Abstract
One hundred thirty-two glycopeptide-resistant Enterococcus faecium (GREF) isolates from different hospitals and pig and poultry farms in Belgium were compared on the basis of (i) their antibiotic susceptibilities, (ii) their SmaI pulsed-field gel electrophoresis (PFGE) patterns, and (iii) the organization of their Tn1546 or related elements in order to detect possible phenotypic and genotypic relationships among both groups of isolates. Human and animal vanA-positive GREF isolates were found to have similar susceptibility patterns; they remained susceptible to gentamicin and were, in general, susceptible to ampicillin. PFGE demonstrated a very high degree of genomic heterogeneity in both groups of isolates. However, indistinguishable isolates were found within different farms or hospitals, and in two instances, epidemiologically unrelated pig and human isolates showed indistinguishable PFGE patterns. In total, eight different transposon types were identified, and all were related to the prototype transposon Tn1546. The two predominant types, Tn1546 and type 2 transposons, which differed at three band positions, were present in both human and animal isolates. Type 2 transposons were significantly associated with pig isolates. The other types were seldom detected. These data suggest a possible exchange of glycopeptide resistance markers between animals and humans.
Enterococci are part of the normal flora of humans and animals (26), and except for endocarditis, their pathogenic role in immunocompetent persons is limited. Recently, enterococci have caused great concern because of their acquisition of resistance to many antimicrobial agents, including glycopeptides.
Glycopeptide-resistant enterococci (GRE) were first noted in Europe in 1986 (22, 38) and were subsequently noted in the United States in 1987 (34). These GRE have rapidly emerged as major nosocomial pathogens throughout the United States (23); however, until now they have not been detected in the community or in livestock (9, 35, 37, 43). In Europe, GRE are widely distributed in the community (12), in farm and pet animals (1, 11), and on raw meat and meat products (5, 6, 41), although their prevalence in hospitals remains very low (39).
In Europe, avoparcin, which is structurally related to vancomycin and teicoplanin, was used as a growth promoter in food animals until it was banned in April 1997. The question of whether the use of avoparcin selected for GRE in animals and whether GRE subsequently spread to humans remains (1, 24). Molecular typing of both human and animal vanA GRE isolates revealed a very high degree of polyclonal variability among isolates in Europe (6, 12, 20, 21), which does not support clonal transmission and expansion. The spread of vanA-encoded resistance may be related to the mobility of Tn1546 or related elements (7, 41). Typing of these highly transferable genetic determinants may provide additional insights into the spread of GRE among humans and animals (17, 44, 45).
To detect phenotypic and genotypic relationships among Belgian human and animal glycopeptide-resistant Enterococcus faecium (GREF) isolates, both groups of isolates were compared on the basis of their antibiotic susceptibilities, their SmaI pulsed-field gel electrophoresis (PFGE) patterns, and the presence and organization of Tn1546 or related elements, as analyzed by restriction fragment length polymorphism (RFLP) analysis of almost the complete transposon.
MATERIALS AND METHODS
Strains studied.
A total of 132 vanA-positive GREF isolates comprising 59 animal isolates and 73 human isolates were investigated (Table 1). All strains were collected during the course of several surveillance studies on vancomycin resistance from 1995 to 1997; they originated from 18 geographically distinct hospitals, from 12 pig and 4 poultry farms distributed over the provinces of East and West Flanders, and from other animals, as indicated in Table 1. No vanB GRE were detected during the course of these studies. All animal strains were isolated from feces, while 16 of the 73 human isolates were from clinical samples.
TABLE 1.
Sources and transposon types of the 132 GREF isolates studied
| Source and sample | Transposon type | No. of strains | Strain no.a |
|---|---|---|---|
| Human | |||
| Feces | 1 | 13 | AUZ012-1, AUZ013-2, BG010-1, BRU084-1, BRU084-2, CR030-4, BSL001-1, LG054-2, LG054-3, GT038-2, SN028-1, ST014-1, VV085-1 |
| Feces | 2 | 42 | 005G, 035G, 156G, 160G, 192G, 201G, 272G1, 382G, 545G, AUZ017-2, AL012-2, AL012-3, AL113-2, AL113-3, BG020-2, BG024-3, BSL006-1, CR030-5, D11, D19, DM014-2, DM022-1, DM037-1, DM040-1, GT010-2, GT010-1, GT038-1, GT039-1, GT039-2, GT051-1, GT081-1, GT085-1, GT085-2, LG034-1, LG044-1, LG071-1, MA039-1, MC023-1, RO038-3, RO055-1, RS046-1, VUB045-1 |
| Feces | 4 | 1 | 091G1 |
| Feces | NTb | 1 | 224G |
| Urine | 7 | 1 | 960047 |
| Urine | NT | 1 | 960005 |
| Surface | 2 | 1 | 960034 |
| Surface | NT | 1 | 960007 |
| Wound | 1 | 1 | 960020 |
| Wound | 7 | 1 | 960049 |
| Blood | 3 | 1 | 960050 |
| Blood | 8 | 1 | 960018 |
| Pus | 3 | 1 | 960051 |
| Pus | 6 | 1 | 960036 |
| —c | 2 | 2 | LAB 1267, LAB 1268 |
| — | 3 | 1 | LAB 1264 |
| — | NT | 3 | LAB 1265, LAB 1266, LAB 1270 |
| Animal | |||
| Pig | 1 | 2 | LMG16267, ZMA5 |
| Pig | 2 | 36 | LMG16297, LMG16196, LMG16268, LMG17124, LMG17180, LMG17181, LMG17182, LMG17183, LMG17184, LMG17185, LMG17188, LMG17200, LMG17203, LMG17204, TZM13, TZM18, TZM19, TZM20, VM115, VM122, VM177, VM178, VM23, VM26, ZH48, ZM42, ZM43, ZM47, ZM80, ZM81, ZMA12, ZMA26, ZMA44, ZMA45, ZMA49, ZMA50 |
| Pig | NT | 3 | LMG16265, LMG16266, LMG17178 |
| Chicken | 1 | 2 | LMG16474t1, LMG16475t1 |
| Chicken | 2 | 1 | LMG16194t1 |
| Chicken | 5 | 1 | LMG16271 |
| Chicken | 1 | 1 | LMG16192 |
| Chicken | NT | 3 | LMG16174, LMG16195, LMG16173 |
| Dog | 1 | 1 | LMG16171 |
| Dog | 2 | 1 | LMG16270 |
| Cat | 2 | 1 | LMG16269 |
| Duck | 1 | 1 | LMG16170 |
| Horse | 2 | 4 | LMG16478, LMG16164, LMG16167, LMG16168 |
| Horse | NT | 2 | LMG16165, LMG16166 |
LMG, Culture Collection Laboratorium voor Microbiologie Universiteit Gent, Ghent, Belgium; LAB, Lactic Acid Bacteria Culture Collection, Laboratorium voor Microbiologie Universiteit Gent, Ghent, Belgium; AUZ, Universitair Ziekenhuis Antwerpen, Culture Collection of the University Hospital Antwerp, Antwerp, Belgium. The strain numbers starting with the acronyms refer to the cities where the strains were isolated: AL, Aalst; BG, Brugge; BSL, Brugge; BRU, Brussels; CR, Chalerloi; D, Aalst; DM, Dendermonde; GT, Ghent; LG, Luik; MA, Malle; MC, Mouscron; RO, Ronse; RS, Roeselaere; SN, Sint-Niklaas; ST, Sint-Truiden; VUB, Brussels; VV, Verviers.
NT, not typeable.
—, exact source unknown.
Antimicrobial susceptibility testing.
MICs were determined by the agar dilution method by using the criteria recommended by the National Committee for Clinical Laboratory Standards (27). The following drugs were tested: vancomycin (Eli Lilly Benelux, Brussels, Belgium), teicoplanin (Hoechst Marion Roussel, Romainville, France), streptomycin (Sigma, Steinheim, Germany), gentamicin (Sigma), ampicillin (Sigma), and ciprofloxacin (Bayer AG, Wuppertal, Germany). Enterococcus faecalis ATCC 29212 and two laboratory strains were used for quality control.
High-level resistance to gentamicin and streptomycin was determined on Mueller-Hinton agar (Becton Dickinson, Cockeysville, Md.) supplemented with streptomycin (2,000 μg/ml) and gentamicin (500 μg/ml), respectively, as described by Sahm and Torres (33).
PFGE.
Clonal relatedness was estimated by PFGE with the restriction enzyme SmaI and GelCompar, version, 4.0 image analysis software (Applied Maths, Kortrijk, Belgium), as described before (10).
RFLP analysis of Tn1546 and related elements.
The structures of the putative Tn1546-related elements were analyzed by amplification of almost the complete transposon with the Long and Accurate-PCR (LA-PCR) technology (4, 8), followed by RFLP analysis of the amplicon.
DNA was prepared by the rapid procedure described by Pitcher et al. (31). On the basis of the sequences of the orf1 and vanY genes of Tn1546 (2, 3), the 25-bp forward primer ORF1-F1 (5′-AAT CTT CAT TAA AGC TAC CTG TCC G-3′), located at position 191 of Tn1546, and the 25-bp reverse primer VanY-R1 (5′-TAT CTC ATA ACG AAG ATT AGT CGG C-3′), located at position 9867 of transposon Tn1546 (GenBank accession no. M97297), were selected.
Amplification was performed in a Perkin-Elmer GeneAmp PCR system 9600 with MicroAmp tubes. The 50-μl PCR mixtures contained 5 μl of 10× ExTaq buffer, 4 μl of a deoxynucleoside triphosphate mixture (each deoxynucleoside triphosphates at a concentration of 2.5 mM), 1 μl of each primer (20 pmol/μl), 0.25 μl of ExTaq (5 U/μl; Takara, Gennevilliers, France), 500 ng of extracted DNA, and sterile distilled water up to 50 μl. The amplification process consisted of 35 cycles of denaturation at 98°C for 20 s, annealing at 68°C for 1 min, and elongation at 72°C for 15 min.
For RFLP analysis the following enzymes were tested: HinfI, DdeI, EcoRI, EcoRV, HaeIII, HindIII, BfaI, BamHI, DraI, NheI, XbaI, BglI, SpeI, SmaI, and SfiI. A total of 5 μl of the amplification product was digested for 2 h at 37°C with 10 U of restriction enzyme and the buffer prescribed by the manufacturer.
The restriction product was mixed with 7 μl of loading buffer (50% glycerol, 0.8 mg of bromophenol blue/ml) and was run in a 1.5% pronarose D1 gel (Sphaero Q, Burgos, Spain) for 2 h at 150 V in 0.5× TBE (Tris-borate-EDTA, which contained 0.05 mg of ethidium bromide/liter). The RFLP patterns that were obtained were digitized with the Gel Doc 1000 documentation system (Bio-Rad Laboratories). Conversion, normalization, and further analysis of the patterns were performed with the GelCompar software, version 4.0 (Applied Maths), as described previously (32, 42).
Sequencing.
The nucleotide sequences of parts of transposon Tn1546 or related elements were determined as described by Willems et al. (44) with the ABI PRISM Dye cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.) on an ABI PRISM 377 DNA Sequencer (Perkin-Elmer). GeneCompar software (Applied Maths) was used for sequence analysis.
RESULTS
Susceptibility testing.
The MICs at which 50% of isolates are inhibited (MIC50s), MIC90s, and the range of the MICs of six antibiotics for the human and animal GREF isolates are listed in Table 2. All strains were resistant to both vancomycin and teicoplanin. No high-level resistance (MICs, ≥500 μg/ml) to gentamicin was detected, while high-level resistance to streptomycin was detected in 69.9% (n = 51) and 47.5% (n = 28) of the human and animal isolates, respectively. Ampicillin resistance (MICs, ≥16 μg/ml) was found in 24.7% (n = 18) and 28.8% (n = 17) of the human and animal isolates, respectively. Finally, resistance to ciprofloxacin (MICs, ≥4 μg/ml) was detected in 47.9% of human isolates and 37.3% of animal isolates.
TABLE 2.
Resistance patterns of 73 human and 59 animal vanA-positive enterococcal isolates
| Isolate source and drug | MIC (μg/ml)
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| Human | |||
| Vancomycin | 256 | 256 | 64–512 |
| Teicoplanin | 32 | 64 | 16–>64 |
| Gentamicin | 8 | 16 | 4–>64 |
| Streptomycin | 512 | >512 | 64–>512 |
| Ampicillin | 8 | 16 | 0.5–>64 |
| Ciprofloxacin | 2 | 4 | 0.25–>64 |
| Animal | |||
| Vancomycin | 256 | 256 | 64–512 |
| Teicoplanin | 32 | 64 | 16–>64 |
| Gentamicin | 8 | 16 | 4–64 |
| Streptomycin | >512 | >512 | 64–>512 |
| Ampicillin | 8 | 64 | 1–>64 |
| Ciprofloxacin | 2 | 8 | 0.5–32 |
Clonal distribution.
All strains were typeable by PFGE with the restriction enzyme SmaI, and the intra- and intergel reproducibilities were >95%, as determined with the Pearson correlation coefficient (data not shown).
Visual and computerized analysis of all SmaI patterns obtained revealed 81 clones when three or fewer band differences was used as the criterion to define a different clone (36). Besides indistinguishable isolates that originated from the same farms or the same hospitals, four clones constituted indistinguishable isolates that originated from two different farms each, and nine other clones constituted indistinguishable isolates that originated from up to four different hospitals. Human isolate GT085-1 was indistinguishable from pig isolates ZH48, ZM43, ZMA45, ZMA49, and ZMA50, which were isolated from the same farm; human isolate LG044-1 and pig isolate TZM20 also showed indistinguishable PFGE patterns. There was no geographical clustering of these human and animal strains, and we found no relationships between their hosts (data not shown).
RFLP analysis of the transposon.
To determine the reproducibility and discriminatory power of each enzyme, we digested the amplification product of the transposon from a selection of 25 strains with 15 restriction enzymes. The restriction enzyme DdeI generated the most discriminatory profiles (data not shown) and was consequently used to study all strains. Combination of the DdeI restriction profile with the restriction profile for the same amplicon but obtained with another restriction enzyme did not increase the discriminatory power of the method (data not shown).
No amplification product could be generated for 10.6% of the GREF isolates studied (Table 3), although in nine isolates the primer binding sites were shown to be present. Indeed, we could amplify the orf1 and vanY genes in which the forward and reverse primers are located, using the primers described by Miele et al. (25) and the PCR conditions described above. In addition, by combining one of the orf1- or vanY-specific primers with one of the primers used to amplify the complete transposon, respectively, we were able to generate an amplification product, which confirmed the presence of both primer binding sites.
TABLE 3.
Distribution of different DdeI RFLP transposon types among human and animal isolates
| Transposon type | % (no.) of isolates
|
|||
|---|---|---|---|---|
| Human | Pig | Other animal species | Total | |
| 1 | 19.2 (14) | 4.9 (2) | 27.8 (5) | 15.9 (21) |
| 2 | 61.6 (45) | 87.8 (36) | 38.9 (7) | 66.6 (88) |
| 3 | 4.1 (3) | 0.0 (0) | 0.0 (0) | 2.3 (3) |
| 4 | 1.4 (1) | 0.0 (0) | 0.0 (0) | 0.8 (1) |
| 5 | 0.0 (0) | 0.0 (0) | 5.6 (1) | 0.8 (1) |
| 6 | 1.4 (1) | 0.0 (0) | 0.0 (0) | 0.8 (1) |
| 7 | 2.7 (2) | 0.0 (0) | 0.0 (0) | 1.5 (2) |
| 8 | 1.4 (1) | 0.0 (0) | 0.0 (0) | 0.8 (1) |
| NTa | 8.2 (6) | 7.3 (3) | 27.8 (5) | 10.6 (14) |
| Total | 73 | 41 | 18 | 132 |
NT, not typeable.
A total of eight different types of transposons were identified (Fig. 1). Type 1 transposons were found in 15.9% of the strains investigated, while 66.6% of the strains harbored type 2 transposons (Table 3). Those two frequently recovered types of transposons were found among both human and animal isolates and were found in isolates that originated from the same farm or hospital. Type 2 transposons were identified in 36 of 41 (87.8%) GREF isolates from pigs but in only 7 of 18 (38.9%) GREF isolates from other animal sources (P < 0.0001) (Table 3). Transposons of the remaining types were less frequently found in human and/or animal isolates. Transposons of types 3, 4, 6, 7, and 8 were found only in human isolates, while transposons of type 5 were detected only in animal isolates, but the number of strains with transposons of type 5 was small. The untypeable transposons were found in both human and animal isolates, and no association with a farm or hospital was found.
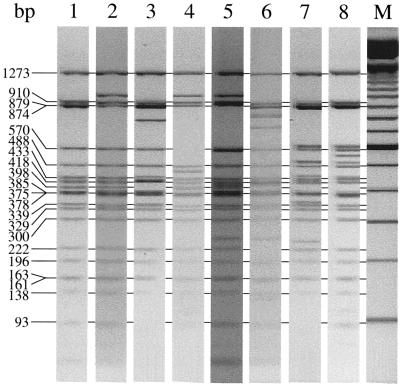
FIG. 1.
Different transposon types (types 1 to 8 in lanes 1 to 8, respectively) detected among human and animal enterococcal isolates. The type 1 transposon is identical to Tn1546; the sizes of the DdeI restriction fragments are indicated. Lane M, 100-bp DNA ladder.
Except for isolates GT085-1, GT081-1, GT038-2, and DM040-1, isolates characterized by indistinguishable PFGE patterns always had the same transposon patterns. These isolates were characterized by indistinguishable PFGE patterns, but isolate GT038-2 harbored the type 1 transposon, while isolates GT085-1, GT081-1, and DM040-1 harbored the type 2 transposon.
Characterization of different transposon types.
The transposons from strains 960020, 960034, 960050, 091G1, LMG16271, 960036, 960049, and 960018, representing transposon types 1 to 8, respectively, were selected for further characterization by sequence analysis of parts of the transposon.
Transposon type 1 showed complete sequence homology to the sequence of transposon Tn1546 (GenBank accession no. M97297) (3), which consequently allowed allocation of each DdeI restriction fragment to a particular gene located on Tn1546 and determination of the exact size of each fragment (Fig. 1, lane 1). The second transposon type differed from type 1 by a single nucleotide variation in the vanX gene (G to T) at position 8234, resulting in the loss of a DdeI restriction site but with the creation of a new fragment of 964 bp from 90- and 874-bp fragments (Fig. 1, lane 2) (18, 44, 45). Type 3 was characterized by a deletion of 27 nucleotides (positions 8804 to 8831) in the vanX-vanY intergenic region and the insertion of IS1216V at that position. IS1216V contains one DdeI restriction site at position 150 of the insertion sequence (IS) element (GenBank accession no. L38972). This results in the disappearance of the 398-bp fragment and the creation of a 415-bp fragment and a 765-bp fragment (Fig. 1, lane 3). Type 4 has a mutation in the vanH gene (position 6908; C to T) and a mutation in the vanX gene (position 8234; G to T). The first mutation creates an extra DdeI restriction site, resulting in the split of the 570-bp fragment into fragments of 460 and 110 bp. The second mutation results in the loss of a DdeI restriction site, with the creation of a new fragment of 964 bp from 90- and 874-bp fragments (Fig. 1, lane 4). Transposon type 5 has an IS16-like element inserted at position 3108 in the orf1-orf2 intergenic region (the complete IS element was not sequenced). The 910-bp fragment is replaced by multiple new DNA fragments (Fig. 1, lane 5). Type 6 has IS1216V at position 8839 (vanX-vanY intergenic region) and a yet unknown IS256-like element at position 3932 in the orf2-vanR intergenic region (the complete IS element was not sequenced). The first IS element results in the disappearance of the 398-bp band and the creation of 800- and 407-bp bands. The second IS element results in the disappearance of the 222-bp band and the creation of multiple new DNA fragments (Fig. 1, lane 6). Type 7 has a 392-bp deletion from positions 9049 to 9441, which deletes the 5′ end of the vanY gene. At the position of the deletion a yet unknown IS element was introduced (the complete IS element was not sequenced) (Fig. 1, lane 7). This results in the loss of 398-, 375-, and 35-bp bands and the creation of multiple extra bands. Type 8 probably has the same IS element as type 7 (the complete IS sequence is not known) at the same position (position 9049), however, without the deletion at the 5′ end of the vanY gene. This also results in the loss of the 398-bp band but the creation of extra bands different from those in type 7 (Fig. 1, lane 8).
DISCUSSION
Resistance to glycopeptides among the GREF isolates included in our study was always due to the presence of the vanA gene. We never isolated vanB enterococci from human or animal sources. In Europe, vanB enterococci have only occasionally been isolated from humans (14), and we are not aware of the detection of any vanB Enterococcus species in isolates from animals. This is in contrast to the situation in the United States, where both vanA and vanB genes are present in GRE (9, 29, 30).
The antibiotic susceptibility profiles of the human and the animal GREF isolates tested in this study were comparable. No GREF isolates with high-level gentamicin resistance were detected, and the majority of the strains were susceptible to ampicillin. This is in contrast to the situation in the United States, where GRE are important nosocomial pathogens and are highly resistant to most of the available antibiotics (16).
Although a high degree of clonal variability was detected among the 132 GREF isolates studied, confirming the results of studies by others (5, 12, 20, 21), four clones were found to have spread over two different farms each, and nine other clones had spread over up to four different hospitals each. In addition, two clones comprising indistinguishable human and animal isolates with no apparent relationship between their hosts were found. Thus, humans and animals can carry indistinguishable GRE, corroborating the observations of Jensen et al. (19) and van den Bogaard et al. (40). In addition, isolates that belonged to the same clone, as defined by indistinguishable PFGE patterns, may have spread geographically between different hosts. This might indicate a possible intrahospital or intrafarm spread of GRE or, alternatively, the existence of a common reservoir of isolates with glycopeptide resistance.
On the basis of the results of a study by van den Bogaard et al. (40), in which one strain from a farmer and one strain from his turkey flock appeared to be indistinguishable by PFGE and transposon typing, it was suggested that animal GRE can colonize the human gut. The same conclusions were made by Jensen et al. (19). Clearly, a generalized deduction from these observations is highly hypothetical. In the present study the unidirectional spread of GRE from animals to humans could not be concluded, and also, the study of van den Braak et al. (41) rejected the direct horizontal transmission of GRE from poultry to humans via the food chain. Indeed, indistinguishable GRE may be widely distributed (19), even over different countries (unpublished data), not necessarily indicating epidemiologically related transmission but, rather, indicating the omnipresence of GRE clones. Only prospective study of the presence and colonization of GRE of animal origin in the human gut, after uptake of this isolate by uncolonized human volunteers, may add evidence to the hypothesis that animal GRE colonize humans (15).
In the present study, eight different transposon types were detected. However, in 82.5% of the strains studied, transposons of only two types were recovered, and these differed only in the loss of a DdeI restriction site in the vanX gene (Fig. 1). Type 1, which is identical to the prototype transposon described by Arthur et al. (3), was less frequently present (15.9%) than type 2 (66.6%). The predominance of these two types is in agreement with the study of Willems et al. (44). However, as reported previously (18), we found a statistically significant association of transposon type 2 with pig GREF isolates. Similarly, all 11 pig GREF isolates in the study of Willems et al. (44) and 5 of the 7 pig isolates of GRE (isolates from feces and raw meat) in the study of Woodford et al. (45) also harbored type 2 transposons. This observation merits further study. Transposons of the remaining types were not detected frequently enough to make conclusions as to their host specificities. Several untypeable GRE may represent isolates with additional transposon types, as was shown in the study of Palepou et al. (28), in which the long PCR technique was not able to amplify the DNAs of isolates with particular transposon types, probably because of mutations in the primer binding sites or deletions of parts of the transposon. Other researchers, however (17, 41, 44, 45), found additional transposon types that were not detected in present study. This variability in the transposon types may be due to multiple factors, including the discriminatory power of the typing technique, the geographic differences in transposon distributions among GRE isolates, and the origins of the strains. In addition, comparison of the transposon types described in different studies necessitates a consensus technique and a consensus manner of describing these transposon types. DNA sequence analysis of the variable parts of the transposon with pinpointing of the altered positions in relation to the position in prototype transposon Tn1546 may be a feasible way. However, most important, all researchers (17, 41, 44, 45) found identical transposon types among human, nonhuman, and animal isolates, which may indicate, although it is hypothetical, a possible exchange of glycopeptide resistance markers between isolates from animals and humans or the existence of a common reservoir of glycopeptide resistance.
The highly polyclonal nature of GRE, the infrequent presence of indistinguishable GRE among human and animal isolates, the predominance of a limited number of transposon types in both groups of isolates, and the finding of different transposon types in indistinguishable isolates (45; present study) corroborate a possible epidemic spread of a limited number of transposon types that carry the vanA gene and that are able to transfer resistance to other species and genera in vivo (13). Indeed, GRE that enter the intestinal tracts of humans or animals via the food chain or via direct contamination may transfer their resistance genes during their passage through the gut, even though they do not necessarily colonize the host. The selective pressure by antibiotics used in patients and as growth promoters in animal husbandry may have stimulated the cross contaminations among and between the different hosts.
In conclusion, we found that human and animal vanA-positive GRE were characterized by comparable susceptibility patterns, although they were genetically heterogeneous. In addition, identical types of transposons were found in both groups of isolates. These data suggest that GRE are not confined to a restricted set of perhaps more virulent clones but that glycopeptide resistance is spread through horizontal transfer of the resistance genes among different hosts. Still, firm conclusions on the transfer of strains or genes from animals to humans await further study.
ACKNOWLEDGMENTS
P.V. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral research fellow.
We thank Rob Willems and Janetta Top, National Institute of Public Health and the Environment, Bilthoven, The Netherlands, for help in sequencing the different transposon types.
REFERENCES
- 1.Aarestrup F M. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;1:255–257. doi: 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Courvalin P. Sequence of the vanY gene required for production of a vancomycin-inducible d,d-carboxypeptidase in Enterococcus faecium BM4147. Gene. 1992;120:111–114. doi: 10.1016/0378-1119(92)90017-j. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates J, Jordens J Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–514. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 6.Bates J. Epidemiology of vancomycin-resistant enterococci in the community and the relevance of farm animals to human infection. J Hosp Infect. 1997;37:89–101. doi: 10.1016/s0195-6701(97)90179-1. [DOI] [PubMed] [Google Scholar]
- 7.Biavasco F, Miele A, Vignaroli C, Manso E, Lupidi R, Varaldo P E. Genotypic characterization of a nosocomial outbreak of vanA Enterococcus faecalis. Microb Drug Resist. 1996;2:231–237. doi: 10.1089/mdr.1996.2.231. [DOI] [PubMed] [Google Scholar]
- 8.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. 1994. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descheemaeker P, Lammens C, Pot B, Vandamme P, Goossens H. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int J Syst Bacteriol. 1997;47:555–561. doi: 10.1099/00207713-47-2-555. [DOI] [PubMed] [Google Scholar]
- 11.Devriese L A, Ieven M, Goossens H, Vandamme P, Pot B, Hommez J, Haesebrouck F. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother. 1996;40:2285–2287. doi: 10.1128/aac.40.10.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endtz H P, Van Den Braak N, Van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weersink A J L, Vandenbroucke-Grauls C M J E, Buiting A G M, Van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana R, Ligozzi M, Pedrotti C, Padovani E M, Cornaglia G. Vancomycin-resistant Bacillus circulans carrying the vanA gene responsible for vancomycin resistance in enterococci. Eur J Clin Microbiol Infect Dis. 1997;16:473–474. doi: 10.1007/BF02471915. [DOI] [PubMed] [Google Scholar]
- 14.Goossens H. Spread of vancomycin-resistant enterococci: differences between the United States and Europe. Infect Control Hosp Epidemiol. 1998;19:546–551. doi: 10.1086/647871. [DOI] [PubMed] [Google Scholar]
- 15.Goossens H. Vancomycin-resistant Enterococcus faecium derived from animals colonizing the human gut: the missing evidence? Clin Microbiol Infect. 1999;5:64–66. doi: 10.1111/j.1469-0691.1999.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 16.Iwen P C, Kelly D M, Linder J, Hinrichs S H, Dominguez E A, Rupp M E, Patil F D. Change in prevalence and antibiotic resistance of Enterococcus species isolated from blood cultures over an 8-year period. Antimicrob Agents Chemother. 1997;41:494–495. doi: 10.1128/aac.41.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolates from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen L B. Differences in the occurrence of two base pair variants of Tn1546 from vancomycin-resistant enterococci from humans, pigs, and poultry. Antimicrob Agents Chemother. 1998;42:2463–2464. doi: 10.1128/aac.42.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen L B, Hammerum A M, Poulsen R L, Westh H. Vancomycin-resistant Enterococcus faecium strains with highly similar pulsed-field gel electrophoresis patterns containing similar Tn1546-like elements isolated from a hospitalized patient and pigs in Denmark. Antimicrob Agents Chemother. 1999;43:724–725. doi: 10.1128/aac.43.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klare I, Heier H, Claus H, Reiibrodt R, Witte W. VanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;125:165–172. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 21.Klare I, Heier H, Claus H, Böhme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 22.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 23.Martone W J. Spread of vancomycin-resistant enterococci: why did it happen in the United States? Infect Control Hosp Epidemiol. 1998;19:539–545. doi: 10.1086/647870. [DOI] [PubMed] [Google Scholar]
- 24.McDonald L C, Kuehnert M J, Tenover F C, Jarvis W R. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miele A, Bandera M, Goldstein B P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in vanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray B E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 28.Palepou M-F I, Adebiyi A-M A, Tremlett C H, Jensen L B, Woodford N. Molecular analysis of diverse elements mediating vanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998;42:605–612. doi: 10.1093/jac/42.5.605. [DOI] [PubMed] [Google Scholar]
- 29.Pegues D A, Pegues C F, Hibberd P L, Ford D S, Hooper D C. Emergence and dissemination of a highly vancomycin-resistant vanA strain of Enterococcus faecium at a large teaching hospital. J Clin Microbiol. 1997;35:1565–1570. doi: 10.1128/jcm.35.6.1565-1570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perlada D E, Smulian A G, Cushion M T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 32.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O’Donnell A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: J. Wiley & Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 33.Sahm D F, Torres C. Effects of medium and inoculum variations on screening of high-level aminoglycoside resistance in Enterococcus faecalis. J Clin Microbiol. 1988;26:250–256. doi: 10.1128/jcm.26.2.250-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman J, Thal L A, Perri M B, Bostic G, Zervos M J. Epidemiologic evaluation of antimicrobial resistance in community-acquired enterococci. J Clin Microbiol. 1998;36:830–832. doi: 10.1128/jcm.36.3.830-832.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thal L A, Chow J W, Mahayni R, Bonilla H, Perri M B, Donabedian S A, Silverman J, Taber S, Zervos M J. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1995;39:2112–2115. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 39.Vandamme P, Vercauteren E, Lammens C, Pensart N, Ieven M, Pot B, Leclercq R, Goossens H. Survey of enterococcal susceptibility patterns in Belgium. J Clin Microbiol. 1996;34:2572–2576. doi: 10.1128/jcm.34.10.2572-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Bogaard A E, Jensen L B, Stobberingh E E. Vancomycin-resistant enterococci in turkeys and farmers. N Engl J Med. 1997;337:1558–1559. doi: 10.1056/NEJM199711203372117. [DOI] [PubMed] [Google Scholar]
- 41.van den Braak N, Van Belkum A, Van Keule M, Vliegenthart J, Verbrugh H A, Endtz H P. Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in The Netherlands. J Clin Microbiol. 1998;36:1927–1932. doi: 10.1128/jcm.36.7.1927-1932.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]
- 43.Welton L A, Thal L A, Perri M B, Donabedian S, McMahon J, Chow J W, Zervos M J. Antimicrobial resistance in enterococci isolated from turkey flocks fed virginiamycin. Antimicrob Agents Chemother. 1998;42:705–708. doi: 10.1128/aac.42.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willems R J L, Top J, Van Den Braak N, Van Belkum A, Mevius D J, Hendriks G, Van Santen-Verheuvel M, Van Embden J D A. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43:483–491. doi: 10.1128/aac.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodford N, Adebiyi A A, Palepou M I, Cookson B D. Diversity of vanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]