Abstract
Tanshinones are the bioactive diterpenoid constituents of the traditional Chinese medicinal herb Danshen (Salvia miltiorrhiza), and are examples of the phenolic abietanes widely found within the Lamiaceae plant family. Due to the significant interest in these labdane-related diterpenoid natural products, their biosynthesis has been intensively investigated. In addition to providing the basis for metabolic engineering efforts, this work further yielded pioneering insights into labdane-related diterpenoid biosynthesis in the Lamiaceae more broadly. This includes stereochemical foreshadowing of aromatization, with novel protein domain loss in the relevant diterpene synthase, as well as broader phylogenetic conservation of the relevant enzymes. Beyond such summary of more widespread metabolism, formation of the furan ring that characterizes the tanshinones also has been recently elucidated. Nevertheless, the biocatalysts for the pair of demethylations remain unknown, and the intriguing potential connection of these reactions to the further aromatization observed in the tanshinones are speculated upon here.
Introduction
Salvia miltiorrhiza is a Lamiaceae species whose rhizome (roots), termed danshen, is one of the most widely used herbs in traditional Chinese medicine (TCM). Its intense coloration led to its original designation as red sage, and danshen is recorded as a supergrade herb in Shennong’s Herbal Classic of Materia Medica (written in 25–220 CE during the East Han Dynasty). Thus, danshen has been used for more than 2000 years and is currently one of the most important herbs for TCM treatment of cardiovascular diseases in China and other Asian countries. Moreover, Compound Danshen Dripping Pills T89, with danshen as one of the major active components, has completed phase III clinical trials in the USA in 2017 (NCT01659580) and is now undergoing outcome research to confirm the anti-anginal effect in a phase III multi-centered clinical trial (NCT03789552). Many pharmacological studies have shown that danshen has various biological activities, including anti-oxidant, anti-inflammatory, anti-tumor and antimicrobial effects [1].
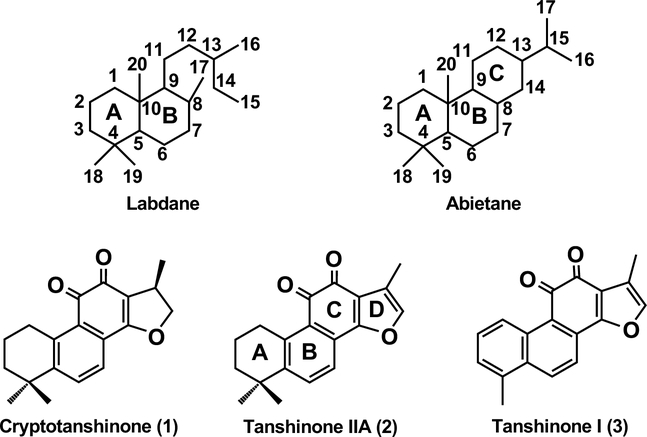
The lipophilic bioactive component of danshen is composed of the tanshinones [2], which also are then among the molecular markers used to evaluate the quality of danshen medicinal materials for use in TCM [3]. These diterpenoid natural products are highly elaborated derivatives of phenolic abietanes, which are widely found in the Lamiaceae [4]. In particular, not only is the phenolic ‘C’ ring further transformed to an ortho-quinone, but carbon-20 (C20) is lost (demethylation) with additional aromatization of the adjacent ‘B’ ring and formation of an additional 14,16-epoxy ‘D’ ring as well [5]. Thus, the tanshinones are minimally norditerpenoids, such as cryptotanshinone (1), with the ‘D’ ring generally further oxidized to a furan, such as in tanshinone IIA (2). Also observed is an additional demethylation (loss of C19) and aromatization of the ‘A’ ring, yielding 19,20-dinor derivatives such as tashinone I (3) (Figure 1). In keeping with their extensively conjugated structures, the tanshinones provide the reddish pigmentation of the rhizome.
Figure 1. Tanshinones and their base structures.
Shown are the base structures for labdanes and abietanes, along with the three most prevalent tanshinones found in S. miltiorrhiza (numbered as described in the text)
Due to their medicinal value, chemical total syntheses of tanshinones have attracted substantial interest, but these have been limited by low yields and are not economically viable [6]. Thus, the commercial supply of tanshinones relies on extraction from S. miltiorrhiza rhizomes. While danshen was originally harvested from the wild, due to increasing demand this is no longer practical. Accordingly, S. miltiorrhiza is now field cultivated, which has become the main source of danshen. With good agricultural practices the resulting danshen is relatively uniform in quality, but can contain heavy metals and pesticide residues [7]. An alternative source is hairy root and cell cultures of S. miltiorrhiza, which produce and secrete tanshinones, particularly in response to elicitation with fungal extracts [8], but the yields from such cultures also does not yet appear to be economically viable.
As with many other natural products of interest, elucidation of tanshinone biosynthesis has been sought to provide insight into the underlying intriguing enzymatic transformations as well as molecular targets for metabolic engineering, both in planta and for synthetic biology approaches in microbial hosts such as yeast (Saccharomyces cerevisiae) [2]. Here the current progress towards this goal is summarized, not least as the recent identification of the cytochrome P450 (CYP) monooxygenases that form the 14,16-epoxy ‘D’ ring provides demarcation between the upstream transformations, which are more widespread in the Lamiaceae, and the downstream more tanshinone specific pathway [9]. Moreover, as also reviewed here, elucidation of the upstream portions of tanshinone biosynthesis provided entry into not only phenolic abietane but also other labdane-related diterpenoid biosynthesis in the Lamiaceae more broadly. Indeed, even beyond the prevalent phenolic abietanes, the Lamiaceae plant family is a particularly rich source of diverse labdane-related diterpenoids [10].
Precursor supply
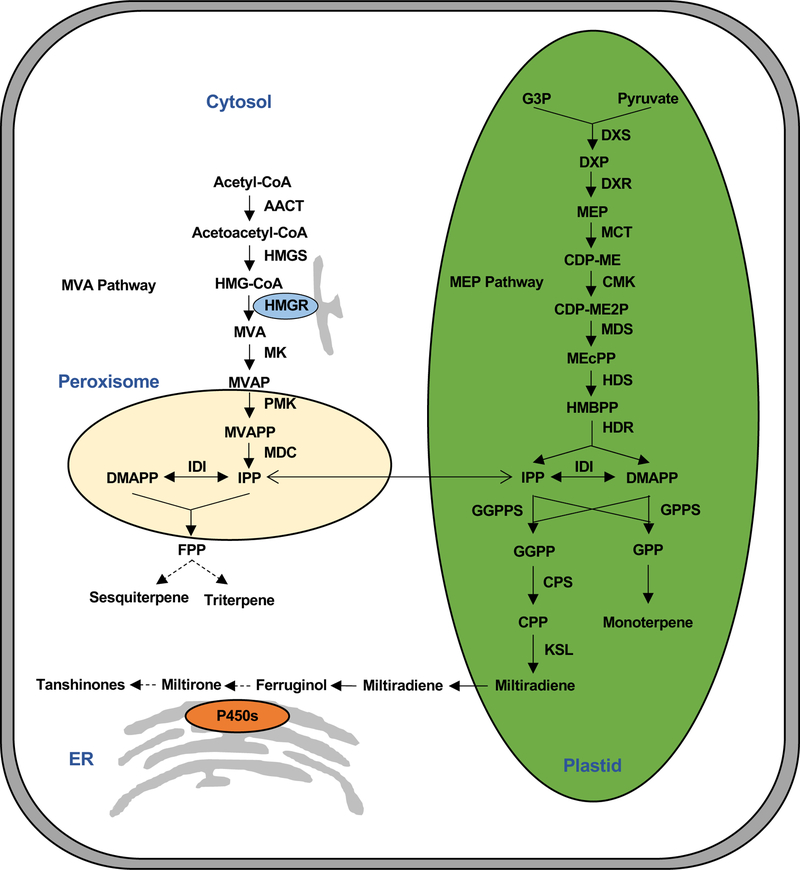
As isoprenoids, tanshinones originate from the universal precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). In plants, these precursors are produced via distinct pathways in the cytoplasm and plastid (Figure 1), which are termed the mevalonate (MVA) and 2-C-methyl-D-erythritol 4-phosphate (MEP) dependent pathways, respectively [11]. As diterpenoids, the tanshinones are largely derived from the plastid MEP pathway, although cross-talk between these sub-cellular compartments has been observed. Indeed, cloning and overexpression of a hydroxymethylglutaryl-CoA reductase (HMGR) from the MVA dependent pathway in S. miltiorrhiza increased tanshinone production in hairy root cultures [12]. Nevertheless, overexpression of a 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from the MEP dependent pathway increases tanshinone production significantly more than does overexpression of HMGR [13].
Following production of the universal isoprenoid precursors, DMAPP and three units of IPP must then be condensed to form the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP, 4) by a GGPP synthase (GGPPS). As with many plant species [14], S. miltiorrhiza contains a family of genes encoding GGPPS, as well as for the various enzymes from both MVA and MEP dependent isoprenoid precursor pathways [15]. Also as seen in other plant species [14], overexpression of an S. miltiorrhiza GGPPS significantly increases tanshinone production, even more than overexpression of either HMGR or DXS [13].
Expression pattern matters
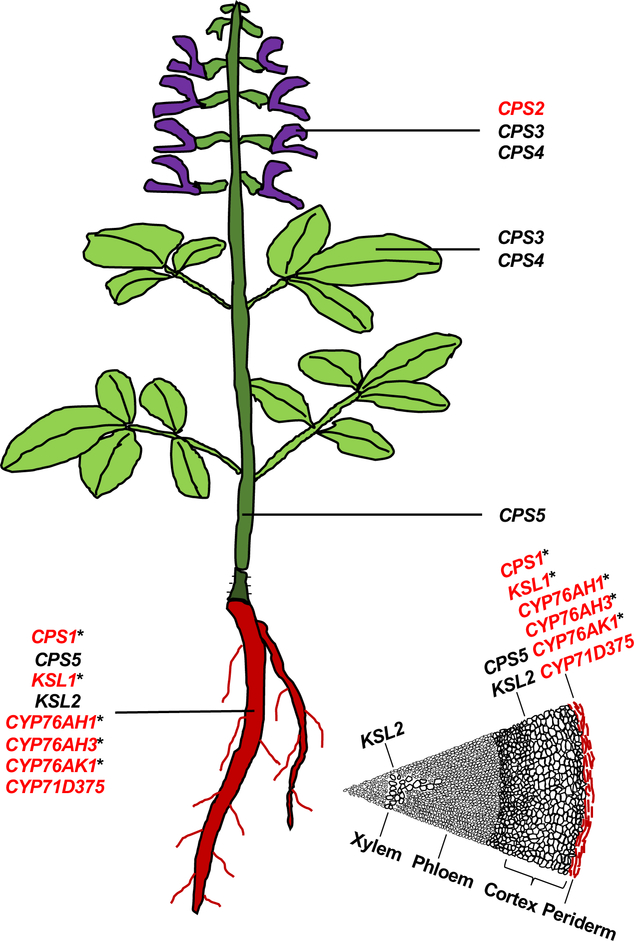
In the case of the GGPPS family, as well as those for the enzymes from both the MVA and MEP dependent isoprenoid precursor pathways, members of each family exhibit essentially identical catalytic activity. The individual members then generally differ in expression pattern and/or regulatory mechanism. Accordingly, while overexpression of any family member is usually sufficient to increase metabolic flux, at least through the corresponding reaction, the family member(s) most relevant to tanshinone production are presumed to be those with matching expression pattern. For example, the age-dependent production of tanshinones in S. miltiorrhiza hairy root cultures was used to propose the relevant genes from analysis of a cDNA-array with ~4,350 genes [16]. Similarly, an expressed sequence tag (EST) approach to root and leaf tissues was used to generate a more comprehensive list [17]. A combination of metabolomics and transcriptomics was used in combination with induction of such hairy root cultures to provide a more comprehensive view of the relevant members from the various enzymatic families expected to play roles in tanshinone biosynthesis [18]. More precise localization of tanshinone accumulation to the rhizome/root periderm provided further specificity for identification of the relevant genes (Figure 3) [19]. Beyond those involved in assembly of the general diterpenoid precursor GGPP, these include members of the terpene synthase (TPS) family and those for oxygenases/oxidases, particularly from the cytochrome P450 (CYP), 2-oxo-glutarate dependent dioxygenase (2ODD) and short chain alcohol dehydrogenase/reductase (SDR) families. The members of these latter families differ in catalytic activity and, thus, require biochemical characterization for identification of the relevant enzyme(s), which are then more specific to tanshinone biosynthesis as described below.
Figure 3. Tissue and cell-type specific CPS, KSL and CYP gene expression in S. miltiorrhiza.
Key enzymatic genes highly expressed in root (colored red, consistent with the accumulation of the pigmented tanshinones in this organ), leaf and flower. In addition, the root was further divided into four cross-section cell-types, roughly correlated to the periderm (colored red, consistent with the more specific localization of tanshinones in this cell-type), cortex, phloem and xylem, and those genes known to have higher expression in these cell-types also are indicated. Red text indicates genes with known roles in tanshinone biosynthesis and those known to exhibit inducible expression with an asterisk (*)
Formation of the abietane backbone
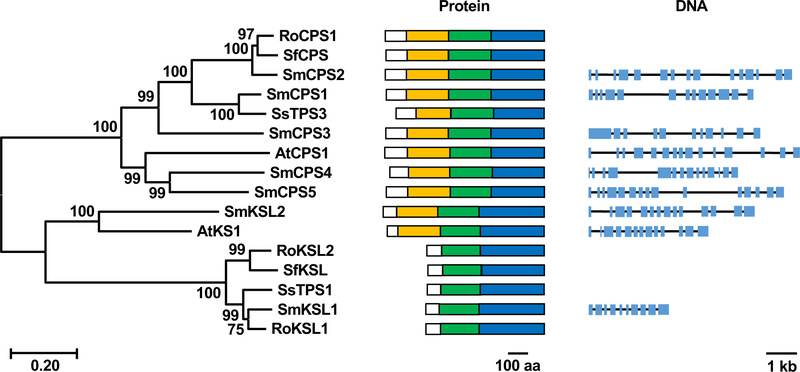
As abietanes, the tanshinones fall within the labdane-related diterpenoid superfamily, which is characterized by initial cyclization of GGPP (4) catalyzed by a class II diterpene cyclase [20]. These enzymes generate a trans-decalin bicycle via protonation of the terminal alkene in 4, with sequential anti addition of the two internal alkenes yielding the eponymous labda-13-en-8-yl+ diphosphate carbocation intermediate, which is most commonly immediately deprotonated at the adjacent methyl to produce copalyl diphosphate (CPP). The resulting bicyclic backbone typically undergoes further cyclization and/or rearrangement catalyzed by a more prototypical (class I) TPS, which initiates its reaction by ionization of the allylic diphosphate ester. In angiosperms, these diterpene synthases generally fall into the TPS-e subfamily, which is derived from the ent-kaurene synthase (KS) required for gibberellin phytohormone biosynthesis and, hence, these TPSs are often termed kaurene synthase-like (KSL) [21].
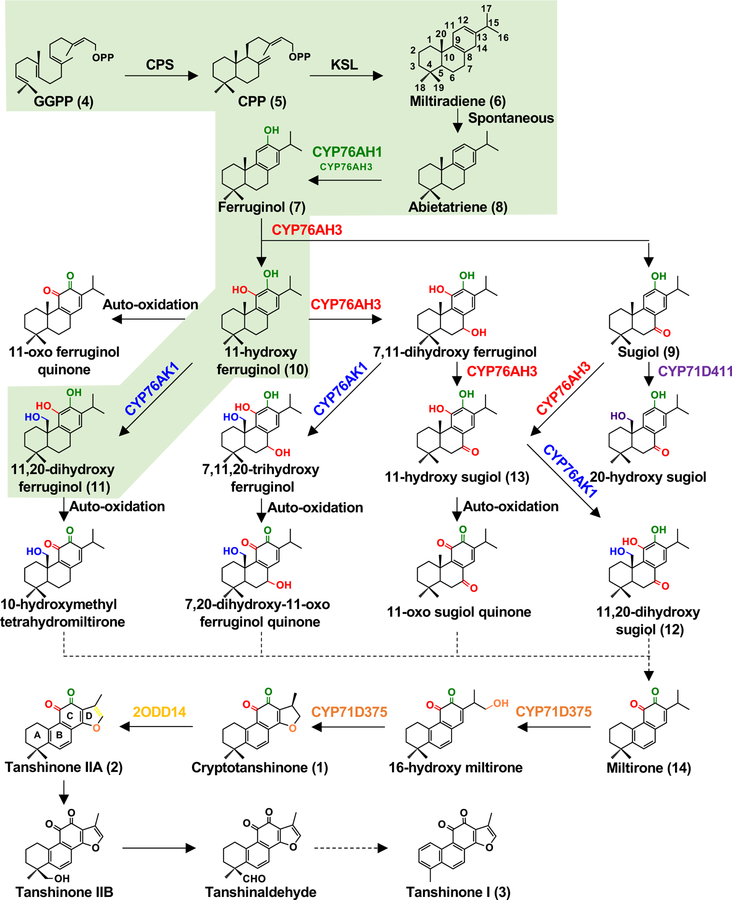
While the tanshinones are clearly derived from CPP, this metabolite can be produced in any of four distinct stereochemical configurations [20], and the aromatic nature of the tanshinones obscured which of these is relevant. Even assuming that the tanshinones are derived from the base phenolic abietane commonly found in the Lamiaceae (ferruginol) only provides orientation of the bridgehead methyl substituent at C10, which could stem from either normal or syn- CPP – i.e., those with the corresponding 10S configuration. To identify the relevant CPP synthase (CPS), as well as subsequently acting KSL, a cDNA-array with ~8,700 genes was used to analyze elicitation of S. miltiorrhiza hairy root cultures [22]. Only a single class II diterpene cyclase was found to exhibit inducible transcription, and biochemical characterization of the encoded SmCPS1 demonstrated that this produced normal (9S,10S) CPP (5) (Figure 4). Similarly, only a single KSL exhibited inducible transcription, and the encoded SmKSL1 was found to readily convert 5 to an abietane, termed miltiradiene (abieta-8,12-diene, 6), which also was readily found in induced S. miltiorrhiza hair root cultures. Notably, the cyclohexa-1,4-diene arrangement of the ‘C’-ring in 6 imposes a planar configuration, leaving it poised for aromatization [22].
Figure 4. Proposed tanshinone biosynthesis.
Miltiradiene is produced from the general diterpenoid precursor GGPP by the successive action of SmCPS1 and SmKSL1, and undergoes presumably spontaneous oxidation to abietatriene. The ferruginol synthase (CYP76AH1, green), 11-hydroxyferruginol synthase (CYP76AH3, red), and 20-hydroxylase (CYP76AK1, blue) then catalyze successive hydroxylation reactions at C12, C11, C7, and C20, respectively. CYP71D375 (orange) is known to generate the 14,16-epoxy D-ring, which is then further oxidized to a furan ring by Sm2ODD14 (yellow). Compounds numbered as described in the text, while the shaded area indicates the linear pathway that seems to predominate in planta biosynthesis
Unusually, while KS(L)s typically exhibit a tridomain architecture stemming from their evolutionary origins in bifunctional (class I and II activity) CPS/KS [23], SmKSL1 instead has a bidomain architecture, with loss of the relictual, class II activity associated, N-terminal γ domain [24]. Later studies demonstrated that both SmCPS1 and SmKSL1 are expressed in the rhizome/root periderm where tanshinones accumulate, as is strikingly evident from the coloration of root cross-sections (Figure 3). S. miltiorrhiza was further found to contain a number of other CPSs and KS(L)s, with the normal CPP production exhibited by SmCPS2, along with its expression pattern, leading to the surprising finding of tanshinone production in aerial tissues [19]. Nevertheless, RNA interference (RNAi) knock-down of SmCPS1 expression led to decreased tanshinone accumulation in hairy root cultures [25], as well as in the root/rhizome of whole plants [19]. By contrast, SmCPS5 produces ent-CPP and RNAi of this gene leads to a dwarf phenotype, indicating its relevance to gibberellin biosynthesis, consistent with phylogenetic grouping of this with other such CPSs. Similarly, SmKS(L2) reacts with ent-CPP to produce ent-kaurene, exhibits tridomain architecture and phylogenetically groups with other KSs that act in gibberellin biosynthesis [19].
Oxygenation and oxidation to the base phenolic abietane ferruginol
As with other diterpene olefins, miltiradiene (6) is highly hydrophobic and presumably partitions into membranes, necessitating the addition of oxygen to increase solubility, which must then be catalyzed by the membrane-associated CYPs [21], which also play a major role in generating (di)terpenoid structural diversity [26]. However, in order for 6 to be acted upon by CYPs, it first must be transported from the plastid where it is formed to the endoplasmic reticulum (ER) where CYPs are localized (Figure 2) [27]. While the underlying mechanism remains uncertain, it is possible that this occurs via simple diffusion, as it has been reported that the plastid and ER may share their nonpolar metabolites (such as miltiradiene), presumably via hemi-fusion of the outer leaflets of their outer membrane lipid bilayers [28]. Alternatively, such inter-organelle transport might be accomplished by carriers such as lipid-transport proteins.
Figure 2. The subcellular compartmentalization of isoprenoid and tanshinone biosynthesis more specifically.
AACT, acetoacetyl-CoA thiolase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, HMG-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDC, pyrophosphate decarboxylase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; G3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-Methyl-D-erythritol 4-phosphate; DXR, DXP reductoisomerase; CDP-ME, 4-diphosphocytidyl-2-C-methyl D-erythritol; MCT, 4-diphosphocytidyl-2-C-methyl D-erythritol synthase; CMK, 4-diphosphocytidyl-2-C-methyl D-erythritol kinase; CDP-ME2P, 4-diphosphocytidyl-2-C-methyl D-erythritol 2-phosphate; MEcPP, 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IPI, IPP isomerase; GPP, geranyl pyrophosphate; GPPS, geranyl diphosphate synthase; FPP, farnesyl diphosphate; FPPS, farnesyl diphosphate synthase; GGPP, geranyl geranyl pyrophosphate; GGPPS, geranylgeranyl diphosphate synthase
Regardless of transport mechanism, conversion of miltiradiene (6) to the base phenolic abietane ferruginol (7) can be accomplished by CYP76AH1, which was identified from fourteen CYP genes that exhibited inducible transcription in an RNA-Seq analysis of S. miltiorrhiza hairy root cultures by its ability to catalyze this transformation [29]. However, it appears that CYP76AH1 simply hydroxylates C12 of already aromatized abietatriene (8), which spontaneously forms from miltiradiene in the presence of oxygen, as indicated by careful separation and in vitro assays [30]. Consistent with such activity, knocking down CYP76AH1 expression via RNAi led to accumulation of 6 and reduction of 7 in hairy root cultures [31].
Further oxygenation and oxidation
Further oxygenation of ferruginol (7) in S. miltiorrhiza is catalyzed by CYP76AH3 and CYP76AK1, with recombinant CYP76AK1 acting as a 20-hydroxylase, while CYP76AH3 was reported to act as a multi-functional oxidase, carrying out hydroxylation at both C11 and C7, as well as subsequent oxidation of the 7-hydroxy to a ketone [32]. Given both CYPs react with ferruginol, it was suggested that tanshinone biosynthesis proceeds via a bifurcating pathway. However, the same study reported that RNAi of CYP76AH3 significantly reduces accumulation of only 11-hydroxylated metabolites (not 7 or its 7-keto derivative – i.e., sugiol, 9), while RNAi of CYP76AK1 significantly reduces that of all 20-hydroxylated metabolites, suggesting a main linear pathway in planta wherein CYP76AH3 acts as a 11-hydroxylase of 7, producing 11-hydroxy-ferruginol (10), and CYP76AK1 as a subsequently acting 20-hydroxylase, producing 11,20-dihydroxy-ferruginol (11) (Figure 4). Nevertheless, the recombinant activity of these two CYPs is sufficient to convert 7 to 11,20-dihydroxy-sugiol (abietatrien-11,12,20-triol-7-one, 12) in metabolic engineering studies. Intriguingly, it has since been shown that recombinant CYP76AH3 can convert abietatriene (8) to 11-hydroxy-sugiol (13), catalyzing hydroxylation at C12, C11 and C7, as well as further oxidation to form a 7-ketone, such that expression of this single CYP enables an astonishing series of transformations [33].
Formation of the characteristic 14,16-epoxy ‘D’-ring
While it remains unclear how miltirone (14) is formed, requiring demethylation (loss of C20) and aromatization of the ‘B’ ring, production of the 14,16-epoxy (dihydrofuran) ‘D’-ring provides a clear demarcation in production of the first eponymous metabolite – i.e., cryptotanshinone (1). Thus, recent identification of the relevant CYP71D subfamily members provides clarity to this ‘committed’ step of tanshinone biosynthesis [9]. In particular, it was reported that recombinant CYP71D375 can convert 14 to 1, as well as form this 14,16-epoxy (dihydrofuran) ‘D’-ring with the demethylated (19,20-dinor) derivative 4-methylenemiltirone, while both CYP71D375 and the closely related CYP71D373 can do the same with the further aromatized ‘A’ ring derivative to yield 15,16-dihydrotanshinone I. In addition, RNAi of these CYP71D sub-family members led to reduced accumulation of ‘D’ ring containing metabolites and increases in not only 14 and other upstream metabolites (e.g., ferruginol, 7), but also derived metabolites such as 4-methylenemiltirone, indicating that substrate promiscuity of the downstream enzymes enables operation of a metabolic network/grid for tanshinone biosynthesis.
Putting it all together for heterologous microbial production
Identification of the relevant enzymes has enabled heterologous introduction of the upstream portions of tanshinone biosynthesis (i.e., at least for production of 11,20-dihydroxy-sugiol, 12) into microbial hosts, generally yeast. Such efforts began upon identification of SmCPS1 and SmKSL1, leading to metabolic engineering efforts to increase miltiradiene production, including fusion of these two enzymes to enforce physical proximity and enhance metabolic flux [34]. Indeed, identification of CYP76AH1 as the relevant 12-hydroxylase was accompanied by its incorporation into this recombinant miltiradiene (6) producing strain of yeast, which then produces ferruginol (7) as well [29]. Similarly, characterization of the activity of CYP76AH3 and CYP76AK1 also was accompanied by their incorporation into the ferruginol producing strain of yeast, which then produced largely (>70%) 12, among other elaborated phenolic abietane diterpenoids [32]. Building on the crystal structure obtained for CYP76AH1 [35], protein engineering was undertaken to convert this 12-hydroxylase to a multiply reactive mono-oxygenase (equivalent to CYP76AH3), which could be accomplished with switching just two residues, enabling even more efficient production of 11-hydroxy-ferruginol (10) than with CYP76AH3 [36].
Opening the way to Lamiaceae labdane-related diterpenoid biosynthesis
Notably, identification of SmCPS1 and SmKSL1 presaged discovery of orthologous CPS and bidomain KSLs also producing miltiradiene in other Lamiaceae plants, including Rosmarinus officinalis (Rosemary) [37], Salvia pomifera [38] and Salvia fruticosa [39], as well as Isodon rubescens [40] (Figure 5). Indeed, use of these CPSs and KSLs from different plants has been shown to increase miltiradiene (6) yields in yeast metabolic engineering studies [41]. Beyond 6, other close homologs of SmCPS1 and SmKSL1 produce similar labdane-related diterpenoids, including hydroxylated derivatives resulting from the addition of water to carbocationic intermediates by the relevant diterpene synthases and/or cyclases. For example, Salvia sclarea contains a homolog of SmCPS1 that adds water to the labda-13-en-8-yl+ diphosphate carbocation intermediate to yield labda-13-en-8α-ol diphosphate, which is then further hydroxylated to sclareol (labda-14-en-13R,8α-diol) by a subsequently acting KSL, which also is a homolog of SmKSL1 [42,43]. Alternatively, labda-13-en-8α-ol diphosphate can be heterocyclized to manoyl oxide as found with sequentially acting CPSs and KSLs from Coleus forskohlii [44] and other Lamiaceae species [10]. Similarly, 1,2-hydride transfer within the labdadien-8-yl+ diphosphate carbocation intermediate enables the addition of water to C9, forming labda-13-en-9α-ol diphosphate, which then also undergoes heterocyclization to a spiro-9,13-epoxy ring as found in Marrubium vulgare [45] and Vitex agnus-castus [46] as well as other Lamiaceae species [10].
Figure 5. Phylogeny, protein and genomic structure of CPS and KSL genes in S. miltiorrhiza.
Left) Phylogenetic tree of diterpene cyclases and synthases from Salvia miltiorrhiza (Sm), Salvia splendens (Ss), Salvia fruticosa (Sf), Rosemarinus officinalis (Ro) and Arabidopsis thaliana (At), constructed using the neighbor-joining method with MEGA-X. Numbers at nodes indicate bootstrap values (percentage of 1000 replicates). The scale bar at bottom represents the genetic distance. Middle) Domain architecture (box coloring: white, plastid targeting sequence; orange, γ domain; green, β domain; blue, α domain). Right) Exon/intron structure of the indicated genes. Blue boxes indicate exons and black lines indicate introns
The Lamiaceae go beyond miltiradiene
It should be noted that many of these Lamiaceae bidomain KSLs exhibit a certain degree of substrate promiscuity. For example, the miltiradiene synthases readily react with the hydroxylated derivative of their native substrate (i.e., labda-13-en-8α-ol diphosphate) and catalyze heterocyclization to manoyl oxide [37,44,47–49]. Indeed, analogous heterocyclization was reported with even more widely distributed KSs from gibberellin metabolism, demonstrating that class I (di)terpene synthases broadly exhibit such catalytic promiscuity [48]. However, these all exhibit limited substrate promiscuity. By contrast, sclareol synthase was found to react with all known class II diterpene cyclase products, but exhibits catalytic specificity, generally adding water to C13 of the allylic (distributed) carbocation formed by initiating lysis of the diphosphate ester bond to form tertiary alcohol derivatives [49–51]. Intriguingly, sequence comparison to the other (non-hydroxylating) Lamiaceae bidomain KSLs led to identification of a key residue for the addition of water catalyzed by sclareol synthase, offering some insight into evolution of the underlying enzymatic activities [52].
Conservation of CYP activity with miltiradiene in the Lamiaceae
Identification of the relevant CYPs from S. miltiorrhiza similarly presaged discovery of orthologous CYPs from other Lamiaceae plants that act on abietatriene. This included CYP76AH4 [30], as well as CYP76AH22 and CYP76AH23 from rosemary and CYP76AH24 from S. fruticosa [39], which were all initially reported to act as 12-hydroxylases (i.e., ferruginol synthases), but later found to exhibit analogous (multiply reactive) activity as CYP76AH3, being sufficient to convert abietatriene (8) to 11-hydroxy-sugiol (13) [33]. The ability of CYP76AH sub-family members to act at least as bi-functional hydroxylases at C12 and then C11 upon recombinant expression in yeast, enabling the production of 11-hydroxy-ferruginol (10), was first shown for the CYP76AH24 ortholog from S. pomifera (SfCYP76AH24) and then shown for CYP76AH4 from rosemary as well [53]. This same study also reported that CYP76AK sub-family members can act as multiply reactive oxidases at C20, converting this methyl group to a carboxylic acid, as found with both CYP76AK6 from S. pomifera and CYP76AK8 from rosemary. Thus, recombinant co-expression of CYP76AH24 and either CYP76AK6 or CYP76AK8 in yeast enables conversion of 8 to carnosic acid [53]. Similarly, a separate study reported that not only SfCYP76AH24 (from S. fruticosa) but also CYP76AH22 and CYP76AH23 from rosemary analogously act as bifunctional hydroxylases (i.e., 11-hydroxy-ferruginol synthases), along with characterization of SfCYP76AK6 (from S. fruticosa) and CYP76AK7 as well as CYP76AK8 from rosemary as 20-oxidases, again enabling recombinant production of carnosic acid in yeast. This study also probed the basis for the restricted 12-hydroxylase (ferruginol synthase) activity of CYP76AH1, finding that switching three residues converted this to production of 10 [54]. Note that these included both residues later shown to be sufficient to enable CYP76AH1 to catalyze the even more extended series of transformations leading to production of 13 [36]. Again, these studies offer some insight into evolution of the underlying enzymatic activities.
CYP76 family members act in Lamiaceae labdane-related diterpenoid biosynthesis more broadly
Beyond those reacting with abietatriene (8) described above, CYP76AH sub-family members play roles in Lamiaceae labdane-related diterpenoid biosynthesis more broadly. Indeed, the elaboration of manoyl oxide to deacetyl-forskolin in C. forskohlii seems to entirely rely on this CYP76AH sub-family, with CYP76AH15, CYP76AH8 and CYP76AH17 acting as 11-oxidases to produce the 11-keto moiety, with the latter two also able to act as 1α-hydroxylases, as is CYP76AH11, which also additionally can install both 6β and 7β hydroxyl groups, while CYP76AH16 acts as a 9α-hydroxylase, such that recombinant co-expression of CYP76AH15, CYP76AH11 and CYP76AH16 enables production of deacetyl-forskolin in yeast [55]. Perhaps not surprisingly, other CYPs also seem to operate in such biosynthetic pathways, with CYP76BK1 acting as a peregrinol 9α-hydroxylase in V. agnus-castus [46], CYP71BE52 as a ferruginol 2α-hydroxylase in S. pomifera [53], and CYP71AU87 as a hydroxylase targeting the C4 geminal methyls (C18 and C19) in spiro-9,13-epoxy labda-14-ene in M. vulgare [56]. While these also fall within the large CYP71 clan, given the range of CYPs observed to function in labdane-related diterpenoid biosynthesis in plant more generally [26], it can be expected that more divergent CYPs will be found to do so in the Lamiaceae as well.
A tanshinone-associated biosynthetic gene cluster in S. miltiorrhiza
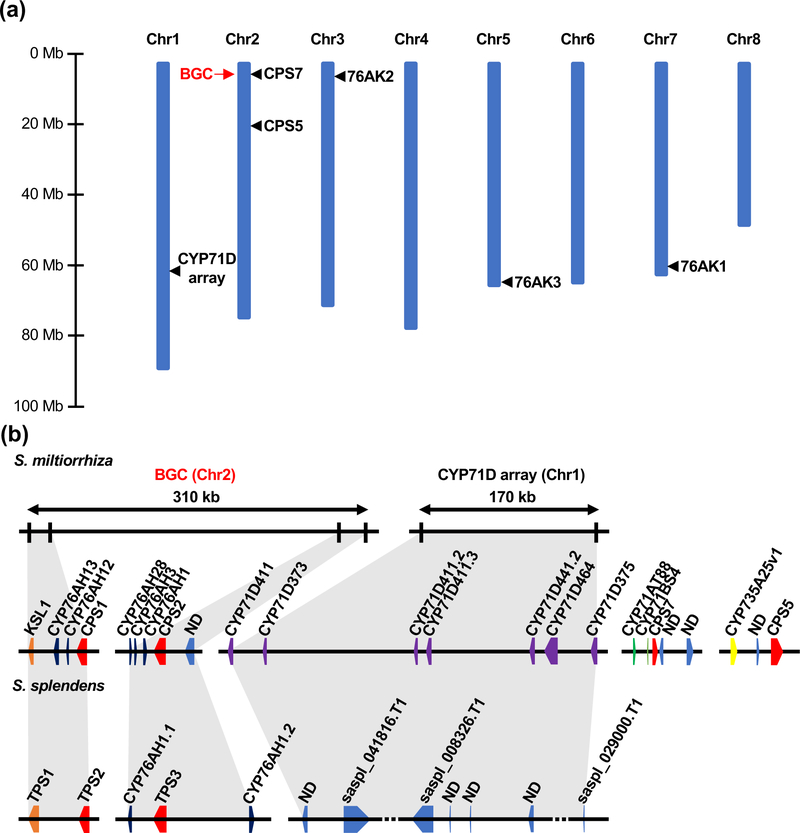
Plant genomes occasionally contain biosynthetic gene clusters (BGCs), composed of proximal unrelated genes involved a common metabolic pathway [57]. Given the interest in danshen, the S. miltiorrhiza genome has been subjected to a number of sequencing efforts, with early draft genomes [58,59] now supplemented with a genetic linkage map based chromosome-scale assembly [60]. Even in the initial draft genome there was a suggestion that there might be a BGC associated with tanshinone biosynthesis, as SmCPS1 was found to neighbor CYP76AH sub-family members (CYP76AH12 and CYP76AH13), albeit these still have not been shown to be involved in such metabolism [58]. Notably, SmKSL1 also has been found to reside nearby, along with CYP76AH1, CYP76AH3 and SmCPS2, all of which act in tanshinone biosynthesis (as described above), as well as the uncharacterized CYP76AH28, defining a 310 kb BGC (Figure 6) [9,60].
Figure 6. Chromosomal location of tanshinone biosynthesis genes in S. miltiorrhiza and comparison of associated BGC with S. splendens.
(a) S. miltiorrhiza chromosomes with position of the tanshinone (ferruginol) associated biosynthetic gene cluster (BGC) as well as that of the CYP71D array and individual genes (CPS and CYP76AK1–3) indicated by a red arrow and black triangles, respectively.
(b) Syntenic relationship of the Danshen BGC and CYP71D tandem array with S. splendens. Gene nomenclature follows that previously assigned [9,58,60,66]
Notably, the expression patterns of the genes within this S. miltiorrhiza BGC indicates roles in tanshinone biosynthesis in both roots and aerial tissues. The putative role for SmCPS2 in tanshinone production in aerial tissues has already been noted above, and while CYP76AH1 and CYP76AH3 are specifically expressed in roots, CYP76AH12 and CYP76AH28 are expressed in aerial tissues [9,60]. It can then be speculated that these might play similar roles for tanshinone production in aerial tissues.
Wider conservation of the (tanshinone) biosynthetic gene cluster
Strikingly, there appears to be an ortholog of the S. miltiorrhiza BGC in Salvia splendens, for which a chromosomal-scale genome sequence was recently reported [61]. This S. splendens BGC contains homologs to not only SmCPS1 and SmKSL1 but CYP76AH1 as well (Figure 6) [9]. Given that the divergence between Salvia splendens and S. miltiorrhiza is estimated to have occurred over 28 million years ago [61,62], this suggests both early assembly and long-standing retention of this BGC in the Salvia genus. From the observed enzymatic activities, this BGC enables production of the base phenolic abietane ferruginol (7) rather than that of tanshinones more specifically. Indeed, the CYP71D sub-family members that form the heterocyclic ‘D’ ring characteristic of the tanshinones are found elsewhere in the S. miltiorrhiza genome as a tandem gene array [9]. On the other hand, the orthologous region in the genome of the more distantly related Scutellaria baicalensis [63], despite also falling within the Lamiaceae, contains only homologs to SmCPS1 and SmCPS2 [9]. Accordingly, the exact age and relevance of this BGC to phenolic abietane diterpenoid production in the Lamiaceae remains uncertain, although it will be of interest to determine if this is present in other Salvia species, as well as rosemary.
Future directions towards elucidating tanshinone biosynthesis
Numerous transformations in tanshinone biosynthesis remain unresolved. Relevant enzymes may be found within not only the CYP but also the 2ODD and SDR super-families, all of which contain over 100 members in S. miltiorrhiza [64]. In order to reduce the number of enzymes to be investigated various co-expression analyses have been applied. For example, an RNA-Seq approach was applied to identify 35 CYP450 genes co-expressed with CYP76AH1 [65], while 16 CYPS (including CYP76AH1) were identified as being most specifically expressed in the periderm [64]. Indeed, knock-out lines of two homologs of CYP76AK1 (CYP76AK2 and CYP76AK3) that were closely co-expressed with this and CYP76AH1 were very recently shown to exhibit reduced tanshinone accumulation, although the relevant reaction catalyzed by these CYPs remains unclear [66]. Periderm specific expression was similarly applied to the 2ODD and SDR super-families, identifying 16 such 2ODDs and 5 SDRs [64]. More comprehensive analysis of the 2ODD super-family has been used to highlight a list of 13 2ODDs that might be involved in tanshinone biosynthesis, with RNAi of the most promising candidate (Sm2ODD5) found to reduce the accumulation of miltirone (14) and downstream tanshinones, although the exact reaction catalyzed by this remains unclear [67]. Just recently, another 2ODD (Sm2ODD14) was found to act as a dehydrogenase, introducing the 15,16-double bond that completes formation of the furan ‘D’ ring to yield tanshinone IIA (2) from cryptotanshinone (1) [68]. Obviously, the potential roles of these enzymes in tanshinone biosynthesis invites further investigation.
More speculatively, conversion of 11,20-dihydroxy-ferruginol (11) to 14 represents a particularly intriguing transformation, requiring demethylation (loss of C20) and aromatization of the ‘B’ ring. In particular as these two modifications may be coupled since essentially all reported diterpenoids from S. miltiorrhiza only contain an aromatic ‘B’ ring in the absence of this C10 methyl substituent and all such norditerpenoids exhibit such aromaticity [1]. Thus, while carnosic acid, with C20 already oxidized to a carboxylic acid, provides an obvious potential intermediate for demethylation, the observed oxidative targeting of C7 by CYP76AH3 lends itself to speculation that formation of a heterocyclic bridge between C20 and C7 (e.g., the 20,7β-olide of carnosol) might enable such cooperative aromatization (formation of 5(10) and 6(7) double bonds) with loss of C20 (originally the C10 methyl substituent). The case for subsequent coupled demethylation (loss of C19) and aromatization of the ‘A’ ring is less compelling, as S. miltiorrhiza contains such 19,20-dinor-diterpenoids in which loss of C19 is not necessarily accompanied by such aromatization, although an aromatic ‘A’ ring is only found in the absence of this C4β methyl substituent. Nevertheless, formation of a heterocyclic bridge between C19 and C2 could be envisioned to similarly enable cooperative aromatization (formation of 1(2) and 3(4) double bonds) with loss of C19. Indeed, precedent for such heterocyclic bridge formation in diterpenoid demethylation can be found in gibberellin biosynthesis, where C20 is lost concurrently with formation of a 19,10α-olide (γ lactone ring), which has been shown to proceed via a key heterocyclic C20 gem-diol anhydride intermediate [69]. It can be expected that coupled demethylation and aromatization reaction(s) in tanshinone biosynthesis also would draw significant mechanistic interest.
Conclusions and Outlook
With the recent identification of the role of CYP71D sub-family members in formation of the 14,16-epoxy (dihydrofuran) ‘D’-ring that characterizes these (di)norditerpenoids [9], it seemed an opportune time to review progress towards elucidation of tanshinone biosynthesis. While upstream transformations for production of the relevant nor-diterpenoid intermediate miltirone remain unknown, those that have been identified enable production of 11,20-dihydroxy-sugiol (12), which has led to significant metabolic engineering efforts, particularly in yeast. Significantly, elucidation of tanshinone production provided pioneering insights into not only phenolic abietane but labdane-related diterpenoid biosynthesis in the Lamiaceae more broadly, enabling identification of the relevant enzymes in a number of these plant species. Building on the discovery of the ferruginol BGC in the S. miltiorrhiza genome, it will be of further interest to determine if this locus has been functionally retained more widely. Regardless, investigation of tanshinone biosynthesis, as well as that of the various labdane-related diterpenoids of interest in the Lamiaceae more generally, remains an active area of study, with more intriguing discoveries sure to be made.
Acknowledgements
We apologize to colleagues whose work was not included in this review due to space constraints. Research in R.J.P.’s laboratory is funded by the National Institutes of Health (GM131885). Z.W. is partly supported by a grant from the National Natural Science Foundation of China (81701083) and a Post-Doctoral Fellowship from the China Scholarship Council (201906555001).
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mei X, Cao Y, Che Y, Li J, Shang Z, Zhao W, Qiao Y, Zhang J: Danshen: a phytochemical and pharmacological overview. Chin J Nat Med 2019, 17:59–80. [DOI] [PubMed] [Google Scholar]
- 2.Ma XH, Ma Y, Tang JF, He YL, Liu YC, Ma XJ, Shen Y, Cui GH, Lin HX, Rong QX, et al. : The Biosynthetic Pathways of Tanshinones and Phenolic Acids in Salvia miltiorrhiza. Molecules 2015, 20:16235–16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinese Pharmacopoeia Commission: Pharmacopoeia of the People’s Republic of China. Vol. 1. 2020. [Google Scholar]
- 4.Gonzalez MA: Aromatic abietane diterpenoids: their biological activity and synthesis. Nat Prod Rep 2015, 32:684–704. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Morris-Natschke SL, Lee KH: New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 2007, 27:133–148. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Morris-Natschke SL, Lee KH: Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat Prod Rep 2011, 28:529–542. [DOI] [PubMed] [Google Scholar]
- 7.Harris ES, Cao S, Littlefield BA, Craycroft JA, Scholten R, Kaptchuk T, Fu Y, Wang W, Liu Y, Chen H, et al. : Heavy metal and pesticide content in commonly prescribed individual raw Chinese Herbal Medicines. Sci Total Environ 2011, 409:4297–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JW, Wu JY: Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures. Appl Microbiol Biotechnol 2010, 88:437–449. [DOI] [PubMed] [Google Scholar]
- 9. Ma Y, Cui G, Chen T, Ma X, Wang R, Jin B, Yang J, Kang L, Tang J, Lai C, et al. : Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat Commun 2021, 12:685. •• This study identified CYP71D subfamily members that seem to function as (dehydro)furan ring synthases in tanshinone biosynthesis. The catalyzed heterocyclization (14,16-ether formation) is of some mechanistic interest. Moreover, this corresponds to the first reaction specific to the tanshinones.
- 10.Johnson SR, Bhat WW, Bibik J, Turmo A, Hamberger B, Evolutionary Mint Genomics C, Hamberger B: A database-driven approach identifies additional diterpene synthase activities in the mint family (Lamiaceae). J Biol Chem 2019, 294:1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vranova E, Coman D, Gruissem W: Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 2013, 64:665–700. [DOI] [PubMed] [Google Scholar]
- 12.Dai Z, Cui G, Zhou SF, Zhang X, Huang L: Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J Plant Physiol 2011, 168:148–157. [DOI] [PubMed] [Google Scholar]
- 13.Kai G, Xu H, Zhou C, Liao P, Xiao J, Luo X, You L, Zhang L: Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab Eng 2011, 13:319–327. [DOI] [PubMed] [Google Scholar]
- 14.Nagel R, Schmidt A, Peters RJ: Isoprenyl diphosphate synthases: the chain length determining step in terpene biosynthesis. Planta 2019, 249:9–20. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y, Yuan L, Wu B, Li X, Chen S, Lu S: Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot 2012, 63:2809–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui G, Huang L, Tang X, Zhao J: Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Mol Biol Rep 2011, 38:2471–2478. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, Fang X, Liu R, Wang L, Chen X, et al. : Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS One 2013, 8:e80464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao W, Sun HX, Xiao H, Cui G, Hillwig ML, Jackson A, Wang X, Shen Y, Zhao N, Zhang L, et al. : Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics 2014, 15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, Snyder JH, Song J, Chen S, Huang L, et al. : Functional Divergence of Diterpene Syntheses in the Medicinal Plant Salvia miltiorrhiza. Plant Physiol 2015, 169:1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters RJ: Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 2010, 27:1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zi J, Mafu S, Peters RJ: To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol 2014, 65:259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ: A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 2009, 11:5170–5173. •• This study provided the first insight into tanshinone biosynthesis, revealing stereochemical foreshadowing of aromatization, a key aspect of these highly conjugated abietane diterpenoids. Specifically, via biochemical identification of the relevant SmCPS1 and SmKSL1, with their combined activity leading to miltiradiene, whose cyclohexa-1,4-diene arrangement imposes a planar configuration on the ’C’ ring, enabling spontaneous aromatization.
- 23.Gao Y, Honzatko RB, Peters RJ: Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat Prod Rep 2012, 29:1153–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G, Huang L, Peters RJ: Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J 2011, 68:1051–1060. • In this study, the surprising discovery that SmKSL1, despite falling within the TPS-e/f (KSL) subfamily that prototypically exhibit the ancestral tri-domain architecture, is missing the N-terminal γ domain, realization of which helped discovery of orthologous class I labdane-related diterpene synthases.
- 25.Cheng Q, Su P, Hu Y, He Y, Gao W, Huang L: RNA interference-mediated repression of SmCPS (copalyldiphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol Lett 2014, 36:363–369. [DOI] [PubMed] [Google Scholar]
- 26.Bathe U, Tissier A: Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry 2019, 161:149–162. [DOI] [PubMed] [Google Scholar]
- 27.Tholl D, Lee S: Terpene Specialized Metabolism in Arabidopsis thaliana. Arabidopsis Book 2011, 9:e0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehrshahi P, Stefano G, Andaloro JM, Brandizzi F, Froehlich JE, DellaPenna D: Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc Natl Acad Sci U S A 2013, 110:12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, et al. : CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci U S A 2013, 110:12108–12113. •• This study biochemically identified the first CYP operating in tanshinone biosynthesis. Discovery of this ferruginol synthase activity helped pave the way for subsequent identification of orthologous CYP76AH subfamily acting in analogous phenolic abietane diterpenoid metabolism more broadly in the Lamiaceae.
- 30. Zi J, Peters RJ: Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem 2013, 11:7650–7652. • In this study it was shown that ferruginol synthases seem to act more simply as 12-hydroxylases of abietatriene generated by (spontaneous) aromatization of miltiradiene.
- 31. Ma Y, Ma XH, Meng FY, Zhan ZL, Guo J, Huang LQ: RNA interference targeting CYP76AH1 in hairy roots of Salvia miltiorrhiza reveals its key role in the biosynthetic pathway of tanshinones. Biochem Biophys Res Commun 2016, 477:155–160. • In this study genetic (RNAi) evidence was provided to bolster the hypothesis that CYP76AH1 acts in S. miltiorrhiza ferruginol biosynthesis.
- 32. Guo J, Ma X, Cai Y, Ma Y, Zhan Z, Zhou YJ, Liu W, Guan M, Yang J, Cui G, et al. : Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol 2016, 210:525–534. •• This study biochemically identified two CYPs that can act on ferruginol, with CYP76AK1 acting as a 20-hydroxylase, while CYP76AH3 is capable of catalyzing hydroxylation at both C11 and C7, as well as further oxidation to form a 7-keto group.
- 33. Bathe U, Frolov A, Porzel A, Tissier A: CYP76 Oxidation Network of Abietane Diterpenes in Lamiaceae Reconstituted in Yeast. J Agric Food Chem 2019, 67:13437–13450. • In this study the multiply reactive nature of the vast majority of Lamiaceae CYP76AH family members was demonstrated.
- 34.Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G, et al. : Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 2012, 134:3234–3241. [DOI] [PubMed] [Google Scholar]
- 35.Gu M, Wang M, Guo J, Shi C, Deng J, Huang L, Huang L, Chang Z: Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza. Biochem Biophys Res Commun 2019, 511:813–819. [DOI] [PubMed] [Google Scholar]
- 36.Mao Y, Ma Y, Chen T, Ma X, Xu Y, Bu J, Li Q, Jin B, Wang Y, Li Y, et al. : Functional Integration of Two CYP450 Genes Involved in Biosynthesis of Tanshinones for Improved Diterpenoid Production by Synthetic Biology. ACS Synth Biol 2020, 9:1763–1770. [DOI] [PubMed] [Google Scholar]
- 37.Brückner K, Bozic D, Manzano D, Papaefthimiou D, Pateraki I, Scheler U, Ferrer A, de Vos RC, Kanellis AK, Tissier A: Characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry 2014, 101:52–64. [DOI] [PubMed] [Google Scholar]
- 38.Trikka FA, Nikolaidis A, Ignea C, Tsaballa A, Tziveleka LA, Ioannou E, Roussis V, Stea EA, Bozic D, Argiriou A, et al. : Combined metabolome and transcriptome profiling provides new insights into diterpene biosynthesis in S. pomifera glandular trichomes. BMC Genomics 2015, 16:935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozic D, Papaefthimiou D, Bruckner K, de Vos RC, Tsoleridis CA, Katsarou D, Papanikolaou A, Pateraki I, Chatzopoulou FM, Dimitriadou E, et al. : Towards Elucidating Carnosic Acid Biosynthesis in Lamiaceae: Functional Characterization of the Three First Steps of the Pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS One 2015, 10:e0124106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelot KA, Hagelthorn LM, Addison JB, Zerbe P: Biosynthesis of the oxygenated diterpene nezukol in the medicinal plant Isodon rubescens is catalyzed by a pair of diterpene synthases. PLoS One 2017, 12:e0176507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu T, Zhou J, Tong Y, Su P, Li X, Liu Y, Liu N, Wu X, Zhang Y, Wang J, et al. : Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab Eng 2020, 60:87–96. [DOI] [PubMed] [Google Scholar]
- 42.Schalk M, Pastore L, Mirata MA, Khim S, Schouwey M, Deguerry F, Pineda V, Rocci L, Daviet L: Toward a biosynthetic route to sclareol and amber odorants. J Am Chem Soc 2012, 134:18900–18903. [DOI] [PubMed] [Google Scholar]
- 43.Caniard A, Zerbe P, Legrand S, Cohade A, Valot N, Magnard JL, Bohlmann J, Legendre L: Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture. BMC Plant Biol 2012, 12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pateraki I, Andersen-Ranberg J, Hamberger B, Heskes AM, Martens HJ, Zerbe P, Bach SS, Moller BL, Bohlmann J, Hamberger B: Manoyl Oxide (13R), the Biosynthetic Precursor of Forskolin, Is Synthesized in Specialized Root Cork Cells in Coleus forskohlii. Plant Physiol 2014, 164:1222–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerbe P, Chiang A, Dullat H, O’Neil-Johnson M, Starks C, Hamberger B, Bohlmann J: Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J 2014, 79:914–927. [DOI] [PubMed] [Google Scholar]
- 46.Heskes AM, Sundram TCM, Boughton BA, Jensen NB, Hansen NL, Crocoll C, Cozzi F, Rasmussen S, Hamberger B, Hamberger B, et al. : Biosynthesis of bioactive diterpenoids in the medicinal plant Vitex agnus-castus. Plant J 2018, 93:943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ignea C, Ioannou E, Georgantea P, Loupassaki S, Trikka FA, Kanellis AK, Makris AM, Roussis V, Kampranis SC: Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast. Metab Eng 2015, 28:91–103. [DOI] [PubMed] [Google Scholar]
- 48.Mafu S, Potter KC, Hillwig ML, Schulte S, Criswell J, Peters RJ: Efficient heterocyclisation by (di)terpene synthases. Chem Commun (Camb) 2015, 51:13485–13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen-Ranberg J, Kongstad KT, Nielsen MT, Jensen NB, Pateraki I, Bach SS, Hamberger B, Zerbe P, Staerk D, Bohlmann J, et al. : Expanding the Landscape of Diterpene Structural Diversity through Stereochemically Controlled Combinatorial Biosynthesis. Angew Chem Int Ed Engl 2016, 55:2142–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia M, Potter KC, Peters RJ: Extreme promiscuity of a bacterial and a plant diterpene synthase enables combinatorial biosynthesis. Metab Eng 2016, 37:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia M, Mishra SK, Tufts S, Jernigan RL, Peters RJ: Combinatorial biosynthesis and the basis for substrate promiscuity in class I diterpene synthases. Metab Eng 2019, 55:44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia M, O’Brien TE, Zhang Y, Siegel JB, Tantillo DJ, Peters RJ: Changing Face: A Key Residue for the Addition of Water by Sclareol Synthase. ACS Catal 2018, 8:3133–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ignea C, Athanasakoglou A, Ioannou E, Georgantea P, Trikka FA, Loupassaki S, Roussis V, Makris AM, Kampranis SC: Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proc Natl Acad Sci U S A 2016, 113:3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheler U, Brandt W, Porzel A, Rothe K, Manzano D, Bozic D, Papaefthimiou D, Balcke GU, Henning A, Lohse S, et al. : Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat Commun 2016, 7:12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pateraki I, Andersen-Ranberg J, Jensen NB, Wubshet SG, Heskes AM, Forman V, Hallstrom B, Hamberger B, Motawia MS, Olsen CE, et al. : Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karunanithi PS, Dhanota P, Addison JB, Tong S, Fiehn O, Zerbe P: Functional characterization of the cytochrome P450 monooxygenase CYP71AU87 indicates a role in marrubiin biosynthesis in the medicinal plant Marrubium vulgare. BMC Plant Biol 2019, 19:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nutzmann HW, Scazzocchio C, Osbourn A: Metabolic Gene Clusters in Eukaryotes. Annu Rev Genet 2018, 52:159–183. [DOI] [PubMed] [Google Scholar]
- 58.Xu H, Song J, Luo H, Zhang Y, Li Q, Zhu Y, Xu J, Li Y, Song C, Wang B, et al. : Analysis of the Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. Mol Plant 2016, 9:949–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang G, Tian Y, Zhang J, Shu L, Yang S, Wang W, Sheng J, Dong Y, Chen W: Hybrid de novo genome assembly of the Chinese herbal plant danshen (Salvia miltiorrhiza Bunge). Gigascience 2015, 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song Z, Lin C, Xing P, Fen Y, Jin H, Zhou C, Gu YQ, Wang J, Li X: A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 2020, 13:e20041. [DOI] [PubMed] [Google Scholar]
- 61.Jia KH, Liu H, Zhang RG, Xu J, Zhou SS, Jiao SQ, Yan XM, Tian XC, Shi TL, Luo H, et al. : Chromosome-scale assembly and evolution of the tetraploid Salvia splendens (Lamiaceae) genome. Hortic Res 2021, 8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong AX, Xin HB, Li ZJ, Liu H, Sun YQ, Nie S, Zhao ZN, Cui RF, Zhang RG, Yun QZ, et al. : High-quality assembly of the reference genome for scarlet sage, Salvia splendens, an economically important ornamental plant. Gigascience 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Q, Yang J, Cui MY, Liu J, Fang Y, Yan M, Qiu W, Shang H, Xu Z, Yidiresi R, et al. : The Reference Genome Sequence of Scutellaria baicalensis Provides Insights into the Evolution of Wogonin Biosynthesis. Mol Plant 2019, 12:935–950. [DOI] [PubMed] [Google Scholar]
- 64. Xu Z, Peters RJ, Weirather J, Luo H, Liao B, Zhang X, Zhu Y, Ji A, Zhang B, Hu S, et al. : Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 2015, 82:951–961. • In this study periderm specific expression was used to highlight CYP, 2ODD and SDR family members that might act in S. miltiorrhiza tanshinone biosynthesis.
- 65.Chen H, Wu B, Nelson DR, Wu K, Liu C: Computational Identification and Systematic Classification of Novel Cytochrome P450 Genes in Salvia miltiorrhiza. PLoS One 2014, 9:e115149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B, Li J, Chai Y, Huang Y, Li L, Wang D, Wang Z: Targeted mutagenesis of CYP76AK2 and CYP76AK3 in Salvia miltiorrhiza reveals their roles in tanshinones biosynthetic pathway. Int J Biol Macromol 2021, 189:455–463. [DOI] [PubMed] [Google Scholar]
- 67.Xu Z, Song J: The 2-oxoglutarate-dependent dioxygenase superfamily participates in tanshinone production in Salvia miltiorrhiza. J Exp Bot 2017, 68:2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Song JJ, Fang X, Li CY, Jiang Y, Li JX, Wu S, Guo J, Liu Y, Fan H, Huang YB, et al. : A 2-oxoglutarate dependent dioxygenase converts dihydrofuran to furan in Salvia diterpenoids. Plant Physiol 2021. (in press). • This study biochemically identified the first 2ODD operating in tanshinone biosynthesis.
- 69.Nagel R, Peters RJ: Diverging Mechanisms: Cytochrome-P450-Catalyzed Demethylation and gamma-Lactone Formation in Bacterial Gibberellin Biosynthesis. Angew Chem Int Ed Engl 2018, 57:6082–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]