Abstract
The Omicron variant is rapidly becoming the dominant SARS-CoV-2 virus circulating globally. It is important to define reductions in virus neutralizing activity in serum of convalescent or vaccinated individuals to understand potential loss of protection against infection by Omicron. We have previously established that a 50% plaque reduction neutralization (PRNT50) antibody titre ≥25.6 in our live virus assay corresponded to the threshold for 50% protection from infection against wild-type (WT) SARS-CoV-2. Here we show markedly reduced serum antibody titres against the Omicron variant (geometric mean titre (GMT) <10) as compared to wild-type virus 3–5 weeks after two doses of BNT162b2 (GMT 218.8) or CoronaVac vaccines (GMT 32.5). A BNT162b2 booster dose elicited Omicron PRNT50 titres ≥25.6 in 88% of individuals (22 of 25) who previously received 2 doses of BNT162b2 and 80% of individuals (24 of 30) who previously received CoronaVac. However, few (3%) previously infected individuals (1 of 30) or those vaccinated with three doses of CoronaVac (1 of 30) met this threshold. Our findings suggest that countries primarily using CoronaVac vaccines should consider mRNA vaccine boosters in response to the spread of Omicron. Studies evaluating the effectiveness of different vaccines against the Omicron variant are urgently needed.
A novel SARS-CoV-2 variant with increased transmissibility was first reported in South Africa in November 20211, classified as a variant of concern and named Omicron (BA.1 sublineage of B.1.1.529)2. This variant has 37 amino acid substitutions in the spike protein of the virus, 15 of them being in the receptor binding domain. It was predicted that some of these amino acid substitutions would enable the evasion of neutralizing antibodies. Virus neutralizing antibodies are a major determinant of protection from infection in humans and in macaques experimentally challenged with virus3,4. Neutralizing antibody thresholds associated with protection from re-infection or severe disease have been reported5,6. Although CD8 T cells have been shown to contribute to protection, quantitative correlates of protection remain elusive. 4
CoronaVac is one of the WHO approved vaccines and over two billion doses have been administered in more than 40 countries. Phase 3 randomized clinical trials of CoronaVac showed vaccine efficacy of 50.7% and higher vaccine effectiveness against severe disease7,8. However, there have been reports of breakthrough infections leading to severe disease and death in CoronaVac vaccinated adults9. Data on the immunogenicity of current COVID-19 vaccines against the Omicron variant is urgently needed.
We have previously demonstrated that those vaccinated with BNT162b2 had markedly higher levels of geometric mean PRNT50 antibody titres against SARS-CoV-2 isolated in Hong Kong in January 2020 compared to those vaccinated with CoronaVac vaccines at 3–5 weeks post-second vaccine dose 10. Allowing for antibody waning, we estimated that only 16% of the CoronaVac vaccinated individuals would retain PRNT50 antibody titres above protective thresholds against the WT virus while 79.6% of BNT162b2 vaccinees would, by six months after second dose of vaccine10. Subsequently, we randomized the cohort receiving CoronaVac vaccine to receive booster doses of CoronaVac or BNT162b2 and showed a marked increase in neutralizing antibodies to WT SARS-CoV-2 following boosting with BNT162b2, but less of an increase with CoronaVac11.
Here we compare PRNT50 and PRNT90 geometric mean antibody titres (GMTs) to WT SARS-CoV-2 and Omicron BA.1 variant in subsets of sera from 7 groups of vaccinated individuals, convalescent individuals and individuals with breakthrough infections (Table 1, Extended Data Table 1, Extended Data Table 2). We evaluated sera from a) vaccinated individuals with no evidence of prior COVID-19 infection (see methods) 3–5 weeks after receiving two doses of BNT162b2 (n=31) or two doses of CoronaVac (n=30), randomly selected from a previous study, 10 and b) individuals 3–5 weeks after receiving a 3rd dose of CoronaVac (n=30) or a heterologous booster dose of BNT162b2 after two prior doses of CoronaVac (n=30), randomly selected from a previous study (ClinicalTrials.Gov NCT04611243),11 and c) those receiving 3rd dose of BNT162b2 (n=25). We also evaluated sera from the following groups of previously infected individuals: (a) individuals 143–196 days post-infection who had recovered from COVID-19 (pre-omicron emergence) and had not yet received vaccine (n=30),12 (b) COVID-19 convalescent individuals who had received one dose of BNT162b2 (n=30) or (c) COVID-19 convalescent individuals who had received one dose of CoronaVac (n=28). Sera collected during acute infection and during convalescence from six Omicron-infected individuals identified in Hong Kong in November and December 2021 were also profiled. Plaque reduction neutralization tests were carried out as previously described (Methods).12,13 The highest serum dilutions neutralizing ≥50% or ≥90% of plaques were regarded as the PRNT50 and PRNT90, respectively. SARS-CoV-2 viruses used were a WT virus isolated in January 2020, an Omicron variant isolated on November 13, 2021 and a Delta lineage virus isolated in June 2021.
Table:
Age, sex and geometric mean 50% plaque reduction neutralization test (PRNT50) titres vs. Wild-type (WT) or Omicron BA.1 variant 3–5 weeks post vaccine
| Exposure group | n | Mean Age (SD) | Age range | Male: female | WT GMT | Omicron GMT | WT: Omicron Fold GMT reduction | No (%) of sera with detectable antibody to Omicron | No. above PRNT50 threshold of protection (1:25.6) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Omicron | Wild-type | |||||||||
| BNT162b2 (2 doses) | 31 | 51.7(14.4) | 25–76 | 17:14 | 218.8 | 7.0 | 31.3 | 8/31 (25.8%) | 2/31 | 30/31 |
| BN162b2 (3 doses) | 25 | 50.6(14.9) | 22–72 | 14:11 | 320 | 77.8 | 4.1 | 25/25 (100.0%) | 22/25 | 25/25 |
| CoronaVac (2 doses) | 30 | 52.1(8.2) | 39–73 | 10:20 | 32.5 | 5 | 6.5 | 0/ 30 (0.0%) | 0/30 | 14/30 |
| CoronaVac (2 dose+CoronaVac booster) | 30 | 50.5(8.8) | 36–73 | 8:22 | 65 | 8.9 | 7.3 | 19/30 (63.3%) | 1/30 | 25/30 |
| CoronaVac (2 dose+BNT162b2 booster) | 30 | 50.4(9.3) | 31–66 | 13:17 | 305.5 | 59.2 | 5.2 | 30/30 (100.0%) | 24/30 | 30/30 |
| SARS-CoV-2 Convalescent (day 143–196 days) | 30 | 48.9(15.8) | 20–72 | 13:17 | 85.7 | 8.1 | 10.6 | 12/30 (40.0%) | 1/30 | 29/30 |
| SARS-CoV-2 convalescent + BNT162b2 (1 dose) | 30 | 49.8(14.8) | 20–71 | 15:15 | 320 | 130 | 2.5 | 30/30 (100.0%) | 29/30 | 30/30 |
| SARS-CoV-2 convalescent + CoronaVac (1 dose) | 28 | 55.0(10.6) | 29–81 | 14:14 | 237.8 | 29.7 | 8.0 | 27/28 (96.4%) | 18/28 | 28/28 |
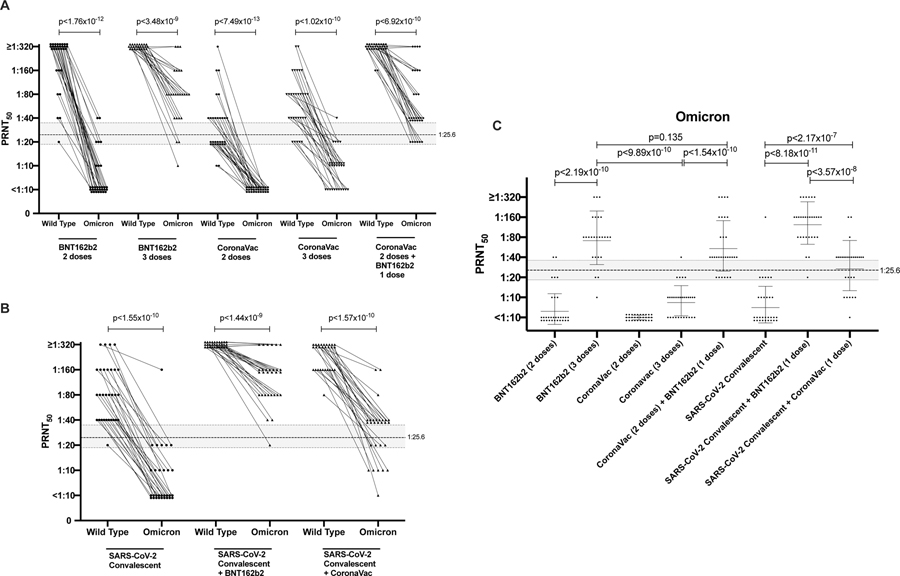
The PRNT50 titres with the wild-type SARS-CoV-2 or Omicron variant in these cohorts are shown in Figure 1. PRNT90 data are shown in Extended data figure 1. PRNT50 titres in all COVID-19 vaccinated or convalescent cohorts were markedly lower against Omicron variant than against wild-type virus (Figure 1; Table 1). Relative to GMT against wild-type virus, there was a 31-fold reduction post-two doses of BNT162b2 vaccine, a 6.5-fold reduction post-two doses of CoronaVac vaccine and a 10.6-fold reduction in COVID-19 convalescent sera (Figure 1A, Table1). Other studies have reported comparable reductions in sera neutralizing titres in mRNA vaccinated or COVID-19 convalescent individuals14,15. However, it is important to note that comparison of fold-difference in GMT to the two viruses is confounded when some vaccine groups have low neutralizing antibody GMT to the wild-type virus, because only a smaller fold-reduction is possible before titres reach the detection threshold. Booster doses of BNT162b2 and CoronaVac vaccines increased Omicron PRNT50 GMT from 7 to 77.8 and from 5 to 8.9, respectively but CoronaVac vaccinees boosted with BNT162b2 had GMT of 59.2. COVID-19 convalescent individuals had Omicron GMT of 8.1 and one dose of BNT162b2 or CoronaVac increased these GMT to 130 and 29.7, respectively.
Figure 1: 50% plaque reduction neutralization test (PRNT50) antibody titres to wild-type (WT) virus and Omicron BA.1 variant.
A. Individuals with 2 or 3 doses of BNT162b2 or CoronVac vaccines, as indicated. B. SARS-CoV-2 convalescent individuals with or without BNT162b2 or CoronaVac vaccine (one dose). C. PRNT50 titres to Omicron in the different groups with geometric mean +/− SD are denoted. See Table for numbers of individuals in each group. Mann-Whitney test (two tailed) was used for significance testing. P values for each comparison are denoted. Dotted line indicates PRNT50 threshold of protection and shading indicates 95% confidence intervals (see text).
Using methods described by Khoury and colleagues5, we had previously determined that the threshold for 50% protection from symptomatic WT SARS-CoV-2 infection was a PRNT50 titre of 25.6 with 95% confidence intervals of 18.3–36.012. Using this threshold for protection, we observed that 30 of 31 BNT162b2 vaccinated individuals retained protective PRNT50 antibody titres to wild-type virus at 3–5 weeks after the second BNT162b2 dose but only 2 in 31 met this threshold with Omicron variant. None of those vaccinated with CoronaVac met this protection threshold with the Omicron variant (Table1). A homologous booster dose of BNT162b2 in individuals who had been previously vaccinated with BNT162b2 restored protective thresholds of Omicron variant PRNT50 antibody in 22 of 25 vaccinees but a homologous booster dose of CoronaVac in CoronaVac vaccinees did not (one in 30). However, following a heterologous BNT162b2 booster, 24 of 30 individuals who had previously received two doses of CoronaVac had protective levels of PRNT50 antibodies to the Omicron variant. In previously infected individuals who were subsequently vaccinated with 1 dose of either BNT162b2 or CoronaVac, 29 of 30 and 18 of 28 respectively had protective levels of PRNT50 antibody to the Omicron variant, 3–5 weeks after the vaccine dose (Table 1).
It was estimated that neutralizing antibodies were reduced by 4.7-fold within 5 to 5.8 months after the second dose of BNT162b2 vaccine and declined 6.4-fold within 6 months following two doses of CoronaVac vaccination16,17. It is not clear if antibody waning is similar after a booster dose nor whether waning of cross-neutralization antibodies against Omicron would follow similar kinetics. There is, however, evidence that waning of protection against Delta variant following a booster dose of BNT162b2 is relatively rapid.18 Allowing for a hypothetical 4.7-fold decline of PRNT50 antibody levels to Omicron variant, we estimated that 7of 25 (28%) of those vaccinated and boosted with BTN162b2, and 8 of 30 (27%) CoronaVac vaccinated individuals boosted with BNT162b2 would retain PRNT50 antibody levels above this threshold. However, 0 of 30 (0%) of individuals who received 3 doses of CoronaVac would.
Our COVID-19 convalescent cohort was already 4.8 to 6.5 months post-symptom onset and only one of 30 individuals met the protective antibody threshold for Omicron variant. Vaccinating COVID-19 convalescent individuals with one dose of BNT162b2 or CoronaVac vaccine boosted PRNT50 antibody levels to above the protective threshold against Omicron in 29 of 30 (97%) and 18 of 28 (64%) individuals respectively, at 3–5 weeks post-vaccination. However, the levels achieved in CoronaVac vaccinated individuals (GMT 29.7) was lower than for BNT162b2 (GMT 130), and assuming a 4.7-fold reduction in antibodies, only 2 of 28 would have PRNT50 titres above 25.6 by 5–5.8 months post-vaccination, although 19 of 30 of those receiving BNT162b2 would.
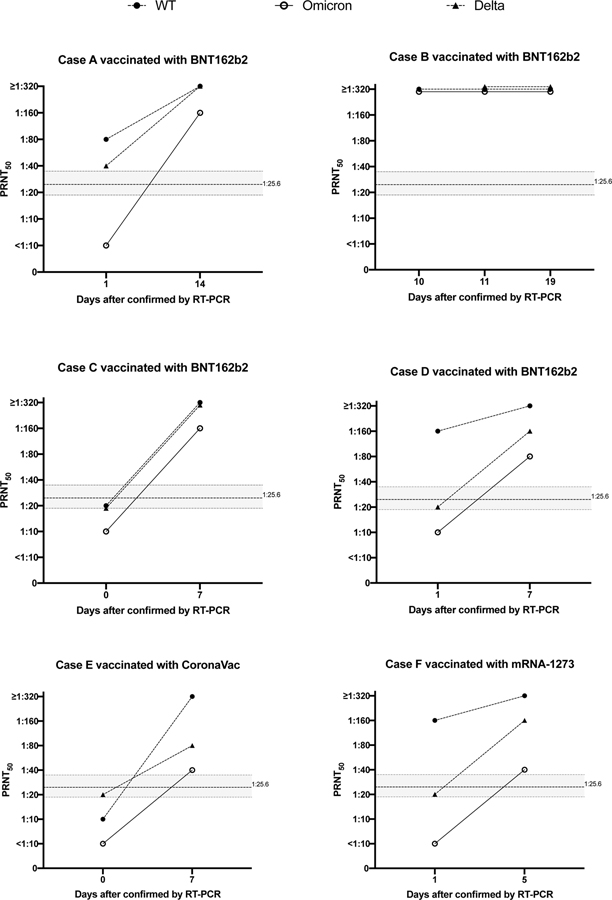
Six individuals with travel-associated vaccine breakthrough Omicron variant infection were also studied, three with mildly symptomatic illness (Cases B, D, E) and three with asymptomatic infection (Cases A, C, F). Five were males and one female, ranging in age from 22–62 years, vaccinated with two doses of BNT162b2 (n=4), mRNA-1273 (n=1) and CoronaVac (n=1) (Extended data Table 2). Five of them had a first serum sample collected within 1 day of RT-PCR diagnosis with Omicron infection and they all had Omicron PRNT50 titres of 10 or lower, possibly explaining why they had Omicron breakthrough infections even though three (A, D, F) had PRNT50 of 80–160 to WT virus (Extended data figure 2). All five showed a rise in antibody titres to Omicron as well as to WT and Delta viruses. None of these individuals were previously diagnosed with SARS-CoV-2 infection. Acquiring Delta in infection in Hong Kong would be unlikely as there have been only 6 locally acquired Delta infections in Hong Kong in 2021. The duration of travel away from Hong Kong for five of these individuals was 18–31 days (Extended Data Table 2) and it would be unlikely that they recovered from Delta and got re-infected with Omicron within that time. Three of them (A,D,F) had higher titres to WT than to Delta in the acute serum, more compatible with vaccine immunity than past infection with Delta. The consistent finding of boosting of antibody responses to Delta virus in these individuals may indicate that Omicron infection in people vaccinated with mRNA or inactivated vaccines may lead to broadening immunity to a range of virus variants, even ones they have not been previously exposed to. A similar finding of increased neutralizing antibodies to the Delta variant in previously vaccinated individuals with Omicron infection has been reported by others.19
This study had some limitations. There were relatively small numbers in the individual groups, but as estimated from our power calculations, the sample size was adequate to demonstrate significant difference in neutralizing antibody responses between the groups studied. We have not investigated other antibody activities, such as binding to spike protein or binding to Fc receptors, which may enable antibody-dependent cell cytotoxicity. In a previous study with the cohorts vaccinated with two doses of BNT-162b2 or two doses of CoronaVac, we showed that both spike binding antibodies and FcgRIIIa binding antibodies correlated with the PRNT antibody responses.10 However, it is possible that these non-neutralizing antibodies may play more important roles in protection from Omicron because they may be less affected by the spike mutations found in the Omicron variant virus. We have also not investigated T cell responses elicited by these vaccines, which may provide protection.
In summary, we demonstrate that two doses of BNT162b2 or CoronaVac vaccines elicit low neutralizing antibodies against the Omicron variant, even at 3–5 weeks post-vaccination. Homologous or heterologous BNT162b2 booster doses following two doses of either BNT162b2 or CoronaVac improve neutralizing antibody levels against Omicron variant at 3–5 weeks post-booster dose. Three doses of CoronaVac failed to elicit neutralizing antibody responses to Omicron in most recipients. Our findings suggest that countries that primarily use CoronaVac vaccines may need to consider mRNA vaccine boosters in response to the spread of Omicron. However, It must be noted that CoronaVac has previously been shown to elicit a wider range of virus-specific T cell responses, which may compensate for some loss of neutralizing antibody protection.10 Immunogenicity studies provide rapid assessments of potential protective effects of vaccines against novel variants but vaccine effectiveness studies in the field are urgently required.
Methods:
Clinical specimens:
Sera were randomly selected from two previously studied cohorts investigating immunogenicity of two doses10 or booster doses11 of COVID-19 vaccines. The first study had sera collected 3–5 weeks after the second dose of CoronaVac (28 days between first and second dose) or BNT162b2 vaccines (21 days between first and second dose).10 The second cohort had sera collected 3–5 weeks after booster vaccination of CoronaVac vaccinated individuals who were given a third dose of CoronaVac (interval between second and third vaccine dose was 61–160 days)(17 of them also included in the previous two dose CoronaVac study) or BNT162b2 (interval between second and third vaccine dose was 51–141 days)(9 of them also included in the previous two dose CoronaVac study) (ClinicalTrials.gov with identifier NCT04611243).11 A third group comprised of BNT162b2 vaccinated individuals receiving a third dose of BNT162b2 (180–234 days after second dose) (4 of them included in the previous two dose BNT162b2 study) with serum collected 3–5 weeks after the third vaccine dose. None of these individuals had previous history of SARS-CoV-2 infection confirmed with negative IgG to SARS-CoV-2 receptor binding domain antibody15 in the pre-first vaccine dose serum.
Separately, sera were obtained from an ongoing longitudinal study of virologically confirmed SARS-CoV-2 convalescent individuals. Sera from non-vaccinated COVID-19 convalescent individuals were selected from day 143–196 days (4.7–6.5 months) post-infection to represent waning antibody titres in convalescence (n=30).12 Sera were also collected 3–5 weeks post vaccination from COVID-19 convalescent individuals who received either BNT162b2 (n=30) or CoronaVac (n=28) vaccines.
These studies were carried out in Hong Kong during the period 21st February 2020 to November 20th 2021 and was approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (Ref no: 2020.229). All participants provided informed written consent. Sample sizes for each group are indicated in Table. See statistical methods for the basis for sample size calculations.
Sequential paired sera from six patients diagnosed with Omicron variant infection in November-December 2021 were included as positive controls for comparison (See Extended data table 2 for clinical details).
Virology:
Vero-E6 cells (ATCC CRL-1586) and Vero E6 cells overexpressing TMPRSS2 (kindly provided by Dr S Matsuyama and colleagues)20 were used. Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) medium (ThermoFisher Scientific), Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (ThermoFisher Scientific), Waltham, MA, USA) and 100 U/ml of penicillin–streptomycin (ThermoFisher Scientific), Waltham, MA, USA). The cells used in the study were regularly tested for mycoplasma contamination at the Core Facility, Centre for PanorOmic Sciences, The University of Hong Kong. We used a strain of Omicron BA.1 variant isolated from the nose & throat swab of a returning traveler diagnosed with Pango lineage B.1.1.529 Omicron variant infection on November 13, 2021, while in hotel quarantine in Hong Kong. The virus, designated hCoV-19/Hong Kong/VM21044713_WHP5047-S5/2021 was isolated in Vero-E6 TMRSS2 cells and the passage level 3 virus aliquots were used in these studies. The virus had amino acid R346 in the spike protein, as do the majority of Omicron variant viruses to date. The wild type SARS-CoV-2 BetaCoV/Hong Kong/VM20001061/2020 (WT) isolated in Hong Kong in January 2020 was used for comparison. Delta lineage virus isolated from a clinical specimen collected in June 2021 in Vero-E6 TMPRSS2 cells designated hCoV-19/Hong Kong/21TM280310_HKUVOC0013/2021 was used for comparison. The original clinical swab and the virus isolates were genetically sequenced and it was confirmed that the isolates had identical amino acid sequence to the virus from the original swab. The virus sequences reported are available in GISAID as EPI_ISL_412028, EPI_ISL_6716902 and EPI_ISL_6716902.
Plaque reduction neutralization tests were carried out in duplicate using 24-well tissue culture plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) in a biosafety level 3 facility using Vero E6 TMRESS2 cells.12,21 Serial two-fold dilutions from 1:10 to 1:320 of each serum sample was incubated with 30–40 plaque-forming units of virus for 1 h at 37 °C. The virus-serum mixtures were added onto pre-formed cell monolayers and incubated for 1 h at 37° C in 5% CO2 incubator. The cell monolayer was then overlaid with 1% agarose in cell culture medium and incubated for 3 days, at which time the plates were fixed and stained. Antibody titers were defined as the highest serum dilution that resulted in ≥90% (PRNT90) reduction or >50% (PRNT50) in the number of virus plaques. This method has been extensively validated on SARS-CoV-2 infected and control sera previously. 12,20
The highest serum dilution neutralizing ≥50% and ≥90% of input plaques was regarded as the 50% plaque reduction neutralization (PRNT50) and PRNT90 titres, respectively. A virus back titration and a positive control serum was included in every experiment. The WHO control serum NIBSC 20/136 was also included in two titrations and gave PRNT50 titres of 20 and 40 with Omicron virus and 320 and 320 with wild-type virus. Virus titres are designated as reciprocal of the serum dilution throughout the text.
Statistical analysis.
Sample size calculations: The maximum standard deviation (SD) of log titers for the uninfected vaccinated groups were 1.37 and 1.77 for PRNT50 and PRNT90 respectively. Assuming a 3-fold difference in GMT, a sample size of 10 in each group would have statistical power of >0.99 and 0.94 for PRNT50 and PRNT90 respectively, for detecting a difference between groups using two tailed Mann-Whatney U test. Comparisons between groups with larger sample size or smaller within-group variation would have larger statistical power.
Continuous variables were summarized as geometric mean with standard deviation (SD) while categorical variables were summarized as proportions or percentage. Sera with undetectable (<10) antibody titres were assigned an antibody titre of 5, for purposes of geometric mean titre calculations or statistical comparisons. Comparison between antibody titres to wild type and Omicron variant viruses was done using the two tailed Mann-Whitney U test. Absolute P values were provided. P values < 0.05 were considered statistically significant.
Extended Data
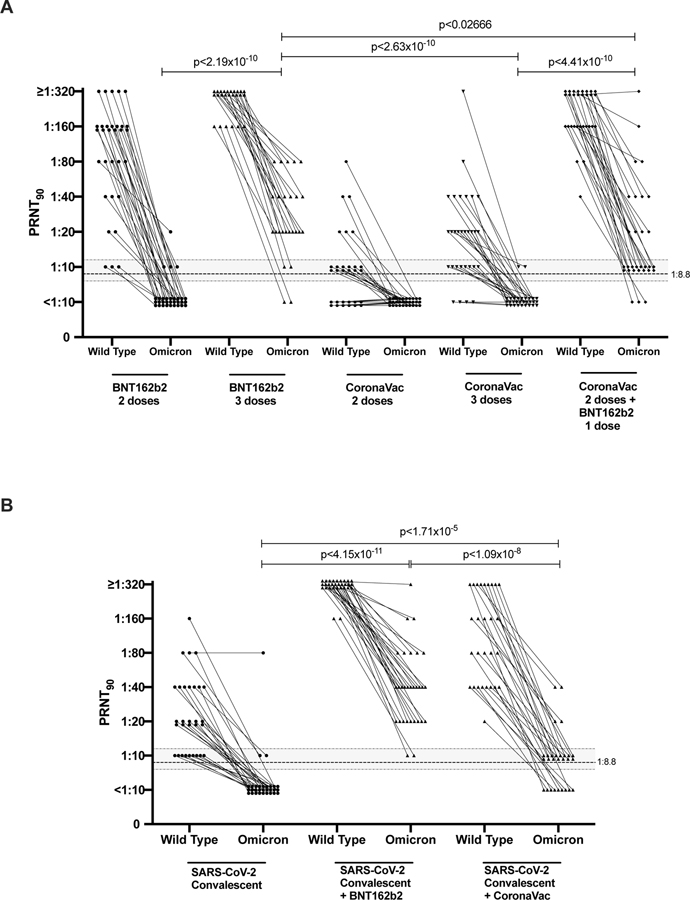
Extended data Fig 1. 90% plaque reduction neutralization test (PRNT90) antibody titres to wild-type virus and Omicron BA.1 variant.
PRNT90 antibody titres to Wild-type and Omicron Variant A. Individuals with 2 or 3 doses of BNT162b2 or CoronaVac vaccines, as indicated. B. SARS-CoV-2 convalescent individuals with or without BNT162b2 or CoronaVac vaccines (one dose). See table for numbers of individuals in each group. Mann-Whitney test (two tailed) was used for significance testing. P values are denoted. Dotted line indicates the threshold of protection and the shading indicates 95% confidence intervals (see text).
Extended data Fig2. 50% plaque reduction neutralization test (PRNT50) antibody titres to wild-type virus, Omicron BA.1 variant and Delta variant in paired sera in six vaccinated individuals with breakthrough Omicron infections.
50% plaque reduction neutralization test (PRNT50) antibody titres wild type virus, Omicron variant and Delta variant in paired sera in six vaccinated individuals with breakthrough Omicron infections. The vaccine used in each individual is provided in the panel heading and clinical data provided in extended data table 2. Dotted line indicates PRNT50 threshold of protection and shading indicates 95% confidence intervals (see text).
Extended data table 1:
Co-morbidities in the cohorts investigated.
| Exposure group | N | Co-morbidity | ||||
|---|---|---|---|---|---|---|
| None | Diabetes | Hypertension | Others | Not known | ||
| BNT162b2 2 doses | 31 | 18 | 2 | 7 | 10 | 0 |
| BNT162b2 3 doses | 25 | 13 | 0 | 3 | 2 | 8 |
| CoronaVac 2 doses | 30 | 18 | 1 | 5 | 9 | 0 |
| CoronaVac 3 doses | 30 | 20 | 3 | 3 | 8 | 0 |
| CoronaVac 2 doses + BNT162b2 | 30 | 23 | 2 | 2 | 6 | 0 |
| SARS-CoV-2 convalescent | 30 | 11 | 5 | 8 | 19 | 0 |
| SARS-CoV-2 convalescent + 1 dose BNT162b2 | 30 | 14 | 5 | 6 | 15 | 0 |
| SARS-CoV-2 convalescent + 1 dose CoronaVac | 28 | 14 | 4 | 4 | 14 | 0 |
Individuals may have more than one co-morbidity.
Other: includes ischemic heart disease, hyperlipidaemia, renal disease, stroke, Thrombocytopenia, hepatitis B, hepatitis C, post renal transplant (n=1), hyperthyroidism, hypothyroidism, asthma, gout, dermatitis, TB, depression.
Extended data table 2:
Clinical information on Omicron infections in returning travellers
| Case number | Age | Sex | Date of vaccine second dose | Type of vaccine | Date of RT-PCR diagnosis | Days since second dose to first RT-PCR positive | Symptoms | Period of travel |
|---|---|---|---|---|---|---|---|---|
| A | 36 | M | 4/6/2021 | BNT162b2 | 13/11/2021 | 163 | Asymptomatic | 18 days |
| B | 62 | M | 25/5/2021 | BNT162b2 | 20/11/2021 | 187 | Mild | 31 days |
| C | 37 | M | 25/6/2021 | BNT162b2 | 7/12/2021 | 165 | Asymptomatic | 19 days |
| D | 22 | M | 29/5/2021 | BNT162b2 | 19/12/2021 | 204 | Mild | 18 days |
| E | 28 | F | 10/2/2021 | CoronaVac | 20/12/2021 | 312 | Mild | 20 days |
| F | 37 | M | 5/10/2021 | mRNA-1273 | 26/11/2021 | 53 | Asymptomatic | 3 years |
Supplementary Material
Acknowledgements
This research was supported by grants from the Health and Medical Research Fund Commissioned Research on the Novel Coronavirus Disease (COVID-19), Hong Kong SAR (COVID1903003; COVID190126) (CKPM, DSH and MP), US National Institutes of Health (contract no. U01-Grant AI151810) (MP), National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme (N_HKU737/18) (CKPM and MP), RGC theme-based research schemes (T11-712/19-N and T11-705/21-N) (DSH, MP), Guangdong Province International Scientific and Technological Cooperation Projects (2020A0505100063) (CKPM), the National Research Foundation of Korea (NRF) grant funded through the Korea government (NRF-2018M3A9H4055203) (CKPM), AIR@InnoHK (EHYL) and C2i (LLMP, MP) administered by Innovation and Technology Commission of Hong Kong.
Footnotes
Competing interests: None of the authors had competing financial or non-financial interests.
Data availability
Source data (individual anonymized patient and linked laboratory data) and raw antibody data are provided with this paper. The viruses used can be obtained on request to Malik Peiris, malik@hku.hk
References:
- 1.Viana R, et al. 2021. https://krisp.org.za/manuscripts/ZHTOWa-MEDRXIV-2021-268028v1-deOliveira.pdf
- 2.World Health Organization. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 3.Addetia A, et al. J Clin Microbiol. 58(11):e02107–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahan K, et al. Nature. 590:630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoury DS, et al. Nat Med. 27:1205–1211 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Cromer D, et al. Lancet Microbe. (2021) doi: 10.1016/S2666-5247(21)00267-6. [DOI] [Google Scholar]
- 7.Zhang Y, et al. Lancet Infect Dis. 21:181–192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jara A, et al. N Engl J Med. In press. [Google Scholar]
- 9.Bangkok Post, 2021. (https://www.bangkokpost.com/world/2133987/hundreds-of-vaccinated-indonesian-health-workers-infected).
- 10.Mok CKP, et al. Respirology Epub ahead of print. [Google Scholar]
- 11.Mok CKP, Am J Resp and Crit Care Med -in press. [Google Scholar]
- 12.Lau EH, et al. EClinicalMedicine. 41:101174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuyama S, et al. Proc Natl Acad Sci U S A. 117:7001–7003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cele S, et al. medRxiv [Preprint]. 2021. doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 15.Schmidt F et al. N Engl J Med. Online (2022). [Google Scholar]
- 16.Levin EG, et al. N Engl J Med. 385:e84. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng G, et al. Lancet Infect Dis. 7:S1473–3099(21)00681–2. (2021). [Google Scholar]
- 18.Levine-Tiefenbrun M et al. medRxiv. Doi: 10.1101/2021.12.27.21268424 (2021). [DOI] [Google Scholar]
- 19.Khan K, et al. medRxiv [Preprint]. 2021. Dec 27:2021.12.27.21268439. doi: 10.1101/2021.12.27.21268439. [DOI] [Google Scholar]
- 20.Perera RA et al. Euro Surveill. 25(16):2000421 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data (individual anonymized patient and linked laboratory data) and raw antibody data are provided with this paper. The viruses used can be obtained on request to Malik Peiris, malik@hku.hk