Abstract
Objectives
The antimicrobial resistance of Helicobacter pylori (H. pylori) in most countries and regions has increased significantly. It has not been fully confirmed whether the detection of H. pylori resistance gene mutation can replace antibiotic drug sensitivity test to guide the clinical personalized treatment. The objective of this study was to assess and compare the efficacy of different antimicrobial resistance-guided quadruple therapies in refractory H. pylori-infected individuals who had undergone unsuccessful prior eradication treatments.
Methods
From January 2019 to February 2020, genotypic and phenotypic resistances were determined by polymerase chain reaction (PCR), whole genome sequencing (WGS) and broth microdilution test, respectively, in 39 H. pylori-infected patients who have failed eradication for at least twice. The patients were retreated with bismuth quadruple therapy for 14 days according to individual antibiotic resistance results. Eradication status was determined by the 13C-urea breath test.
Results
The overall eradication rate was 79.5% (31/39, 95% CI 64.2–89.5%) in the intention-to-treat (ITT) analysis and 88.6% (31/35, 95% CI 73.5–96.1%) in the per- protocol analysis (PP) analysis. The presence of amoxicillin resistance (OR, 15.60; 95% CI, 1.34–182.09; p = 0.028), female sex (OR, 12.50; 95% CI, 1.10–142.31; p = 0.042) and no less than 3 prior eradication treatments (OR, 20.25; 95% CI, 1.67–245.44; p = 0.018), but not the methods for guiding therapy (p > 0.05) were associated with treatment failure. Resistance-guided therapy achieved eradication rates of more than 80% in these patients. The eradication rate of H. pylori in the phenotypic resistance-guided group was correlated well with genotype resistance-guided groups, including PCR and WGS.
Conclusion
Culture or molecular method guiding therapy can enable personalized, promise salvage treatments, and achieve comparably high eradication rates in patients with refractory H. pylori infection. The detection of H. pylori resistance mutations has a good clinical application prospect.
Protocol Study Register
[clinicaltrials.gov], identifier [ChiCTR1800020009].
Keywords: Helicobacter pylori, antibiotic resistance, mutations, whole-genome sequencing, eradication
Introduction
Helicobacter pylori (H. pylori) is a Gram-negative microaerophilic bacterium that persistently colonizes the stomach of approximately 50% of the world’s population, equivalent to approximately 4.4 billion people (O’Connor et al., 2017). This infection establishes lifelong chronic progressive gastric inflammation, leading to a stepwise progression through gastric atrophy, intestinal metaplasia, and dysplasia, to the development of carcinoma (McColl, 2010). In the absence of an effective vaccine, treatment of chronic H. pylori infection has emerged as the main strategy for preventing subsequent gastric cancer development (Ford et al., 2014; Choi et al., 2018; Doorakkers et al., 2018).
Bismuth quadruple therapy, containing two antibiotics, plus bismuth and proton pump inhibitor (PPI) for 14 days, is recommended as a first-line empirical treatment for H. pylori infections by the Masstricht V Consensus (Malfertheiner et al., 2017). However, curing H. pylori infection has been proved remarkably difficult as the cure rates of empirical treatments are often <70% due to the increasing antimicrobial resistance (Malfertheiner et al., 2012; López-Góngora et al., 2015). And personalized treatment could be the main direction in the future, as it can significantly enhance the eradication rate, diminish the abuse of antibiotics, and avoid secondary antibiotic resistance (Fallone et al., 2016; Chey et al., 2017). Consequently, evaluating the resistance of H. pylori to drugs is of major clinical importance for guiding decisions about appropriate therapies in individuals and treatment policies in populations.
Susceptibility of H. pylori to antibiotics can be assessed by culture-based or molecular-based drug susceptibility testing (DST) techniques. Culture-based techniques are the standard DST for H. pylori and provide in vitro phenotypic susceptibly information (Miura and Hokari, 2012). However, this method is hampered by a relatively high rate of false negative, ranging from 45 to 90% (Mégraud and Lehours, 2007). Growth of H. pylori can be affected by many environmental factors and determination of the minimum inhibitory concentration (MIC) for H. pylori is cumbersome with a long turn-around-time, which limits their clinical application (Kjøller et al., 1991; Giorgio et al., 2016). Molecular-based methods rely mainly on the detection of specific H. pylori mutations encoding resistance. Globally, polymerase chain reaction (PCR)-based techniques have been developed to shorten the turn-around time and provide rapid detection of genotypic resistance in H. pylori (Cattoir et al., 2007). Besides, next-generation sequencing (NGS) refers to technologies that enable massively parallel sequencing (of DNA or RNA) to provide high-throughput genetic data at relatively low cost. Consequently, NGS would be applied in combination with culture for whole-genome sequencing (WGS), which is a powerful tool for antibiotic resistance prediction in H. pylori (Nezami et al., 2019).
However, in view of the correlation between genotypic resistance and phenotypic resistance and clinical treatment effect have not yet been fully elucidated, especially in China. Hence, we tried to further verify their consistency and the clinical application prospect of molecular techniques (PCR and WGS) guided eradication therapy. We conducted this trial by testing the resistance of antibiotics in vitro, detecting the mutation in the antibiotic resistance site by PCR and WGS, following up the efficacy of resistance-guided clinical eradication therapy, and analyzing the general data (demography, clinical diagnosis, and medical history, etc.) of the patients with refractory H. pylori infection.
Materials and Methods
Study Design and Population
This prospective, open-label trial was conducted in Wuhan Union Hospital and Pathogen Microbiology Laboratory, Tongji Medical College, Huazhong University of Science and Technology (clinicaltrials.gov identifier: ChiCTR1800020009). Between January 2019 and February 2020, a total of 39 H. pylori-infected patients who failed at least twice were prospectively enrolled in this study. The study protocol was approved by the Institutional Review Board of Tongji Medical College, Huazhong University of Science and Technology. Written informed consent was obtained from all patients prior to enrollment. We searched published works from PubMed, MEDLINE and the Cochrane Library for the terms “H. pylori,” “quadruple therapy,” “third-line,” “phenotypic resistance,” “genotypic resistance,” “polymerase chain reaction,” and “next generation sequencing” with no language or date limits. No publication of clinical trials that assessed and compared the efficacy of phenotypic resistance-guided and genotypic resistance-guided quadruple therapy in the at least third-line treatment of H. pylori infection was identified.
Eligibility Criteria and Randomization
Patients were excluded from the study if any of the following criteria was present: (i) children and teenagers aged <18 years old; (ii) history of gastrectomy or non-curable malignancy; (iii) contraindication or previous allergic reaction to proton pump inhibitors (PPI), bismuth, or antibiotics (clarithromycin, amoxicillin, metronidazole, levofloxacin, furazolidone, and tetracycline); (iv) pregnant or lactating women or severe concurrent diseases; (v) any of the three methods for detecting antibiotic resistance failed. Before H. pylori eradication, patients were randomly assigned to three groups using a random-number chart: (i) treated according to culture-based antibiotic susceptibility testing; (ii) treated according to PCR-based testing; (iii) treated according to WGS.
Determination of Helicobacter pylori Status
Before enrollment, the status of H. pylori infection was determined by the 13C-urea breath test (13C-UBT) or 14C-urea breath test (14C-UBT). Patients with either positive 13C-UBT or positive 14C-UBT at least twice were defined as refractory to previous treatment and were eligible for enrollment. Oesophago-gastro-duodenoscopy with biopsies from gastric antrum (two pieces for H. pylori culture, one pieces for PCR test and necessary number of pieces for histology) were performed for all patients. After treatment, H. pylori status was determined by 13C-UBT or 14C-UBT ≥4 weeks after the completion of eradication therapy. Successful eradication of H. pylori was defined as a negative 13C-UBT or 14C-UBT result. All patients were asked to stop proton pump inhibitors (PPI) for ≥2 weeks and antibiotics or bismuth for ≥4 weeks before endoscopy examination. Positive and negative results of 13C-UBT were defined according to the results of previous study as a cut-off value of ≥5 and <2.5‰, respectively (Chen et al., 2003). Patients with uncertain results received another 13C-UBT until the result was conclusive.
Determination of Phenotypic and Genotypic Resistance
Phenotypic Resistance: Culture-Based Drug Susceptibility Testing
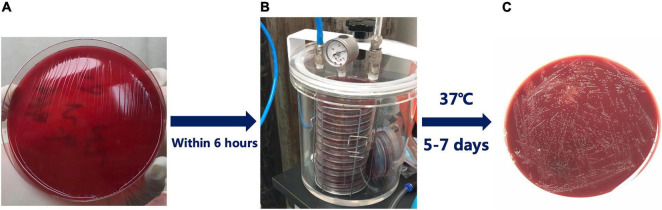
The biopsy specimens were cultured on chocolate agar plates containing 10% sheep blood and incubated under microaerobic conditions (5% O2, 10% CO2, and 85% N2) for 5–7 days (Figure 1). Phenotypic resistance was determined by the broth microdilution test if strains were available. H. pylori was inoculated onto antibiotic-containing broth medium supplemented with 5% defibrinated sheep blood. H. pylori ATCC 26695 was used as the quality control strain. The MIC of each antibiotic was determined after 72 h of incubation. The breakpoints for amoxicillin, clarithromycin, levofloxacin, tetracycline, metronidazole, and furazolidone resistance were defined as ≥0.5, ≥1, ≥1, ≥1, ≥8, and ≥2 μg/mL, respectively (Midolo et al., 1997; Liou et al., 2010; Chung et al., 2017). Each experiment was performed in triplicate, and experiments were repeated at least three times per strain.
FIGURE 1.
Isolation of Helicobacter pylori (H. pylori) from clinical biopsy specimens. (A) The biopsy specimens were applied to chocolate agar plates containing 10% sheep blood; (B) Sealing device for culturing H. pylori; (C) The transparent color colonies of H. pylori after successful culture.
Genotypic Resistance: Polymerase Chain Reaction-Based Assays
The DNA of H. pylori was extracted from gastric biopsy tissues using DNA extraction kit (Gentra DNA Purification Kit, QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The gene fragment of H. pylori correlated with different antibiotics was amplified by PCR with specific primers and then sequenced (Table 1). As established by previous studies, the presence of the specific variants is listed (Supplementary Table 1). All PCR mixtures were prepared in a final volume of 25 μL containing 50 ng DNA from the samples served as the template for PCR performed in a thermal cycler (Master cycler gradient, Eppendorf, Germany), 10 μM of each primer, 8.5 μL ddH2O, 2 μL template DNA, 12.5 μL Mix of Taq DNA polymerase. Thermal amplification of PCR products was performed at 94°C for 4 min and then for 35 cycles of 94°C for 1 min, 55°C for 40 s, 72°C for 1 min, and a final extension at 72°C for 7 min, with a final hold at 10°C in a PCR thermal cycler (Master cycler gradient). The PCR amplified products (10 μL) were subjected to electrophoresis in 1.5% agarose gel in 1 × TBE buffer at 80 V for 30 min stained with a solution of ethidium bromide (EMD Millipore, Billerica, MA, United States). And examined under ultraviolet illumination (Cleaver Scientific Ltd., Rugby, United Kingdom). The genotypic information of H. pylori ATCC 26695 was used as reference.
TABLE 1.
Polymerase chain reaction primers used for detecting mutations of H. pylori strains in this study.
| Antibiotics | Genes | Primer sequence (5′–3′) |
| clarithromycin | 23S rRNA | F: ACAGCACTTTGCCAACTCGTAA R: GCTTGTGCCATTACACTCAACTTG |
| amoxicillin | PBP-1A | F: GCCATTCTTATCGCTCAAGTTTGG R: ATCGCTAAAATGTTACGCATGAAATACG |
| levofloxacin | gyrA | F: TGGGGATTGATTCTTCTATTGAAGA R: TGCACTAAAGCGTCTATGATTTCA |
| Tetracycline | 16S rRNA | F: TCCGTAGAGATCAAGAGGAATACTCATTG R: TCACCGCAACATGGCTGATTTG |
| furazolidone | oorD | F: GGCTTGCCTGGAAATCCTGTAG R: AACCGATTTGCTCCACTTTCAATGA |
| porD | F: GCAAGAAGTCATTGACGC R: GGGGTGATAGGATAGGCT |
|
| metronidazole | rdxA | F: GCAGGAGCATCAGATAGT R: GGGTGATTTCTTGGTTGC |
Genotypic Resistance: Library Preparation and Whole Genome Sequencing of Helicobacter pylori Strains
Library preparation was performed using the Qiagen® QIAseq FX DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Furthermore, next-generation sequencing (MiSeq next-generation sequencer; Illumina, San Diego, CA, United States) was used to analyze all strains for mutation status of 23S rRNA, PBP-1A, gyrA, 16S rRNA, oorD, and porD genotypes. The BLAST algorithm implemented in the CLC Genomics Workbench software (ver. 11; Qiagen NV, Venlo, Netherlands) was used for the analysis. The sequences of hp0425, hp0597, hp0701, hp0374, hp0954, oorD, and porD of the strain 26695 (GenBank accession number AE000511.1) were used as queries to obtain the 23S rRNA, PBP-1A, gyrA, 16S rRNA, rdxA, oorD, and porD sequences, respectively, from the next-generation sequencing data. Subsequently, each codon of the strains was compared to the reference sequence hp 26699 using our original PERL script and confirmed by visual inspection. Discrepancies found between the strains and reference sequence, which was included in the variants of Supplementary Table 1, was considered as variants related to antibiotic resistance.
Susceptibility-Guided Intervention and Assessment of Adverse Effects
Treatment recommendations based on susceptibility testing were given on request in line with guideline from the fifth national consensus report on the treatment of H. pylori infection (Liu et al., 2018; Table 2). Six antibiotics are available for use in combined therapies in H. pylori eradication regimes: clarithromycin, amoxicillin, levofloxacin, metronidazole, tetracycline, and furazolidone. Patients were treated with quadruple therapy containing 30 mg of lansoprazole, 200 mg of pectin bismuth and 2 combined antibiotics for 14 days, according to the phenotypic or genotypic resistance determined using biopsy specimens or isolated strains. If the determined antibiotics for H. pylori sensitivity were less than 2 or not included in the regimens (Table 2), the patients would be treated empirically according to their previous medication history to avoid the reuse of unnecessary antibiotics. Patients were informed of the common adverse effects and were asked to record their symptoms during treatment.
TABLE 2.
Recommended combined regimens of H. pylori in China.†
| Regimen‡ | Drug 1 | Drug 2 |
| 1 | AMX, 1000 mg, bid | CLR, 500 mg, bid |
| 2 | AMX, 1000 mg, bid | LVX, 500 mg, qd/200 mg, bid |
| 3 | AMX, 1000 mg, bid | FZD, 100 mg, bid |
| 4 | TET, 500 mg, tid/qid | MTZ,400 mg, tid/qid |
| 5 | TET, 500 mg, tid/qid | FZD, 100 mg, bid |
| 6 | AMX, 1000 mg, bid | MTZ,400 mg, tid/qid |
| 7 | AMX, 1000 mg, bid | TET, 500 mg, tid/qid |
CLR, clarithromycin; AMX, amoxicillin; LVX, levofloxacin; MTZ, metronidazole; TET, tetracycline; FZD, furazolidone. Qd, once daily; bid, twice daily; tid, three-times daily; qid, four-times daily.
†Table adapted from the fifth national consensus report on the treatment of Helicobacter pylori infection, Chin J. Gastroenterol., 2017, Vol. 22, No. 6.
‡Standard dose (PPI + bismuth) (bid, orally half an hour before a meal) + 2 antibacterial drugs (orally after a meal). The standard dose of PPI is lansoprazole 30 mg; the standard dose of bismuth is 200 mg of pectin bismuth.
Statistical Analysis
The statistical analyses were performed using the SPSS 26.0 statistical software for Windows. Categorical data were compared using the Chi-square test or Fisher’s exact test, as appropriate. Continuous data were compared with Student’s t-test and expressed as the mean (SD). The kappa coefficient was used to assess the agreement between genotypic resistance and phenotypic resistance. Logistic regression analysis was performed to analyze factors affecting the eradication rates. The statistical significance level was set at 0.05. And odds ratios (ORs) and 95% confidence intervals (CIs) were calculated.
Results
Baseline Characteristics of Patients
Thirty-six H. pylori strains were isolated from 39 patients, and eventually 35 patients were analyzed for H. pylori eradication as a result of 1 follow-up loss (Figure 2). The culture-based DST was successfully determined in all of them with broth microdilution testing. The genotypic resistance was successfully determined using PCR tests as well as WGS in the same thirty-six patients above. There were 17 male patients and 19 female patients. The average age of the patients was 47 years old, ranging from 27 to 62 years old. Of the 36 patients with adequate information regarding their medication history, 100% (36/36) received clarithromycin and amoxicillin in their previous eradication regimens, compared with 11.1% (4/36), 8.3% (3/36), 2.8% (1/36), and 16.7% (6/36) for levofloxacin, metronidazole, tetracycline, and furazolidone, respectively. The baseline characteristics of patients included in the study are summarized (Table 3). Among three groups, all of baseline characteristics are of no significance.
FIGURE 2.
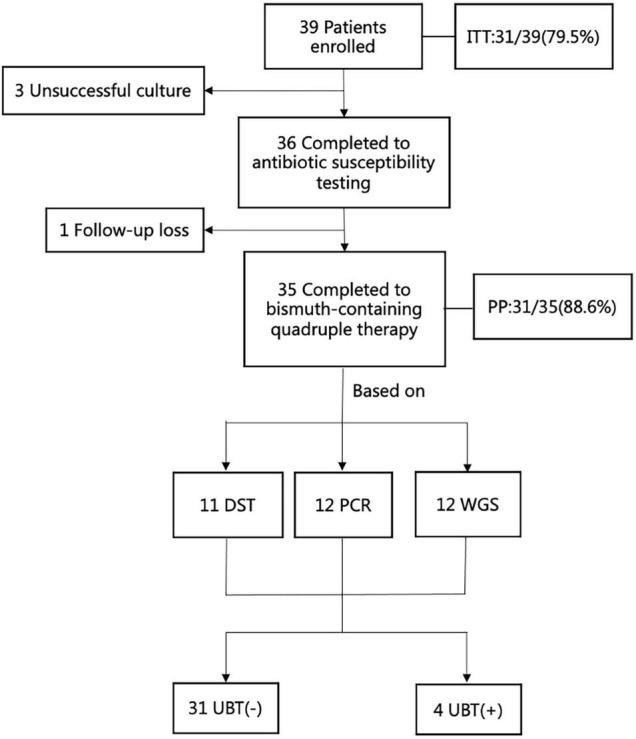
The flow chart of enrollment in this study. ITT, intention -to-treat; PP, per-protocol; UBT, urea breath test; DST, drug susceptibility testing; PCR, polymerase chain reaction; WGS, whole genome sequencing.
TABLE 3.
Baseline characteristics of patients included in the study (n = 36).
| Characteristic | Basis for treatment |
P value* | ||
| DST (n = 12), n (%) | PCR (n = 12), n (%) | WGS (n = 12), n (%) | ||
| Age (years), mean (range) | 42 (27–62) | 43 (26–60) | 45 (29–63) | 0.137 |
| Gender, male | 5 (41.67) | 7 (58.33) | 5 (41.67) | 0.640 |
| Current smoker | 3 (25.00) | 5 (41.67) | 2 (16.67) | 0.381 |
| Endoscopic findings | 0.449 | |||
| peptic ulcer | 2 (16.67) | 3 (25.00) | 0 (0.00) | |
| intestinal metaplasia | 4 (33.33) | 3 (25.00) | 4 (33.33) | |
| atrophic gastritis | 3 (25.00) | 4 (33.33) | 6 (50.00) | |
| others | 3 (25.00) | 2 (16.67) | 2 (16.67) | |
| Number of prior eradication treatments | 0.074 | |||
| 2 | 10 (83.33) | 9 (75.00) | 5 (41.67) | |
| ≥3 | 2 (16.67) | 3 (25.00) | 7 (58.33) | |
| Prior treatment | ||||
| clarithromycin | 12 (100.0) | 12 (100.0) | 12 (100.0) | NA |
| amoxicillin | 12 (100.0) | 12 (100.0) | 12 (100.0) | NA |
| levofloxacin | 1 (8.33) | 2 (16.67) | 1 (8.33) | 0.766 |
| metronidazole | 1 (8.33) | 0 (0.00) | 2 (16.67) | 0.336 |
| tetracycline | 1 (8.33) | 0 (0.00) | 0 (0.00) | 0.324 |
| furazolidone | 1 (8.33) | 4 (33.33) | 1 (8.33) | 0.183 |
| Virulence factors | ||||
| cagA positive | 12 (100.0) | 11 (91.67) | 11 (91.67) | 0.432 |
| ureA/ureB positive | 12 (100.0) | 12 (100.0) | 12 (100.0) | NA |
DST, drug susceptibility testing; PCR, polymerase chain reaction; WGS, whole genome sequencing; cagA, cytotoxin-associated gene A; vacA, gene encoding vacuolating cytotoxin A.
*All P values were for testing the difference between DST, PCR, and WGS subgroups and were calculated by Student’s t test or Fisher’s exact test.
Prevalence of Antibiotic Resistance
As all patients had undergone prior unsuccessful eradication treatments, the proportions of H. pylori antimicrobial resistant to clarithromycin, levofloxacin and metronidazole were markedly high and associated with the number of treatment failures (Table 4). However, none of the strains tested was resistant to furazolidone or tetracycline. And the prevalence of resistance to clarithromycin, levofloxacin, amoxicillin, and metronidazole was 91.7% (33/36), 50.0% (18/36), 25.8 (8/36), 41.7 (15/36), respectively. Markedly, more double resistant H. pylori isolates were observed in these patients (91.7%) (Table 4).
TABLE 4.
Antimicrobial resistance of H. pylori isolated from 36 patients with unsuccessful prior eradication treatments.
| Resistant to | Number of prior eradication treatments |
Overall (n = 36) | ||||
| 2 (n = 24) |
≥3 (n = 12) |
|
|
|||
| n | % | n | % | n | % | |
| CLR* | 21 | 87.5 | 12 | 100.0 | 33 | 91.7 |
| LVX** | 11 | 45.8 | 7 | 58.3 | 18 | 50.0 |
| MTZ*** | 9 | 37.5 | 6 | 50.0 | 15 | 41.7 |
| AMX** | 5 | 20.8 | 3 | 25.0 | 8 | 25.8 |
| CLR/LVX† | 10 | 41.7 | 6 | 50.0 | 16 | 44.4 |
| CLR/MTZ† | 7 | 29.2 | 4 | 33.3 | 11 | 30.6 |
| CLR/AMX† | 1 | 4.2 | 2 | 16.7 | 3 | 8.3 |
| AMX/MTZ† | 2 | 8.3 | 1 | 8.3 | 3 | 8.3 |
| CLR/LVX/AMX†† | 0 | 0.0 | 1 | 100.0 | 1 | 2.8 |
| CLR/LVX/MTZ/AMX††† | 0 | 0.0 | 1 | 100.0 | 1 | 2.8 |
CLR, clarithromycin; AMX, amoxicillin; LVX, levofloxacin; MTZ, metronidazole.
*Resistance included monoresistance, double resistance, triple resistance, and quadruple resistance.
**Resistance included double resistance, triple resistance, and quadruple resistance.
***Resistance included double resistance and quadruple resistance.
†Double resistance.
††Triple resistance.
†††Quadruple resistance.
The mutant genes of H. pylori isolated from 36 patients with unsuccessful prior eradication treatments according to PCR or WGS were shown (Table 5). The proportions of mutant 23S rRNA, PBP-1A, gyrA, and rdxA using PCR were 88.9% (32/36), 22.2% (8/36), 47.2% (17/36), and 44.4% (16/36), respectively. While the proportions of mutant 23S rRNA, PBP-1A, gyrA, and rdxA using whole genome sequencing were 81.7% (33/36), 22.2% (8/36), 50.0% (18/36), and 50.0% (18/36), respectively. None of mutations were found either in 16S rRNA gene or oorD/porD gene (Table 5).
TABLE 5.
Mutant genes of H. pylori isolated from 36 patients with unsuccessful prior eradication treatments.
| Antibiotic | Mutant genes | Genotypic resistance testing | |
|
| |||
| PCR, n (%) | WGS, n (%) | ||
| Clarithromycin | 23S rRNA | 32 (88.9) | 33 (91.7) |
| Amoxicillin | PBP-1A | 8 (22.2) | 8 (22.2) |
| Levofloxacin | gyrA | 17 (47.2) | 18 (50.0) |
| Metronidazole | rdxA | 16 (44.4) | 18 (50.0) |
| Tetracycline | 16S rRNA | 0 (NA) | 0 (NA) |
| Furazolidone | oorD/porD | 0 (NA) | 0 (NA) |
PCR, polymerase chain reaction; WGS, whole genome sequencing.
The agreement between culture-based DST results and identified point mutations by PCR or WGS was shown (Table 6), conferring resistance to CLR, LVX, AMX, and MTZ. The genotypic resistance determined using PCR in biopsy specimens correlated well with the phenotypic DST determined using strains for both clarithromycin and levofloxacin, amoxicillin, as well as metronidazole (k > 0.80) (Table 6). Overall, there was a comparatively high congruence of >90% between phenotypic DST results for clarithromycin, levofloxacin, amoxicillin, and metronidazole and SNPs identified by WGS in the 23S rRNA, gyrA, PBP-1A, and rdxA genes.
TABLE 6.
Correlations between phenotypic resistance by DST and genotypic resistance determined by PCR and WGS.
| Phenotypic | Genotypic |
|||||||
|
|
23S rRNA (PCR) |
P * | k coefficient | 23S rRNA (WGS) |
P * | k coefficient | ||
| CLR MIC | W | M | W | M | ||||
| S | 3 | 0 | <0.001 | 0.842 | 3 | 0 | <0.001 | 1.000 |
| R | 1 | 32 | 0 | 33 | ||||
|
|
||||||||
| LVX MIC |
gyrA (PCR) |
P * | k coefficient |
gyrA (WGS) |
P * | k coefficient | ||
| W | M | W | M | |||||
|
| ||||||||
| S | 17 | 1 | <0.001 | 0.833 | 18 | 0 | <0.001 | 0.944 |
| R | 2 | 16 | 1 | 17 | ||||
|
|
||||||||
| AMX MIC |
PBP-1A (PCR) |
P * | k coefficient |
PBP-1A (WGS) |
P * | k coefficient | ||
| S | R | W | M | |||||
|
| ||||||||
| S | 27 | 1 | <0.001 | 0.839 | 28 | 0 | <0.001 | 0.916 |
| R | 1 | 7 | 1 | 7 | ||||
|
|
||||||||
| MTZ MIC |
rdxA (PCR) |
P * | k coefficient |
rdxA (WGS) |
P * | k coefficient | ||
| S | R | S | R | |||||
|
| ||||||||
| S | 20 | 1 | <0.001 | 0.943 | 18 | 3 | <0.001 | 0.833 |
| R | 0 | 15 | 0 | 15 | ||||
DST, drug susceptibility testing; PCR, polymerase chain reaction; WGS, whole genome sequencing; MIC, minimum inhibitory concentration; CLR, clarithromycin; AMX, amoxicillin; LVX, levofloxacin; MTZ, metronidazole; S, susceptible; R, resistant; W, wild type; M, mutant.
*All P values were calculated by Fisher’s exact test.
Eradication Rates and Factors Affecting the Efficacy
In the present study, four antibiotic combination regimens were applied in the quadruple therapy, TET + FZD, TET + MTZ, AMX + FZD, and AMX + LVX, respectively. Moreover, AMX + FZD was the main regimen used in the quadruple therapy for patients in the DST and WGS groups, while AMX + LVX was the main regimen used in the quadruple therapy for patients in the PCR group (Table 7). The overall eradication rate was 79.5% (31/39, 95% CI 64.2–89.5%) in the ITT analysis and 88.6% (31/35, 95% CI 73.5–96.1%) in the PP analysis. Factors possibly related to eradication failure are summarized (Table 8). Furthermore, the four patients with failed eradication consisted of one from DST group, two from PCR group and one from WGS group with treatment regimens of TET + FZD, TET + MTZ, AMX + LVX, and TET + MTZ, respectively. In univariate analysis, there were statistically significant clinical factors associated with eradication failure, including male: female ratio (p = 0.042), number of prior eradication treatments (p = 0.018) and AMX resistance (p = 0.028). Besides, we also found no statistical difference in eradication failure rates related to age, cigarette smoking, alcohol intake, diabetes, hypertension, high salt diet, family history of GC, method for guiding therapy, endoscopic findings, CLR resistance, LVX resistance, MTZ resistance, CagA, and VacA (p > 0.05). It is note-worthy that the eradication rate based on DST, PCR and WGS was 90.9% (10/11), 83.3% (10/12), and 91.7% (11/12), respectively, which indicated that genotypic resistance-guided therapy might achieve satisfactory results as same as phenotypic resistance-guided therapy. Furthermore, logistic regression analysis revealed that female sex (OR, 12.50; 95% CI, 1.10–142.31; p = 0.042), not less than 3 prior eradication treatments (OR, 20.25; 95% CI, 1.67–245.44; p = 0.018) and AMX resistance (OR, 15.60; 95% CI, 1.34–182.09; p = 0.028) were significantly associated with eradication failure (Table 9).
TABLE 7.
Antibiotic combination regimens in three groups of patients in the quadruple therapy.
| Regimen | Basis for treatment | Overall (n = 36), n (%) | ||
|
| ||||
| DST (n = 12), n (%) | PCR (n = 12), n (%) | WGS (n = 12), n (%) | ||
| TET + FZD | 1 (8.3) | 2 (16.7) | 1 (8.3) | 4 (11.1) |
| TET + MTZ | 1 (8.3) | 1 (8.3) | 2 (16.7) | 4 (11.1) |
| AMX + FZD | 7 (58.3) | 3 (25.0) | 6 (50.0) | 16 (44.4) |
| AMX + LVX | 3 (25.0) | 6 (50.0) | 3 (25.0) | 12 (33.3) |
AMX, amoxicillin; LVX, levofloxacin; MTZ, metronidazole; TET, tetracycline; FZD, furazolidone. DST, drug susceptibility testing; PCR, polymerase chain reaction; WGS, whole genome sequencing.
TABLE 8.
Clinical characteristics in eradication success and failure groups (n = 35).
| Factor | Success group (n = 31), n (%) | Failure group (n = 4), n (%) | P value* |
| Age, yr. | 42.6 ± 11.0 | 42.5 ± 9.7 | 0.993 |
| Male: Female ratio | 25:6 (80.6:19.4) | 1:3 (25.0:75.0) | 0.044 |
| Cigarette smoking | 9 (29.0) | 1 (25.0) | 0.681 |
| Alcohol intake | 19 (38.7) | 1 (25.0) | 0.522 |
| Diabetes | 3 (9.7) | 0 (0.0) | 0.687 |
| Hypertension | 8 (25.8) | 1 (25.0) | 0.732 |
| High salt diet | 19 (61.3) | 2 (50.0) | 0.530 |
| Family history of GC | 2 (6.5) | 0 (0.0) | 0.782 |
| Method for guiding therapy | 0.788 | ||
| DST | 10 (32.3) | 1 (25.0) | |
| PCR | 10 (32.3) | 2 (50.0) | |
| WGS | 11 (31.4) | 1 (25.0) | |
| Number of prior eradication treatments | 0.019 | ||
| 2 | 27 (87.1) | 1 (25.0) | |
| ≥3 | 4 (12.9) | 3 (75.0) | |
| Endoscopic findings | 0.478 | ||
| Non-atrophic | 11 (35.5) | 2 (50.0) | |
| Atrophic | 20 (64.5) | 2 (50.0) | |
| CLR resistance† | 29 (93.5) | 3 (75.0) | 0.313 |
| AMX resistance† | 5 (16.1) | 3 (75.0) | 0.030 |
| LVX resistance† | 10 (55.6) | 2 (50.0) | 0.632 |
| MTZ resistance† | 9 (60.0) | 3 (75.0) | 0.525 |
| CagA | 0.218 | ||
| Negative | 1 (3.2) | 1 (25.0) | |
| Positive | 30 (96.8) | 3 (75.0) | |
| VacA | 0.732 | ||
| m1 | 8 (25.8) | 1 (25.0) | |
| m2/m1 + m2 | 23 (74.2) | 3 (75.0) |
GC, gastric cancer; DST, drug susceptibility testing; PCR, Polymerase chain reaction; WGS, whole genome sequencing; CLR, clarithromycin; AMX, amoxicillin; LVX, levofloxacin; MTZ, metronidazole; cagA, cytotoxin-associated gene A; vacA, gene encoding vacuolating cytotoxin A.
*All P values were for testing the difference between Success group and failure group and were calculated by Student’s t test or Fisher’s exact test.
†The resistance determined by antibiotic susceptibility testing.
TABLE 9.
Logistic regression analysis for Helicobacter pylori eradication failure.
| Variable | Unadjusted OR | 95% CI | P value* |
| Female sex | 12.50 | 1.10–142.31 | 0.042 |
| Not less than 3 prior eradication treatments | 20.25 | 1.67–245.44 | 0.018 |
| AMX resistance† | 15.60 | 1.34–182.09 | 0.028 |
OR, odds ratio; CI, confidence interval; AMX, amoxicillin.
*All P values were calculated by logistic regression analysis.
†The resistance determined by antibiotic susceptibility testing.
The Side Effects of Helicobacter pylori Eradication
Of the 35 participants, 15 participants (42.9%) complained of adverse events after 14 days of quadruple therapy. The side effects were not significantly different between the success group and failure group (Supplementary Table 2). In addition, the combination of TET and FZD was significantly associated with side effects of H. pylori eradication (p = 0.026), but the presence of TET and FZD alone was not associated with the development of adverse reactions (TET + MTZ, p = 0.581; AMX + FZD, p = 0.557) (Supplementary Table 3).
Discussion
The work presented above lead to several novel findings relevant to the optimization of refractory H. pylori eradication. This was the first study to compare the prevalence of refractory antibiotic resistance of six antibiotics in H. pylori by using phenotypic DST and PCR-based assays as well as WGS-based assays at the same time. We further showed that resistance-guided modified quadruple therapy was effective (>80%) in the treatment of refractory H. pylori infection. More importantly, the eradication rates appeared to be same in patients treated with genotypic resistance-guided therapy as compared with those treated with phenotypic resistance-guided therapy. Besides, we found that not less than 3 prior eradication treatments, the presence of amoxicillin resistance and female sex were associated with treatment failure in patients treated with resistance-guided quadruple therapy.
In the present study, we have demonstrated that the genotypic resistance determined using gastric biopsy specimens as well as WGS correlated well with the phenotypic resistance determined in H. pylori strains. We could show a clear correlation between the occurrence of mutations in the 23S rRNA, gyrA, PBP-1A, and rdxA genes of H. pylori and clarithromycin, fluoroquinolone, amoxicillin, and metronidazole resistance, respectively. In a prospective study, Konrad also found that clarithromycin and levofloxacin gene resistance is consistent with the phenotypic resistance of H. pylori, which was consistent with our results (Egli et al., 2020). Few molecular diagnostic tests were generally performed for detecting the tetracycline or furazolidone resistance on gastric tissue samples because this may be of a little significance for initial antibiotic selection at the beginning of therapy, owing to the well documented low resistance rate to tetracycline/furazolidone (Fiorini et al., 2018; Palmitessa et al., 2020). And in this study, interestingly, neither phenotypic resistance nor genotypic resistance appeared in tetracycline and furazolidone, which demonstrated that it might be reasonable to use tetracycline and furazolidone empirically even in patients who have failed multiple treatment.
During this study, we realized that the efficacies appeared to be similar in patients treated with therapies guided by three types of testing as p value was of no significance among them. The successful cultivation from gastric biopsy specimens and DST are the gold standards for the phenotypic sensitivity test of H. pylori, which provide reliable information for the development of personalized clinical programs (Gerrits et al., 2006). However, this method is challenging due to the pathogen’s fastidious growth requirements for laboratory environment, taking up to at least 10 days to obtain results (Gerrits et al., 2006). Besides, culture-based methods could be hampered by a few technical factors, such as quality of the clinical specimen, occurrence of microbial commensal flora and inappropriate transport conditions (Cuadrado-Lavín et al., 2012). And most clinical laboratories, especially small and medium-sized hospitals in China, could not carry out this clinical project on a large scale, and most patients are not willing to wait for 2 weeks to be treated. Therefore, more rapid, and convenient molecular detection technology is ready. The determination of genotypic resistance using biopsy specimens is more expedient (culture not needed) and speedier, and it is easier to transfer the specimen (Cui et al., 2021) and has a higher success rate compared with traditional susceptibility tests (100.0 versus 92.3% in the present study).
Moreover, WGS delivers a more complete picture of resistance determinants present in a clinical isolate than PCR that can only examine a limited number of nucleotide positions (Binh et al., 2015). And the relevance of new polymorphisms can easily be assessed by later retrospective analysis of WGS data (Binh et al., 2014). However, WGS is obviously costlier than qPCRs. Anyway, genotypic resistance-guided quadruple therapies might be practical strategies in the treatment of refractory H. pylori infection in future clinical practice.
We demonstrated that female sex, not less than 3 prior eradication treatments and the presence of AMX resistance were associated with treatment failure in patients treated with resistance-guided quadruple therapy. However, the use of AMX under resistance guidelines remains acceptable in the third-line treatment of AMX-sensitive patients, as the rate of resistance to AMX remained relatively low in patients who have failed at least two eradication therapies. Besides, patients with AMX resistant can use a combination regimen containing TET with a lower resistance rate under resistance guidance. Recommended antimicrobial combinations for bismuth quadruple regimens include: (i) TET + MTZ; (ii) TET + FZD; (iii) TET + LVX. In addition, Vonoprazan, a new potassium competitive acid blocker, can be used to replace PPI. Vonoprazan is stable, highly efficacious, and less affected by CYP2C19 gene polymorphism, which is beneficial to improve H. pylori eradication rate (Akazawa et al., 2016). There are several reports that female gender can influence H. pylori eradication (Binh et al., 2014, 2015). One study suggested that there might be a difference in gastric physiology between males and females (Akazawa et al., 2016). Our study showed that the female gender is an unfavorable factor affecting eradication. However, the cause of gender differences in the eradication rate needs further research. Besides, data has shown that after just one unsuccessful therapy, resistance to clarithromycin rose to 60%, and further vain treatment attempts resulted in resistance as high as 80% (Wüppenhorst et al., 2014; Yahaghi et al., 2014; Thung et al., 2016). Although we found the efficacy of eradication was affected by these three factors, the results should be validated in further studies because of the wide CIs, which indicated small case numbers for these three variables.
However, this study had some limitations. First, it was a single-arm prospective research which did not include the relatively large sample size in third-line therapy. Second, the prevalence of CLR and AMX resistance is higher than average status because of nearly all of patients enrolled had prior treatments in Wuhan Union Hospital, where the empirically first-line regimen included antibiotic combination of CLR and AMX. Third, we should focus on predicating new point mutations of drug resistance in H. pylori based on the bacterium’s genome using next generation sequencing (NGS) technology in the future.
In conclusion, the results from this study show that genotypic resistance-guided quadruple therapy can achieve comparably high eradication rates as compared with phenotypic resistance-guided therapy in the treatment of refractory H. pylori infection. The detection of H. pylori resistance genes could be a good clinical application in the eradication of H. pylori.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Tongji Medical College, Huazhong University of Science and Technology. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
RL and ZG designed the project. ST and WW supplied samples. ZG performed all experiments, collected, and analyzed data. YZ and JL followed the participants. ZG, ST, and WW drafted and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our special thanks to Institute of Microbiology, Chinese Academy of Sciences for supporting this study. We thank all participants in this study, staff at the Endoscopy Center of Wuhan Union Hospital and without their support, this work would not have been possible.
Abbreviations
- H. pylori
Helicobacter pylori
- PPI
proton pump inhibitor
- DST
drug susceptibility testing
- MIC
minimum inhibitory concentration
- PCR
polymerase chain reaction
- NGS
next-generation sequencing
- WGS
whole-genome sequencing
- 13C-UBT
13C-urea breath test
- 14C-UBT
14C-urea breath test
- ORs
odds ratios
- CIs
confidence intervals
- CLR
clarithromycin
- AMX
amoxicillin
- LVX
levofloxacin
- TET
tetracycline
- FZD
furazolidone.
Funding
This study was supported in part by the National Natural Science Foundation of China (No. 81272656, 81974068, and 81572428), National Key Research and Development Program of China (No. 2017YFC0110003), and Natural Science Foundation of Hubei Province of China for Distinguished Young Scholar (No. 2017CFA061).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.861626/full#supplementary-material
References
- Akazawa Y., Fukuda D., Fukuda Y. (2016). Vonoprazan-based therapy for Helicobacter pylori eradication: experience and clinical evidence. Therap Adv. Gastroenterol. 9 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binh T. T., Shiota S., Suzuki R., Matsuda M., Trang T. T., Kwon D. H., et al. (2014). Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J. Antimicrob Chemother. 69 1796–1803. 10.1093/jac/dku050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binh T. T., Suzuki R., Trang T. T., Kwon D. H., Yamaoka Y. (2015). Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother. 59 2343–2348. 10.1128/AAC.04852-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V., Nectoux J., Lascols C., Deforges L., Delchier J. C., Megraud F., et al. (2007). Update on fluoroquinolone resistance in Helicobacter pylori: new mutations leading to resistance and first description of a gyrA polymorphism associated with hypersusceptibility. Int. J. Antimicrob Agents 29 389–396. 10.1016/j.ijantimicag.2006.11.007 [DOI] [PubMed] [Google Scholar]
- Chen T. S., Chang F. Y., Chen P. C., Huang T. W., Ou J. T., Tsai M. H., et al. (2003). Simplified 13C-urea breath test with a new infrared spectrometer for diagnosis of Helicobacter pylori infection. J. Gastroenterol. Hepatol. 18 1237–1243. 10.1046/j.1440-1746.2003.03139.x [DOI] [PubMed] [Google Scholar]
- Chey W. D., Leontiadis G. I., Howden C. W., Moss S. F. (2017). ACG clinical guideline: treatment of Helicobacter pylori infection. Am. J. Gastroenterol. 112 212–239. 10.1038/ajg.2016.563 [DOI] [PubMed] [Google Scholar]
- Choi I. J., Kook M. C., Kim Y. I., Cho S. J., Lee J. Y., Kim C. G., et al. (2018). Helicobacter pylori therapy for the prevention of metachronous gastric Cancer. N. Engl. J. Med. 378 1085–1095. 10.1056/NEJMoa1708423 [DOI] [PubMed] [Google Scholar]
- Chung J. W., Kim S. Y., Park H. J., Chung C. S., Lee H. W., Lee S. M., et al. (2017). In vitro activity of diphenyleneiodonium toward multidrug-resistant Helicobacter pylori strains. Gut Liver 11 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado-Lavín A., Salcines-Caviedes J. R., Carrascosa M. F., Mellado P., Monteagudo I., Llorca J., et al. (2012). Antimicrobial susceptibility of Helicobacter pylori to six antibiotics currently used in Spain. J. Antimicrob Chemother. 67 170–173. 10.1093/jac/dkr410 [DOI] [PubMed] [Google Scholar]
- Cui R., Song Z., Suo B., Tian X., Xue Y., Meng L., et al. (2021). Correlation analysis among genotype resistance, phenotype resistance and eradication effect of Helicobacter pylori. Infect Drug Resist. 14 1747–1756. 10.2147/IDR.S305996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorakkers E., Lagergren J., Engstrand L., Brusselaers N. (2018). Helicobacter pylori eradication treatment and the risk of gastric adenocarcinoma in a Western population. Gut 67 2092–2096. [DOI] [PubMed] [Google Scholar]
- Egli K., Wagner K., Keller P. M., Risch L., Risch M., Bodmer T. (2020). Comparison of the diagnostic performance of qPCR, sanger sequencing, and whole-genome sequencing in determining clarithromycin and levofloxacin resistance in Helicobacter pylori. Front. Cell Infect Microbiol. 10:596371. 10.3389/fcimb.2020.596371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallone C. A., Chiba N., van Zanten S. V., Fischbach L., Gisbert J. P., Hunt R. H., et al. (2016). The toronto consensus for the treatment of Helicobacter pylori infection in adults. Gastroenterology 151 51–69.e14. [DOI] [PubMed] [Google Scholar]
- Fiorini G., Zullo A., Saracino I. M., Pavoni M., Vaira D. (2018). Antibiotic resistance pattern of Helicobacter pylori strains isolated in Italy during 2010-2016. Scand. J. Gastroenterol. 53 661–664. [DOI] [PubMed] [Google Scholar]
- Ford A. C., Forman D., Hunt R. H., Yuan Y., Moayyedi P. (2014). Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. BMJ 348:g3174. 10.1136/bmj.g3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits M. M., van Vliet A. H., Kuipers E. J., Kusters J. G. (2006). Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 6 699–709. 10.1016/S1473-3099(06)70627-2 [DOI] [PubMed] [Google Scholar]
- Giorgio F., Ierardi E., Sorrentino C., Principi M., Barone M., Losurdo G., et al. (2016). Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand. J. Gastroenterol. 51 1429–1432. 10.1080/00365521.2016.1216592 [DOI] [PubMed] [Google Scholar]
- Kjøller M., Fischer A., Justesen T. (1991). Transport conditions and number of biopsies necessary for culture of Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 10 166–167. 10.1007/BF01964450 [DOI] [PubMed] [Google Scholar]
- Liou J. M., Lin J. T., Chang C. Y., Chen M. J., Cheng T. Y., Lee Y. C., et al. (2010). Levofloxacin-based and clarithromycin-based triple therapies as first-line and second-line treatments for Helicobacter pylori infection: a randomised comparative trial with crossover design. Gut 59 572–578. 10.1136/gut.2009.198309 [DOI] [PubMed] [Google Scholar]
- Liu W. Z., Xie Y., Lu H., Cheng H., Zeng Z. R., Zhou L. Y., et al. (2018). Fifth chinese national consensus report on the management of Helicobacter pylori infection. Helicobacter 23:e12475. 10.1111/hel.12475 [DOI] [PubMed] [Google Scholar]
- López-Góngora S., Puig I., Calvet X., Villoria A., Baylina M., Muñoz N., et al. (2015). Systematic review and meta-analysis: susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J. Antimicrob Chemother. 70 2447–2455. 10.1093/jac/dkv155 [DOI] [PubMed] [Google Scholar]
- Malfertheiner P., Megraud F., O’Morain C. A., Atherton J., Axon A. T., Bazzoli F., et al. (2012). Management of Helicobacter pylori infection–the Maastricht IV/florence consensus report. Gut 61 646–664. 10.1136/gutjnl-2012-302084 [DOI] [PubMed] [Google Scholar]
- Malfertheiner P., Megraud F., O’Morain C. A., Gisbert J. P., Kuipers E. J., Axon A. T., et al. (2017). Management of Helicobacter pylori infection-the maastricht V/Florence consensus report. Gut 66 6–30. 10.1136/gutjnl-2016-312288 [DOI] [PubMed] [Google Scholar]
- McColl K. E. (2010). Clinical practice. Helicobacter pylori infection. N. Engl. J. Med. 362 1597–1604. [DOI] [PubMed] [Google Scholar]
- Mégraud F., Lehours P. (2007). Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 20 280–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midolo P. D., Bell J. M., Lambert J. R., Turnidge J. D., Grayson M. L. (1997). Antimicrobial resistance testing of Helicobacter pylori: a comparison of Etest and disk diffusion methods. Pathology 29 411–414. 10.1080/00313029700169415 [DOI] [PubMed] [Google Scholar]
- Miura S., Hokari R. (2012). Seeking an optimal eradication therapy for Helicobacter pylori infection. J. Gastroenterol. Hepatol. 27 7–9. 10.1111/j.1440-1746.2011.06953.x [DOI] [PubMed] [Google Scholar]
- Nezami B. G., Jani M., Alouani D., Rhoads D. D., Sadri N. (2019). Helicobacter pylori mutations detected by next-generation sequencing in formalin-fixed, paraffin-embedded gastric biopsy specimens are associated with treatment failure. J. Clin. Microbiol. 57:e01834-18. 10.1128/JCM.01834-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor A., O’Morain C. A., Ford A. C. (2017). Population screening and treatment of Helicobacter pylori infection. Nat. Rev. Gastroenterol. Hepatol. 14 230–240. [DOI] [PubMed] [Google Scholar]
- Palmitessa V., Monno R., Panarese A., Cuppone R., Burattini O., Marangi S., et al. (2020). Evaluation of antibiotic resistance of Helicobacter pylori strains isolated in bari, southern italy, in 2017-2018 by phenotypic and genotyping methods. Microb Drug Resist. 26 909–917. 10.1089/mdr.2019.0262 [DOI] [PubMed] [Google Scholar]
- Thung I., Aramin H., Vavinskaya V., Gupta S., Park J. Y., Crowe S. E., et al. (2016). Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol. Ther. 43 514–533. 10.1111/apt.13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüppenhorst N., Draeger S., Stüger H. P., Hobmaier B., Vorreiter J., Kist M., et al. (2014). Prospective multicentre study on antimicrobial resistance of Helicobacter pylori in Germany. J. Antimicrob Chemother. 69 3127–3133. 10.1093/jac/dku243 [DOI] [PubMed] [Google Scholar]
- Yahaghi E., Khamesipour F., Mashayekhi F., Safarpoor Dehkordi F., Sakhaei M. H., Masoudimanesh M., et al. (2014). Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed. Res. Int. 2014:757941. 10.1155/2014/757941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.