of the agent to an exponentially growing culture of a sensitive organism at concentrations up to 20× MIC completely inhibits the synthesis of DNA with little or no effect on the synthesis of RNA or other macromolecules (5, 6, 8). The third important characteristic of HPUra is its target within the DNA replication machinery. It is absolutely specific for the polC-encoded pol III enzyme that is unique to low G+C content gram-positive eubacteria (10, 11, 13, 25). The active form of HPUra has no inhibitory effect on a wide variety of prokaryotic and eukaryotic polymerases—including the mammalian pol alpha and the pol IIIs of gram-negative and high G+C content gram-positive bacteria (4–6, 10, 11).
Considering their potential for further development as antibiotics (see below), we sought to determine whether the latest generation of soluble EMAUs (i.e., HE-EMAU, ME-EMAU, HP-EMAU, and MP-EMAU) faithfully retained the HPUra-like specificity for DNA replication and pol III target. We assessed their pol III specificities by exploiting E. coli pol III and calf thymus DNA pol alpha as relevant control enzymes. As the results shown in the rightmost columns of Table 4 indicate, the four compounds displayed HPUra-like specificity; none of them displayed significant activity against either of these test enzymes.
We next sought to determine if the N-3 hydroxyalkyl- and N-3 methoxyalkyl-EMAUs also conserve HPUra’s specificity for DNA replication in the intact cell. To this end, we exploited an experimental approach that we used previously to assess the pol III and DNA replication specificities of N2-(3,4-dichlorobenzyl)guanine, an even more distant guanine derivative of the HPUra pharmacophore (4). The experiment compared the effect of each agent at 100 μM with that of 100 μM HPUra on the incorporation of [3H]adenine into RNA and DNA (see the Materials and Methods section for experimental details). The results, expressed as percent inhibition of 3H incorporation into DNA:percent inhibition of 3H incorporation into RNA, were as follows: HPUra control, 88%:6%; HE-EMAU, 85%:5%; ME-EMAU, 88%:5%; HP-EMAU, 86%:8%; and MP-EMAU, 89%:4%. In sum, these results strongly suggest that the four hydrophilic N-3-substituted EMAUs and the HPUra molecule from which they evolved share the same level of target selectivity in vivo in the intact cell.
Effects of MP-EMAU and HP-EMAU on murine staphylococcal infection.
Given their relatively high aqueous solubilities and the potency of their bactericidal effects against the Smith strain of S. aureus (Fig. 2B), we selected the two propyl derivatives, MP-EMAU and HP-EMAU, for testing in a simple in vivo lethal infection model employing the same organism (details of model appear in the Materials and Methods section). Specifically, mice were infected by intraperitoneal injection of a fixed, lethal dose of bacteria, and 15 min thereafter, groups of 10 mice each were injected by the same route with (i) the agent to be tested, (ii) vehicle, or (iii) vancomycin as a positive control agent of known efficacy. Thereafter, each treatment group was monitored over a 3-day period. The results, expressed as the number of mice surviving at 3 days after infection, are summarized in Table 5.
TABLE 5.
Effects of MP-EMAU and HP-EMAU on lethal infection of mice by S. aureusa
| Treatment used (mg/kg) | No. of survivors at 3 days postinfection (total no. treated) |
|---|---|
| Vehicleb only | 1 (10) |
| Vancomycin (20) | 10 (10) |
| HP-EMAU (10) | 10 (10) |
| HP-EMAU (5) | 7 (10) |
| MP-EMAU (10) | 5 (10) |
| MP-EMAU (5) | 2 (10) |
Intraperitoneal infection followed by a single intraperitoneal injection of drug or drug vehicle; see the Materials and Methods section for experimental details.
Saline for vancomycin and 10% DMSO:90% peanut oil for EMAU derivatives.
As expected, all of the animals that received 20 mg of vancomycin per kg survived throughout the 3-day period, while 9 of the 10 control mice that received only vehicle died. Similarly, all animals treated with HP-EMAU at a dose of 10 mg/kg survived, and 7 of 10 animals treated with half that dose (5 mg/kg) survived. After they were treated with MP-EMAU at 10 mg/kg, only 5 of 10 animals survived, and for those treated at 5 mg/kg, survival (20%) was barely distinguishable from that seen for animals in the vehicle (control) group (10%).
What is the significance of this in vivo protective effect of HP-EMAU and MP-EMAU? This so-called “furry test tube” infection model in which animals are infected and treated in the same, intraperitoneal, compartment, is one of the simplest infection models available for use in antibiotic testing. We selected this specific model in an effort simply to “prove principle” of antibiotic potential, reasoning that any candidate agent which failed to display efficacy in this system was unlikely to show efficacy in a more valid, pharmacokinetically complex model exploiting infection at one site and administration of agent at a different enteral or parenteral site. These preliminary results strongly suggest that the N-3-substituted AUs, indeed, have considerable potential as antiinfectives—a potential which we intend to probe further with experiments employing more relevant infection models. To achieve this goal we presently are evaluating, in mice, the basic pharmacokinetics of HP-EMAU to permit its rational parenteral and oral application, and we are continuing to devise even-more-soluble and -potent AU derivatives through further manipulation of both the ring N-3 substituent of the base-pairing domain and substituents of the aryl ring.
Potential of pol III as an antibiotic target.
Like DNA topoisomerase, the target of the highly effective fluoroquinolones, pol III is an essential, rate-limiting component of the bacterial DNA replication machinery (10, 15, 22). Given the essential, replication-specific function of pol III and the results presented above for the AU class of pol III-specific inhibitors, we propose that the development of pol III-specific antibiotics is feasible. We are presently pursuing this proposition with two approaches. The first, which is noted above, is to continue pursuit of the anti-infective potential of our pol III-specific AU platform compounds. The second is to identify and characterize novel pol III-targeted molecules by exploiting high-throughput screening of other, unrelated compounds for activity in vitro against both the polC-specific pol III of the low G+C content gram-positive organisms and the dnaE-specific pol III of relevant gram-negative pathogens.
ACKNOWLEDGMENT
This work was supported by STTR phase I grant AI41260 from the National Institutes of Health.
REFERENCES
- 1.Barnes M H, Brown N C. Antibody to B. subtilis DNA polymerase III: use in the purification of enzyme and the study of its structure. Nucleic Acids Res. 1979;6:1203–1219. doi: 10.1093/nar/6.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes M H, Leo C, Brown N C. DNA polymerase III of gram-positive eubacteria is a zinc metalloprotein conserving an essential finger-like domain. Biochemistry. 1998;37:15254–15260. doi: 10.1021/bi981113m. [DOI] [PubMed] [Google Scholar]
- 3.Barnes M H, Tarantino P M, Spacciapoli P, Yu H, Brown N C, Dybvig K. DNA polymerase III of Mycoplasma pulmonis: isolation and characterization of the enzyme and its structural gene, polC. Mol Microbiol. 1994;13:843–854. doi: 10.1111/j.1365-2958.1994.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown, N. C. Unpublished data.
- 5.Brown N C, Dudycz L W, Wright G E. Rational design of substrate analogues targeted to selectively inhibit replication-specific DNA polymerases. Drugs Exp Clin Res. 1986;12:555–564. [PubMed] [Google Scholar]
- 6.Brown N C, Handschumacher R E. Inhibition of the synthesis of deoxyribonucleic acid in bacteria by 6-(p-hydroxyphenylazo)-2,4-dihydroxypyrimidine: metabolic studies in Streptococcus faecalis. J Biol Chem. 1966;241:3083–3089. [PubMed] [Google Scholar]
- 7.Brown N C, Gambino J J, Wright G E. Inhibitors of Bacillus subtilis DNA polymerase III. 6-(Arylalkylamino)uracils and 6-anilinouracils. J Med Chem. 1977;20:1186–1189. doi: 10.1021/jm00219a015. [DOI] [PubMed] [Google Scholar]
- 8.Butler M M, Dudycz L W, Khan N N, Wright G E, Brown N C. Development of novel inhibitor probes of DNA polymerase III based on dGTP analogs of the HPUra type: base, nucleoside, and nucleotide derivatives of N2-(3,4-dichlorobenzyl)guanine. Nucleic Acids Res. 1990;18:7381–7387. doi: 10.1093/nar/18.24.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements J E, D’Ambrosio J, Brown N C. Inhibition of Bacillus subtilis DNA polymerase III by phenylhydrazinopyrimidines: demonstration of a drug-induced DNA:enzyme complex. J Biol Chem. 1975;250:522–526. [PubMed] [Google Scholar]
- 10.Cozzarelli N R. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977;46:641–668. doi: 10.1146/annurev.bi.46.070177.003233. [DOI] [PubMed] [Google Scholar]
- 11.Flett F, Jungmann-Campello D, Mersinias V, Koh S, Godden R, Smith C. A ‘Gram-negative type’ DNA polymerase III is essential for replication of the linear chromosome of Streptomyces coelicolor A3(2) Mol Microbiol. 1999;31:949–958. doi: 10.1046/j.1365-2958.1999.01237.x. [DOI] [PubMed] [Google Scholar]
- 12.Hammond R A, Brown N C. Overproduction and purification of Bacillus subtilis DNA polymerase III. Protein Expr Purif. 1992;3:65–70. doi: 10.1016/1046-5928(92)90057-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y-P, Ito J. The hyperthermophilic bacterium Thermotoga maritima has two different classes of family C DNA polymerases: evolutionary implications. Nucleic Acids Res. 1998;26:5300–5309. doi: 10.1093/nar/26.23.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D R, McHenry C S. In vivo assembly of overproduced DNA polymerase III. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg A, Baker T. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 16.Love E, D’Ambrosio J, Dubnau D, Brown N C. Mapping of the gene specifying DNA polymerase III of Bacillus subtilis. Mol Gen Genet. 1976;144:313–321. doi: 10.1007/BF00341730. [DOI] [PubMed] [Google Scholar]
- 17.Mackenzie J M, Neville M N, Wright G E, Brown N C. Hydroxyphenylazopyrimidines: characterization of the active forms and their inhibitory action on a DNA polymerase from Bacillus subtilis. Proc Natl Acad Sci USA. 1973;70:512–516. doi: 10.1073/pnas.70.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacitti D F, Barnes M H, Li D H, Brown N C. Characterization and expression of Staphylococcus aureus polC, the structural gene for DNA polymerase III. Gene. 1995;165:51–56. doi: 10.1016/0378-1119(95)00377-i. [DOI] [PubMed] [Google Scholar]
- 19.Tarantino P M, Zhi C, Gambino J J, Wright G E, Brown N C. 6-Anilinouracil-based inhibitors of Bacillus subtilis DNA polymerase III: antipolymerase and antimicrobial structure-activity relationships based on substitution at uracil N-3. J Med Chem. 1999;42:2035–2040. doi: 10.1021/jm980693i. [DOI] [PubMed] [Google Scholar]
- 20.Tomasz A. Multiple-antibiotic-resistant pathogenic bacteria. N Engl J Med. 1994;330:1247–1251. doi: 10.1056/NEJM199404283301725. [DOI] [PubMed] [Google Scholar]
- 21.Trantolo D J, Wright G E, Brown N C. Inhibitors of Bacillus subtilis DNA polymerase III. Influence of modifications in the pyrimidine ring of anilino- and (benzylamino)uracils. J Med Chem. 1986;29:676–681. doi: 10.1021/jm00155a016. [DOI] [PubMed] [Google Scholar]
- 22.Wolfson J S, Hooper D C. Fluoroquinolone antimicrobial agents. Clin Microbiol Rev. 1989;2:378–424. doi: 10.1128/cmr.2.4.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright G E, Brown N C. The mechanism of inhibition of B. subtilis DNA polymerase III by 6-(arylhydrazino)pyrimidines: novel properties of 2-thiouracil derivatives. Biochim Biophys Acta. 1976;432:37–48. doi: 10.1016/0005-2787(76)90039-3. [DOI] [PubMed] [Google Scholar]
- 24.Wright G E, Brown N C. Inhibitors of Bacillus subtilis DNA polymerase III. 6-Anilinouracils and 6-(alkylamino)uracils. J Med Chem. 1980;23:34–38. doi: 10.1021/jm00175a007. [DOI] [PubMed] [Google Scholar]
- 25.Wright G E, Brown N C. Deoxyribonucleotide analogs as inhibitors and substrates of DNA polymerases. Pharmacol Ther. 1990;47:447–497. doi: 10.1016/0163-7258(90)90066-b. [DOI] [PubMed] [Google Scholar]
- 26.Wright G E, Gambino J J. Quantitative structure-activity relationships of 6-anilinouracils as inhibitors of Bacillus subtilis DNA polymerase III. J Med Chem. 1984;27:181–185. doi: 10.1021/jm00368a013. [DOI] [PubMed] [Google Scholar]