Abstract
Biological systems with intrinsic complexity require multiplexing techniques to comprehensively describe the phenotype, interaction, and heterogeneity. Recent years have witnessed the development of supermultiplexed vibrational microscopy, overcoming the ‘color barrier’ of fluorescence-based optical techniques. Here, we will review the recent progress in the design and applications of super-multiplexed vibrational probes. We hope to illustrate how rainbow-like vibrational colors can be generated from systematic studies on structure-spectroscopy relationships and how being colorful makes a difference to various biomedical applications.
Introduction
Working with systems of intrinsic complexity, biologists have a long-lasting demand for simultaneous detection of multiple targets of interest. Optical techniques, such as fluorescence microscopy, have superb biocompatibility and have become essential in biological studies. Regular fluorescence microscopy usually enables two to five resolvable colors for simultaneous detection, which already gives rise to vast biomedical applications. There are now powerful multiplexed fluorescence methods in basic life science for various cell sorting and phenotyping1–2, imaging-based in situ multiplexed nucleic acid and epitope profiling3–8, which fuel the understanding of biological heterogeneity. Assays are also developed for fluorescence-based multiplexed detection of antigens and nucleic acids with high throughput and sensitivity9–10. Such techniques present outstanding utility in medical diagnoses and give birth to a thriving market.
However, fluorescence microscopy runs into a ‘color barrier’ to further expand the level of multiplexing. Imaging more than five targets simultaneously is typically forbidden without specialized instrumentation due to the broad fluorescence spectrum resulting from the fundamentally fast electronic dephasing11. Unlike fluorescence spectroscopy, vibrational spectroscopy involves molecular vibrational transitions with much narrower spectral resonance. Consider a typical fluorescent dye, Cy5, for example. The full width at half maximum (FWHM) of its fluorescence spectrum is about 50 times larger than that of its Raman peak corresponding to the conjugated double bonds. Such sharp spectral resonance gives vibrational microscopy intrinsic advantages in multiplexing. Many multiplexed bioassays and bioimaging, mostly based on surface-enhanced Raman scattering (SERS), have already been developed to explore this potential in multiplexing12–13. However, as most works use the already-existing fingerprint signatures of Raman probes, the achievable level of multiplexing is still hindered by the inevitable overlapping of multiple peaks crowding in the fingerprint region. The tipping point came for vibrational microscopy to ultimately overcome the color barrier when two super-multiplexed vibrational palettes were designed and synthesized in the cell-silent spectral region for stimulated Raman scattering (SRS) microscopy (Figure 1)14–15.
Figure 1|. Super-multiplexed vibrational palettes in the cell-silent window for SRS microscopy.
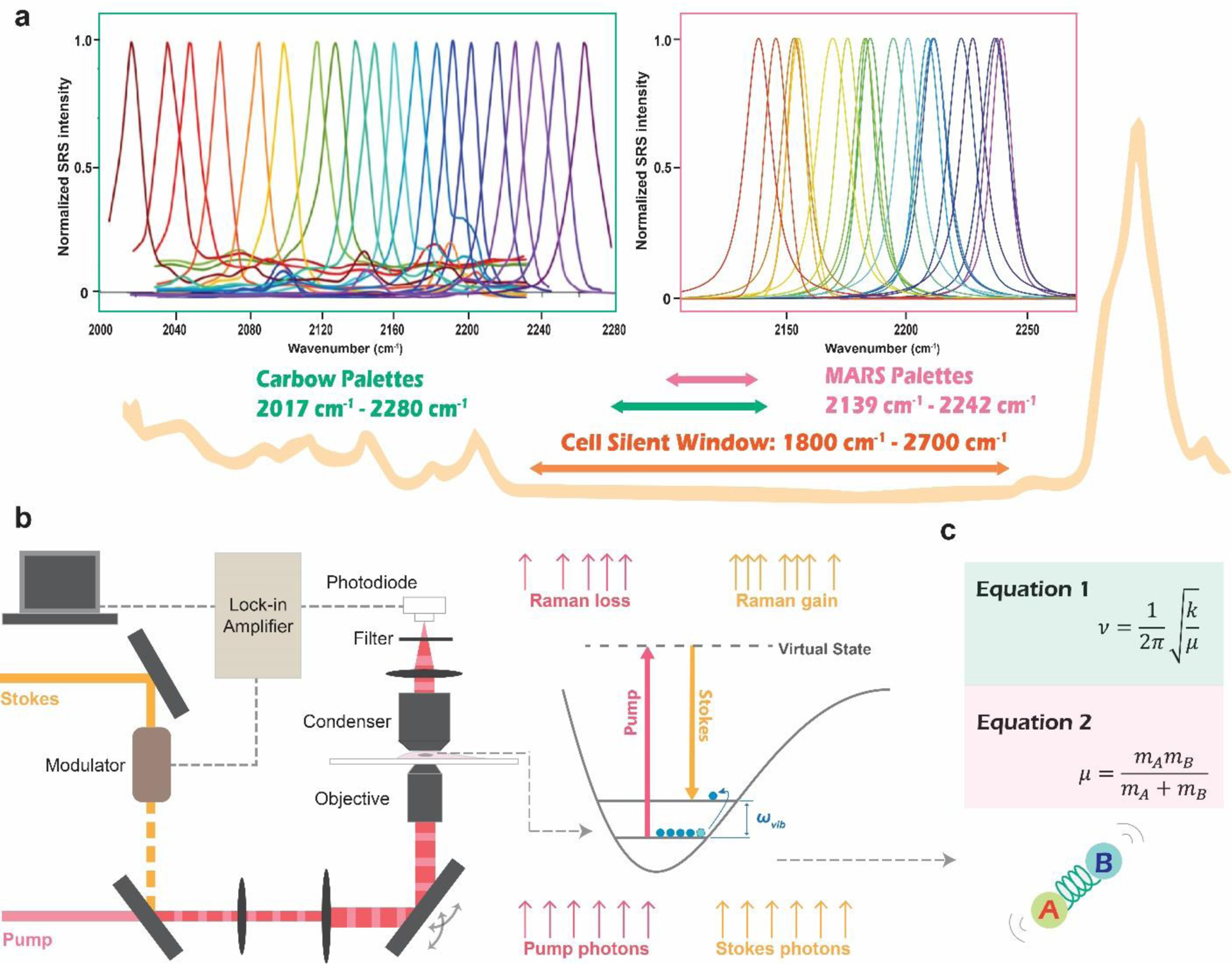
a. The typical Raman spectrum of cells contains a spectral window from 1800 cm−1 to 2700 cm−1, with no Raman peaks from endogenous biomolecules. Both of the super-multiplexed vibrational palettes, MARS and Carbow, have been developed within this spectral window. b. SRS microscopy illustration. Pump laser beam and modulated Stokes laser beam are combined and focused onto the sample. When the energy difference between the pump and the Stokes laser matches the vibrational energy(ωvib) of the chemical bond of interest, vibrational excitation happens. Each event is accompanied by one photon loss in the pump beam (stimulated Raman loss, SRL) and one photon gain in the Stokes beam (stimulated Raman gain, SRG). SRS signal is detected as the relative intensity change of the pump beam, which is extracted by the lock-in amplifier to achieve sensitive detection above the low-frequency laser background noise. c. Classical spring model to account for the tuning of vibrational frequency. ν (Hz) is the vibrational frequency determined by force constant k and reduced mass μ, while mA and mB are the atomic mass of atom A and B from the chemical bond.
SRS microscopy involves two synchronized pulsed lasers. The pump and the Stokes beams are focused collinearly onto the same focal spot and coherently excite the target molecular vibration (Figure 1b). The stimulated Raman scattering effect, analogous to stimulated emission in quantum mechanics, happens when the energy difference between the pump and the Stokes laser matches the vibrational energy(ωvib) of the target chemical bond (Figure 1b). The vibrational activation rates can be drastically accelerated by 108 than that of the inherently weak spontaneous Raman scattering, providing the ultra sensitivity of SRS microscopy for vibrational bio-imaging in the far-field16. Although SRS microscopy was initially developed as a label-free technique17, its utility has gone through an astonishing expansion with the development of various vibrational probes to transform the unique chemical specificity and sharp resonance of vibrational spectroscopy into valuable biological information18. Especially, the latest biomedical applications, demonstrated by super-multiplexed vibrational imaging, suggest that more colors for simultaneous observation is not only a simple accumulation of quantities but can bring qualitative transformation as well. This review will focus on how the super-multiplexed vibrational palettes are systematically designed and what unique applications such multiplexing capability has brought to reality during the past several years.
Development of super-multiplexed vibrational palettes
The early attempts exploring the systematic strategies to tune the vibrational frequencies can trace back to the studies on structure−Raman shift/intensity relationships for alkynes. Alkyne molecules caught researchers’ attention due to their unique sharp Raman single peak in a spectral region known as ‘cell-silent (1800 cm−1 - 2700 cm−1, Figure 1a)19–20. This region in the Raman spectrum is particularly favorable, avoiding interference from biomass’s complicated endogenous Raman signals and rendering vibrational bioimaging with good contrast. From the early studies on alkynes, researchers have accumulated the two key building blocks to systematically tune vibrational frequencies. One is changing substitution pattern, which affects the force constant k (Figure 1c, Equation 1) in the classical spring model of molecular vibrations21. The other is the isotope-editing, which manipulates reduced mass (Figure 1c, Equation 1, 2) to give different vibrational frequencies22. It turns out to be a general strategy that works for various vibrational probes to expand the vibrational colors without changing any chemical properties. Inspired by these successes, many small biomolecules were developed with Raman tags of distinct vibrational colors for multiplexed vibrational bioimaging.23–24 However, small Raman tags have little room for chemical structural modifications, resulting in fundamental restrictions on generating new vibrational colors or boosting detection sensitivity. With Raman multiplicity of around three and detection limit of mM to a few hundred μM, the actual utility of multiplexed vibrational imaging is unfortunately confined. The breakthrough finally comes when two super-multiplexed vibrational palettes, MARS14 and Carbow15, were developed from systematically modifying a group of Raman-active molecules of characteristic chemical structures. The two palettes take advantage of the nitrile or alkyne triple bonds in the cell-silent region to ensure the tuning of vibrational resonance without running into complicated spectral overlapping in the fingerprint region (Figure 1a). Nevertheless, each work provides original solutions to systematically design vibrational probes for the ultimate realization of super-multiplexed vibrational imaging.
Manhattan Raman scattering (MARS) dyes developed in 2017 were the first super-multiplexed vibrational palettes for SRS microscopy14. By coupling bond vibration with the electronic transition of particular chromophores, the electronic pre-resonance (epr) effect was introduced into SRS microscopy to further enhance the sensitivity by more than four orders of magnitude. Obtaining such detection limit down to a few hundred nanomolar is crucial to image specific molecular targets, which expands territories for biomedical applications. The designing intuition for MARS palettes is to conjugate the alkyne or nitrile group onto the 9-position of xanthene scaffolds. This structural feature not only ensures the coupling enhancement but also allows adjustment on force constant k for vibrational frequency tuning by modifying the electron density on the xanthene ring. With the additional isotope-editing strategy, the single sharp Raman peak of the triple bond can be appropriately tuned to give a full palette, which enabled the first super-multiplexed vibrational imaging of cells labeled by 16 colors simultaneously. A more recent paper did a comprehensive investigation on 9-Cyanopyronins as typical MARS structures, providing solutions for chemical synthesis, vibrational frequency tuning, and versatile labeling modalities25. Such probe development work establishes robust synthetic methods to account for essential structural features of a MARS probe (Figure 2a). The systematically synthesized palettes allow quantitative evaluation of the tuning effects from isotope editing, core atom substitution, and ring expansion (Figure 2a). The contribution from the three tuning strategies turned out to be additive, which provides a quantification model for structural-frequency prediction. Finally, functionalization is another critical issue to address by chemical synthesis for turning the premature vibrational colors into actual target-specific probes to convey biological information. Super-multiplexed biological imaging naturally requires multiple compatible labeling modalities integrated into the vibrational palettes to encode different information detected simultaneously. Up to now, abundant biological probing functionalities have been explored taking the 9-Cyanopyronins as structural templates, including epitope immunostaining (Figure 3b), organelle targeting (Figure 3d), metabolic imaging through click chemistry (Figure 3i,j), as well as enzymatic activity profiling (Figure 3k,l)25–26.
Figure 2|. Structural features and structure-spectroscopy relationship for MARS and Carbow palettes.
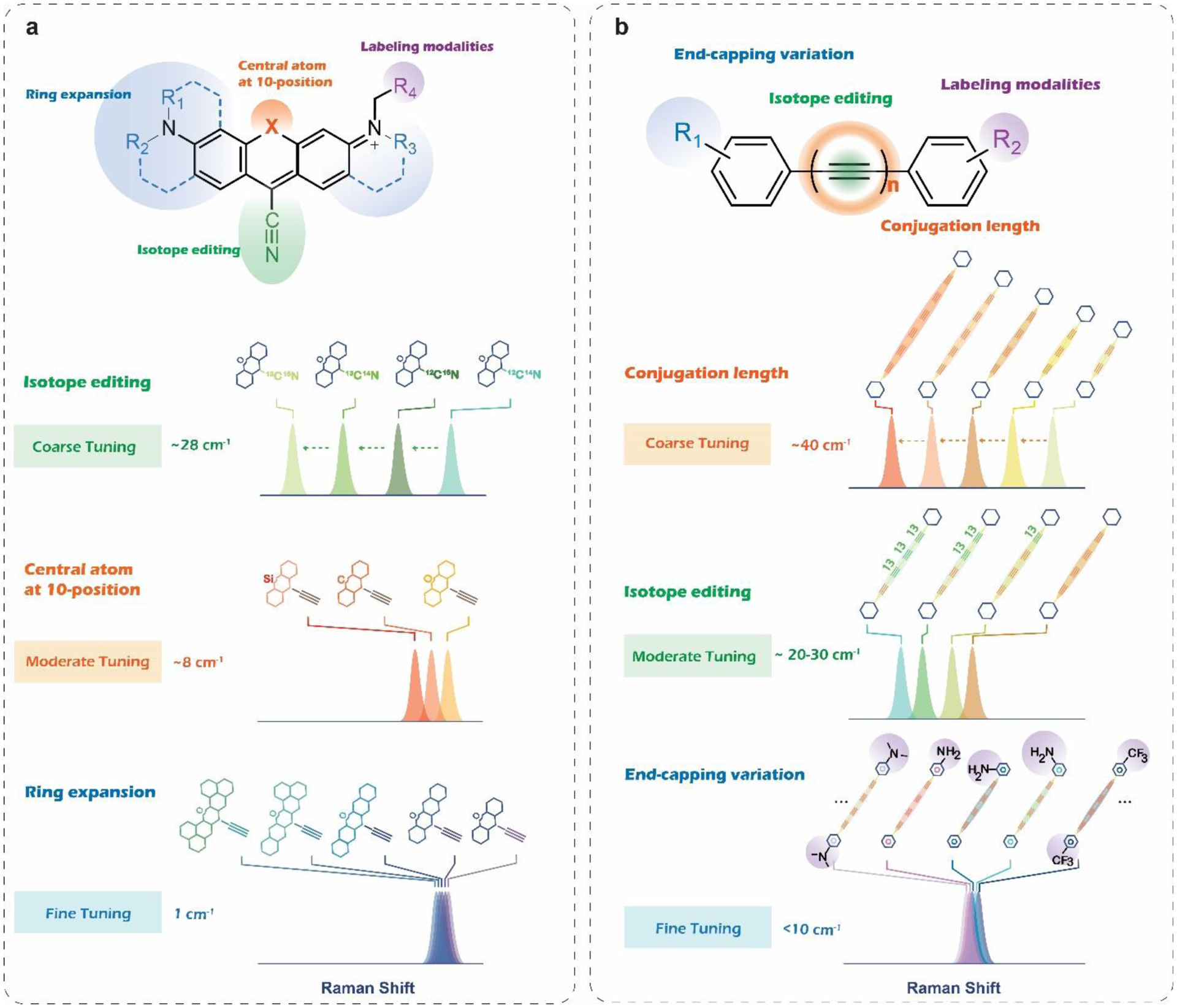
a. Schematic illustration of 9-Cyanopyronin based MARS dyes. From coarse tuning to fine tuning of the vibrational frequencies, isotope editing, central atom variation, and ring expansion can be used. The latter two rely on the specific chemical modification to xanthene rings to influence the k of the vibrational mode. b. Schematic illustration of polyyne-based Carbow dyes. Other than common isotope editing, two other tuning strategies on modifying conjugation length and end-capping variation are identified to be characteristic in engineering the Raman frequencies of polyynes.
Figure 3|. Functionalized library of vibrational probes.
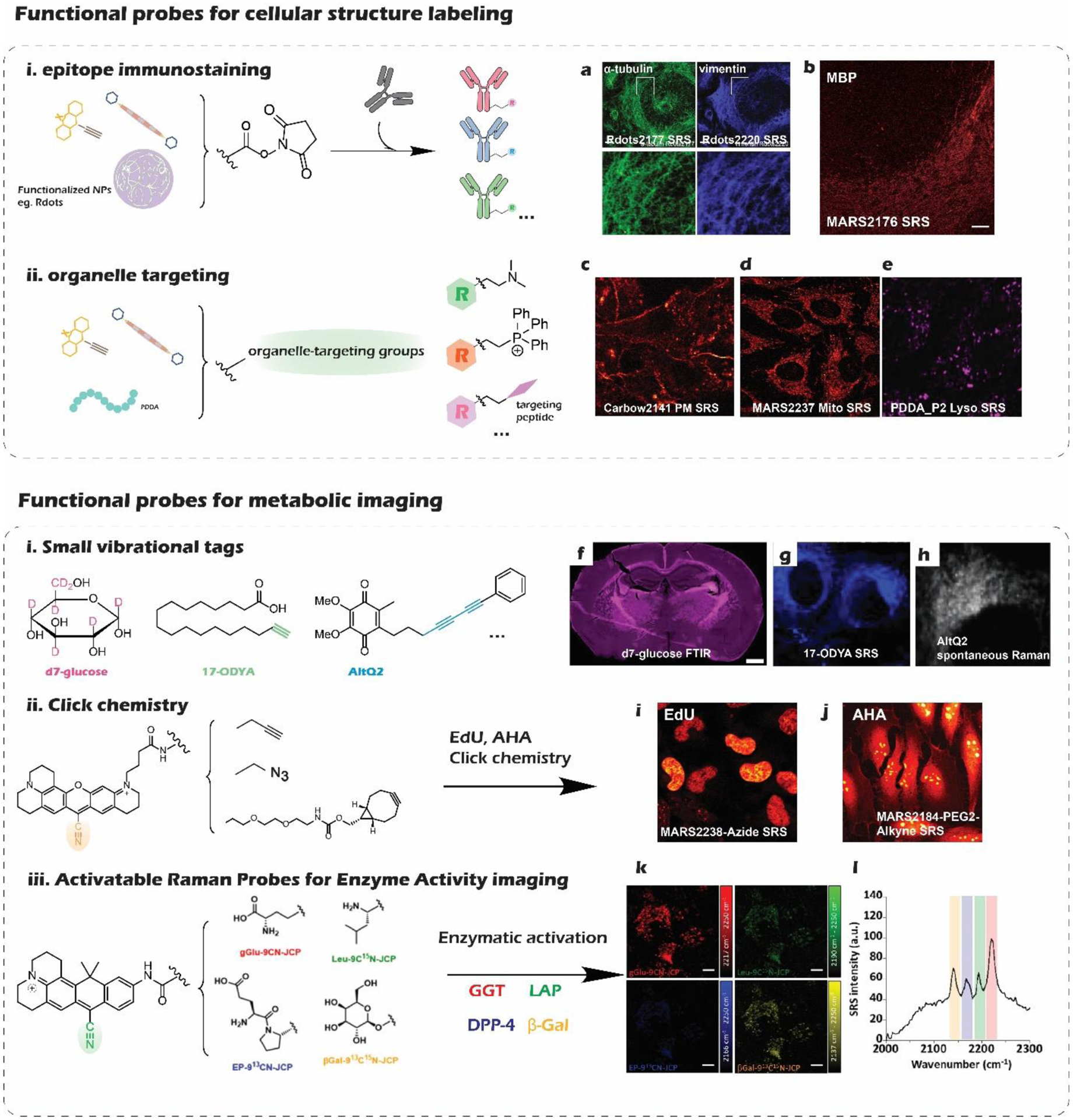
a. α-tubulin immunostaining with functionalized Rdots2177. b. Vimentin immunostaining with functionalized Rdots2220. c. MBP (myelin basic protein) in mouse cerebellum labeled by immunostaining with MARS2176 antibody conjugate. Scale bar, 50 μm. c. SRS imaging of plasma membrane (PM) in live Hela cells labeled by Carbow2141 PM. d. Imaging mitochondria with MARS2237 in live cells. e. SRS imaging of lysosomes in Hela cells labeled by PDDAP2. f. IR imaging of glucose anabolic activity in the whole brain with d7-glucose, integrating the C–D band as 2,060–2,220 cm−1. Scale bar, 1 mm. g. SRS imaging of fatty acid metabolism with fatty acyl derivatives 17-ODYA. h. Spontaneous Raman imaging of alkyne-labeled coenzyme Q (AltQ2) in live cells. i. SRS imaging of DNA synthesis in Hela cells with EdU and MARS2238-Azide through click reaction. j. SRS imaging of protein synthesis in Hela cells with AHA and MARS2184-PEG2-Alkyne through click reaction. k, Simultaneous detection of four enzyme activities in live A594 cells with SRS microscopy using four activable probes: gGlu-9CN-JCP, Leu-9C15N-JCP, EP-913CN-JCP, and βGal-913C15N-JCP. Scale bar, 10 μm. l. SRS spectra of enzyme activities obtained from A549 cells (left)
The other super-multiplexed vibrational palettes, termed Carbon rainbow (Carbow), are based on polyynes, which were brought to the field in 201815. Polyynes contain a special linear structure of an sphybridized carbon chain with alternating single and triple bonds, presenting unique properties on Raman spectroscopy. A thorough investigation of the structure-spectroscopy relationship, founded by chemical synthesis and modification of the various polyyne molecules, allows rational engineering to create the most extensive super-multiplexed vibrational palette on record now. Polyynes are particularly preferential in Raman detection for their intrinsically strong peak with narrow linewidth (13 cm−1) in the cell-silent window. Both Raman intensity and frequency vary according to different conjugation lengths of different polyynes. The Raman intensity also grows superlinearly as the triple bonds increase from two to six, rendering the detection limit down to sub-uM for polyynes beyond 4-ynes. Other than the intrinsic coarse tuning capability from various conjugation lengths, the general isotope-editing strategy also works to tune the vibrational frequency in a relatively moderate way. Substituents on the phenyl cap of the polyyne molecules can further be modified to influence the force constant, providing capacities for frequency finetuning. With such rational frequency-tuning strategies(Figure 2b), Carbow palettes contain 20 well-resolved vibrational colors, perfectly filling the spectral window from 2000 cm−1 to 2280 cm−1 (Figure 1 a). The further functionalization of organelle-targeting side chains on the Carbow molecules turn these vibrational colors into organelle-specific probes(Figure 3c). The neutral and non-polar polyyne backbones turn out to be preferential for the probe’s superb biocompatibility and labeling specificity in live-cell organelle imaging (Figure 4a).
Figure 4. Biomedical applications enabled by super-multiplexed vibrational microscopy.
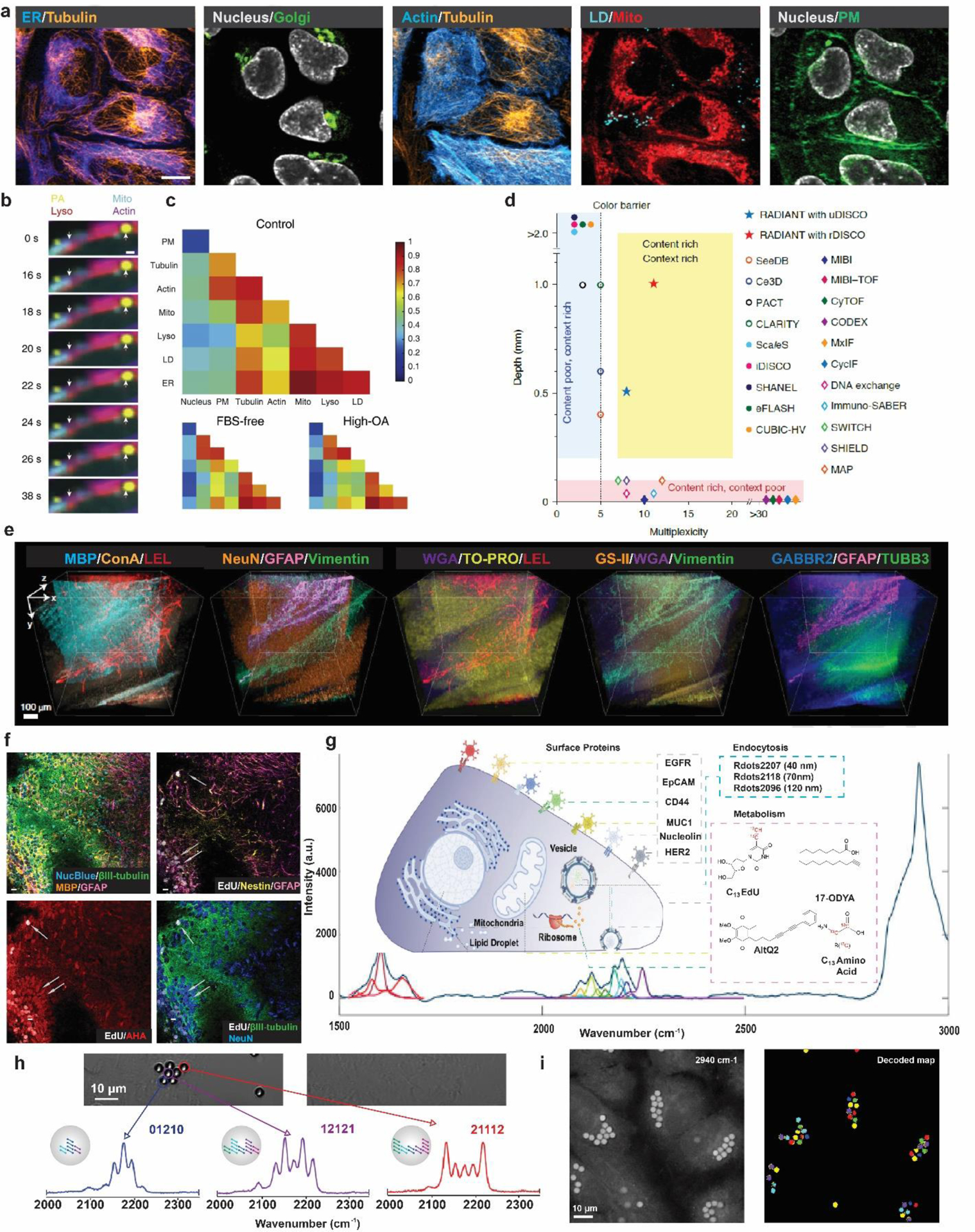
a. Ten-color optical imaging of PM (Carbow2141), ER (Carbow2226), Golgi (BODIPY TR), Mito (Carbow2262), LD (Carbow2202), Lyso (Carbow2086), nucleus (NucBlue), tubulin (SiR650), actin (GFP) and FM 4–64 in living Hela cells. Scale bar, 10 μm. b. Super-multiplex time-lapse imaging of live cells simultaneously showing shortening of Mito along the actin and small movement of large LD. The arrows indicate the initial positions of the mitochondria edge and the large LD. Scale bar, 1 μm. c. Single-cell analysis of organelle interactome by super-multiplex image-based cytometry of live cells. The matrix representation of medians of organelle correlation coefficients here reveals cell-to-cell heterogeneity for live HeLa cells grown in different culture conditions. d. Illustration on the content-context trade-off in immunohistochemistry-based protein imaging for current techniques. e. One-shot eleven-target volumetric imaging of 1-mm thick mouse cerebellum sections by RADIANT. Fluorescence: ConA (Concanavalin A), GS-II (Griffonia simplicifolia lectin), TUBB3, TO-PRO-3 (cell nuclei); epr-SRS: NeuN, LEL (Lycopersicon Esculentum lectin), MBP, GABBR2, WGA, Vimentin, GFAP. Z-step: 5 μm. Scale bars, 50 μm. f. Eight-color epr-SRS imaging of DNA replication and protein synthesis in hippocampal neuronal cultures epr-SRS: β-III-tubulin(neurons, green), myelin basic protein (MBP; oligodendrocytes, orange), glial fibrillary acidic protein (GFAP, astrocytes and neural stem cells, magenta), EdU(newly synthesized DNA, gray), AHA(newly synthesized proteins, red). Fluorescence: Nucblue (total DNA, cyan), NeuN (neurons, blue) and nestin (neural stem cells and astrocytes, yellow). Scale bars, 10 μm. g. Diagram illustrating of 14 Raman probes for multiparameter live-cell profiling and their subcellular targets and the resulting 14-plexed live-cell Raman spectra with unmixing processing. h. Typical Carbow barcoded cell IDs read out by spontaneous Raman measurement. i. Decoding and spatial visualization of barcoded beads in live cells with SRS microscopy. Scale bars, 10 μm.
However, sub-uM sensitivity is still not enough to visualize low-abundance molecular targets in the biological system. Thus, a recent study introduced a nano method, providing a simple intuition to overcome the long-lasting sensitivity bottleneck for Raman microscopy. The study makes use of Carbow dyes and incorporates them into small polymeric nanoparticles to generate ultrabright Raman dots (Rdots) as new imaging reagents for SRS microscopy27. 20 nm polystyrene nanoparticles turned out to concentrate the hydrophobic Carbow dyes through a simple swell-diffusion strategy so effectively that single-particle sensitivity was accomplished under the SRS microscope. With negligible spectral shift and broadening, this method serves as a general and robust way to boost the sensitivity down to the sub-nM level while preserving the full super-multiplexed capability of Carbow palettes. Labeling functionality can also be realized by conjugating PEGs and antibodies with the abundant carboxyl groups on the particle surface, allowing further biomedical applications such as multiplexed immunostaining protein imaging (Figure 3a).
In general, building successful super-multiplexed vibrational palettes have three critical criteria to consider: superb detection sensitivity, tunable and resolvable vibrational signatures, and labeling functionalities. Superb sensitivity sometimes originates from intrinsic large Raman cross-sections of certain vibrational bonds that are susceptible to polarization. Coupling vibrational modes with the electronic transition will further boost the sensitivity by orders of magnitude depending on electronic resonance enhancement28–30. Even single molecules can be observed with vibrational signature when fluorescence signals are induced for detection in Stimulated Raman excited fluorescence spectroscopy (SREF)31–33. Nano strategy provides an alternative solution to complement sensitivity by increasing the local concentration of the Raman active bonds in the focal volume. Polymer chemistry also makes notable demonstration on its profound utility in making Raman-active bonds condensed imaging probes with devised functional modalities for biomedical applications, such as live-cell organelle imaging (Figure 3e) and multiplexed tumor detection34–35. Distinct and resolvable Raman signatures, fundamental to supermultiplexed palettes development, are more accessible in the cell-silent window, free from complicated peak interferences. Investigation on the structure-spectroscopy relationship is helpful to rationalize frequency tuning on different molecular templates (Figure 2). Despite some modifications unique to certain structural features, adjusting the force constant and the reduced mass provides a relative general intuition. Isotope-editing can serve as a universal strategy to expand the vibrational palettes in a relative coarse-tuning manner with negligible alternation on chemical properties. Targeting functionalities to label with specificity is the ultimate step for vibrational colors to become real imaging probes, whose intrinsic properties in the biological system render specific applications under different biological scenarios (Figure 3). We also want to note some recent efforts on developing photo-switchable Raman probes, which greatly enrich the Raman toolbox for bio-imaging and potentially expand the utility for vibrational microscopy to decipher complex biological systems36–38.
Biomedical applications
Super-multiplexed vibrational probe development lays a solid foundation for various interesting biomedical applications. From structural imaging to metabolic profiling, super-multiplexed vibrational microscopy fosters powerful new techniques to visualize and understand complex biological systems. The unique optical multiplexing capability of vibrational microscopy outstands current multiplexed imaging techniques. Especially when it comes to tricky yet valuable scenarios, such as working with living cells or thick tissues, one-shot imaging becomes essential as iterative or destructive methods no longer apply7–8, 39–40 (Figure 4d). The expanded available multiplexing colors also give rise to barcoding applications with impressive potential in optical data storage and identification for high-throughput biomedical analysis. Here, we highlight some latest super-multiplexed biomedical applications based on vibrational microscopy, hoping to illustrate the idea of how advances in optical multiplexing capabilities can bring a revolutionary difference.
Supermultiplexed structural imaging of biological system
On establishing the Carbow palette, researchers functionalized five Carbow dyes with different organelle targeting groups for specific organelle imaging. Combining with another five compatible fluorescent reporters, researchers first achieve 10-color optical imaging of specific subcellular structures in vivo(Figure 4a)15. Simultaneous visualization of multiple organelles and skeleton proteins in living Hela cells was demonstrated with high quality, which is particularly favorable in studying organelle interactome. Organelle interactome regarding cellular organization and dynamics has been recognized as an important question in cell biology41. With recent instrumental advances of integrated stimulated Raman and fluorescence microscopy, visualization of dynamic organelle interactomes are enabled using Carbow probes at seconds level temporal resolution for image-based cytometry (Figure 4b), unveiling the dynamics and heterogeneity in live cells (Figure 4c)42.
Highly multiplexed-epitope imaging, which provides comprehensive, spatially resolved information on cell types and cell states in tissues, is pivotal to comprehending the complex biological system7. With the organization being 3D by its nature, structural mapping of the biological system, especially for the nervous system, where cells extend over large distances43, requires volumetric imaging techniques44–45. However, there is a general trade-off between contents (high multiplexing) and context (thick tissues)(Figure 4d), leaving highly multiplexed protein imaging in large 3D volumes out of reach. Recently, researchers addressed this challenge with SRS microscopy by integrating multiplexed imaging enabled by well-engineered MARS palettes and inspiration for large context imaging from Raman-tailored tissue clearing. They developed a method of Raman Dye Imaging and Tissue Clearing (RADIANT) optimized for immunostaining based on MARS labeled antibodies46. Combining with fluorescence microscopy, up to 11 targets in millimeter-thick brain slices were imaged in one shot(Figure 4e), which outperformed prior multiplexed protein imaging methods by 10- to 100- fold on imaging depths40, 47–50. Valuable system information extracted from such multiplexed volumetric data can reveal region-specific correlation network and their topology during life development, facilitating the comprehensive understanding of 3D protein interaction in the complex biological system.
Supermultiplexed metabolic imaging and profiling
Metabolism is also one of the defining characters of life, which fundamentally requires multiplexing techniques for a thorough description of the synthesis, transformation, and degradation of various biomolecules inside cells. There have long been small metabolic Raman probes developed from stable isotope labels (C-D) or triple-bond vibrational tags (C≡C, C≡N) to image the metabolic activity with minimized perturbation23 (Figure 3). Multiplexed application of metabolic probes is beneficial in single-cell profiling on metabolic activity to disclose cell heterogeneities. Integrating instrumental advances, a technique, metabolic activity phenotyping (MAP), was developed to quantitatively measure lipid and protein synthesis at high throughput (6000 cells/ hour) for single-cell profiling with micro-Raman spectroscopy51. Much higher throughput (over 10,000 cells in several minutes) can be achieved when applying IR-active metabolic vibrational probes to mid-infrared microscopy52. Multiplexed imaging of metabolic activity was demonstrated with large field of view and short data acquisition time, reporting the intricate heterogeneity in various biological systems (Figure 3f).
Moving from small vibrational tags, specific functionalization of the current super multiplexed vibrational palettes paves another way for more multiplexed detection and better imaging sensitivity. Click chemistry combined with metabolic labeling reagents (e.g., AHA and EdU) enable MARS dyes to visualize metabolic activities, such as DNA replication and protein synthesis25 (Figure 3 i,j). With other biomarkers stained simultaneously, super-multiplexed vibrational imaging is prone to give a multi-omics view of the molecular heterogeneity in situ (Figure 4f)14. Harnessing the unique epr-SRS spectroscopy, a new study invented a set of activatable Raman probes for multiplexed sensing of enzymatic activities. Appropriate enzyme-substrate moieties were conjugated onto 9-Cyanopyronin via an amide bond, giving switchable SRS contrasts. After enzymatic bond cleavage, the absorption of the fluorophore red-shifts from electronic non-resonance condition to electronic pre-resonance condition. Isotope-editing helps expand the vibrational colors and enables multiplexed SRS imaging of enzymatic activities (Figure 3k,l)26.
Multiplexed Raman techniques have been widely used to detect specific biomarkers and endocytosis for live-cell imaging using nanoparticles12, 35, 53–55. To fulfill the powerful multiplexing potential, a live-cell profiling technology based on Raman micro-spectroscopy came to place this year, combining the surface marker detection and metabolic activities evaluation of individual live cells56. In total 14-plexed live-cell profiling was demonstrated, which includes ten modified super-multiplexed Rdots quantifying surface protein markers and probing the endocytosis activities plus four compatible Raman-tagged metabolites revealing cellular metabolism simultaneously (Figure 4g). The single-cell multiparameter measurement platform on a simple instrument enables informative clustering, correlation, and network analysis regarding single-cell phenotyping of general biological interests.
Supermultiplexed information barcoding
Multiplexing capability also promotes creating barcodes for data storage, which set platforms for high throughput biomedical analysis to inventory biological molecules, cells, or even spatial relationships. With Carbow palettes, the researchers have explored its utility in optical barcoding by incorporating specified polyyne mixtures into micro-size polystyrene beads via swell-diffusion. The researchers showcased that using ten distinct vibrational colors at three intensity levels (digitized levels 0, 1, 2) can generate up to 59,048 (310 − 1) unique spectral barcodes to tag individual cells with unique IDs (Figure 4h,i)15. Recently, such color-intensity coding strategies have also been applied to covalent polymer nanoparticles to effectively expand the Raman-active codes for multiplexed SRS imaging of the biological system57. More ambitious intensity-coding and more mindful Raman frequency combination can further leverage vibrational barcoding capability, yielding the largest capacity of optical barcodes to date (524,288)58.
Conclusion and outlook
Super-multiplexed vibrational imaging has unique advantages working with biological systems with intrinsic complexity. Microscopy is intrinsically complementary to the popular approaches for “omics” which have massive multiplexing ability but poor spatiotemporal resolution. The recent two years have witnessed blooming applications with super-multiplexed vibrational microscopy, coming after the development MARS and Carbow palettes. Nevertheless, potential challenges are to be addressed for further expanding the vibrational palettes. Vibrational probes with frequencies in the uncharted region of the cell-silent window are waiting for exploration, which might require specialties in chemical synthesis to cope with the relatively uncommon chemical bonds59. New vibrational color alone does not readily become an imaging probe without sufficient sensitivity and proper functionalities. As typical chemical bonds have detection limits at millimolar magnitude with SRS microscopy, strategies enhancing the sensitivity might be necessary to visualize specific targets. Finally, the unique utility of super-multiplexed vibrational microscopy for biological systems is to visualize multiple aspects of the same sample simultaneously. Proper functionalization of the probes are substantial to ensure the multi-parameter measurement is bio-compatible, and the detection results together provide meaningful information to resolve true biological questions. We envision that with the field’s effort being dedicated towards vibrational probe development, the ultimate super-multiplexed vibrational techniques will present their power in biomedical applications far beyond our imagination.
Acknowledgment:
W.M. acknowledges support from NIH (R01 EB029523, R01 GM132860 and R01 GM128214).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Krutzik PO; Nolan GP, Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 2006, 3 (5), 361–8. [DOI] [PubMed] [Google Scholar]
- 2.Nitta N; Sugimura T; Isozaki A; Mikami H; Hiraki K; Sakuma S; Iino T; Arai F; Endo T; Fujiwaki Y, Intelligent image-activated cell sorting. Cell 2018, 175 (1), 266–276. e13. [DOI] [PubMed] [Google Scholar]
- 3.Chen KH; Boettiger AN; Moffitt JR; Wang S; Zhuang X, Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348 (6233). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su JH; Zheng P; Kinrot SS; Bintu B; Zhuang X, Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 2020, 182 (6), 1641–1659 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang X, Spatially resolved single-cell genomics and transcriptomics by imaging. Nature Methods 2021, 18 (1), 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goltsev Y; Samusik N; Kennedy-Darling J; Bhate S; Hale M; Vazquez G; Black S; Nolan GP, Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 2018, 174 (4), 968–981. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]; The following two reviews provide comprehensive accounts for prevalent multiplexed imaging techniques at current.
- 7.Bodenmiller B, Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst 2016, 2 (4), 225–38. [DOI] [PubMed] [Google Scholar]
- 8.Lewis SM; Asselin-Labat ML; Nguyen Q; Berthelet J; Tan X; Wimmer VC; Merino D; Rogers KL; Naik SH, Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods 2021. [DOI] [PubMed] [Google Scholar]
- 9.Ackerman CM; Myhrvold C; Thakku SG; Freije CA; Metsky HC; Yang DK; Ye SH; Boehm CK; Kosoko-Thoroddsen TF; Kehe J; Nguyen TG; Carter A; Kulesa A; Barnes JR; Dugan VG; Hung DT; Blainey PC; Sabeti PC, Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582 (7811), 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M; Gao X; Su JZ; Nie S, Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nature biotechnology 2001, 19 (7), 631–635. [DOI] [PubMed] [Google Scholar]
- 11.Niehorster T; Loschberger A; Gregor I; Kramer B; Rahn HJ; Patting M; Koberling F; Enderlein J; Sauer M, Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat Methods 2016, 13 (3), 257–62. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z; Zong S; Wu L; Zhu D; Cui Y, SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem Rev 2017, 117 (12), 7910–7963. [DOI] [PubMed] [Google Scholar]
- 13.Wang YW; Doerksen JD; Kang S; Walsh D; Yang Q; Hong D; Liu JT, Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection - Enhanced Topical Staining with SERS - Coded Nanoparticles. Small 2016, 12 (40), 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported the first super-multiplexed vibrational palettes, MARS.
- 14.Wei L; Chen Z; Shi L; Long R; Anzalone AV; Zhang L; Hu F; Yuste R; Cornish VW; Min W, Super-multiplex vibrational imaging. Nature 2017, 544 (7651), 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work came slightly later than MARS, presenting another super-multiplxed vibrational palette, Carbow. Having 20 well-resolved vibrational colors, Carbow is the most extensive palette up to now.
- 15.Hu F; Zeng C; Long R; Miao Y; Wei L; Xu Q; Min W, Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat Methods 2018, 15 (3), 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min W; Freudiger CW; Lu S; Xie XS, Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annual review of physical chemistry 2011, 62, 507–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freudiger CW; Min W; Saar BG; Lu S; Holtom GR; He C; Tsai JC; Kang JX; Xie XS, Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 2008, 322 (5909), 1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu F; Shi L; Min W, Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nat Methods 2019, 16 (9), 830–842. [DOI] [PubMed] [Google Scholar]
- 19.Wei L; Hu F; Shen Y; Chen Z; Yu Y; Lin C-C; Wang MC; Min W, Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nature methods 2014, 11 (4), 410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamakoshi H; Dodo K; Okada M; Ando J; Palonpon A; Fujita K; Kawata S; Sodeoka M, Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J Am Chem Soc 2011, 133 (16), 6102–5. [DOI] [PubMed] [Google Scholar]
- 21.Yamakoshi H; Dodo K; Palonpon A; Ando J; Fujita K; Kawata S; Sodeoka M, Alkyne-tag Raman imaging for visualization of mobile small molecules in live cells. J Am Chem Soc 2012, 134 (51), 20681–9. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z; Paley DW; Wei L; Weisman AL; Friesner RA; Nuckolls C; Min W, Multicolor live-cell chemical imaging by isotopically edited alkyne vibrational palette. J Am Chem Soc 2014, 136 (22), 8027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y; Hu F; Min W, Raman Imaging of Small Biomolecules. Annu Rev Biophys 2019, 48, 347–369. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Z; Shen Y; Hu F; Min W, Applications of vibrational tags in biological imaging by Raman microscopy. Analyst 2017, 142 (21), 4018–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]; This follow-up work on MARS palettes provides a solid structure-spectroscopy study of 9-Cyanopyronin based MARS dyes and built a probe library with expanded functionalities.
- 25.Miao Y; Qian N; Shi L; Hu F; Min W, 9-Cyanopyronin probe palette for super-multiplexed vibrational imaging. Nat Commun 2021, 12 (1), 4518. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrated the first activatable Raman sensors for multiplexed visualization of live-cell enzymatic activities.
- 26.Fujioka H; Shou J; Kojima R; Urano Y; Ozeki Y; Kamiya M, Multicolor Activatable Raman Probes for Simultaneous Detection of Plural Enzyme Activities. J Am Chem Soc 2020, 142 (49), 20701–20707. [DOI] [PubMed] [Google Scholar]; This work introduces a novel nano-strategy to boost the detection sensitivity with Carbow dyes for high-quality multiplexed epitope imaging.
- 27.Zhao Z; Chen C; Wei S; Xiong H; Hu F; Miao Y; Jin T; Min W, Ultra-bright Raman dots for multiplexed optical imaging. Nat Commun 2021, 12 (1), 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi L; Xiong H; Shen Y; Long R; Wei L; Min W, Electronic Resonant Stimulated Raman Scattering Micro-Spectroscopy. J Phys Chem B 2018, 122 (39), 9218–9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L; Min W, Electronic Preresonance Stimulated Raman Scattering Microscopy. J Phys Chem Lett 2018, 9 (15), 4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y; Zhuang Y; Zhang S; Smith ZJ; Li Y; Mu X; Li M; He C; Zheng X; Pan F, AzoEnhanced Raman Scattering for Enhancing the Sensitivity and Tuning the Frequency of Molecular Vibrations. ACS Central Science 2021, 7 (5), 768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong H; Shi L; Wei L; Shen Y; Long R; Zhao Z; Min W, Stimulated Raman Excited Fluorescence Spectroscopy and Imaging. Nat Photonics 2019, 13 (6), 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong H; Qian N; Miao Y; Zhao Z; Min W, Stimulated Raman Excited Fluorescence Spectroscopy of Visible Dyes. J Phys Chem Lett 2019, 10 (13), 3563–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong H; Min W, Combining the best of two worlds: Stimulated Raman excited fluorescence. The Journal of Chemical Physics 2020, 153 (21), 210901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian S; Li H; Li Z; Tang H; Yin M; Chen Y; Wang S; Gao Y; Yang X; Meng F; Lauher JW; Wang P; Luo L, Polydiacetylene-based ultrastrong bioorthogonal Raman probes for targeted live-cell Raman imaging. Nat Commun 2020, 11 (1), 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Q; Fan X; Chen C; Huang L; Wang J; Tang X, Multicolor Raman Beads for Multiplexed Tumor Cell and Tissue Imaging and in Vivo Tumor Spectral Detection. Anal Chem 2019, 91 (6), 3784–3789. [DOI] [PubMed] [Google Scholar]
- 36.Ao J; Fang X; Miao X; Ling J; Kang H; Park S; Wu C; Ji M, Switchable stimulated Raman scattering microscopy with photochromic vibrational probes. Nat Commun 2021, 12 (1), 3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee D; Qian C; Wang H; Li L; Miao K; Du J; Shcherbakova DM; Verkhusha VV; Wang LV; Wei L, Toward photoswitchable electronic pre-resonance stimulated Raman probes. The Journal of Chemical Physics 2021, 154 (13), 135102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shou J; Ozeki Y, Photoswitchable stimulated Raman scattering spectroscopy and microscopy. Optics Letters 2021, 46 (9), 2176–2179. [DOI] [PubMed] [Google Scholar]
- 39.Gut G; Herrmann MD; Pelkmans L, Multiplexed protein maps link subcellular organization to cellular states. Science 2018, 361 (6401). [DOI] [PubMed] [Google Scholar]
- 40.Giesen C; Wang HA; Schapiro D; Zivanovic N; Jacobs A; Hattendorf B; Schüffler PJ; Grolimund D; Buhmann JM; Brandt S, Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature methods 2014, 11 (4), 417–422. [DOI] [PubMed] [Google Scholar]
- 41.Valm AM; Cohen S; Legant WR; Melunis J; Hershberg U; Wait E; Cohen AR; Davidson MW; Betzig E; Lippincott-Schwartz J, Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 2017, 546 (7656), 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shou J; Oda R; Hu F; Karasawa K; Nuriya M; Yasui M; Shiramizu B; Min W; Ozeki Y, Super-multiplex imaging of cellular dynamics and heterogeneity by integrated stimulated Raman and fluorescence microscopy. iScience 2021, 24 (8), 102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichtman JW; Denk W, The big and the small: challenges of imaging the brain’s circuits. Science 2011, 334 (6056), 618–623. [DOI] [PubMed] [Google Scholar]
- 44.Ueda HR; Erturk A; Chung K; Gradinaru V; Chedotal A; Tomancak P; Keller PJ, Tissue clearing and its applications in neuroscience. Nat Rev Neurosci 2020, 21 (2), 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueda HR; Dodt HU; Osten P; Economo MN; Chandrashekar J; Keller PJ, Whole-Brain Profiling of Cells and Circuits in Mammals by Tissue Clearing and Light-Sheet Microscopy. Neuron 2020, 106 (3), 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrated the highly multiplexed volumetric epitope imaging. Mapping up to 11 targets in a millimeter-thick brain slice, this work overcame the typical trade-off between content and context from current multiplexed imaging techniques.
- 46.Shi L; Wei M; Miao Y; Qian N; Shi L; Singer R; Benninger R; Min W, Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nature Biotechnology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray E; Cho JH; Goodwin D; Ku T; Swaney J; Kim S-Y; Choi H; Park Y-G; Park J-Y; Hubbert A, Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 2015, 163 (6), 1500–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ku T; Swaney J; Park J-Y; Albanese A; Murray E; Cho JH; Park Y-G; Mangena V; Chen J; Chung K, Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature biotechnology 2016, 34 (9), 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park Y-G; Sohn CH; Chen R; McCue M; Yun DH; Drummond GT; Ku T; Evans NB; Oak HC; Trieu W, Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nature biotechnology 2019, 37 (1), 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelo M; Bendall SC; Finck R; Hale MB; Hitzman C; Borowsky AD; Levenson RM; Lowe JB; Liu SD; Zhao S; Natkunam Y; Nolan GP, Multiplexed ion beam imaging of human breast tumors. Nat Med 2014, 20 (4), 436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Z; Chen C; Xiong H; Ji J; Min W, Metabolic Activity Phenotyping of Single Cells with Multiplexed Vibrational Probes. Anal Chem 2020, 92 (14), 9603–9612. [DOI] [PubMed] [Google Scholar]; This work first introduced labeled metabolic probes into mid-IR microscopy, achieving high-throughput metabolic imaging under vibrational microscopy.
- 52.Shi L; Liu X; Shi L; Stinson HT; Rowlette J; Kahl LJ; Evans CR; Zheng C; Dietrich LEP; Min W, Mid-infrared metabolic imaging with vibrational probes. Nature Methods 2020, 17 (8), 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S; Chen T; Wang Y; Liu L; Lv F; Li Z; Huang Y; Schanze KS; Wang S, Conjugated Polymer with Intrinsic Alkyne Units for Synergistically Enhanced Raman Imaging in Living Cells. Angew Chem Int Ed Engl 2017, 56 (43), 13455–13458. [DOI] [PubMed] [Google Scholar]
- 54.Hu F; Brucks SD; Lambert TH; Campos LM; Min W, Stimulated Raman scattering of polymer nanoparticles for multiplexed live-cell imaging. Chem Commun (Camb) 2017, 53 (46), 6187–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis RM; Kiss B; Trivedi DR; Metzner TJ; Liao JC; Gambhir SS, Surface-enhanced Raman scattering nanoparticles for multiplexed imaging of bladder cancer tissue permeability and molecular phenotype. Acs Nano 2018, 12 (10), 9669–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work showcase the utilities in live-cell profiling with 14-plexed Raman probe panel, quantifying cell surface proteins, endocytosis activities, and metabolic dynamics of individual cells simultaneously.
- 56.Chen C; Zhao Z; Qian N; Wei S; Hu F; Min W, Multiplexed live-cell profiling with Raman probes. Nat Commun 2021, 12 (1), 3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu W; Cai EL; Li HZ; Wang P; Shen AG; Popp J; Hu JM, Precise Encoding of TripleBond Raman Scattering of Single Polymer Nanoparticles for Multiplexed Imaging Application. Angew Chem Int Ed Engl 2021. [DOI] [PubMed] [Google Scholar]
- 58.Tang Y; He C; Zheng X; Chen X; Gao T, Super-capacity information-carrying systems encoded with spontaneous Raman scattering. Chem Sci 2020, 11 (11), 3096–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mochizuki M; Sato S; Asatyas S; Leśnikowski ZJ; Hayashi T; Nakamura H, Raman cell imaging with boron cluster molecules conjugated with biomolecules. RSC Advances 2019, 9 (41), 23973–23978. [DOI] [PMC free article] [PubMed] [Google Scholar]


