Abstract
Background.
Throughout the animal kingdom, GABA is the principal inhibitory neurotransmitter of the nervous system. It is essential for maintaining the homeostatic balance between excitation and inhibition required for the brain to operate normally. Identification of GABAergic neurons and their GABA release sites are thus essential for understanding how the brain regulates the excitability of neurons and the activity of neural circuits responsible for numerous aspects of brain function including information processing, locomotion, learning, memory, and synaptic plasticity, among others.
New method.
Since the structure and features of GABA synapses are critical to understanding their function within specific neural circuits of interest, here we developed and characterized a conditional marker of GABAergic synaptic vesicles for Drosophila, 9XV5-vGAT.
Results.
9XV5-vGAT is validated for conditionality of expression, specificity for localization to synaptic vesicles, specificity for expression in GABAergic neurons, and functionality. Its utility for GABAergic neurotransmitter phenoltyping and identification of GABA release sites was verified for ellipsoid body neurons of the central complex. In combination with previously reported conditional SV markers for acetylcholine and glutamate, 9XV5-vGAT was used to demonstrate fast neurotransmitter phenotyping of subesophageal ganglion neurons.
Comparison with Existing Methods.
This method is an alternative to single cell transcriptomics for neurotransmitter phenotyping and can be applied to any neurons of interest represented by a binary transcription system driver.
Conclusion.
A conditional GABAergic synaptic vesicle marker has been developed and validated for GABA neurotransmitter phenotyping and subcellular localization of GABAergic synaptic vesicles.
Keywords: Drosophila, vGAT, GABAergic, synaptic vesicle, epitope tag
1. Introduction
Gamma amino butyric acid (GABA) is the primary inhibitory neurotransmitter in the nervous system of both vertebrates and invertebrates (Smart and Stephenson 2019). It is essential for maintaining homeostasis in the brain by counterbalancing the effects of the excitatory neurotransmitters glutamate and acetylcholine. In humans, GABA dysfunction has been linked to numerous neurological conditions including epilepsy (Ben-Ari and Holmes 2005), schizophrenia (Taylor and Tso 2015), and autism spectrum disorder (Marotta et al. 2020) (Schur et al. 2016). The ability to identify GABAergic neurons and sites of GABA release are thus critical for understanding how GABAergic neurons contribute to the plethora of functions accomplished by the brains of humans and organisms throughout the animal kingdom by facilitating assessment of their development and function within circuits that control behavior.
An established marker for GABAergic neurons is the vesicular GABA neurotransmitter transporter vGAT (Gasnier 2004) (Schioth et al. 2013). vGAT also serves as a marker for GABAergic synaptic vesicles (SVs) and designates sites of GABA release in the presynaptic terminals of GABAergic neurons. However, due to its abundance in the brain and subcellular localization in presynaptic terminals far from the cell bodies of GABAergic neurons, it is problematic to use vGAT immunostaining for identifying GABAergic neurons. To restrict vGAT expression to neuronal subsets of interest and make it possible to identify GABAergic neurons based on expression of vGAT, we have developed a conditional epitope-tagged variant of vGAT for the Drosophila model system via CRISPR/Cas9 genome editing.
In this report the development and characterization of RSRT-STOP-RSRT-9XV5-vGAT is described. This genome edited chromosome contains a translation STOP cassette flanked by R recombinase target sites (RSRTs) inserted into the vGAT intron to provide conditionality. Additionally, it contains nine copies of the V5 epitope tag (9XV5) to distinguish it from endogenous vGAT and provide high sensitivity detection. RSRT-STOP-RSRT-9XV5-vGAT is designed to be used in combination with any binary transcription driver that expresses in neuronal subsets, but especially with split-GAL4 drivers that express in individual neuron types or single neurons of which there is a large and growing collection (Aso et al. 2014; Robie et al. 2017; Wolff and Rubin 2018; Dolan et al. 2019; Davis et al. 2020; Sterne et al. 2021), among others https://splitgal4.janelia.org/cgi-bin/splitgal4.cgi. The conditionality and specificity of RSRT-STOP-RSRT-9XV5-vGAT is first described and subsequently its utility for GABAergic neurotransmitter phenotyping and identification of GABA release sites is demonstrated using split-GAL4 drivers that express in neuronal subsets of the ellipsoid body. Lastly, RSRT-STOP-RSRT-9XV5-vGAT is used with existing conditional cholinergic and glutamatergic SV reporters to demonstrate the utility of conditional neurotransmitter-specific SV markers for fast neurotransmitter phenotyping of subesophageal ganglion neurons.
2. Materials and Methods
2.1. Plasmid construction
The pCFD4-vGAT1 and pCFD4-vGAT2 double guide RNA plasmids were generated as previously described (Port et al. 2014). pCFD4-vGAT1 contains guide RNA sequences ggtgggtcattctcatta and aatccatcttccgcaggc. pCFD4-vGAT2 contains guide RNA sequences and cgaaccggatccggcaac and cagggctcgttcgactcg. The RSRT-STOP-RSRT-9XV5-vGAT donor plasmid was assembled in vector pHSG298 (Takara Biosciences) using NEBuilder HiFi (New England Biolabs). The 9XV5 epitope tag was generated using the epitope multimerization strategy previously described (Tison et al. 2020). The complete annotated sequence of the donor plasmid is shown in Supplemental Information Figure S1.
2.2. Genome editing
The pCFD4-vGAT1 and pCFD4-vGAT2 double guide RNA plasmids were co-injected with the RSRT-STOP-RSRT-9XV5-vGAT donor plasmid into embryos of strain nos-Cas9 TH_attP2 (Ren et al. 2013) by Bestgene, Inc. Surviving adults injected as embryos were crossed to the balancer stock yw: Sp/CyO. ~100 male CyO progeny were crossed individually to yw; vGAT1/CyO and screened for failed complementation on the expectation the desired genome edit would result in a lethal vGAT allele. Males from each cross that yielded failed complementation were subsequently crossed to strain yw; 20XUAS-DSCP-R; n-syb-GAL4 and third instar larval progeny of the appropriate genotype were assessed for anti-V5 immunostaining. A stable stock of RSRT-STOP-RSRT-9XV5-vGAT was established from males showing positive immunostaining.
2.3. Germline excision
Germline excision of the STOP cassette of RSRT-STOP-RSRT-9XV5-vGAT was accomplished by crossing it to yw; nos-GAL4; 20XUAS-DSCP-R. Progeny males of the appropriate genotype were crossed to a 2nd chromosome balancer stock to recover potential germline excision chromosomes. Candidate germline excision progeny males from the balancer cross were individually crossed to the 2nd chromosome balancer stock and third instar larva were screened by anti-V5 immunofluorescence. A stable stock of the RSRT-9XV5-vGAT germline excision chromosome was established from a male whose progeny exhibited positive anti-V5 immunostaining in the second balancer cross.
2.4. Immunostaining
Larval and adult immunostaining were performed as previously described (Certel and Thor 2004; Petersen and Stowers 2011). Briefly, dissected brains were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 minutes. Brains were then blocked with PBSTBSA (2% Triton-X100, 2% bovine serum albumin in PBS) for three hours before overnight incubation with primary antibodies in PBSTBSA at 4°C. Brains were then washed 3X in PBSTBSA for 20 minutes before a three-hour incubation with secondary antibodies in PBSTBSA at room temperature. Brains were then washed 3X in PBSTBSA for 20 minutes before mounting in VECTASHIELD PLUS (H-1900) on microscope slides for imaging. All steps were performed in microfuge tubes with rotation. Primary antibodies and dilution factors: The SYN (3C11) mAb 1:10 developed by E. Buchner (Klagges et al. 1996) was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Rabbit anti-vGAT (Fei et al. 2010) 1:200, Mouse anti-mCherry (Biorbyt orb256058) 1:200; Rabbit anti-mCherry (Abcam ab213511) 1:500, Rat anti-mCherry 16D7 (ThermoFisher-M11217) 1:200, Rabbit anti-V5 SV5-P-K (Novus NBP2-52653) 1:200, Mouse anti-HA C5 (Covalab mab90002-P) 1:200, Rabbit Abfinity anti-GFP (ThermoFisher) 1:400, Mouse anti-GFP 3E6 (ThermoFisher A-11120) 1:200, Rat anti-FLAG (Novus NBP1-06712) 1:200, Rabbit anti-MYC 9E10 (Novus NBP2-52636) 1:200. Secondary antibodies and dilution factors: Donkey anti-Rat Alexa 488 (Jackson Immunoresearch 712-546-153) 1:400, Donkey anti-Mouse Alexa 488 (Jackson Immunoresearch 715-545-151) 1:400, Goat anti-Rabbit Alexa 488 (Thermo-Fisher A32731) 1:200, Donkey anti-Mouse JF549 (Novus NBP1-75119JF549) 1:200, Goat anti-Rabbit JF549 (Novus NBP1-72732JF549) 1:200, Donkey anti-Rat JF549 (Novus NBP1-72743JF549) 1:200.
2.5. Fly strains
Stocks from the Bloomington Drosophila Stock Certer (NIH P40OD018537) were used in this study. Previously described fly strains: 20XUAS-DSCP-B2 and B2RT-STOP-B2RT-GFP-Rab3 (Williams et al. 2019); FRT-STOP-FRT-7XMYC-vAChT and 20XUAS-DSCP-FLP (Tison et al. 2020); 20XUAS-DSCP-R (Sherer et al. 2020); B2RT-STOP-B2RT-smFLAG-vGlut (Certel et al. 2022); vGAT1 (Fei et al. 2010); UAS-CD8-mCherry (BDSC #27392); nos-GAL4 (Tracey et al. 2000) (BDSC #4442); MBON-6/MB434B (Aso et al. 2014); LH2094 (Dolan et al. 2019); 3XUAS-Syt-sm-HA (Wolff and Rubin 2018); SS02704, SS02714, SS02766, SS02705, SS02767, SS02702, SS07206, SS02710, SS02709, SS02712, SS02765, and SS02769 (Robie et al. 2017); SS00238 (Turner-Evans et al. 2020); SS46209, SS47082, SS30377 (Sterne et al. 2021). The 20XUAS-DSCP-R plasmid (Sherer et al. 2020) was inserted at landing site JMK66B (Knapp et al. 2015).
2.6. Data availability
The complete annotated sequence of the RSRT-STOP-RSRT-9XV5-vGAT donor plasmid is shown in Supplemental Information (Figure S1). The RSRT-STOP-RSRT-9XV5-vGAT donor plasmid, and pCFD4-vGAT1 and pCFD4-vGAT2 guide RNA plasmids will be made available upon request. Fly strains original to this publication will be deposited at the Bloomington Drosophila stock center or will be made available upon request.
3. Results
3.1. Strategic design of a conditional epitope-tagged GABAergic SV marker
To develop a conditional GABAergic synaptic vesicle (SV) marker for Drosophila, the gene encoding the vesicular GABA transporter (vGAT) was subjected to CRISPR/Cas9 genome editing. The Drosophila genome encodes a single vGAT homolog that is accepted as utilized by all GABAergic neurons for packaging GABA into SVs (Adams et al. 2000; Deshpande et al. 2020). Thus, conditional epitope tagging of the sole Drosophila vGAT gene is sufficient as a GABAergic SV marker for all Drosophila GABAergic neurons. vGAT was genome edited at its endogenous chromosomal locus to ensure the entirety of the vGAT regulatory is intact and the neuronal expression of vGAT is thus complete across the nervous system. Furthermore, expression of epitope-tagged vGAT under its endogenous promoter is expected to result in expression at native vGAT levels and thereby precise subcellular localization to GABAergic SVs that recapitulates that of endogenous vGAT for accurate identification of GABAergic release sites. This avoids potential artifactual mis-localization due to ectopic overexpression from binary transcription systems (GAL4, LexA, Q). However, it should be noted that insertion of the 9XV5 epitope tag creates the potential caveat that vGAT trafficking, localization, or function could be altered.
An internal site between amino acids 134 and 135 located 29 amino acids upstream of the first transmembrane domain of the vGAT protein was chosen for insertion of a multimerized V5 epitope tag. This region of the vGAT protein prior to the first transmembrane domain exhibits low evolutionary conservation and was selected on the basis that it would likely tolerate insertion of additional amino acids without altering the SV localization or vesicular GABA transporter function of the vGAT protein. The vGAT gene is composed of two exons and a 72bp intron (Figure 1A). Genome editing included insertion of a FlpStop cassette (hereafter STOP cassette) (Fisher et al. 2017) flanked by R recombinase target sites (RSRTs) into the intron and an in-frame insertion of nine tandem copies of a V5 epitope tag (9XV5) near the beginning of the second exon (Figure 1B). Nine copies of the V5 epitope tag were inserted to enhance the sensitivity of detection of the conditionally expressed vGAT protein. Prior to excision of the STOP cassette, vGAT expression is prevented and the 9XV5-vGAT protein is not expressed, thus creating a presumptive null allele of vGAT. However, after excision of the STOP cassette in neurons of interest via expression of the R recombinase, 9XV5-vGAT will be expressed in GABAergic neurons (Figure 1C).
Figure 1.
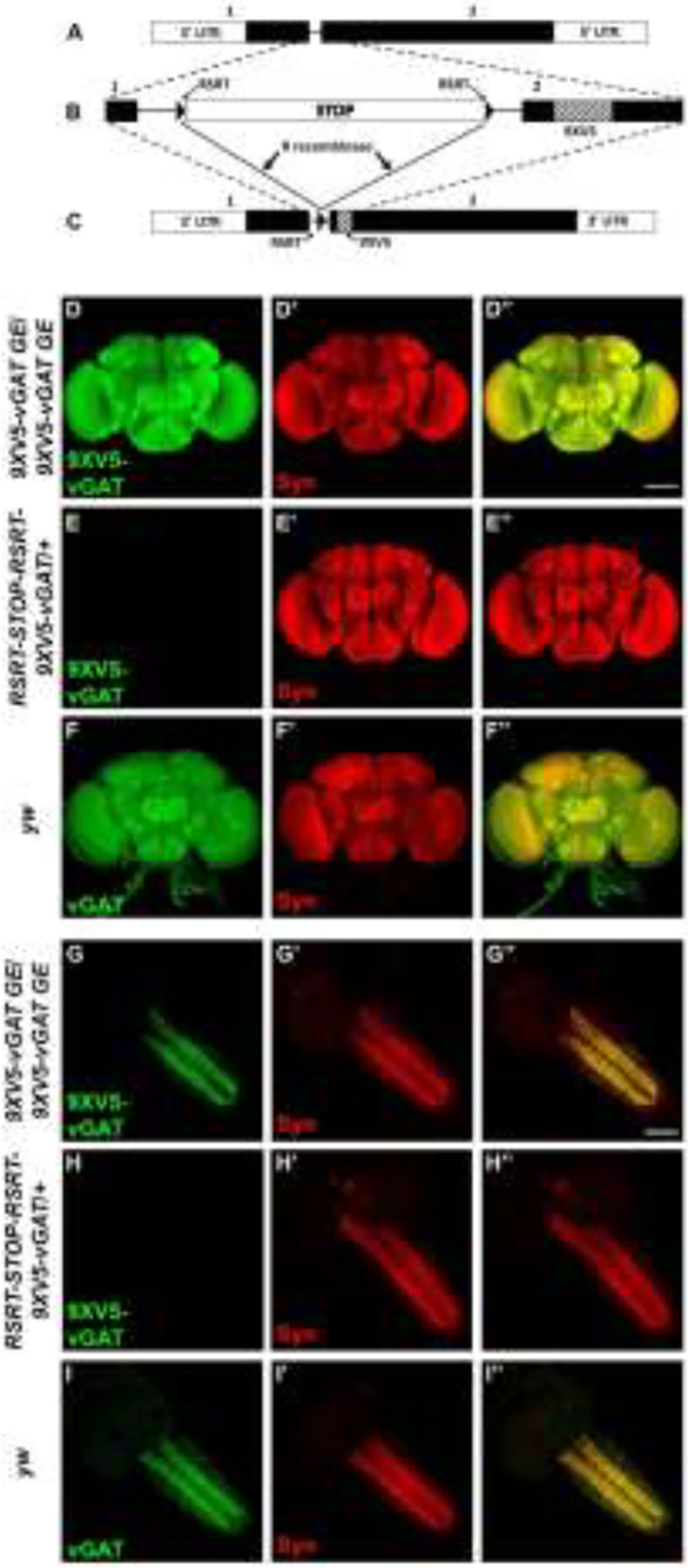
Strategic design of the conditional GABAergic synaptic vesicle marker RSRT-STOP-RSRT-9XV5-vGAT and conditional neuropil expression in adult brain and larval ventral nerve cord. A) Genomic exon structure of Drosophila vGAT. B) Genome editing at the endogenous vGAT genomic locus included insertion of a FlpStop cassette flanked by R recombinase target sites into the vGAT intron and insertion of a 9XV5 epitope tag near the beginning of exon 2. The FlpStop cassette includes two transcription termination sequences and an upstream splice acceptor sequence followed by translation STOP codons in all three reading frames. Prior to excision of the STOP cassette, 9XV5-vGAT is not expressed. C) After expression of the R recombinase using a binary transcription system driver and a corresponding R recombinase responder transgene, the STOP cassette is excised and 9XV5-vGAT is expressed in GABAergic neurons. D-F) Conditional expression in adult brain. D-D”) 9XV5-vGAT germline excision/9XV5-germline excision. D) 9XV5-vGAT; D’) Syn; D”) overlay. E-E”) RSRT-STOP-RSRT-9XV5-vGAT/+. E) 9XV5-vGAT; E’) Syn; E”) overlay. F-F”) yw. F) vGAT; F’) Syn; F”) overlay. Images in panels D and E were collected and processed identically. G-I) Conditional expression in larval ventral nerve cord). G-G”) 9XV5-vGAT germline excision/9XV5-germline excision. G) 9XV5-vGAT; G’) Syn; G”) overlay. H-H”) RSRT-STOP-RSRT-9XV5-vGAT/+. H) 9XV5-vGAT; H’) Syn; H”) overlay. I-I”) yw. I) vGAT; I’) Syn; I”) overlay. Images in panels G and H were collected and processed identically. Germline excision of the RSRT-STOP-RSRT-9XV5-vGAT STOP cassette results in strong neuropil-specific 9XV5-vGAT immunostaining in the adult brain and larval ventral nerve cord that is not observed prior to STOP cassette excision. The 9XV5-vGAT distribution is nearly indistinguishable from endogenous vGAT. Syn-Synapsin. Scale bar: 100μm.
3.2. Assessment of conditionality and synaptic vesicle specificity
To evaluate the conditionality of expression of RSRT-STOP-RSRT-9XV5-vGAT, its expression was assessed with and without germline excision of the STOP cassette in adult brains and in the third instar larval ventral nerve cord (VNC). Anti-V5 immunostaining of 9XV5-vGAT in adult brains containing a germline excision of the STOP cassette reveals robust signal broadly distributed across the neuropil of the adult brain (Figure 1D). This is in sharp contrast to anti-V5 immunostaining in RSRT-STOP-RSRT-9XV5-vGAT with the STOP cassette intact in which no signal was detectable (Figure 1E). Similarly, robust signal was observed in the neuropil of the third instar larval VNC in the 9XV5-vGAT germline excision (Figure 1G) but no signal was detectable with the STOP cassette intact (Figure 1H). These results indicate the STOP cassette is highly effective in suppressing expression of 9XV5-vGAT prior to excision and thus that there is no detectable “leaky” expression when the STOP cassette is intact.
To evaluate the specificity of 9XV5-vGAT for localization to SVs, co-labeling with the pan-SV-specific protein Synapsin (Syn) was incorporated into the above experiments. Syn signal in both the adult brain (Figure 1D’) and larval VNC (Figure 1G’) exhibited a nearly precise co-localization with 9XV5-vGAT (Figures 1D” and 1G”, respectively). Additional evidence supporting the SV localization of 9XV5-vGAT is the close resemblance of its expression pattern to that of endogenous vGAT in both adult brains (Figure 1F) and in the larval VNC (Figure 1I).
Genetic data also supports the immunolabeling experiments above as to the effectiveness of the STOP cassette in preventing 9XV5-vGAT expression. Complementation tests of RSRT-STOP-RSRT-9XV5-vGAT with the STOP cassette intact in heterozygous combination with the previously characterized vGAT null allele vGAT1 revealed a failure of complementation. Moreover, the embryonic lethal phase of this failed complementation is the same as the known embryonic lethal phase of the vGAT1 null mutant (Fei et al. 2010). This result suggests RSRT-STOP-RSRT-9XV5-vGAT is also a null/complete loss-of-function vGAT allele. Thus, both immunolabeling and genetic data are consistent in establishing the high level of effectiveness of the STOP cassette in preventing 9XV5-vGAT expression.
Genetic results also support the idea that the vGAT protein retains functionality as a vesicular GABA transporter despite insertion of the 9XV5 epitope tag. The 9XV5-vGAT germline excision chromosome, in which the STOP cassette has been removed and 9XV5-vGAT expression occurs normally in GABAergic neurons, is viable in heterozygous combination with the vGAT1 null allele and as a homozygote. Neither genotype exhibited developmental delay nor showed any obvious external morphological phenotypes. Additionally, 9XV5-vGAT germline excision homozygotes are fertile and a homozygous stock can be maintained for an indefinite number of generations.
3.3. Single neuron assessment of 9XV5-vGAT for SV and neurotransmitter specificity
To determine if 9XV5-vGAT localizes specifically to SVs and whether its expression is restricted to GABAergic neurons, the STOP cassette of RSRT-STOP-RSRT-9XV5-vGAT was excised using a 20XUAS-DSCP-R recombinase transgene in combination with split-GAL4 drivers that express in single neuron types with known neurotransmitter usage previously demonstrated using independent approaches.
The ellipsoid body ring neuron R4d represented by the split-GAL4 driver SS00238 (Turner-Evans et al. 2020) was selected as the positive control for GABAergic neurons. In this and subsequent experiments, the plasma membrane marker CD8-mCherry was used to visualize the entire anatomy of neurons of interest. R4d neurons are symmetric with lateral cell bodies (arrowheads) and processes that project medially to a central ring (arrow, Figure 2A) that includes the presynaptic terminals (Turner-Evans et al. 2020). In contrast to the uniform distribution of CD8-mCherry throughout R4d neurons, 9XV5-vGAT expression is highly polarized and is not detected in the cell bodies or processes but is restricted to the ring (Figure 2A’). The distribution of 9XV5-vGAT to the ring of R4d neurons is highly similar to the conditionally-expressible pan-SV marker GFP-Rab3 (Figure 2C’). Additionally, high resolution (100X) images of the presynaptic ring region of R4d neurons in which 9XV5-vGAT (Figure 2B) is conditionally co-expressed with the pan-SV marker Syt-smHA (Figure 2B’) reveals near precise co-localization (Figure 2B”).
Figure 2.
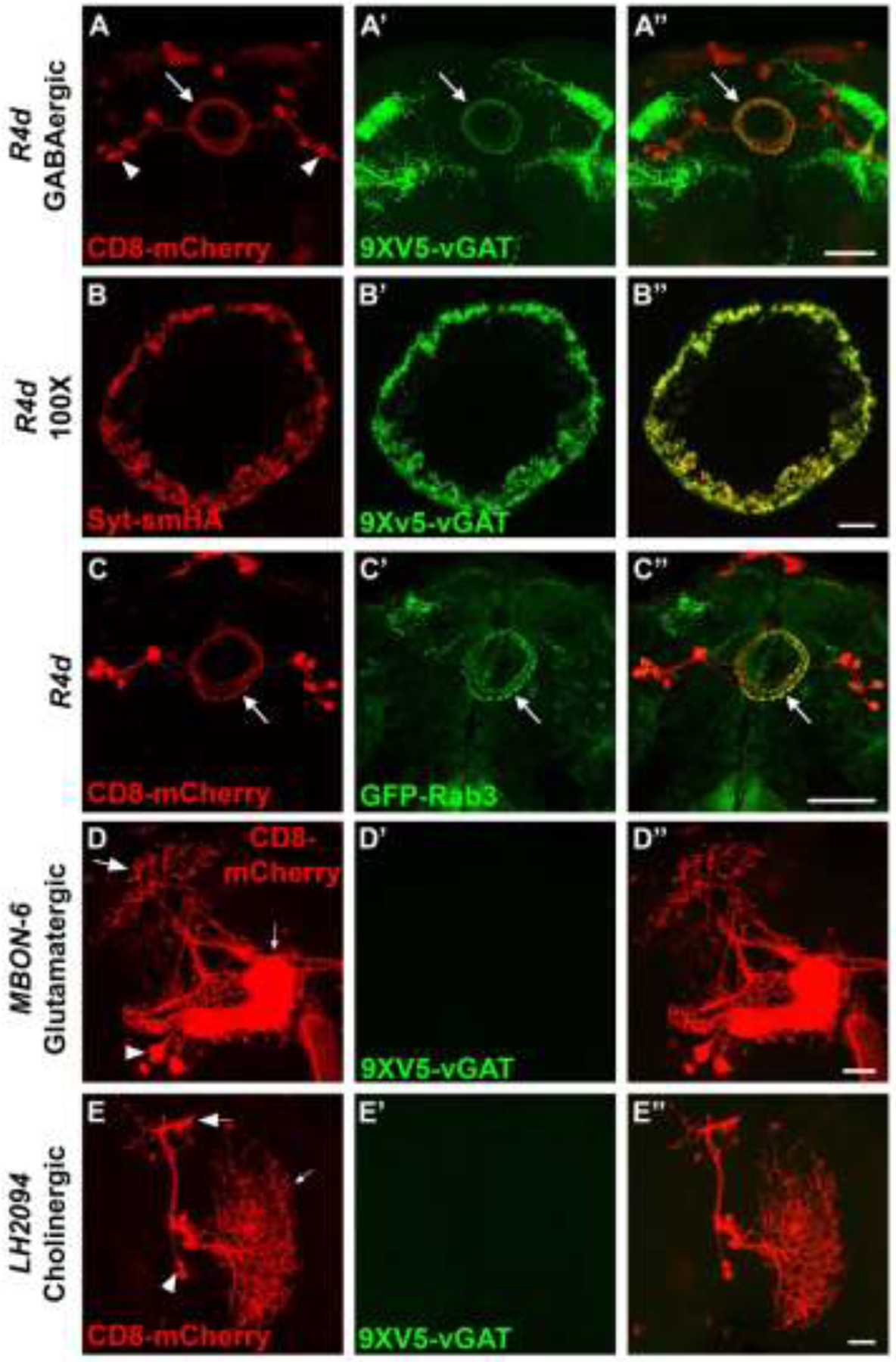
Assessment of neurotransmitter and synaptic vesicle specificity of RSRT-STOP-RSRT-9XV5-vGAT in single neuron types. Neuron anatomy is visualized with the plasma membrane marker CD8-mCherry. A-A”) GABAergic neuron R4d. A) CD8-mCherry; A’) 9XV5-vGAT; A”) overlay. 9XV5-vGAT expression exhibits highly restricted localization to the ring region of R4d neurons. B-B”) High resolution 100X images of pre-synaptic terminals of GABAergic neuron R4d. B) Syt-smHA; B’) 9XV5-vGAT; B”) overlay. High resolution images of R4d pre-synaptic terminals reveals near precise overlap of 9XV5-vGAT with the synaptic vesicle marker Syt-smHA. C-C”) GABAergic neuron R4d. C) CD8-mCherry; C’) GFP-Rab3; C”) overlay. GFP-Rab3 is expressed in GABAergic neuron R4d and distributes predominantly to pre-synaptic terminals in a pattern similar to that of 9XV5-vGAT. D-D”) Glutamatergic neuron MBON-6. D) CD8-mCherry; D’) 9XV5-vGAT; D”) overlay. E-E”) Cholinergic neuron LH2094. E) CD8-mCherry; E’) 9XV5-vGAT; E’) overlay. 9XV5-vGAT expression was not detected in the glutamatergic neuron MBON-6 or the cholinergic neuron LH2094. Large arrows-presynaptic terminals; small arrows-dendrites; arrowheads-cell bodies. Scale bars: A, C-50μm; B-10μm; D, E-20μm.
The expression of 9XV5-vGAT was also assessed in the known glutamatergic neuron MBON-6 (Aso et al. 2014) and known cholinergic neuron LH2094 (Dolan et al. 2019). MBON-6 is a highly polarized neuron with distinct cell bodies (arrowhead), dendrites (small arrow), and presynaptic regions (large arrow). LH2094 is also a highly polarized neuron with distinct cell bodies (arrowhead), dendrites (small arrow), and presynaptic regions (large arrow). 9XV5-vGAT expression was not detected in any region of either MBON-6 (Figure 2D’) or LH2094 (Figure 2E’) neurons.
Taken together, these data establish 9XV5-vGAT as a conditional marker of GABAergic SVs by: 1) establishing that 9XV5-vGAT distributes to the presynaptic region of R4d neurons in a pattern highly reminiscent of the pan-SV marker GFP-Rab3; 2) precise co-localization of 9XV5-vGAT with the pan-SV marker Syt-smHA to the presynaptic region of R4d neurons; and 3) the absence of detectable expression of 9XV5-vGAT in known glutamatergic and cholinergic neurons.
3.4. GABAergic neurotransmitter phenotyping of ellipsoid body neurons
With the conditionality and specificity for GABAergic SVs of RSRT-STOP-RSRT-9XV5-vGAT thus established, its utility for GABAergic neurotransmitter phenotyping and identifying sites of GABA release was demonstrated for 12 ellipsoid body neuron subsets of unreported neurotransmitter usage represented by split-GAL4 drivers (Robie et al. 2017). The ellipsoid body is a centrally located region of the adult Drosophila brain especially known for its role in locomotion control (Martin-Pena et al. 2014; Robie et al. 2017; Kottler et al. 2019). Knowledge of their GABAergic neurotransmitter usage would augment our understanding of how these ellipsoid body neurons regulate locomotive behavior.
The ellipsoid body split-GAL4 drivers were combined with 20XUAS-DSCP-R to induce excision of the STOP cassette in RSRT-STOP-RSRT-9XV5-vGAT and subjected to anti-V5 immunostaining. In all of these experiments the CD8-mCherry plasma membrane marker is used for visualization of the neuron of interest (red, left column) and the presynaptic regions where GABAergic SVs accumulate are indicated by 9XV5-vGAT (green, indicated with arrows, middle column). The 12 ellipsoid body neuron types assessed exhibit 9XV5-vGAT labeling indicating they are GABAergic including SS02704 (Figure 3A–A”), SS02714 (Figure 3B–B”), SS02766 (Figure 3C–C”), SS02705 (Figure 3D–D”), SS02767 (Figure 3E–E”), SS02702 (Figure 3F–F”), SS02706 (Figure S2A–A”), SS02710 (Figure S2B–B”), SS02709 (Figure S2C’C”), SS02712 (Figure S2D–D”), SS02765 (Figure S2E–E”), and SS02769 (Figure S2F’–F”). While this result would be statistically unlikely for 12 randomly chosen neurons, it is possible less than 12 distinct neuron types are represented by the 12 split-GAL4 drivers examined (for instance the neurons expressing in SS02706, SS02710, SS02709, and SS02712 look very similar and may well all represent the same neuronal subset). Additionally, it has recently been shown that the neurotransmitter identity of Drosophila neurons is determined during the stem cell stage of development (Lacin et al. 2019). As all of the neurons tested show significant neuroanatomical similarity, they may well have developed from the same GABAergic stem cell lineage and differentiated late in development. Nevertheless, the ellipsoid body neurons represented by these 12 split-GAL4 drivers exhibit robust, highly polarized 9XV5-vGAT signal restricted to their ring regions where it is known from connectome data that their presynaptic terminals are located (Scheffer et al. 2020; Turner-Evans et al. 2020; Hulse et al. 2021). These neurons thus all have an inhibitory valence that can be incorporated into the neural circuit modeling of how they regulate locomotive behavior.
Figure 3.
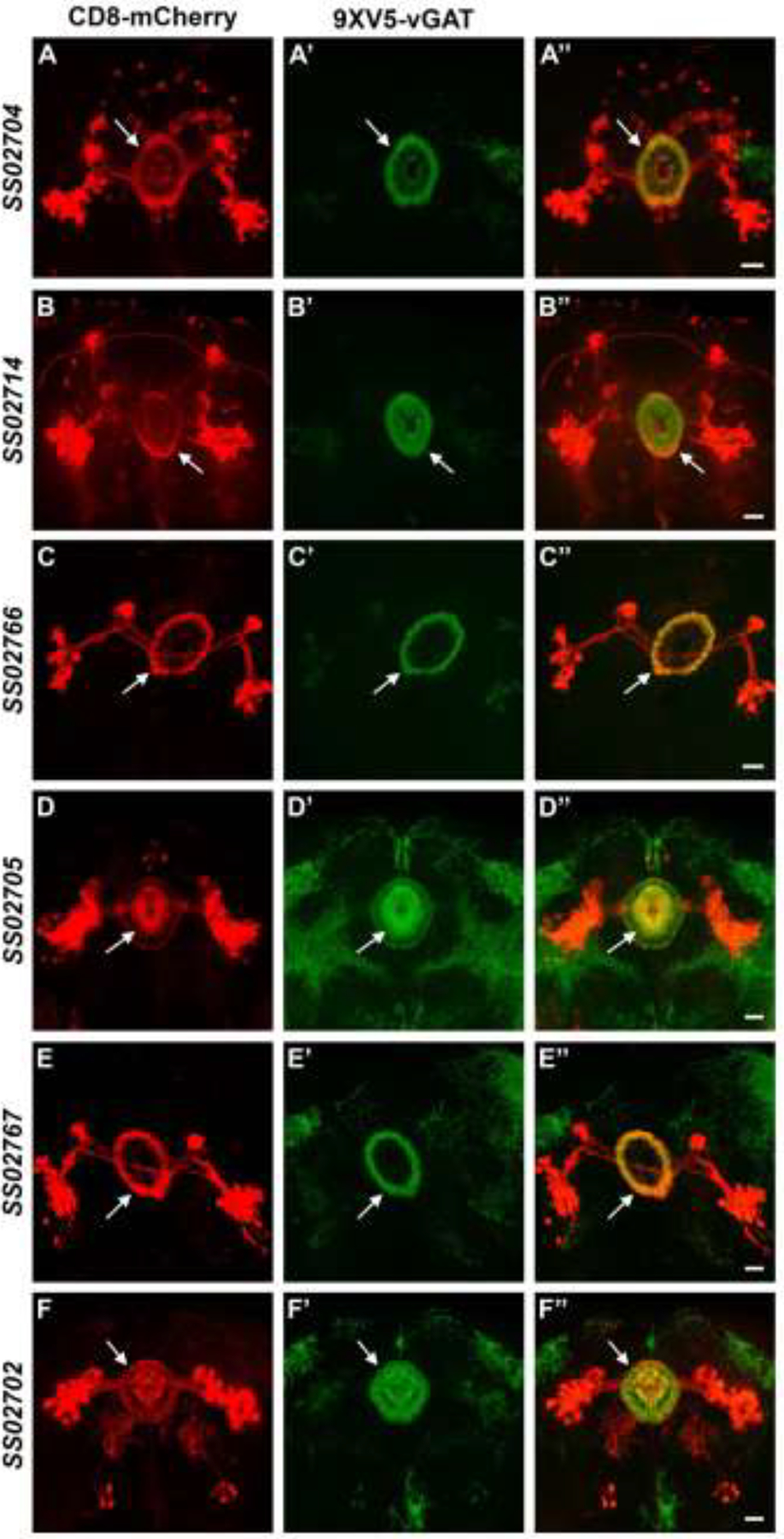
GABAergic neurotransmitter phenotyping of ellipsoid body neurons. A-A”) SS02704. B-B”) SS02714. C-C”) SS02766. D-D”) SS02705. E-E”) SS02767. F-F”) SS02702. The plasma membrane marker CD8-mCherry (red, left column) allows visualization of the neuroanatomy of each ellipsoid body neuron. The presence of 9XV5-vGAT (arrows, middle column) indicates each ellipsoid body neuron is GABAergic and its subcellular localization reveals the site of presynaptic terminals where SV fusion and GABA release occurs. Scale bars: 20μm.
It should be noted in these experiments that there is a variable amount of 9XV5-vGAT signal observed outside the neurons of interest that are marked with CD8-mCherry. This is most likely due to variable levels of expression of each individual split-GAL4 driver in other GABAergic neurons during development. Thus, even though a given split-GAL4 driver may be transiently expressed during development in neurons in which its expression is not maintained into adulthood, STOP cassette excisions during development are permanent.
It should also be noted that although all 12 ellipsoid body neurons neurotransmitter phenotyped for GABA in these experiments were positive, and thus confidence is high that they are GABAergic, the confidence in a negative result that a neuron is not GABAergic is less certain. If the strength of a given split-GAL4 driver is insufficient to drive expression of the R recombinase at the level required to elicit excision of the STOP cassette, it is possible a GABAergic neuron will not exhibit labeling with 9XV5-vGAT and thus the potential of false negatives should be kept in mind.
3.5. Conditional neurotransmitter-specific SV markers for fast neurotransmitter phenotyping
With the addition of the conditional GABAergic SV reporter described here to previously developed conditional reporters of cholinergic (Tison et al. 2020) and glutamatergic (Certel 2022, in press) SVs, there now exists conditional SV reporters for all three Drosophila fast neurotransmitters. This makes it possible to perform fast neurotransmitter phenotyping for any neuron(s) represented by a binary transcription system driver. This is most feasible with neurons represented by split-GAL4 drivers expressed in single neuron types as the sparser the expression of the driver the easier it is to discern overlapping expression of the fast neurotransmitter SV reporters with individual neurons of interest from the background expression of other neurons.
To demonstrate the use of conditional neurotransmitter-specific SV markers for neurotransmitter phenotyping of GABA, acetylcholine, and glutamate neurotransmitter identity, three neuron types of unknown neurotransmitter usage represented by recently developed split-GAL4 drivers for the subesophageal region of the brain were assessed for expression of all three conditional fast neurotransmitter SV reporters. Fast neurotransmitter phenotyping of trident neurons represented by the SS46209 split-GAL4 driver revealed overlapping expression with 9XV5-vGAT (arrows, Figure 4A–A”) but not 7XMYC-vAChT (Figure 4B–B”) or smFLAG-vGlut (Figure 4C–C”), thus establishing trident neurons as GABAergic. This experiment also reveals that SS46209 neurons exhibit a high degree of polarization with clearly discernible axonal and dendritic regions. Fast neurotransmitter phenotyping of G2N-1 neurons represented by the SS47082 split-GAL4 driver demonstrated overlapping expression with 7XMYC-vAChT (arrows, Figure 5B–B”), but no detectable expression of 9XV5-vGAT (Figure 5A–A”) or smFLAG-vGlut (Figure 5C–C”), thus establishing G2N-1 neurons as cholinergic. In this experiment 7XMYC-vAChT is distributed broadly in the processes of G2N-1 neurons without clearly separable axonal and dendritic regions indicating these are mixed polarity neurons. Fast neurotransmitter phenotyping of genie neurons represented by the SS30377 split-GAL4 driver showed overlap with smFLAG-vGlut (arrows, Figure 6C–C”) but no detectable expression of 9XV5-vGAT (Figure 6A–A”) or 7XMYC-vAChT (Figure 6B–B”), thus establishing genie neurons as glutamatergic. smFLAG-vGlut is distributed throughout genie neurons without clearly separable axonal and dendritic regions and are thus also mixed polarity neurons.
Figure 4.
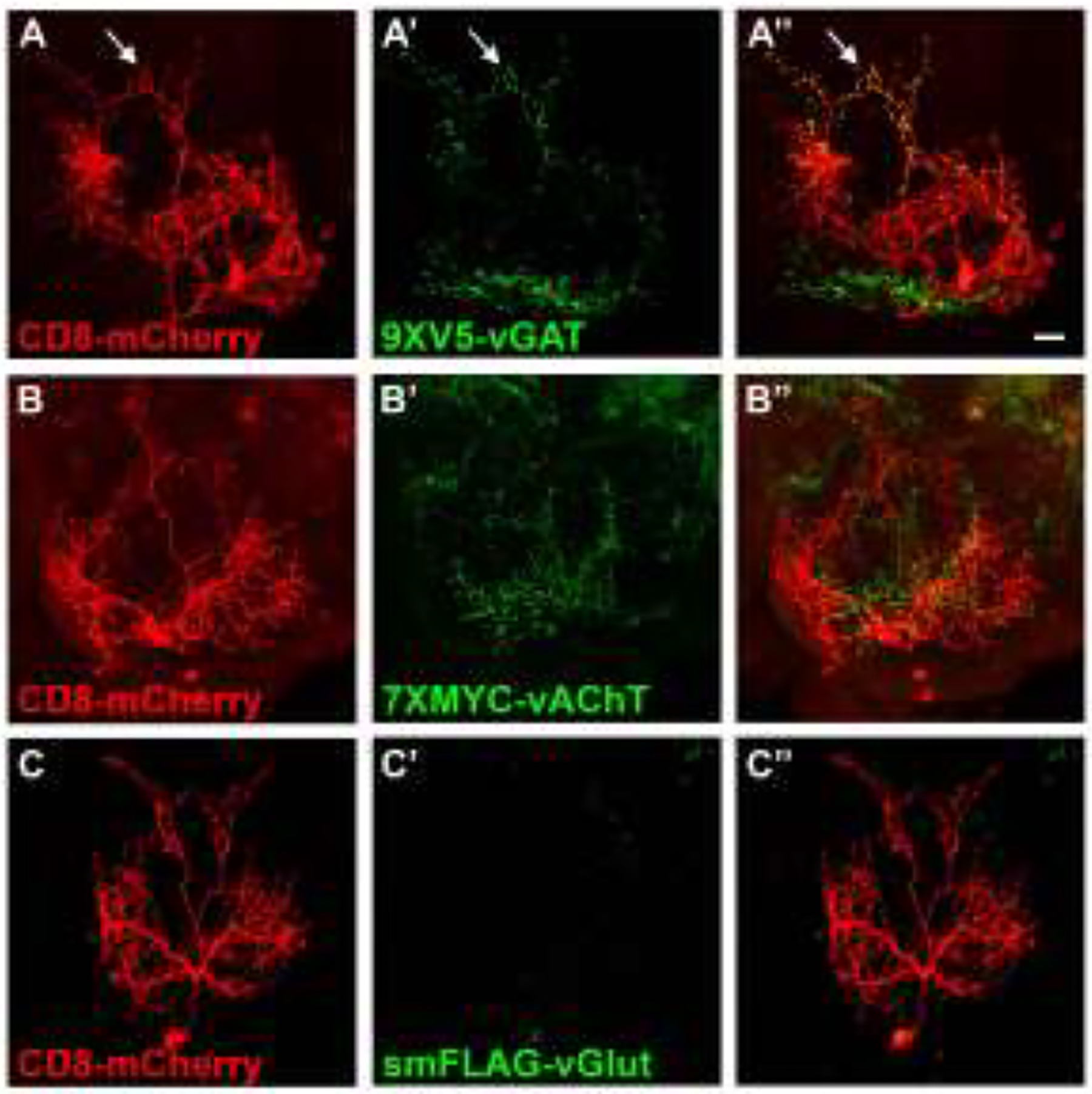
Fast neurotransmitter phenotyping of the subesophageal neuron trident/SS46209. A) CD8-mCherry; A’) 9XV5-vGAT); A”) overlay. 9XV5-vGAT expression in trident neurons indicates a GABAergic neurotransmitter phenotype. B) CD8-mCherry; B’) 7XMYC-vAChT; B”) overlay. C) CD8-mCherry; C’) smFLAG-vGlut; C”) overlay. 7XMYC-vAChT and smFLAG-vGlut were not detected in trident neurons. Scale bar: 20μm.
Figure 5.
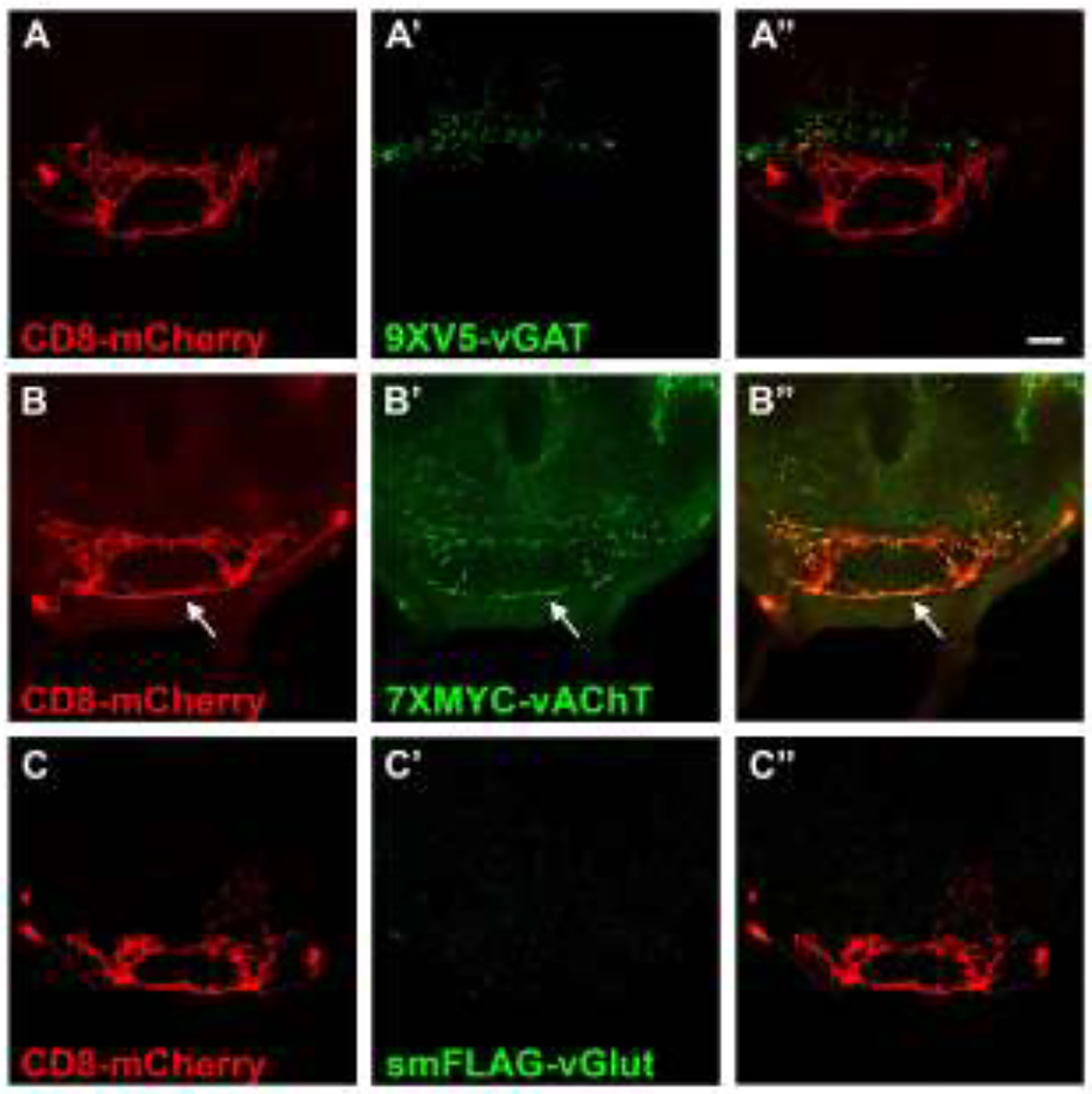
Fast neurotransmitter phenotyping of the subesophageal neuron G2N-1/SS47082. A) CD8-mCherry; A’) 9XV5-vGAT); A”) overlay. B) CD8-mCherry; B’) 7XMYC-vAChT; B”) overlay. 7XMYC-vAChT expression in G2N-1 neurons indicates a cholinergic neurotransmitter phenotype.C) CD8-mCherry; C’) smFLAG-vGlut; C”) overlay. 9XV5-vGAT and smFLAG-vGlut were not detected G2N-1 neurons. Scale bar: 20μm.
Figure 6.
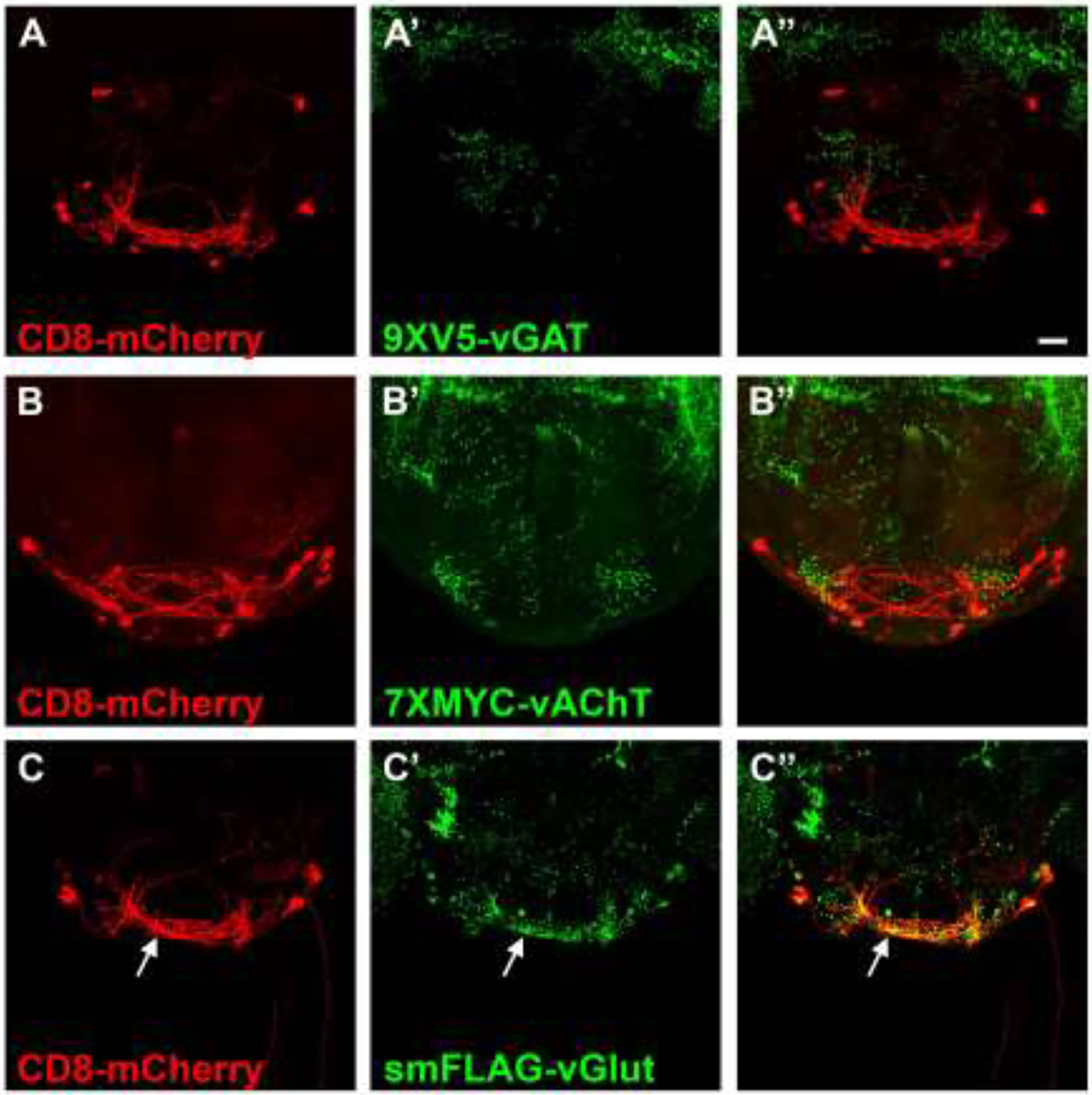
Fast neurotransmitter phenotyping of the subesophageal neuron genie/SS30377. A) CD8-mCherry; A’) 9XV5-vGAT); A”) overlay. B) CD8-mCherry; B’) 7XMYC-vAChT; B”) overlay. C) CD8-mCherry; C’) smFLAG-vGlut; C”) overlay. smFLAG-vGlut expression in genie neurons indicates a GABAergic neurotransmitter phenotype. 9XV5-vGAT and 7XMYC-vAChT were not detected in genie neurons. Scale bar: 20μm.
4. Discussion
The development and characterization of a conditional epitope tagged GABAergic SV marker for Drosophila, RSRT-STOP-RSRT-9XV5-vGAT, has been described. RSRT-STOP-RSRT-9XV5-vGAT was genome-edited at the endogenous locus of the sole Drosophila vesicular GABA transporter gene vGAT, thus making it a SV reporter for all GABAergic neurons. Data demonstrating its conditionality, specificity for GABAergic neurons, and specificity for SVs has been presented. The multimerized 9XV5 epitope tag provides robust signal and high sensitivity that makes it easily detectable in individual neurons. Evidence that the 9XV5-vGAT protein retains its function as a vesicular GABA transporter includes the viability of the 9XV5-vGAT germline excision chromosome in heterozygous combination with a null allele of vGAT and in homozygous condition.
Knowledge of the neurotransmitter identity of a neuron is critical to understand its contribution to neural circuit function and the generation of behavior. The utility of RSRT-STOP-RSRT-9XV5-vGAT for GABAergic neurotransmitter phenotyping and identifying sites of GABA release was demonstrated using 12 ellipsoid body neurons represented by split-GAL4 drivers. Many of these neurons have been implicated in the regulation of walking behavior on the observation that their activation increases the fraction of time spent walking, especially the outer ring neurons R2/R4 represented by split-GAL4 drivers SS0269, SS0267, and SS0266 (Robie et al. 2017). Knowledge that these ellipsoid body ring neurons are GABAergic provided by RSRT-STOP-RSRT-9XV5-vGAT suggests a model whereby they are part of a neural circuit that contains neurons downstream of them that inhibit walking. Thus, by this model GABAergic ellipsoid body neurons inhibit downstream neurons that themselves function to inhibit walking behavior with the net effect that the amount of time spent walking is enhanced. A completely different model of the function of these neurons in walking behavior would have emerged if these neurons were instead determined to be excitatory. This example with ellipsoid body neurons of the central complex thus demonstrates the value of the knowledge of the neurotransmitter identity of individual neurons for modeling how neural circuits regulate behavior.
Now that a connectome that includes the majority of neurons comprising the adult Drosophila brain has been completed (Scheffer et al. 2020), an important next step in understanding how these neurons work in concert to accomplish the multitude of functions performed by the brain will be to determine the neurotransmitter identity of each and every neuron. The principal method used heretofore for neurotransmitter phenotyping Drosophila brain neurons has been single cell transcriptomics in one of its variations (Henry et al. 2012; Croset et al. 2018; Davis et al. 2020). With this report of a GABAergic SV reporter there are now available conditional vesicular neurotransmitter reporters for all three Drosophila fast neurotransmitters GABA, acetylcholine (Tison et al. 2020), and glutamate (Certel 2022, in press), as well as a conditional vesicular monoamine transporter, 6XV5-vMAT (Sherer et al. 2020) for the monoamine neurotransmitters dopamine, octopamine, and serotonin (Chen et al. 2013). This makes it possible to perform neurotransmitter phenotyping for all of these neurotransmitters for any neurons represented by a binary transcription system driver. Use of conditional neurotransmitter-specific SV markers is thus an alternative method for neurotransmitter phenotyping of Drosophila brain neurons that can be employed on its own or as a complement to single cell transcriptomics to enhance confidence in the results of each method. It should be noted though that the conditional neurotransmitter-specific SV strategy has the advantages of technological simplicity and providing not just neurotransmitter identity but also knowledge of the spatial location of sites of neurotransmitter release.
As proof of principle, fast neurotransmitter phenotyping was demonstrated for three neuron types of the supesophageal ganglion with the result that determination of fast neurotransmitter identity was clear and unambiguous in each case. Thus, the addition of RSRT-STOP-RSRT-9XV5-vGAT described herein makes the use of conditional neurotransmitter-specific SV reporters for fast neurotransmitter phenotyping a valid alternative to single cell transcriptomics. Given there now exists split-GAL4 drivers representing many hundreds of neuron types of the adult Drosophila brain this same strategy can now be applied extensively for assessment of neurotransmitter identity and thereby facilitate our understanding of Drosophila brain function.
Supplementary Material
Highlights.
Conditionally expressible GABAergic synaptic vesicle marker for Drosophila
Robust visualization in individual neurons
Applicable for GABAergic neurotransmitter phenotyping
Identifies GABAergic synaptic vesicles and presynaptic sites of GABA release
Alternative to single cell transcriptomics for GABA neurotransmitter phenotyping
Acknowledgements:
We thank the Bloomington Drosophila Stock Center and the Janelia Farm Research Campus for fly strains, and the Developmental Studies Hybridoma Bank and David Krantz for antibodies.
Funding:
This work was supported by National Institutes of Health (NIH) grant R01GM115510 to S.J.C. and R.S.S and Swiss National Science Foundation grant 31003A_179587 to B.D.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: None.
CRediT authorship contribution statement
S.J. Certel-writing-review and editing, acquisition.
B.D. McCabe- writing-review and editing, acquisition.
R.S. Stowers-conceptualization, methodology, validation, investigation, resources, writing-original draft, visualization, acquisition.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA et al. , 2014. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3: e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y, and Holmes GL, 2005. The multiple facets of gamma-aminobutyric acid dysfunction in epilepsy. Curr Opin Neurol 18: 141–145. [DOI] [PubMed] [Google Scholar]
- Certel SJ, Ruchti E, McCabe BD and Stowers RS, 2022. A conditional glutamatergic synaptic vesicle marker for Drosophila. G3 (Bethesda). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, and Thor S, 2004. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development 131: 5429–5439. [DOI] [PubMed] [Google Scholar]
- Chen A, Ng F, Lebestky T, Grygoruk A, Djapri C et al. , 2013. Dispensable, redundant, complementary, and cooperative roles of dopamine, octopamine, and serotonin in Drosophila melanogaster. Genetics 193: 159–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Treiber CD and Waddell S, 2018. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Nern A, Picard S, Reiser MB, Rubin GM et al. , 2020. A genetic, genomic, and computational resource for exploring neural circuit function. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande SA, Freyberg Z, Lawal HO and Krantz DE, 2020. Vesicular neurotransmitter transporters in Drosophila melanogaster. Biochim Biophys Acta Biomembr 1862: 183308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MJ, Frechter S, Bates AS, Dan C, Huoviala P et al. , 2019. Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Chow DM, Chen A, Romero-Calderon R, Ong WS et al. , 2010. Mutation of the Drosophila vesicular GABA transporter disrupts visual figure detection. J Exp Biol 213: 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher YE, Yang HH, Isaacman-Beck J, Xie M, Gohl DM et al. , 2017. FlpStop, a tool for conditional gene control in Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasnier B, 2004. The SLC32 transporter, a key protein for the synaptic release of inhibitory amino acids. Pflugers Arch 447: 756–759. [DOI] [PubMed] [Google Scholar]
- Henry GL, Davis FP, Picard S and Eddy SR, 2012. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res 40: 9691–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse BK, Haberkern H, Franconville R, Turner-Evans DB, Takemura SY et al. , 2021. A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO et al. , 1996. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci 16: 3154–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp JM, Chung P and Simpson JH, 2015. Generating customized transgene landing sites and multi-transgene arrays in Drosophila using phiC31 integrase. Genetics 199: 919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler B, Faville R, Bridi JC and Hirth F, 2019. Inverse Control of Turning Behavior by Dopamine D1 Receptor Signaling in Columnar and Ring Neurons of the Central Complex in Drosophila. Curr Biol 29: 567–577 e566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H, Chen HM, Long X, Singer RH, Lee T et al. , 2019. Neurotransmitter identity is acquired in a lineage-restricted manner in the Drosophila CNS. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta R, Risoleo MC, Messina G, Parisi L, Carotenuto M et al. , 2020. The Neurochemistry of Autism. Brain Sci 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Pena A, Acebes A, Rodriguez JR, Chevalier V, Casas-Tinto S et al. , 2014. Cell types and coincident synapses in the ellipsoid body of Drosophila. Eur J Neurosci 39: 1586–1601. [DOI] [PubMed] [Google Scholar]
- Petersen LK, and Stowers RS, 2011. A Gateway MultiSite recombination cloning toolkit. PLoS One 6: e24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T and Bullock SL, 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 111: E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robie AA, Hirokawa J, Edwards AW, Umayam LA, Lee A et al. , 2017. Mapping the Neural Substrates of Behavior. Cell 170: 393–406 e328. [DOI] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY et al. , 2020. A connectome and analysis of the adult Drosophila central brain. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schioth HB, Roshanbin S, Hagglund MG and Fredriksson R, 2013. Evolutionary origin of amino acid transporter families SLC32, SLC36 and SLC38 and physiological, pathological and therapeutic aspects. Mol Aspects Med 34: 571–585. [DOI] [PubMed] [Google Scholar]
- Schur RR, Draisma LW, Wijnen JP, Boks MP, Koevoets MG et al. , 2016. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Hum Brain Mapp 37: 3337–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer LM, Catudio Garrett E, Morgan HR, Brewer ED, Sirrs LA et al. , 2020. Octopamine neuron dependent aggression requires dVGLUT from dual-transmitting neurons. PLoS Genet 16: e1008609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart TG, and Stephenson FA, 2019. A half century of gamma-aminobutyric acid. Brain Neurosci Adv 3: 2398212819858249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne GR, Otsuna H, Dickson BJ and Scott K, 2021. Classification and genetic targeting of cell types in the primary taste and premotor center of the adult Drosophila brain. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SF, and Tso IF, 2015. GABA abnormalities in schizophrenia: a methodological review of in vivo studies. Schizophr Res 167: 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tison KV, McKinney HM and Stowers RS, 2020. Demonstration of a Simple Epitope Tag Multimerization Strategy for Enhancing the Sensitivity of Protein Detection Using Drosophila vAChT. G3 (Bethesda) 10: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD Jr., Ning X, Klingler M, Kramer SG and Gergen JP, 2000. Quantitative analysis of gene function in the Drosophila embryo. Genetics 154: 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner-Evans DB, Jensen KT, Ali S, Paterson T, Sheridan A et al. , 2020. The Neuroanatomical Ultrastructure and Function of a Biological Ring Attractor. Neuron 108: 145–163 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Shearin HK and Stowers RS, 2019. Conditional Synaptic Vesicle Markers for Drosophila. G3 (Bethesda) 9: 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, and Rubin GM, 2018. Neuroarchitecture of the Drosophila central complex: A catalog of nodulus and asymmetrical body neurons and a revision of the protocerebral bridge catalog. J Comp Neurol 526: 2585–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete annotated sequence of the RSRT-STOP-RSRT-9XV5-vGAT donor plasmid is shown in Supplemental Information (Figure S1). The RSRT-STOP-RSRT-9XV5-vGAT donor plasmid, and pCFD4-vGAT1 and pCFD4-vGAT2 guide RNA plasmids will be made available upon request. Fly strains original to this publication will be deposited at the Bloomington Drosophila stock center or will be made available upon request.


