Abstract
Background:
Although PD-1 inhibitors have revolutionized treatment for advanced melanoma, not all patients respond. We previously showed that inhibition of the flavoprotein renalase (RNLS) in pre-clinical melanoma models decreases tumor growth. We hypothesized that RNLS inhibition promotes tumor rejection by effects on the tumor micro-environment (TME).
Methods:
We employed two distinct murine melanoma models, studied in RNLS knock-out (KO) or wild-type (WT) mice. WT mice were treated with the anti-RNLS antibody, m28, with or without anti-PD-1. 10X single-cell RNA-sequencing was used to identify transcriptional differences between treatment groups, and tumor cell content was interrogated by flow cytometry. Samples from patients treated with immunotherapy were examined for RNLS expression by quantitative immunofluorescence.
Results:
RNLS knock-out mice injected with wild-type melanoma cells reject their tumors, supporting the importance of RNLS in cells in the TME. This effect was blunted by anti-CD3. However, macrophage-specific RNLS ablation was insufficient to abrogate tumor formation. Anti- RNLS antibody treatment of melanoma-bearing mice resulted in enhanced T cell infiltration and activation, and resulted in immune memory upon rechallenging mice with injection of melanoma cells. At the single cell level treatment with anti-RNLS antibodies resulted in increased tumor density of macrophages, neutrophils, and lymphocytes, and increased expression of IFNγ and granzyme B in NK cells and T cells. Intra-tumoral FOXP3+ CD4 cells were decreased. In two distint murine melanoma models, we showed that melanoma-bearing mice treated with anti-RNLS antibodies plus anti-PD-1 had superior tumor shrinkage and survival than either with either treatment alone. Importantly, in pre-treatment samples from patients treated with PD-1 inhibitors, high RNLS expression was associated with decreased survival (Log-rank P=0.006), independent of other prognostic variables.
Conclusions:
RNLS knock-out results in melanoma tumor regression in a T-cell dependent fashion. Anti-RNLS antibodies enhance anti-PD-1 activity in two distinct aggressive murine melanoma models resistant to PD-1 inhibitors, supporting development of anti-RNLS-antibodies with PD-1 inhibitors as a novel approach for melanomas poorly responsive to anti-PD-1.
Keywords: Renalase, anti-PD-1, tumor immune microenvironment
INTRODUCTION
Tumor cells evade the immune system by using inhibitory signals to exhaust T cells or suppress their effector functions. Several molecules that mediate these inhibitory signals (immune checkpoints) have been identified. Program death 1 (PD-1), its ligand (PD-L1), and cytotoxic T- lymphocytes antigen 4 are the most extensively studied immune checkpoint molecules. They are expressed on chronically activated T cells and their blockade using specific antibodies activates tumor reactive T cells, enabling them to induce an anti-tumor response. Such therapies have achieved remarkable responses in subsets of patients with multiple tumor types, starting with melanoma, and are approved for clinical use for various indications. (1)
Although the incidence of melanoma continues to rise, the death rate from melanoma is finally falling, from 9320 in 2018 to 7230 in 2019 in the United States, due to approval of contemporary systemic therapie.(2, 3) Melanoma is relatively resistant to chemotherapy, and targeted therapy is only effective in the subset of melanomas (~40%) that harbor Braf mutations, particularly when combinations of BRAF and MEK inhibitors are used. (4, 5) Notably, while BRAF/MEK inhibitors are highly active in melanomas harboring BrafV600E mutations, responses are rarely durable. The major breakthrough in treating unresectable melanoma in terms of prolonging survival is due to immune checkpoint inhibitors (anti-CTLA4 or anti-PD-1, or the combination thereof), which stimulate cytotoxic T cells, resulting in tumor regression in subsets of melanoma patients, regardless of whether they harbor Braf mutations (4, 6–12). While responses to immune checkpoint inhibitors are more durable, they are less frequent than responses to targeted therapy. In the frontline setting, approximately 19% of patients respond to CTLA-4 inhibitors and 40% to PD-1 inhibitors (6–10, 13). PD-1 or PD-L1 inhibitors cause less toxicities than CTLA-4 inhibitors and are therefore now widely viewed as the standard of care in the frontline setting, alone or in combination with other drugs. At 36 months of follow-up only 37% of patients treated with anti-PD-1 monotherapy remain in response and the majority develop resistance requiring additional therapy.(14) The combination of the CTLA-4 and PD-1 inhibitors results in a higher response rate (57.6%) and a 52% five-year survival rate, but is associated with a grade 3– 4 toxicity rate >50% (9) and (15). Therefore, less toxic and equally (or more) effective alternative therapeutic approaches are still needed for melanomas resistant to anti-PD-1 monotherapy.
To identify additional drug targets in melanoma, in our previous work we studied the role of the secreted flavoprotein renalase (RNLS) in melanoma (16). Many cells, including melanocytes, express RNLS (17–19). RNLS functions as a general survival factor that protects cells and organs against toxic and ischemic injury (20, 21). It promotes survival in cells exposed to ischemia or toxic injury by activating PI3K/AKT, JAK/STAT, and MAPK pathways (20). Its expression is higher in melanomas than nevi, and higher in metastatic than primary melanoma (16). Knock-down of RNLS by siRNA inhibits human melanoma cell survival in vitro. Inhibition of RNLS signaling using an anti-RNLS monoclonal antibody developed in our lab (m28-RNLS) in murine melanoma models led to dramatic decrease in tumor burden, both in melanoma and in pancreatic cancer (16, 22). Our mechanistic studies showed that RNLS is produced both by tumor cells and by tumor associated macrophages (TAM), and suggest that increased RNLS production by tumor promoting CD163+ TAM facilitates melanoma growth by activating STAT3 in tumor cells (16).
To expand on these findings, with the future goal of developing drugs to inhibit RNLS in patients with advanced melanoma resistant to PD-1 inhibitors, our purpose was to determine how RNLS inhibition promotes tumor rejection. We hypothesized that RNLS inhibition results in tumor rejection primarily via its effects on the tumor immune-microenvironment, and sought to determine whether RNLS inhibition might enhance anti-tumor effects of immune-checkpoint inhibitors in murine melanomas resistant to these therapies.
MATERIALS AND METHODS
Cell lines:
B16-F10 cells were purchased from ATCC. YUMM 1.7 cells were generated in house. (23) Both were shown to express RNLS using the Taqman method. Relative to GAPDH, levels of RNLS were 8.26±0.37 fold higher in B16F10 cells probing for exons 3–4 and 11.19±0.28 probing for exons 4–5. The levels relative to GAPDH in YUMM1.7 for these probes were 3.69±0.19 and 5.25±0.23, respectively (data not shown). shRNA knock-down of RNLS: Wild-type (WT) B16F10 cells were seeded in 12-well plates with complete medium on day 1. Medium was removed, and complete medium plus polybrene (5ug/ml) was added to the wells. Control and RNLS shRNA lentiviral particles were added to the mixtures, and incubated with the cells overnight. The medium mixture was replaced with complete medium on day 3, and the cells were again incubated overnight. On day 4, the cells were split 1:5 and incubated for 48 before undergoing two weeks of puromycin (10ug/ml) stable clone colony selection. The selected colonies were expanded, and evaluated using RTqPCR to determine the effect of shRNA on gene expression.
Murine studies:
RNLS KO models:
WT C57B6 and RNLS-KO (backcrossed to C57B6 >20 generations) mice were bred and housed in the Veterans Affairs of Connecticut Healthcare System (VACHS) facility, and treated with approval of the Institutional Animal Care and Use Committee of VACHS.
Tumor cell injections and treatments:
We established allograft tumors by subcutaneous injection of B16-F10 or YUMM1.7 (5×105 in 100μl PBS). Control mice were treated with rabbit IgG, 40 μg by intraperitoneal injection (IP) once weekly, and 40 μg subcutaneously (SQ) around the tumor site every 3 days,. Experimental groups received the anti-RNLS antibody m28-RNLS (40 μg IP, once weekly, and 40 μg SQ, every 3 days) (16). In experiments using immne checkpoint inhibitors, mice received 60 μg m28-RNLS, 100 μg anti-PD1 (Bio X Cell, Cat # BE0146), or 70 μg anti-CTLA4 (Bio X Cell, Cat # BP0032) every 3 days. Tumor size was measured with digital calipers and volume calculated by (length width2)xπ/2. A syngeneic metastatic model was established by tail vein injection of B16-F10 or YUMM1.7 melanoma cells into C57B6 or RNLS- KO mice. Two weeks later, gross metastatic disease burden was evaluated.
A syngeneic metastatic model was established by injecting into the tail vein of the control mouse B16-F10 melanoma cell line or Sh-RNLS B16F10 into either C57B6 or RNLS KO mice. Two weeks later, the gross metastatic disease burden evaluated, and lung tissue was collected for further study.
Toxicity studies:
Blood pressure and heart rate meaturements: A blood pressure transducer (TA11PA-C10; commercially available through Data Sciences International) was implanted into the carotid artery of mice in order to measure blood pressure, heart rate, and diurnal patterns on a chronic basis. Male C57BL/6J mice 8–10 weeks of age were anesthetized with isoflurane (1– 3% in oxygen) and the blood pressure transducer catheter inserted into the left carotid artery under sterile surgical conditions. A subcutaneous tunnel was made to generate a pocket in the right lateral flank to accommodate the body of the transducer. Approximately 7 days after the surgery, once normal diurnal patterns were re-established, mice were singly housed, and the cages placed on receiver units connected to a computer to record blood pressure. Blood pressure was measured over a 10 second interval and recorded every minute. Mice (n=5 for each group) received either control rabbit IgG or m28 antibody (120 μg per mouse by IP injection) 3 times per week for 3 weeks. The average blood pressure over a 3-day period was calculated at baseline prior to treatment and after 3 weeks of antibody treatment.
At sacrifice, kidneys, livers, hearts, lungs and spleens of mice treated with rabbit IgG or m28 were harvested and assessed for histologic abnormalities.
MØ-specific RNLS-KO:
The RNLS conditional allele is based on the ‘knockout-first’ design, which combines advantages of a reporter-tagged and conditional mutation (24). Trapping cassettes include mouse En2 splice acceptor and SV40 polyadenylation sequences. The cassette is composed of an FRT site followed by a lacZ sequence and loxP site. The loxP site is followed by neomycin under control of the human β-actin promoter, SV40 polyA, a second FRT site and a second loxP site. A third loxP site is inserted downstream of exon 3 on RNLS. The RNLS-KO-first allele (C57BL/6N-Rnlstm1a(EUCOMM)Wtsi/Ieg) was modified by crossing to transgenic FLP and cre mice. Conditional alleles were generated by removal of the gene-trap cassette by Flp recombinase (crossing with 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J, lso Known As: ROSA26::FLPe knock), which reverts the mutation to wild type, leaving loxP sites on either side of exon 3. Exon 3 was deleted by crossing with mice that express Cre in myeloid cells due to targeted insertion of the cre cDNA into their endogenous M lysozyme locus(25).
Statistical analyses for murine studies:
R software was used. Differences between cell content in experimental models were assessed using the Chi-Square test for binarized variables or Student’s t-test for continuous variables. Tukey’s test was used for multiple comparisons. Log- rank statistics were used to determine associations between treatments and survival in animal experiments. Differences with p values of <0.05 were considered statistically significant.
Single-cell RNA sequencing:
Tumors were collected, weighed, dissected and incubated in 1.48U/mL Liberase DL enzyme mixture (Roche) and 200U/mL DNase I (Roche). Cell suspension was filtered and pelleted. Cells were resuspended in ACK Lysing Buffer (ThermoFischer) and washed in DMEM-2% FBS. Cell debris was removed using the Miltenyi Debris Removal Solution. Live cells were diluted to 106 cells/mL.
scRNA-seq library preparation and sequencing were conducted by the Yale Center for Genome Analysis. Single cells, reagents and a single Gel Bead containing barcoded oligonucleotides were encapsulated into nanoliter-sized Gel Bead in Emulsion (GEM) using GemCodeTM Technology (10X Genomics). cDNA libraries were constructed using the 10x ChromiumTM Single Cell 3’ Library Kit. Qualitative analysis was performed using the Agilent Bioanalyzer High Sensitivity DNA assay. Libraries were sequenced on an Illumina HiSeq-4000 sequencer. Cell Ranger version 3.1.0 was used to process Chromium single cell 3’ RNA-seq output and align the read to the mouse reference transcriptome (mm10–3.0.0).
Data analysis was performed using Seurat v3.1.5 R package (26), including cell type identification and comparative analyses between tumors from WT and RNLS-KO mice or from WT mice treated with m28-RNLS versus control IgG. Poor quality cells with <100 (likely cell fragment) or >6,000 (potentially cell duplet) uniquely expressed genes were excluded. Cells were excluded if mitochondrial gene percentages were >75% in samples of WT and conrol, or 25% in KO and m28-treated samples. Data were normalized and scaled with default settings in Seurat. The 2000 top variable genes were identified, and inputted for principle component analysis (PCA) for dimensionality reduction. We retained 20 leading principal components for further visualization and cell clustering. The t-distributed stochastic neighbor embedding (tSNE) projection (27) was used to visualize single cells on two-dimensional spaces with a perplexity of 100. The share nearest neighbor (SNN) graph was constructed by calculating the Jaccard index between each cell and its 20-nearest neighbors, which was then used for cell clustering based on the Louvain algorithm (with a resolution of 0.7). Each cluster was screened for marker genes by differential expression analysis based on the non-parameteric Wilcoxon rank sum test for all clusters with genes expressed in ≥10% of cells either inside or outside a cluster. Based on melanocyte and immune cell lineage-specific marker expression, cell clusters of melanocytes, B cells, T cells, NK cells, macrophages, and neutrophils were identified and preserved (Supplemental Figure 2), while cell clusters of erythroblast, smooth muscle cells, endothelial cells and keratinocytes were filtered out due to lack of relevance. For both WT and RNLS-KO, or Control and m28, differential expression analysis was performed for each cell type separately and visualized in violin plots.
Immune cell profiling:
Mice were euthanized and tumors excised, minced, and digested. Tumor cells were triturated. Viable cells were labeled with fluorescently conjugated antibodies. Flow cytometry was performed on Stratedigm-13 cytometer and data analyzed by FlowJo software (BD Bioscience).
Human tumor studies:
Patient Cohort and Tissue Microarray (TMA) Construction:
With approval of the Yale University Institutional Review Board, we identified 60 cutaneous or mucosal metastatic melanoma patients serially treated with anti-PD-1 therapy, alone or in combination with CTLA-4 inhibitors, between 2011 and 2017 at Yale Cancer Center with tissue available for coring. Specimens were reviewed by a board-certified pathologist (DR). Tumors were represented by up to four cores per tumor creating four TMA blocks. Clinicopathological data were collected from clinical records and pathology reports. Demographic and clinical information is summarized in Supplemental Table 12. This cohort has been used in previous publictaions.(28–30)
Immunofluorescence:
We determined the specificity of an anti-RNLS plyclonal antibody from R&D) using cells over- or under-expressing RNLS by Western blot (not shown). Immunofluorescent staining has previously been described. (31) Antigen retrieval for RNLS was performed in TRIS-EDTA (pH 9.0) using a pressure cooker. Anti-RNLS polyclonal goat IgG was diluted at 1:50 in 0.3% BSA-TBS and antibody detection was conducted with a secondary donkey anti goat at 1:1200 (SouthernBiotech). The signal was amplified using Cyanine 5 (Cy5)-tyramide (Perkin-Elmer) at 1:50. One slide was stained for the tumor mask using S100 polyclonal rabbit (DAKO) as described. (31) To determine whether RNLS was located in macrophages, a consecutive slide was stained with CD68 monoclonal rabbit antibody (abcam) at 1:600. Nuclei were stained on both slides using 1:50 DAPI (Thermo Fisher 1mg/ml).
Quantitation of immunofluorescent signal:
Methods for quantitating immunofluorescent signal have been extensively described.(32) We previously showed that RNLS is expressed in both tumor cells and macrophages. Seeing that macrophages are dispersed among tumor cells, each histospot was individually evaluated to determine the location of RNLS (primarily macrophages, primarily tumor cells or both), and RNLS expression was assessed in the total tumor (S100) mask. Most spots had insufficient CD68 cells to reliably determine RNLS levels solely within macrophages. Spots with insufficient tumor (<3% of the histospot area) or mechanically damaged were eliminated. Cy5 signal intensity within the mask was divided by the tumor mask area to generate a RNLS intensity score for subsequent analysis.
Statistical analyses:
JMP14 software were used. Associations between RNLS intensities within the tumor or CD68 compartment and clinical data were examined by the Wilcoxon rank test. Overall survival (OS) was defined from the date of initiation of immunotherapy (Figure 6) or date of diagnosis of metastatic disease (supplemental materials) to death or patient censoring. Survival estimates were depicted with the Kaplan-Meier method; differences were assessed with the stratified log-rank test. The Cox Proportional Hazed method was used to estimate the hazard ratio for OS. Results are expressed as means±standard error (SE). Comparisons between groups were made by using the Student’s t-test or f-test, where appropriate. Differences with p values of <0.05 were considered statistically significant.
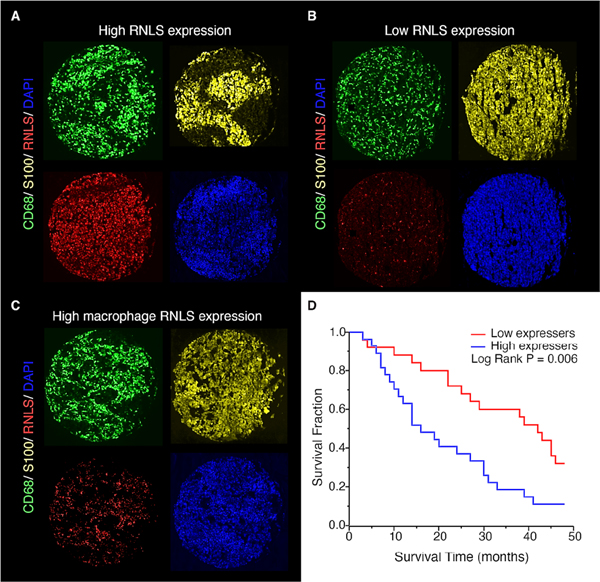
Figure 6.
RNLS expression in human tumors. Immunofluorescent staining showing RNLS expression (red), CD68+ macrophages (green) and S100+ melanoma cells (yellow), with examples from patients with high RNLS expression in both tumor cells and macrophages (A), low expression (B) and RNLS primarily in macrophages (C). High tumor RNLS expression (above the median) in melanoma patients treated with anti-PD-1 alone or with anti-CTLA-1 and anti-PD- 1 was associated with decreased survival (p=0.006, panel D).
RESULTS
RNLS inhibition or elimination leads to tumor regression in syngeneic mouse models of melanoma and promotes long-lasting anti-tumor immunity and memory
In previous work we established direct anti-tumor effects of RNLS inhibition, but our data suggested the primary mechanism of action of the anti-RNLS antibody, m28-RNLS, was via its effects on the TME. While some melanomas have T cell rich tumor microenvironments, others are T cell poor (33). The T-cell inflamed tumor subset contains CD8 T cells, along with macrophages, plasma cells and B cells (33). Importantly, the T-cell poor tumor subset is associated with a less favorable prognosis, and worse response to immunotherapeutic approaches, including immune checkpoint inhibitors (34).
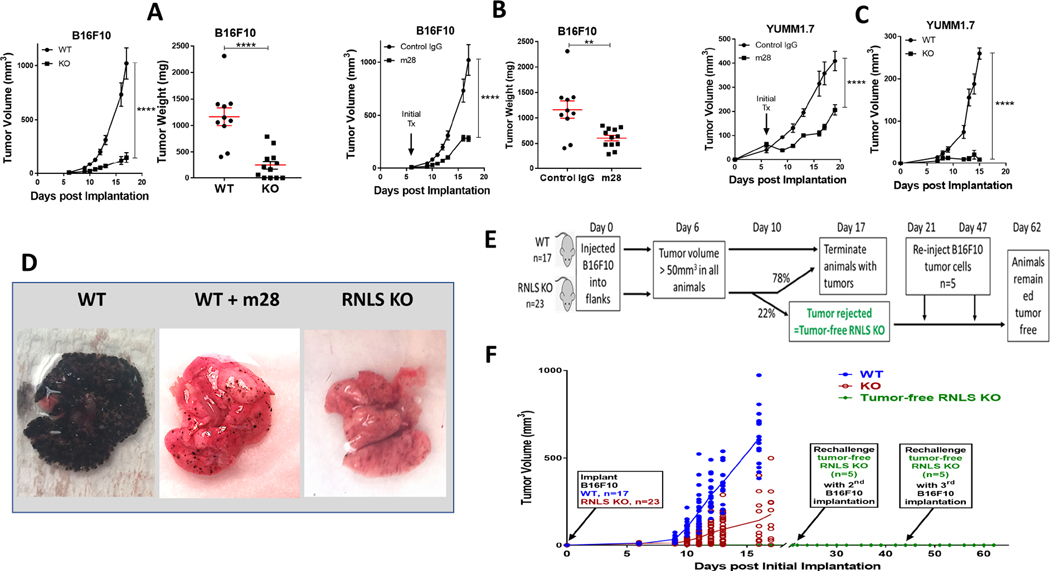
The B16-F10 mouse melanoma cell line is aggressive, highly metastatic, and forms a non- T-cell associated tumor subtype that is poorly immunogenic and resistant to anti-PD-1 (35, 36). To examine how inhibition of RNLS signaling modulates tumor shrinkage, we injected B16-F10 cells subcutaneously into RNLS knock-out (KO) C57B6 mice. We found large differences in tumor volume and weight in B16-F10-injected KO mice compared to wild-type (WT) mice, p<0.0001 (Figure 1A.) We studied effects of partial RNLS inhibition in both tumor and immune cells with m28-RNLS. Once tumors reached 50 mm3, animals were treated every 3 days with either control rabbit IgG or m28-RNLS. Treatment with m28-RNLS resulted in significant decreasea in tumor volume and weight at all points tested, p<0.0001 (Figure 1 B).
Figure 1.
Lack of RNLS and inhibition of RNLS leads to tumor regression and promotes long- lasting anttumor immunity and memory. WT and RNLS-KO mice were injected with B16F10 cells (A-B) or YUMM1.7 cells (C). Starting at Day 6, WT mice were injected with either control IgG or m28-RNLS (B) and tumor volume was measured (A-C). Tumors were isolated and weighed (A and B, right panels), ***denots p<0.0002; **** denotes p<0.0001. D: B16F10 cells were injected intravenously into WT or RNLS-KO mice. WT mice were treated with either PBS (left) or m28- RNLS; lungs were examined for metastasis. E: Schema of experiment demonstrating immune memory; F: Tumor growth curve: B16F10 cells were injected in WT (n=10) or RNLS-KO mice (n=23), tumors in 5 out of 23 RNLS-KO animals were rejected 10d post tumor implantation. These five RNLS-KO mice that had rejected their tumors were reinjected with B16F10 cells on Day 21 and Day 47 after initial B16F10 implantation. Mice were observed every other day for tumor formation and no tumor formed after the two rechallenges with tumor cell injections. WT animals were injected with the same batch of tumor cells at each rechallenge timepoint as a positive control (data not shown) and all tumors in WT mice grew.
YUMM1.7 is a non-immunogenic melanoma cell line derived from immune-competent mice genetically-engineered to develop melanoma through known drivers of human melanoma including Braf activation, Pten loss, Cdkn2a loss (BrafV600ECdkn2a−/−Pten−/−) (23). Similar to B16- F10, YUMM1.7 is insensitive to immune checkpoint inhibition, and remarkably tumorigenic. While the key driver mutations present in B16-F10 are not fully characterized, those of YUMM1.7 are well defined. We therefore verified our results in YUMM1.7 cells. As shown in Figure 1 C, volume of YUMM 1.7 tumors were also lower in either RNLS-KO mice or m28-RNLS treated mice, p<0.0001.
Effects of RNLS inhibition or knock-out were further demonstrated in a metastatic model, using tail vein injection of B16-F10 tumor cells injected into tail veins of RNLS-KO mice or C57B6mice, which received m28-RNLS or rabbit IgG for up to 2 weeks. Evaluation of lung surfaces revealed a major decrease in the burden of pulmonary metastatic disease in m28-RNLS-treated mice compared to control IgG (5.1% vs. 73.4% pulmonary tumor burden), n=5, p=0.01, (Figure 1D left and middle panels). We also observed a marked reduction in extra-pulmonary metastatic tumor burden. Treatment with m28RNLS limited disease spread to 1.25±0.15 non-pulmonary sites, compared to 4.25±.0.41 non-pulmonary sites in the control group (n=5, p=0.001), data not shown. Lung morphology of WT and RNLS-KO were indistinguishable in the absence of tumor. Similar to WT mice treated with m28-RNLS, and in contrast to untreated WT mice, RNLS-KO mice injected with B16-F10 had a dramatic reduction in lung metastases (Figure 1D, right panel).
Importantly, we observed a complete regression rate of 15–30% in both WT mice treated with m28-RNLS and RNLS-KO mice receiving no additional treatment. To test if tumor disappearance conferred durable anti-tumor immunity, RNLS-KO mice with complete tumor elimination after initial tumor formation (n=5) received B16-F10 injections every 2 weeks for two additional implantation cycles, as depicted in Figure 1E. Upon reinjection of tumor cells, none of the five mice developed tumor with either injection (Figure 1F), indicating successful generation of durable anti-tumor immunity.
To determine the relative importance of inhibiting tumor cell RNLS compared to RNLS in the tumor microenvironment, we inhibited RNLS expression in B16F10 cells using shRNA. Cells were then injected into the tail veins of RNLS WT mice, and compared to control cells injected with lentivirus (LV), tumor growth was somewhat impeded (green line, Supplemental Figure 1). However, when either cell line (with or without shRNA) was injected into RNLS KO mice, tumor growth was substantially impeded, and no clear difference was seen between cells containing the shRNA to RNLS versus those without (purple and red lines, Supplemental Figure 1). Images of lungs harvested from these animals are shown in the right hand panel.
RNLS knock-out or inhibition alters quantities and properties of various cellular components of the TME
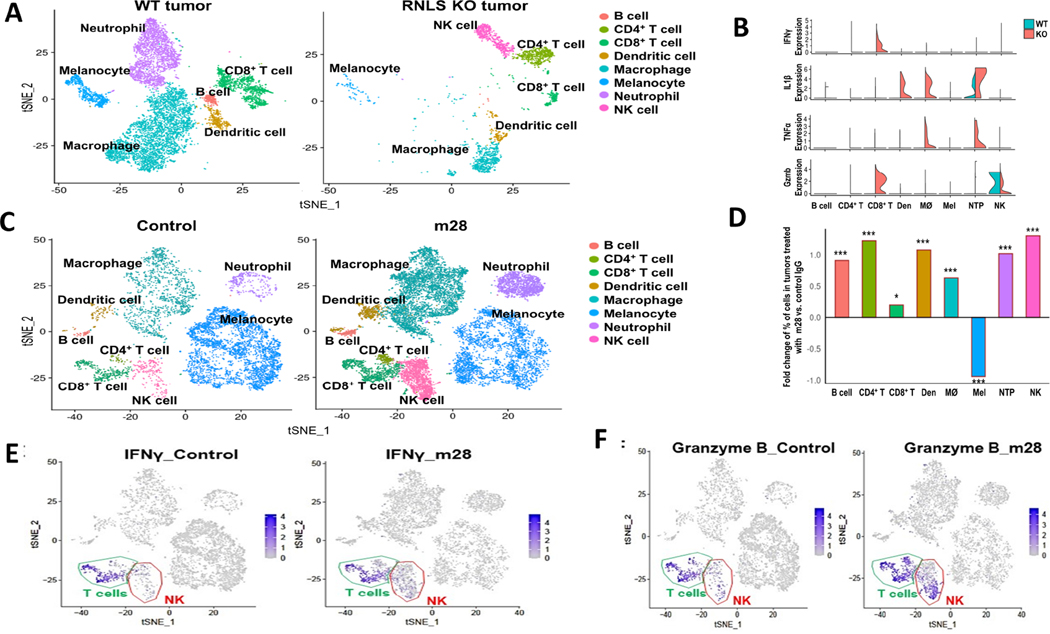
RNLS-KO mice provide an opportunity to specifically study effects of lack of RNLS in host cells, whereas m28-RNLS inhibits RNLS in tumor cells. To differentiate these effects, we conducted single-cell RNA sequencing. We sequenced cells from B16F10 tumors from WT mice and compared them to RNLS-KO mice, combining tumors from three mice due to small tumor size in RNLS-KO mice. The t-distributed stochastic neighbor embedding (t-SNE) plot in Figure 2A shows dramatic differences. RNLS-KO mice had smaller tumors, with less tumor cells and neutrophils. There was a clear increase in natural killer (NK cell content). Properties of macrophages and CD4 cells differ in tumors from RNLS-KO mice compared to WT, as evidenced by change of location on the two-dimensional plot. To further understand differences in these properties, we focused on expression of chemokines, cytokines and molecules involved in inflammation. As demonstrated in the examples in Figure 2B, tumors from RNLS-KO mice had markedly higher levels of granzyme B and interferon-gamma in CD8+ T cells and NK cells compared to tumors implanted in WT mice, supporting a role of T cells in tumor rejection. Macrophages and neutrophils were polarized to an inflammatory phenotype by RNLS-KO, as evidenced by increased TNFα and IL1β. Other examples of differentially expressed genes in RNLS-KO mice compared to WT mice are provided in Supplemental Figures 3–5. These include CD274 (PD-1), which was up-regulated in T cells, macrophages, NK cells and neutrophils in tumors from KO mice, increased CD40 expression, particularly in macrophages, increased PDCD1 and PTPRC (encoding leukocyte common antigen) in T cells and NK cells, increased CD44 expression, particularly in CD4 T cells and increased expression of the leukocyte-homing gene sell in NK cells and CD4 T cells. Complete tables of expressed genes are provided in the Supplemental Tables 1–11.
Figure 2.
Single cell RNA sequencing (scRNA-seq) of murine tumors demonstrates dramatic differences between conditions with and without RNLS. (A) B16F10 tumors from WT mice show decreased neutrophils and macrophages compared to KO mice, which have a relative abundance of NK cells, and different populations of macrophages, CD4+ and CD8+ T cells. (B) Violin plots demonstrating higher levels of granzyme B and IFNγ in CD8+ T cells in RNLS-KO mice compared to WT, increased TNFα and IL1β in macrophages and neutrophils, and activation of NK cells in RNLS-KO mice. (C-D) Treatment of WT mice bearing B16F10 tumors with m28-RNLS results in increased numbers of CD4+ and CD8+ T cells, NK cells, macrophages, neutrophils, dendritic cells (Den) and B cells. m28 treatment results in decreased numbers of melanocytes. Using the Chi-Square text, * < 5e-2, ** < 5e-3, *** < 5e-4. (E-F) tSNE plots showing that IFNγ and Granzyme B expression was primarily in T cells and increased with m28-RNLS treatment.
Employing markers that are commonly applied to determine activation, migration and exhaustion of T cells (37), we compared tumors injected in WT mice to RNLS KO mice, and found much higher levels of Pdcd1, CD44, LAG3 and CD160, all markers of exhaustion, in KO mice compared with WT mice, indicating ongoing CD8 activity and continuous stimulation resulting in exhaustion in the KO mice (Supplemental Figure 3). The scRNAseq data were further modified to include changes in dendritic cells. As shown in the plots in Figure 2A, dendritic cells form a small component of cells in the tumor microenvironment, and appear to include two subsets, which indeed change with RNLS elimination or inhibition. Select qualitative changes are now shown in Figure 2B, demonstrating large increases in IL-1β in tumors harvested from RNLS-KO mice. IL-1β plays a key role in early DC maturation and is important for IL-12 secretion by DCs (38–40). We assessed expression of markers of dendritic cell (DC) activation, and found that IRF7, the major transducer of type 1 interferon production in plasmacytoid DC (41) and Dock 2 expressing activated DCs (42) are more abundant in tumors from RNLS KO mice compared to WT mice, Supplemental Figure 4. To assess effects on CD4 cells, we found large increases in Sell expression, which was essentially absent in CD8 cells, providing evidence of CD4 cell migration and activation in the RNLS-KO mouse, Supplemental Figure 5 (43).
Given our interest in developing anti-RNLS antibodies as a treatment modality for melanomas with limited T-cell infiltration, we assessed effects of m28-RNLS on cells in the TME at the single cell level (Figure 2C). Treatment of mice with m28-RNLS resulted in increases in quantity of multiple immune cell types – macrophages, neutrophils. CD4+ and CD8+ T cells, NK cells and B cells. Increases are demonstrated in the bar graph in Figure 2D, and examples of changes in expression of individual inflammatory markers are provided in Supplemental Figure 6. Interestingly, treatment with m28-RNLS resulted in decreased FOXP3+ CD4 cells (Supplemental Figure 7), while levels of IFNγ and granzyme B were particularly increased in NK cells and T cells, Figure 2 E–F.
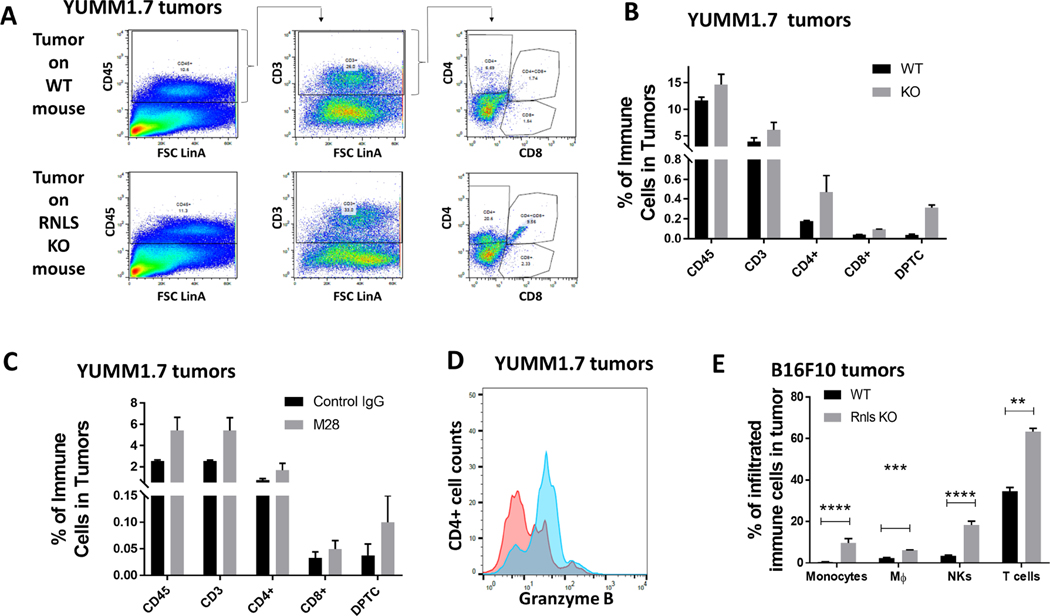
Effects on cells in the TME were confirmed at the protein level by flow cytometry. In YUMM 1.7 tumors harvested from RNLS-KO mice, we found an increase in CD45+ immune cells, with a particular increase in CD4+ cells and CD4/CD8 double positive cells (Figure 3A–B). Similar changes were seen with m28-RNLS treated cells in RNLS WT mice (Figure 3C). In addition to changes in the quantity of CD4+ T cells, we showed that these cells had increased cytotoxic properties, exemplified by increased Granzyme B expression in CD4+ cells (Figure 3D). Similar results were seen in B16F10 implanted tumors, showing increases in a variety of immune cell types, particularly CD4+ T cells and NK cells (Figure 3E).
Figure 3.
Protein level changes induced by RNLS knock-out or inhibition confirm changes seen with scRNA-seq: (A-B) Flow cytometry of T cells in YUMM1.7 tumors implanted on WT or RNLS- KO mice showing increased CD4 and CD8 content. (C) m28-RNLS treatment results in similar increases in T cell content, and CD4 T cell activation as shown by Granzyme B increases (D). Increases in monocytes, macrophages, NK and T cells were confirmed in B16F10 tumors at the protein level (E).
T cells are required for tumor rejection by RNLS elimination
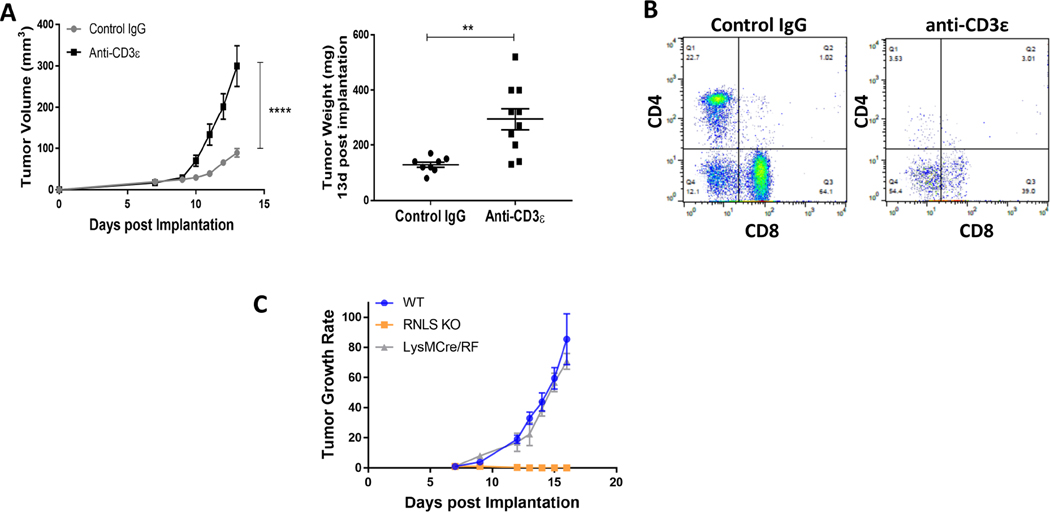
Given the marked increase in T cell content in tumors in RNLS-KO mice compared to wild- type, we hypothesized that T cells are critical for tumor rejection in these mice. Treatment of five RNLS-KO mice bearing B16F10 melanomas with anti-CD-3 antibodies resulted in marked reversal of tumor inhibition compared with five control IgG-treated mice (Figure 4A). As expected, in the presence of anti-CD3 antibodies, quantities of CD4+ and CD8+ T cells were diminished compared to control IgG-treated mice (Figure 4B). We then asked whether deleting RNLS expression in the myeloid lineage alone was sufficient to promote tumor rejection. We created a MØ and neutrophil-specific RNLS-KO and confirmed the absence of RNLS expression in macrophages by qPCR and Western blot (not shown). Growth of B16-F10 tumors in LysMcre- RNLS-KO mice was compared to that in WT and RNLS-KO animals. Global deletion of RNLS expression abolished tumor growth, while that in macrophages conferred no observable protection and tumor size and growth rate in these mice were similar to untreated WT mice (Figure 4C). This indicates that deletion of RNLS in myeloid cells alone is insufficient to confer protection against B16-F10 tumor growth.
Figure 4.
T cells, but not macrophages, are required for anti-tumor activity of RNLS inhibition. (A) Treatment of RNLS-KO mice bearing B16F10 tumors with control IgG results in tumor rejection (grey curve, N=5 mice). This effect is abrogated with anti-CD3 (yellow curve, N=5 mice). (B) Tumors harvested from these mice treated with anti-CD3 have lower CD4 and CD8 cell content, as expected. (C) Macrophage specific RNLS-KO (grey curve) is insufficient to significantly impact tumor growth compared to global RNLS-KO in the host (orange curve), N=5 mice per cohort.
m28-RNLS combined with immune checkpoint inhibitors is superior to either drug alone
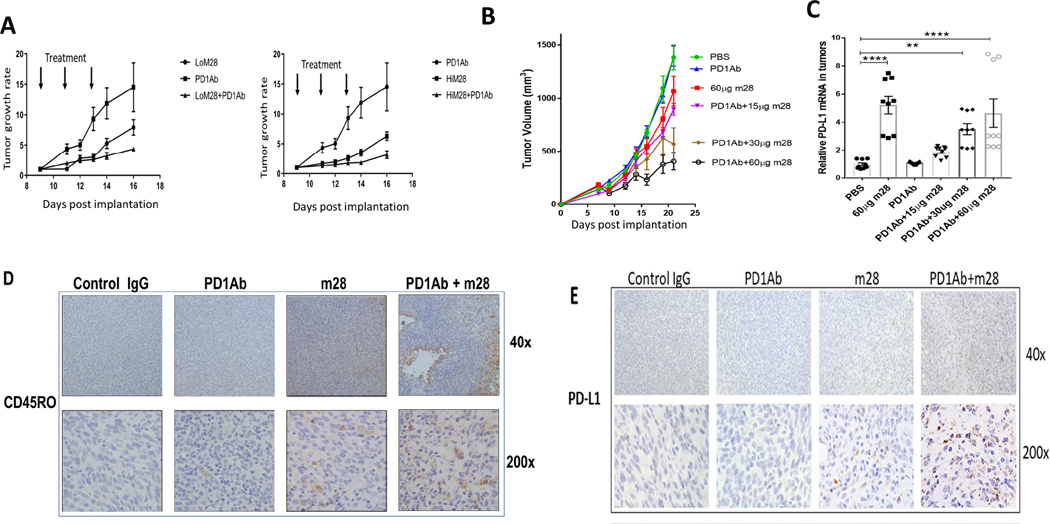
B16-F10 tumors are non-immunogenic and insensitive to immune checkpoint inhibition (36). YUMM 1.7 cells are similarly insensitive. (44) RNLS-KO mice with implanted B16F10 cells tend to reject tumors in the majority of mice (Figure 1A). With the goal of developing anti-RNLS antibodies in combination with anti-PD-1 in melanomas resistant to anti-PD-1, we studied the combination of m28-RNLS and anti-PD-1 in the two melanoma models. As shown in Figure 5 A–B, the combination was superior to either antibody alone, and enhanced activity of the combination was superior with higher doses of m28-RNLS. Tumor-cell PD-L1 expression has been shown in multiple studies to be associated with response to PD-1 inhibitors. (45, 46) We therefore assessed PD-L1 expression by RT-PCR in tumors harvested from mice treated with control IgG, anti-PD-1, m28-RNLS or the combination of anti-PD-1 and m28-RNLS, and found that PD-L1 levels are higher in tumors from m28-RNLS-treated mice, with or without anti-PD-1, particularly at higher doses of m28-RNLS (Figure 5C). By immunohistochemistry we confirmed increased memory T cell content and enhanced increases in PD-L1 in tumors from mice treated with combined anti-PD-1 and m28-RNLS compared to either modality alone (Figure 5D–E). Ipilimumab (anti-CTLA-4) is also used alone or with PD-1 inhibitors to treat advanced melanoma. In groups of 10 mice bearing B16 tumors, the combination of anti-CTLA-4 and anti-RNLS is superior to either drug alone, and this is in fact superior to anti-PD-1 and anti-RNLS. The addition of anti-PD-1 to anti-CTLA-4 and anti-RNLS does not have a significant effect on tumor volume or weight (Supplemental Figure 8).
Figure 5.
Inhibition or elimination of RNLS in combination with anti-PD-1 is superior to either modality alone in two murine melanoma models. (A) Mice bearing B16F10 tumors treated with two doses of m28-RNLS (60ug and 120ug) alone or with anti-PD-1. Combination therapy was superior to either drug alone, N=5 per cohort. (B) Confirmation studies done in mice bearing YUMM1.7 tumors treated with 15 μg, 30 μg or 60 μg m28-RNLS with or without anti-PD-1, demonstrating superior activity of the combination compared to either drug alone (N=5 per cohort). (C) mRNA levels of PD-L1 are higher in tumors from mice treated with m28-RNLS, suggesting a potential mechanism for sensitization of tumors to anti-PD-1 by m28-RNLS. (D-E) Immunohistochemical studies demonstrating increased memory T cell content and increased tumor PD-L1 expression with combined therapy compared to either drug alone. Control IgG treated tumors had a mean of 1.6±0.84 CD45RO positive cells per high power field (400X), anti- PD-1 treated tumors had 3.4±0.9, m28 tumors had 22±1.4, and tumors treated with the combination had 31±3 CD45RO positive cells, markedly more than control or anti-PD-1 alone (P<0.0001, by Tukey’s test for multiple comparisons). We found a mean of 2.2±0.8 PD-L1 positive cells per high power field (400X) in control IgG treated tumors, 2.3±0.5 in anti-PD-1 treated tumors, 19.9±3.4 in m28 treated tumors and tumors treated with the combination had 43.3±3.6 PD-L1 positive cells, markedly more than control or either drug alone (P<0.0001, by Tukey’s test for multiple comparisons).
RNLS knock-out or antibody-driven inhibition does not affect normal organ function
RNLS is known to be expressed in numerous normal organs. (47) Given our desire to develop RNLS inhibiting antibodies as a therapeutic intervention, we determined the effects of RNLS inhibition or knock-down on normal organs and blood pressure. Treatment resulted in slightly lower systolic blood pressures (Supplemental Figure 9). Compared to control IgG, m28 treatment showed slight splenic enlargement due to follicular hyperplasia in the white pulp in two of five mice, and increased neutrophils in the lungs of 4 of 5 mice, without additional pathology in the lungs. A few tiny foci of extramedually hematopoiesis were seen in livers of 5 of 5 mice treated with m28 and in 3 of 4 mice treated with control IgG.
Increased tumor RNLS expression is associated with worse overall survival in melanoma patients receiving immune therapy
We previously showed that RNLS expression is upregulated in human melanoma, and that inhibition of RNLS signaling using anti-RNLS monoclonal antibodies is cytotoxic to human melanoma cells in vitro and in vivo, as evidenced by growth inhibition of human melanoma cells injected in nude mice (16). To further interrogate the importance of RNLS expression in samples from patients treated with immunotherapy, we examined tumors from a cohort of patients with advanced melanoma, treated at Yale University with anti-PD-1 alone or with anti-CTLA-4. Clinical characteristics are shown in Supplemental Table 12. RNLS expression on pre-treatment specimens was variable. Examples of strong and weak RNLS expression in both tumor cells and macrophages are shown in Figure 6A–B. A subset of cases had weak tumor RNLS in the setting of strong macrophage RNLS, as exemplified in Figure 6C. Given that macrophages in many cases were relatively sparse compared with tumor cells, the analyses focus on RNLS expression within the entire tumor.
To evaluate associations between RNLS expression and baseline clinical and pathological parameters, we performed analyses of variance. Age (binarized by median), gender, stage, location of metastasis, level of LDH, site of biopsy, response to therapy and progression-free survival were not associated with RNLS expression. However, high RNLS expression was associated with decreased overall survival by Cox univariate analysis (P=0.039). Continuous RNLS scores were binarized by the median score and Kaplan–Meier survival curves were generated (Figure 6D), confirming the association between high RNLS levels and worse survival (log-rank P=0.006), as measured from time of initiation of immune therapy. As demonstrated in Supplemental Figure 10, when dividing this small cohort into two, the association with survival is maintained in the subset treated with anti-PD-1 alone (P=0.012), while in the subset of patients treated with dual immune checkpoint inhibitors, there is a trend towards worse survival in patients with high RNLS expression. High RNLS levels were similarly associated with worse survival as measured from diagnosis of advanced disease (Supplemental Figure 11). We next sought to determine whether high mRNA expression levels were associated with worse survival in patients treated with anti-PD-1, using data from GSE91061. (48, 49) This data set included tumor RNLS mRNA expression from 51 patients. Using X-Tile (50), we determined the optimal cutpoint at 298, and patients with RNLS levels ≥298 had worse 18 month survival (Supplemental Figure 12).
Finally, we generated a multi-variable Cox model to determine whether RNLS expression has independent prognostic value. Our model included other established clinical prognostic characteristics. On multivariable analysis, high RNLS, high stage (M1c or M1d), elevated LDH, and treatment with anti-PD-1 monotherapy, as opposed to combined therapy, all maintained their independent association with decreased survival (Supplemental Table 13). Taken together with the pre-clinical data in Figure 6 showing upregulation of PD-L1 with RNLS inhibition, these results suggest that co-targeting RNLS and PD-1 might be beneficial for treating patients with tumors resistant to anti-PD-1 expressing high RNLS levels.
DISCUSSION
In this work we demonstrate the importance of RNLS signaling in immune cells in melanoma murine models and human tumors. Specifically, we show that RNLS inhibition in immune cells and host cells is associated with tumor rejection in murine melanoma models, and rechallenging the same mice by repeat injection of viable tumor cells fails to result in subsequent tumor development. Studies were conducted by two methods: RNLS knockout mice were injected with RNLS wild-type melanoma cells, and wild-type mice were injected with anti-RNLS antibodies that inhibit RNLS signaling in both tumor and immune cells. Effects of RNLS inhibition in the TME were demonstrated in two distinct aggressive melanoma murine models, B16F10 and YUMM1.7, both poorly responsive to immune therapy. The fact that the RNLS knockout mice rejected their melanoma tumors upon re-implantation of RNLS wild-type melanoma cells provides strong evidence to support the important role of RNLS signaling in immune infiltrating cells. In our previous studies we showed that anti-RNLS antibodies have some direct anti-tumor effect in vitro and in xenograft models (16), and can therefore conclude that inhibition of RNLS signaling results in tumor shrinkage by two mechanisms. The first, and dominant mechanism, is rooted in inhibition of RNLS signaling in the host that subsequently alters the TME and shapes its response to tumor cells. The second mechanism involves direct, host-independent, cytotoxic and growth inhibitory effect on tumor cells.
Several additional lines of evidence demonstrated in this work suggest that inhibition of RNLS signaling abrogates the immuno-suppressive properties of the TME, and accounts for the majority of the observed therapeutic results. Most importantly, in contrast to WT mice, RNLS-KO mice are protected from metastatic spread when injected with B16-F10 tumors cells which express RNLS. Inhibition of RNLS by m28-RNLS affects all cells, both tumor cells and immune cells, and effects of treatment on lung metastases were similarly impressive, albeit slightly less dramatic than in RNLS-KO mice. The observation that inhibition of TME-derived RNLS results in greater reduction in tumor growth than tumor cell-specific reduction has important implications for clinical development of anti-RNLS antibodies for patients with melanoma.
We showed that the TME of RNLS-KO mice exhibits an inflamed phenotype that differs markedly from WT mice. Single-cell RNA-sequencing studies showed that the TME of RNLS-KO mice compared to RNLS WT mice is characterized by increased Granzyme B in CD8+ T cells and NK cells, increased interferon-gamma in CD8+ T cells and polarization of macrophages to a more inflammatory phenotype, characterized by increased TNFα and IL1β expression. (51) Development of a T-cell inflamed phenotype correlates well, although not perfectly, with tumor rejection. Importantly, treatment of melanoma tumors with m28-RNLS also resulted in increased innate and adaptive immune cells. Taken together, these results strongly suggest that inhibition of RNLS signaling in the host, and therefore in the TME, is a key determinant of whether or not the host’s immune system rejects tumors.
Cells of myeloid lineage that populate TME include granulocytes, dendritic cells, and tumor associated macrophages (TAM). TAM comprise 5–30% of cells in metastatic melanoma deposits (52) and are thought to support tumor growth and suppress immune responses (51). Various mechanisms of TAM immune suppression have been proposed, including (but not limited to) TGF- β, IL10, ARG1, IDO, PGE2, or PD-L1 (53). RNLS expression is markedly increased in human melanoma and inversely correlated with disease-specific survival, and we previously showed that macrophages have high expression of RNLS.(16) Furthermore, RNLS expression appears to be increased dramatically in immunosuppressive tumor-promoting CD163+ macrophages, known to promote tumor invasion and metastatic spread (54–56). Tumor cells appear to secrete factor(s) that modulate RNLS expression by macrophages as evidenced by our studies in which conditioned medium obtained from B16-F10 cells in culture resulted in 8-fold increased RNLS expression in macrophages (16). This likely leads to downstream effects that promote a polarization of macrophages toward a tumor promoting phenotype (57); expression of IL-6 and leukemia inhibitory factor (LIF) along with IL-10, all of which are reported to be important for promotion of the tumor microenvironment, is decreased upon loss of RNLS (16). Despite the importance of macrophage RNLS signaling, our experiments using a macrophage-specific RNLS- KO mouse showed that the effect of RNLS inhibition in macrophages is small and not key or sufficient for tumor rejection.
Conversely, specific T cell depletion using antibodies to CD3 abrogated the effects of RNLS knock-out in host cells. This is further substantiated by the increase in granzyme B in T cells and NK cells and increased expression of interferon-gamma in tumors implanted in RNLS KO mice or WT mice treated with anti-RNLS, while these changes were not seen in myeloid cells (Figure 2 E–F). These data, combined with the major changes in T cell content, particularly CD4+ T cell content with anti-RNLS antibodies, both in terms of number and in terms of T cell activation, suggest that effects of RNLS inhibition on T cells are key for tumor rejection. Importantly, this further supports evaluation of potential combinations of RNLS inhibitors with T cell activating drugs such as immune checkpoint inhibitors in cancer.
Our single-cell RNA-seq studies demonstrate changes in both number and activation status in other cell types, including dendritic cells and NK cells. Further work is being planned to determine the role of these cell types using similar cell depletion approaches to those undertaken for macrophages and CD3+ cells.
Specifically, in melanoma, the striking effect of RNLS inhibition in two different animal melanoma models on both lung and extrapulmonary metastases suggests that evaluation of RNLS inhibiting antibodies is warranted. In our previous studies this was demonstrated in xenograft models,(16) and here we demonstrate strong anti-tumor activity of the m28-RNLS mAb in the highly aggressive B16-F10 immune competent model, both in models of local subcutaneous tumor cell implantation and in a metastatic model. Importantly, we note that RNLS-KO mice do not exhibit abnormal organ function, and mice treated with anti-RNLS antibodies did not exhibit signs of distress, indicating that RNLS inhibition should be feasible in humans with melanoma.
Given the potential for clinical development of anti-RNLS antibodies, it is important to determine the precise mechanism of action of these antibodies. The effect of m28 is not likely related to antibody dependent cellular phagocytosis, as MDSC elimination did not substantially alter the effects of m28. Antibody-dependent cellular cytotoxicity may play a role, and further experiments involving Fab or FcR-deficient mice will be conducted.
Key for clinical development of anti-RNLS antibodies, in two melanoma murine models resistant to PD-1 inhibitors we showed increased tumor reduction and improved survival in mice treated with both anti-PD-1 and anti-RNLS compared with either drug alone or vehicle control. Moreover, melanomas treated with anti-RNLS demonstrated increased PD-L1 expression in tumor cells, which might render them more sensitive to anti-PD-1. This finding supports development of combinations of PD-1 and RNLS targeting drugs in melanomas that are poorly responsive to PD-1 inhibitors, such as PD-L1 negative tumors or tumors with poor T cell infiltration.
In pre-treatment samples from patients with metastatic melanoma treated with anti-PD-1 alone or in combination with anti-CTLA-4, we showed that RNLS is expressed in tumor cells, in cells in the tumor microenvironment (particularly macrophages), or both. High RNLS levels in these patients were associated with decreased overall survival at four years of follow-up. This suggests that high RNLS levels are associated with poor long-term immunity, a hypothesis that is supported by our murine experiments. Treating patients with tumors that have high RNLS and low PD-L1 expression, might enable us to overcome resistance to PD-1 inhibitors.
CONCLUSIONS:
Based on our findings, we conclude that the primary mechanism underlying tumoricidal effects of RNLS signaling inhibition relates to development of a T-cell inflamed microenvironment that promotes rejection and killing of tumor cells. Although inhibition of RNLS signaling decreases tumor cell proliferation in vitro, these effects appear to be less important to the in vivo anti-tumor action of RNLS inhibition than effects on cells in the tumor micro-environment. We also establish the utility of anti-RNLS therapy in syngeneic mouse models of metastatic melanoma, and note that RNLS-KO mice are fully viable, suggesting that targeting RNLS is a promising strategy for treating melanoma. Moreover, our studies suggest that RNLS inhibition might be most useful for treating melanomas with high tumor promoting macrophage content, melanomas that have poor T-cell infiltration, or tumors with low or absent PD-L1 expression. These melanomas are the very melanomas that tend to be resistant to immune checkpoint inhibitors and represent an unmet clinical need, and studies of combinations of anti-RNLS antibodies and immune checkpoint inhibitors in patients with anti-PD-1 resistant melanoma are warranted.
Supplementary Material
HIGHLIGHTS.
RNLS knock-out in immune cells substantially inhibits tumor growth.
Anti-RNLS antibodies enhance effects of anti-PD-1 in melanoma bearing mice.
T cells (but not macrophages) are required for anti-RNLS effects on tumors.
Immunotherapy treated melanoma patients with high tumor RNLS have worse survival.
Anti-RNLS antibodies might have therapeutic value in anti-PD-1 resistant melanomas.
Acknowledgements:
We thank C. J. Booth, DVM, PhD, Director of the Yale Comparative Pathology Research Core in the Department of Comparative Medicine for her expertise in examination of the HE-stained slides.
Funding: This work was supported in part by VA Connecticut (R. Safirstein. and G.V. Desir), National Institute of Health grants RC1DK086465, RC1DK086402, and R01DK081037, (G.V. Desir), and RO1CA216846 (G.V. Desir and H. Kluger), the Yale SPORE in Skin Cancer, P50 CA121974 (M. Bosenberg and H. Kluger) and the Lampman surgical award from Yale University Department of Surgery (C. Cha).
ABBREVIATIONS
- ACK
Ammonium-Chloride-Potassium
- AKT
serine/threonine kinase 1
- ARG1
Arginase 1
- B16-F10
a mouse melanoma cell line
- BRAF
proto-oncogene B-Raf and v-Raf murine sarcoma viral oncogene homolog B
- BSA
bovine serum albumin
- CD (CD3, CD4, CD8, CD163, CD274, etc)
cluster of differentiation 3, 4, 8, 163, 274, etc
- Cdkn2a
cyclin-dependent kinase inhibitor 2a
- Cre
Cre recombinase
- CTLA4
cytotoxic T-lymphocyte-associated protein 4
- Cy5
cyanine 5
- DAPI
4′,6-diamidino-2-phenylindole
- FLP
Flp recombinase
- FOXP3
Forkhead Box P3
- GEM
gel bead in emulsion
- HI-FBS
heat inactivated fetal bovine serum
- IDO
indoleamine 2,3-dioxygenase
- IFNγ
interferon γ
- IgG
immunoglobulin G
- IL
interleukin
- IP
intraperitoneal injection
- JAK
Janus kinase
- KO
knockout
- LDH
lactate dehydrogenase
- LIF
leukemia inhibitoy factor
- LysMcre
Lysozyme 2 driven myeloid cells specific Cre recombinase
- m28
a monoclonal antibody against renalase, clone m28
- MØ
macrophages
- MAPK
mitogen-activated protein kinase
- MDSC
myeloid derived suppressor cells
- MEK
MAPK/ERK Kinase
- NaOH
sodium hydroxide
- NK
natural killer
- OS
overall survival
- PBS
phosphate-buffered saline
- PCA
principal component analysis
- PD-1
programmed cell death protein 1
- PD-L1
programmed cell death protein ligand 1
- PGE2
Prostaglandin E2
- PI3K
phosphoinositide 3-kinase
- Pten
phosphatase and tensin homolog
- QC
quality control
- RECIST
Response Evaluation Criteria in Solid Tumours
- RNLS
renalase
- RT-qPCR
real-time reverse transcription polymerase chain reaction
- scRNAseq
single cell ribonucleic acid sequencing
- SE
standard error
- siRNA
small interfering RNA
- SNN
share nearest neighbor
- SQ
subcutaneous injection
- STAT
signal transducer and activator of transcsription
- t-SNE
t-distributed stochastic neighbor embedding
- TAM
tumor associated macrophages
- TBS
Tris-buffered saline
- TGF-β
transforming growth factor beta
- TMA
tissue microarray
- TME
tumor micro-enviroment
- TNFα
tumor necrosis factor alpha
- TRIS-EDTA
trisaminomethane-ethylenediaminetetraacetic acid
- YCGA
Yale Center for Genome Analysis
- YUMM1.7
Yale University Mouse Melanoma 1.7
- VACHS
Veterans Affairs Connecticut Health System
- WT
wild type
Footnotes
DECLARATIONS
Ethics approval and consent to participate: Collection of human tissues was done with approval of a Yale University Institutional Review Board. Patients provided consent for use of excess tissue at the time of surgery.
Consent for publication: All authors approved the final version of the manuscript and have agreed to publication of the results.
Availability of data and material: All data, including the single-cell RNA sequencing data sets supporting the conclusions are included within the manuscript or the supplemental materials available online.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kluger HM, Tawbi HA, Ascierto ML, Bowden M, Callahan MK, Cha E, et al. Defining tumor resistance to PD-1 pathway blockade: recommendations from the first meeting of the SITC Immunotherapy Resistance Taskforce. J Immunother Cancer 2020;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367(18):1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371(20):1867–76. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol 2015;16(8):908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 2015;372(26):2521–32. [DOI] [PubMed] [Google Scholar]
- 8.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribas A, Kefford R, Marshall MA, Punt CJ, Haanen JB, Marmol M, et al. Phase III randomized clinical trial comparing tremelimumab with standard-of-care chemotherapy in patients with advanced melanoma. J Clin Oncol 2013;31(5):616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364(26):2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367(2):107–14. [DOI] [PubMed] [Google Scholar]
- 13.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2015;16(4):375–84. [DOI] [PubMed] [Google Scholar]
- 14.Regan MM, Werner L, Rao S, Gupte-Singh K, Hodi FS, Kirkwood JM, et al. Treatment- Free Survival: A Novel Outcome Measure of the Effects of Immune Checkpoint Inhibition-A Pooled Analysis of Patients With Advanced Melanoma. J Clin Oncol 2019;37(35):3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med 2019;381(16):1535–1546. [DOI] [PubMed] [Google Scholar]
- 16.Hollander L, Guo X, Velazquez H, Chang J, Safirstein R, Kluger H, et al. Renalase Expression by Melanoma and Tumor-Associated Macrophages Promotes Tumor Growth through a STAT3-Mediated Mechanism. Cancer Research 2016;76(13):3884–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Li G, Wang P, Velazquez H, Yao X, Li Y, et al. Renalase is a novel, soluble monoamine oxidase that regulates cardiac function and blood pressure. J Clin Invest 2005;115(5):1275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X, Wang L, Velazquez H, Safirstein R, Desir GV. Renalase: its role as a cytokine, and an update on its association with type 1 diabetes and ischemic stroke. Curr Opin Nephrol Hypertens 2014;23(5):513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinman EJ, Biswas R, Steplock D, Wang P, Lau YS, Desir GV, et al. Increased renal dopamine and acute renal adaptation to a high-phosphate diet. Am J Physiol Renal Physiol 2011;300(5):F1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Velazquez H, Moeckel G, Chang J, Ham A, Lee HT, et al. Renalase Prevents AKI Independent of Amine Oxidase Activity. Journal of the American Society of Nephrology 2014;25(6):1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HT, Kim JY, Kim M, Wang P, Tang L, Baroni S, et al. Renalase protects against ischemic AKI. J Am Soc Nephrol 2013;24(3):445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Hollander L, MacPherson D, Wang L, Velazquez H, Chang J, et al. Inhibition of renalase expression and signaling has antitumor activity in pancreatic cancer. Scientific Reports 2016;6:22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meeth K, wang J, Micevic G, Damsky WE Jr, Bosenberg M. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment cell & melanoma research 2016;29(5):590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011;474(7351):337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research 1999;8(4):265–277. [DOI] [PubMed] [Google Scholar]
- 26.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177(7):1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linderman GC, Rachh M, Hoskins JG, Steinerberger S, Kluger Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods 2019;16(3):243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta S, McCann L, Chan YGY, Lai EW, Wei W, Wong PF, et al. Closed system RT-qPCR as a potential companion diagnostic test for immunotherapy outcome in metastatic melanoma. J Immunother Cancer 2019;7(1):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Morilla S, Villarroel-Espindola F, Wong PF, Toki MI, Aung TN, Pelekanou V, et al. Biomarker Discovery in Patients with Immunotherapy-Treated Melanoma with Imaging Mass Cytometry. Clin Cancer Res 2021;27(7):1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Morilla S, Zugazagoitia J, Wong PF, Kluger HM, Rimm DL. Quantitative analysis of CMTM6 expression in tumor microenvironment in metastatic melanoma and association with outcome on immunotherapy. Oncoimmunology 2020;10(1):1864909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kluger HM, Zito CR, Barr ML, Baine MK, Chiang VL, Sznol M, et al. Characterization of PD- L1 Expression and Associated T-cell Infiltrates in Metastatic Melanoma Samples from Variable Anatomic Sites. Clin Cancer Res 2015;21(13):3052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res 2004;64(23):8773–7. [DOI] [PubMed] [Google Scholar]
- 33.Woo S-R, Corrales L, Gajewski TF. The STING pathway and the T cell-inflamed tumor microenvironment. Trends in Immunology 2015;36(4):250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji R-R, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunology, Immunotherapy 2012;61(7):1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fidler IJ. Biological Behavior of Malignant Melanoma Cells Correlated to Their Survival in Vivo. Cancer Research 1975;35(1):218–224. [PubMed] [Google Scholar]
- 36.Pan Z-K, Weiskirch LM, Paterson Y. Regression of Established B16F10 Melanoma with a Recombinant Listeria monocytogenes Vaccine. Cancer Research 1999;59(20):5264–5269. [PubMed] [Google Scholar]
- 37.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15(8):486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makino M, Maeda Y, Mukai T, Kaufmann SH. Impaired maturation and function of dendritic cells by mycobacteria through IL-1beta. Eur J Immunol 2006;36(6):1443–52. [DOI] [PubMed] [Google Scholar]
- 39.Kaka AS, Foster AE, Weiss HL, Rooney CM, Leen AM. Using dendritic cell maturation and IL-12 producing capacity as markers of function: a cautionary tale. J Immunother 2008;31(4):359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wesa AK, Galy A. IL-1 beta induces dendritic cells to produce IL-12. Int Immunol 2001;13(8):1053–61. [DOI] [PubMed] [Google Scholar]
- 41.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005;434(7034):772–7. [DOI] [PubMed] [Google Scholar]
- 42.Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology 2018;154(1):3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buhrer C, Berlin C, Jablonski-Westrich D, Holzmann B, Thiele HG, Hamann A. Lymphocyte activation and regulation of three adhesion molecules with supposed function in homing: LECAM-1 (MEL-14 antigen), LPAM-1/2 (alpha 4-integrin) and CD44 (Pgp-1). Scand J Immunol 1992;35(1):107–20. [DOI] [PubMed] [Google Scholar]
- 44.Perry CJ, Munoz-Rojas AR, Meeth KM, Kellman LN, Amezquita RA, Thakral D, et al. Myeloid-targeted immunotherapies act in synergy to induce inflammation and antitumor immunity. J Exp Med 2018;215(3):877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kluger HM, Chiang V, Mahajan A, Zito CR, Sznol M, Tran T, et al. Long-Term Survival of Patients With Melanoma With Active Brain Metastases Treated With Pembrolizumab on a Phase II Trial. J Clin Oncol 2019;37(1):52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kluger HM, Zito CR, Turcu G, Baine MK, Zhang H, Adeniran A, et al. PD-L1 Studies Across Tumor Types, Its Differential Expression and Predictive Value in Patients Treated with Immune Checkpoint Inhibitors. Clin Cancer Res 2017;23(15):4270–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Safirstein R, Velazquez H, Guo XJ, Hollander L, Chang J, et al. Extracellular renalase protects cells and organs by outside-in signalling. J Cell Mol Med 2017;21(7):1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Chai Z, Li Y, Long F, Hao Y, Pan G, et al. Identification of Potential Biomarkers for Anti-PD-1 Therapy in Melanoma by Weighted Correlation Network Analysis. Genes (Basel) 2020;11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017;171(4):934–949 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10(21):7252–9. [DOI] [PubMed] [Google Scholar]
- 51.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496(7446):445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hussein MR. Tumour-associated macrophages and melanoma tumourigenesis: integrating the complexity. Int J Exp Pathol 2006;87(3):163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz-Valdes N, Basagoiti M, Dotor J, Aranda F, Monreal I, Riezu-Boj JI, et al. Induction of monocyte chemoattractant protein-1 and interleukin-10 by TGFbeta1 in melanoma enhances tumor infiltration and immunosuppression. Cancer Res 2011;71(3):812–21. [DOI] [PubMed] [Google Scholar]
- 54.Ruhrberg C, De Palma M. A double agent in cancer: Deciphering macrophage roles in human tumors. Nat Med 2010;16(8):861–862. [DOI] [PubMed] [Google Scholar]
- 55.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer 2004;4(1):71–78. [DOI] [PubMed] [Google Scholar]
- 56.Hao N-B, Lü M-H, Fan Y-H, Cao Y-L, Zhang Z-R, Yang S-M. Macrophages in Tumor Microenvironments and the Progression of Tumors. Clinical and Developmental Immunology 2012;2012:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duluc D, Delneste Y, Tan F, Moles M-P, Grimaud L, Lenoir J, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 2007;110(13):4319–4330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.