Abstract
Saporin, a type I ribosome-inactivating protein from soapwort plant, is a potent protein synthesis inhibitor. Catalytically, saporin is a characteristic N-glycosidase, and it depurinates a specific adenine residue from a universally conserved loop of the major ribosomal RNA (rRNA) of eukaryotic cells. It is well-known that saporin induces apoptosis through different pathways, including ribotoxic stress response, cell signal transduction, genomic DNA fragmentation and RNA abasic lyase (RAlyase) activity, and NAD+ depletion by poly-(ADP)-ribose polymerase hyperactivation. Saporin’s high enzymatic activity, high stability, and resistance to conjugation procedures make it a well-suited tool for immunotherapy approaches.
In the present study, we focus on saporin-based targeted toxins that may be efficacious therapeutic agents for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Our discussed points suggest that saporin may be a strategic molecule for therapeutic knockout treatments and a powerful candidate for novel drugs in the struggle against coronavirus 2019 (COVID-19).
Keywords: SARS-CoV-2, saporin, abasic site, rRNA, N-glycosidase
Introduction
Due to a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a major outbreak of extraordinary viral pneumonia [coronavirus 2019 (COVID-19)] started in Wuhan, China, in late December 2019. The World Health Organization (WHO) declared the epidemic of COVID-19 as a pandemic on March 11, 2020.1 Coronaviruses are pathogenic in humans and other mammals (e.g., bats and pangolins). As the virus has spread around the world, countries have reached different stages of their outbreaks at different times. Across the globe, COVID-19 remains a global leading cause of death, with a still-rising incidence.
The main reason for this situation is that despite the well-known genetic and molecular features of SARS-CoV-2, there are still no therapeutic knockout treatments or vaccines. To date, even though various clinical trials are ongoing, no licensed and effective antiviral vaccine or drug is available against COVID-19. A recent paper reported that understanding the genetic and phenotypic basics of SARS-CoV-2 in pathogenesis is considerably important for drug discovery. The genomes of coronaviruses (CoVs) are single-stranded positive-sense RNA (+ssRNA) with a 5′-cap structure and 3′-poly-A tails. The SARS-CoV-2 genome of approximately 30 kilobase pairs (kb) includes a minimum of six open reading frames (ORFs). The first ORF (ORF1a/b) is approximately two-thirds of the whole genome length and encodes 16 nonstructural proteins. ORFs near the 3′-end of the genome encode four major structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins ( Fig. 1).2
Figure 1.
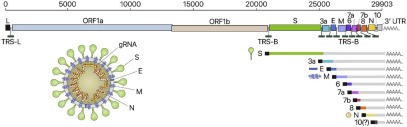
Coronaviruses include the largest genomes among all RNA viruses (26–32 kb).2
Saporin is a toxic RNA N-glycosidase [Enzyme Commission no. (EC): 3.2.2.22] that depurinates eukaryotic ribosomal RNAs (rRNAs), thereby arresting protein synthesis irreversibly during translation. It is one of the type I ribosome-inactivating proteins (RIPs), and it is synthesized from Saponaria officinalis L. (family Caryophyllaceae, common name: soapwort).3 Several isoforms of saporin have been isolated from various parts of the plant, such as saporin-L1 (SAP) and saporin SO6 (Sap-SO6) purified from soapwort leaves and seeds, respectively. The crystal structure of Sap-SO6 [Protein Data Bank (PDB) code: 1QI7] has revealed that this enzyme contains two main domains: an N-terminal domain with a predominantly β-sheet, and a C-terminal domain with a prevalent α-helix structure ( Fig. 2).4 , 5
Figure 2.
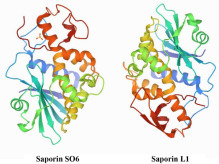
Protein structures of saporin isoforms from the Protein Data Bank (PDB): Saporin O6 (Sap-SO6; PDB code: 1QI7) and saporin-L1 (SAP; PDB code: 3HIS).
RIPs are potent toxins and can cause irreversible cell damage leading to apoptosis and necrosis.5 RIPs were first found in higher plants, and many RIPs have been identified from natural sources (e.g., higher plants, fungi, algae, and bacteria). Examples of plant RIPs include ricin, abrin, gelonin, momardin, and saporin.6, 7, 8
Saporin catalyzes the depurination of invariant adenosine residue (A4324) from the GA4234GA tetraloop motif within the universally conserved α-sarcin–ricin recognition loop of the eukaryotic 28S rRNA molecule.9 Saporin includes a single catalytic chain (domain A) and lacks a lectin-like binding domain; therefore, it is classified as a type I RIP.
Depurination of adenine residue causes a permanent inhibition of the 28S rRNA subunit, arresting the recognition and binding of elongation factor-1 (EF1) and the following creation of the elongation factor-2–guanosine triphosphate (EF2–GTP)–ribosome complex, thereby prohibiting translocation of the transfer RNA (tRNA) from the A site to the P site of the ribosomal complex, thus irreversibly blocking protein synthesis.10
Mode of Action of Saporin
Endo et al. (1987) first discovered the enzymatic activity of RIPs, and all types of RIPs were called rRNA N-glycosidase (EC 3.2.2.22) ( Fig. 3). Studies showed that isoforms of saporin removed more than one adenine residue from rRNA, unlike most RIPs.11 In addition, saporin-R1 depurinated adenine not only from rRNA but also from messenger RNA (mRNA), tRNA, and poly(A) and other cellular DNAs, but not from adenosine triphosphate (ATP) or deoxyadenosine triphosphate (dATP).12 Saporin removes multiple adenines from rRNAs ( Fig. 4), whereas other RIPs exhibit exquisite specificity. This is undoubtedly one of the most important characteristics of saporin proteins. Therefore, it was suggested that saporin proteins would be more appropriately called polynucleotide–adenosine nucleosidases.13
Figure 3.
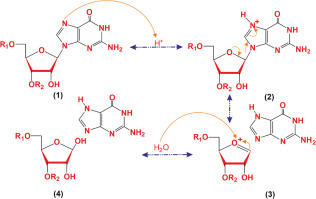
Schematic overview of construction of an abasic site in ribosomal RNA (rRNA).
Figure 4.
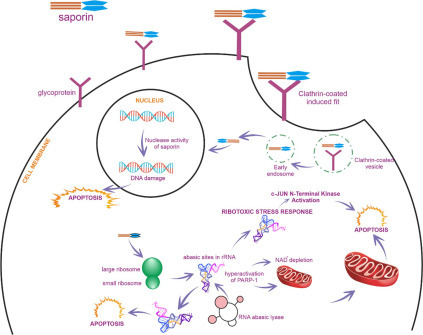
Schematic overview of different mechanisms of action of saporin isoforms.
Basics of Depurination Mechanism
Abasic sites are the lack of a purine or pyrimidine base in a DNA/RNA and arise from spontaneous damage or toxins (e.g., RIPs). The glycosidic bond is mostly unstable and susceptible to hydrolysis by diluted acids and enzymes (e.g., glucosidases). The N-glycosidic bond, particularly that created by nitrogenous bases, is one of the most reactive covalent bonds in DNA and RNA. Saporin-based depurination is a chemical reaction of releasing an adenine residue from nucleic acids and includes the hydrolysis of the β-N-glycosidic bond between the ribose sugar and nitrogenous base (Fig. 3). Hydrolysis reaction commences the protonation of the N3-position of the adenine base that forms a positive charge on the ring backbone and destabilizes the N-glycosidic bond. This leads to β-elimination of the nitrogenous base and creation of a ribofuranosyl oxocarbenium ion intermediate. Ultimately, it builds a ribofuranose hemiacetal via nucleophilic attack of H2O at the position of C1′, and then the hydrolysis process is finalized.14 , 15 Abasic sites as a consequence of base depurination on nucleic acids are vital if unrepaired. Abasic RNA structures were found to be more resistant to β-elimination than abasic DNAs,16suggesting that abasic sites may modify the 3D structure of an RNA molecule and all its interactions in a cell. Abasic RNA molecules can also be substrate for RNA abasic lyase (RAlyase), which is thought to be concerned with apoptosis.17 Saporin induces apoptosis through different pathways, including ribotoxic stress response, cell signal transduction, RAlyase activity, and NAD+ depletion by poly-[adenosine diphosphte (ADP)-ribose] polymerase hyperactivation.
Ribotoxic Stress Response and Cell Signal Transduction
RIPs are a family of highly potent toxins that inhibit protein synthesis by blocking ribosomes. In addition, recent studies suggest that RIPs are also capable of stimulating cell death by apoptosis. Apoptosis is an innate biochemical mechanism by which organisms exterminate undesired cells to maintain healthy cells.18 One of the earliest reports that RIPs are capable of stimulating cell death came from Griffiths et al. (1987).19 They discovered that intramuscular injection of abrin and ricin into rats resulted in the formation of apoptotic bodies and cell deaths.
Researchers discovered that the depurination of rRNA by RIP toxins triggered a ribotoxic stress response that activates c-Jun N-terminal kinase (JNK) (apoptosis) and can lead to cell death.20 Not all forms of RIPs induce a kinase cascade; however, α-sarcin and ricin do, whereas emetine derivatives do not.
Mitogen-activated protein kinases (MAPKs) are a special group of protein kinases that regulate many cellular events, including transcription, translation, proliferation, differentiation, and apoptosis.21 In humans, members of the MAPK superfamily can be divided into six groups: the extracellular signal-regulated protein kinases (ERK1, 2), JNKs (JNK1, 2, 3), p38s (α, β, γ, and δ), ERK5s, ERK3s, and ERK7s. Each member of the MAPKs can be stimulated by different signal transduction cascades concerning environmental stimulants such as ultraviolet radiation, inflammatory signals, and reactive oxygen species.
A recent study demonstrated that destruction to the 3′ end of large rRNA mediates a stress signal that is transduced to a JNK through keeping the ribosome in the pre-translocational state.22 For example, trichosanthin was reported to inhibit HIV replication in T-lymphoblastoid cells and also decrease HIV p24 amounts in HIV-infected macrophages.23 A recent paper discovered that CEP-11004, an effective inhibitor of JNKs, could restrain the antiviral activity of trichosanthin in C8166 cells. The inhibitor alone had no effect on HIV replication, however, and trichosanthin alone significantly inhibited the replication. These results revealed that the anti-HIV action of trichosanthin might be related to MAPK signal transduction downstream from the point of CEP inhibition.24 In contrast, the antiviral activities of RIPs without specific depurination in rRNA also have been reported. Specifically, trichosanthin was shown to bind to chemokine receptors CCR5, CXCR4, CCR1, CCR2B, CCR3, and CCR4 on the surface of HEK293 cells. Thus, these data showed that the RIP enzyme synergizes the chemokines to trigger chemotaxis and G protein action without specific depurination.25
Saporin has been shown to stimulate cell death via apoptosis in a variety of different cell lines, including human peripheral blood B lymphocytes and neutrophils, the Daudi B-cell line, and hematopoietic cell lines HL60 and TF1.12 Similarly, saporin has been linked to monoclonal antibodies against the CD30 antigen of human lymphocytes to form targeted toxins, all of which induced apoptosis in the CD30+ L540 cell lines.26 As highlighted in Ref. 27, the cytotoxicity of saporin-6 is administered by its RNA N-glycosidase and apoptotic properties and the residues tyrosine-16 and tryptophan-208, respectively.
Apoptosis Induction via NAD+ Depletion by PARP [Poly-(ADP-ribose) Polymerase] Hyperactivation
Various studies on RIP-induced apoptosis include alternative pathways of apoptosis. Studies showed that hyperactivation of PARP1 enzymes can result in depletion of NAD+ levels and then can induce mitochondrial damage and apoptosis.28
A recent study found that saporin-L2 and saporin-S6 have transforming activity that involves auto-ADP-ribosylation of the PARP enzyme. Moreover, they also revealed that saporins directly depurinate automodified PARP, removing adenine residue from the ADP-ribosyl group.29 , 30 These results showed that saporin-L2 and saporin-S6 induced apoptosis via NAD+ depletion by PARP hyperactivation, an alternative pathway of apoptosis.
Apoptosis Induction via Nuclease Activity
Another pathway of apoptosis stimulated by saporin is nuclease enzyme activity that leads to DNA damage and apoptosis. Saporin-6 has been shown to have deoxyribonuclease (DNase) activity by which it causes DNA damage and, indirectly, apoptosis.31 In this study, saporin-6 possesses two catalytic activities: RNA N-glycosidase and genomic DNA fragmentation activity. These results suggest that saporin is also active on DNA and induces apoptosis.
Apoptosis Induction via RNA Abasic Lyase (RAlyase) Activity
Abasic RNA molecules can also be substrate for RNA abasic lyase (RAlyase), which is thought to be concerned with apoptosis. RAlyase catalyzes a β-elimination reaction, producing a 5′-phosphate end of the 3′ fragment (α-fragment) of the 28S rRNA and α-hydroxy-α, an β-unsaturated aldehyde end of the 5′ fragment. It is likely that RAlyase constitutes a part of a molecular system that leads the apoptosis.32 Saporin catalyzes the depurination of adenine residue (A4324) from the GA4234GA tetraloop motif within the universally conserved α-sarcin–ricin recognition loop of the eukaryotic 28S rRNA molecule, indicating that saporin may indirectly induce apoptosis by RNA abasic lyase enzyme activity.9
Discussion
COVID-19 remains a global leading cause of death, with a still-rising incidence. This is basically caused by the fact that despite the well-known structural properties of SARS-CoV-2, there are still no therapeutic knockout treatments or vaccines. Scientists are racing, however, to discover the best drugs or vaccines to treat COVID-19 disease.
Antiviral activities of RIPs against many animal, human, and plant viruses are well-documented phenomena. Specifically, several RIPs were tested clinically for therapeutic use; for example, trichosanthin from the Chinese medicinal herb Trichosanthes kirilowii has been applied in clinical treatment of AIDS.25
Saporin, a type I RIP from the soapwort plant, is a potent protein synthesis inhibitor. Catalytically, saporin is a specific N-glycosidase and depurinates a specific adenine residue from a universally conserved loop of the major rRNA of eukaryotic cells.3
Ribotoxic stress response upregulates the transcription and expression of many proteins through its activation of MAPK in cells, suggesting that saporin may be a regulatory agent in the expression of target viral proteins. Saporin may also inhibit specifically the development of viruses in one or more points in their lifecycle, including infection, integration, replication, and translation. One of the most important features that makes saporin a powerful therapeutic agent against SARS-CoV-2 infection is the unusual stability of the protein and its ability to keep its enzymatic activity. The roles of putative active site residues Tyr72, Tyr120, Glu176, Arg179, and Trp208, and two invariant residues Tyr16 and Arg24, are proposed to be important for the structural stability of saporin.31
Scientists revealed that saporin has high coil content (>50%), and therefore it is extremely resistant to high temperature, to denaturation by urea or guanidine, to attack by proteolytic enzymes,33 and to chemical modifications such as derivatization and conjugation procedures.34
Due to saporin lacking the cell-binding lectin domain, saporin is much less toxic than most type II RIPs. Throughout the past decade, our research group has been studying quillaic acid or gypsogenin-based Gypsophila saponins showing their toxicity-enhancing effects on Sap-SO6 without causing toxicity by themselves (up to 100,000-fold).35, 36, 37A recent paper reported that soapwort saponins trigger clathrin-mediated endocytosis of both saporin and saporin-associated immunotoxins.38 Its extraordinary stability and its ability to get into cells by clathrin-mediated endocytosis of saporin or saporin-associated immunotoxins are extremely important for drug delivery.
Saporin’s high enzymatic activity, high stability, and resistance to conjugation procedures make it a well-suited tool for immunotherapy approaches. Previous clinical study on saporin-based immunotoxins has shown that several critical issues must be taken into deeper consideration to fully exploit their therapeutic potential.5
Saporin-based immunotoxins might be used in the treatment of infectious diseases within the scope of targeted therapy. Here, we focus on how saporin-based targeted toxins may be efficacious therapeutic agents for SARS-CoV-2 infection. Our discussed points suggest that saporin might be a strategic molecule for therapeutic knockout treatments and a powerful candidate of novel drugs for the struggle against SARS-CoV-2 infection.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Supplemental Material
Supplemental Material
References
- 1.World Health Organization (WHO). WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020.
- 2.Kim D., Lee J.Y., Yang Y.S., et al. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stirpe F., Gasperi-Campani A., Barbieri L., et al. Ribosome-Inactivating Proteins from the Seeds of Saponaria officinalis L. (Soapwort), of Agrostemma githago L. (Corn Cockle) and of Asparagus officinalis L. (Asparagus), and from the Latex of Hura crepitans L. (Sandbox Tree) Biochem. J. 1983;216:617–625. doi: 10.1042/bj2160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Protein Data Bank. https://www.rcsb.org.
- 5.Giansanti F., Flavell D.J., Angelucci F., et al. Strategies to Improve the Clinical Utility of Saporin-Based Targeted Toxins. Toxins (Basel) 2018;10 doi: 10.3390/toxins10020082. PMID: 29438358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narayanan S., Surolia A., Karande A.A. Ribosome-Inactivating Protein and Apoptosis: Abrin Causes Cell Death via Mitochondrial Pathway in Jurkat Cells. Biochem J. 2004;377:233–240. doi: 10.1042/BJ20030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H.X., Ng T.B. Flammulin: A Novel Ribosome-Inactivating Protein from Fruiting Bodies of the Winter Mushroom. Flammulina velutipes. Biochem. Cell Biol. 2000;78:699–702. doi: 10.1139/o00-087. [DOI] [PubMed] [Google Scholar]
- 8.Tran M., Henry R.E., Siefker D., et al. Production of Anti-Cancer Immunotoxins in Algae: Ribosome Inactivating Proteins as Fusion Partners. Biotechnol. Bioeng. 2013;110:2826–2835. doi: 10.1002/bit.24966. [DOI] [PubMed] [Google Scholar]
- 9.Stirpe F. Ribosome-Inactivating Proteins. Toxicon. 2004;44:371–383. doi: 10.1016/j.toxicon.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Endo Y., Tsurugi K. The RNA N-Glycosidase Activity of Ricin A-Chain: The Characteristics of the Enzymatic Activity of Ricin A-Chain with Ribosomes and with rRNA. J. Biol. Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 11.Barbieri L., Ferreras J.M., Barraco A., et al. Some Ribosome-Inactivating Proteins Depurinate Ribosomal RNA at Multiple Sites. Biochem. J. 1992;286:1–4. doi: 10.1042/bj2860001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergamaschi G., Perfetti V., Tonon L., et al. Saporin, a Ribosome-Inactivating Protein Used to Prepare Immunotoxins Induces Cell Death via Apoptosis. Br. J. Haematol. 1996;93:789–794. doi: 10.1046/j.1365-2141.1996.d01-1730.x. [DOI] [PubMed] [Google Scholar]
- 13.Barbieri L., Gorini P., Valbonesi P., et al. Unexpected Activity of Saporins. Nature. 1994;372:6507–6524. doi: 10.1038/372624a0. [DOI] [PubMed] [Google Scholar]
- 14.An R., Jia Y., Wan B., et al. Non-Enzymatic Depurination of Nucleic Acids: Factors and Mechanisms. PLoS One. 2014;9:e115950. doi: 10.1371/journal.pone.0115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro R., Danzig M. Acidic Hydrolysis of Deoxycytidine and Deoxyuridine Derivatives: The General Mechanism of Deoxyribonucleoside Hydrolysis. Biochemistry. 1972;11:23–29. doi: 10.1021/bi00751a005. [DOI] [PubMed] [Google Scholar]
- 16.Küpfer P.A., Crey-Desbiolles C., Leumann C.J. Trans-Lesion Synthesis and RNaseH Activity by Reverse Transcriptases on a True Abasic RNA Template. Nucleic Acids Res. 2017;35:6846–6853. doi: 10.1093/nar/gkm767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawasaki T., Nishihara M., Endo Y. RIP and RALyase Cleave the Sarcin/Ricin Domain, a Critical Domain for Ribosome Function, during Senescence of Wheat Coleoptiles. Biochem. Biophys. Res. Commun. 2008;370:561–565. doi: 10.1016/j.bbrc.2008.03.124. [DOI] [PubMed] [Google Scholar]
- 18.Adams J.M. Ways of Dying: Multiple Pathways to Apoptosis. Genes Dev. 2003;17:2481–2495. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths D., Leek M.D., Gee D.J. The Toxic Plant Proteins Ricin and Abrin Induce Apoptotic Changes in Mammalian Lymphoid Tissues and Intestine. J. Pathol. 1987;151:221–229. doi: 10.1002/path.1711510310. [DOI] [PubMed] [Google Scholar]
- 20.Jandhyala D.M., Thorpe C.M., Magun B. Ricin and Shiga Toxins: Effects on Host Cell Signal Transduction. Curr. Top. Microbiol. Immunol. 2012;357:41–65. doi: 10.1007/82_2011_181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teramoto H., Gutkind J.S. In: Encyclopedia of Biological Chemistry. 2nd ed. Lennarz W.J., Lane M.D., editors. Academic Press; London: 2013. Mitogen-Activated Proteins Kinase Family. [Google Scholar]
- 22.Iordanov M.S., Pribnow D., Magun J.L., et al. Ribotoxic Stress Response: Activation of the Stress-Activated Protein Kinase JNK1 by Inhibitors of the Peptidyl Transferase Reaction and by Sequence-Specific RNA Damage to the Sarcin/Ricin Loop in the 28S rRNA. Mol. Cell. Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byers V.S., Baldwin P.W. Trichosanthin Treatment of HIV Disease. AIDS. 1991;5:1150–1151. [PubMed] [Google Scholar]
- 24.Ouyang D.Y., Chan H., Wang Y.Y., et al. An Inhibitor of c-Jun N-Terminal Kinases (CEP-11004) Counteracts the Anti-HIV-1 Action of Trichosanthin. Biochem. Biophys. Res. Commun. 2006;339:25–29. doi: 10.1016/j.bbrc.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J., Ben L.H., Wu Y.L. Anti-HIV Agent Trichosanthin Enhances the Capabilities of Chemokines to Stimulate Chemotaxis and G Protein Activation, and This Is Mediated through Interaction of Trichosanthin and Chemokine Receptors. J. Exp. Med. 1999;190:101–111. doi: 10.1084/jem.190.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolognesi A., Tazzari P.L., Tassi C. A Comparison of Anti-Lymphocyte Immunotoxins Containing Different Ribosome-Inactivating Proteins and Antibodies. Clin. Exp. Immunol. 1992;89:341–346. doi: 10.1111/j.1365-2249.1992.tb06959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagga S., Divya S., Batra J.K. The Cytotoxic Activity of Ribosome-Inactivating Protein Saporin-6 Is Attributed to Its rRNA N-Glycosidase and Internucleosomal DNA Fragmentation Activities. J. Biol. Chem. 2003;278:4813–4820. doi: 10.1074/jbc.M207389200. [DOI] [PubMed] [Google Scholar]
- 28.Chiarugi A., Moskowitz M.A. Cell Biology. PARP-1 a Perpetrator of Apoptotic Cell Death? Science. 2002;297:200–201. doi: 10.1126/science.1074592. [DOI] [PubMed] [Google Scholar]
- 29.Barbieri L., Brigotti M., Perocco P., et al. Ribosome-Inactivating Proteins Depurinate Poly-(ADP-Ribosyl)ated Poly-(ADP-Ribose) Polymerase and Have Transforming Activity for 3T3 Fibroblasts. FEBS Lett. 2003;538:178–182. doi: 10.1016/s0014-5793(03)00176-5. [DOI] [PubMed] [Google Scholar]
- 30.Yu S.W., Wang H.W., Poitras M.F., et al. Mediation of Poly-(ADP-Ribose) Polymerase-1-Dependent Cell Death by Apoptosis-Inducing Factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 31.Roncuzzi L., Gasperi-Campani A. DNA-Nuclease Activity of the Single-Chain Ribosome-Inactivating Proteins Dianthin 30, Saporin 6 and Gelonin. FEBS Lett. 1996;392:6–20. doi: 10.1016/0014-5793(96)00776-4. [DOI] [PubMed] [Google Scholar]
- 32.Santanché S., Bellelli A., Brunori M. The Unusual Stability of Saporin, a Candidate for the Synthesis of Immunotoxins. Biochem. Biophys. Res. Commun. 1997;234:129–132. doi: 10.1006/bbrc.1997.6597. [DOI] [PubMed] [Google Scholar]
- 33.Endo Y. In: Ribosome Inactivating Proteins. Stirpe F., Lappi D.A., editors. Wiley-Blackwell; Hoboken (NJ): 2014. Enzymology of the Ribosome-Inactivating Proteins. [Google Scholar]
- 34.Bolognesi A., Tazzari P.L., Olivieri F. Induction of Apoptosis by Ribosome-Inactivating Proteins and Related Immunotoxins. Int. J. Cancer. 1996;68:349–355. doi: 10.1002/(SICI)1097-0215(19961104)68:3<349::AID-IJC13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Arslan I., Celik A., Melzig M.F. Nebulosides A-B, Novel Triterpene Saponins from Underground Parts of Gypsophila arrostii Guss. var. nebulosi. Bioorgan. Med. Chem. 2013;21:1279–1283. doi: 10.1016/j.bmc.2012.12.036. [DOI] [PubMed] [Google Scholar]
- 36.Arslan I. Simenoside A, a New Triterpenoid Saponin from Gypsophila simonii Hub.-Mor. Chem. Biodiv. 2014;11:445–450. doi: 10.1002/cbdv.201300118. [DOI] [PubMed] [Google Scholar]
- 37.Arslan I., Ili P. Genotoxicological Assessment of Nebuloside-A a Triterpenoid Saponin Compound on Whole Blood DNA. Int. J. Food Prop. 2015;18:2374–2379. [Google Scholar]
- 38.Weng A., Bachran C., Fuchs H., et al. Soapwort Saponins Trigger Clathrin-Mediated Endocytosis of Saporin, a Type I Ribosome-Inactivating Protein. Chem. Biol. Interact. 2008;176:204–211. doi: 10.1016/j.cbi.2008.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material


