Abstract
Nanotopography presents an effective physical approach for biomaterial cell manipulation mediated through material-extracellular matrix interactions. The extracellular matrix that exists in the cellular microenvironment is crucial for guiding cell behaviours, such as determination of integrin ligation and interaction with growth factors. These interactions with the extracellular matrix regulate downstream mechanotransductive pathways, such as rearrangements in the cytoskeleton and activation of signal cascades. Protein adsorption onto nanotopography strongly influences the conformation and distribution density of extracellular matrix and, therefore, subsequent cell responses. In this review, we first discuss the interactive mechanisms of protein physical adsorption on nanotopography. Secondly, we summarise advances in creating nanotopographical features to instruct desired cell behaviours. Lastly, we focus on the cellular mechanotransductive pathways initiated by nanotopography. This review provides an overview of the current state-of-the-art designs of nanotopography aiming to provide better biomedical materials for the future.
Keywords: Nanotopography, Cell-material interaction, Protein adsorption, Biomaterials
Highlights
-
●
A comprehensive overview of nanotopography fabrication, and nanotopography regulates various cell behaviours.
-
●
The interactive physical adsorption between nanotopography and extracellular matrix.
-
●
Nanotopography initiates the cellular mechanotransductive pathways and downstream signalling cascades.
1. Introduction
The cellular microenvironment is fundamental to the regulation of cell behaviours. Cells sense physical and functional properties of the external environment through adhesion formation, which is critical to cell decision making and behaviour. Cell membrane receptors, such as integrins, transduce physical information from the microenvironment into intracellular signalling pathways, leading to changes in cell proliferation, differentiation, migration or apoptosis [[1], [2], [3], [4], [5]]. Properties such as chemistry [2,6], stiffness [7,8] and topography [9,10] can be manipulated to control and guide cell behaviour.
Topography refers to specific morphological features in the cellular microenvironment. The scale of these topographical features scale is important, and leads to changes in cell response. At the macroscale (>100 μm), topographical features influence the cells arrangement at colony level [11]. At the microscale, topographies are in the size range of the cell itself (0.1–100 μm), and thus effect cells at the single cell level. Cell alignment to microtopographical features is termed ‘contact guidance’, which was coined around 1964 when Curtis and Varde demonstrated that fibroblasts can position themselves parallel to microstructures with diameters of 10–30 μm [[12], [13], [14]]. Nanotopography refers to specific morphological features that are fabricated at the nanoscopic scale (1–100 nm), and are therefore within the same order of magnitude as cell receptors, such as integrins. Since the contact of the implant surface with the biological environment occurs as soon as the implant is introduced, the implant surface plays an important role in influencing the sequence of initial protein adsorption, interactions with blood such as platelet adhesion and haemostasis, inflammation, and osteogenic cell responses [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24]].
In vivo, cells are surrounded by the extracellular matrix (ECM), which is present in all organs and tissues. The ECM is a complex structure composed of many types of proteins such as collagens, proteoglycans, glycoproteins and glycosaminoglycans [25]. ECM elastin or fibronectin (FN) are highly elastic giving tissue the capability to deform reversibly. Further to this, FN also provides multivalent cellular binding sites that enable cross-linking with other ECM proteins [26,27]. Collagen can form fibers or networks that contribute to the mechanical stiffness of tissues [28]. The ECM acts like a ‘reservoir’ storing and presenting growth factors to cells while also modulating cell-cell interactions and allowing cushioning and pressure resistance [29]. ECM-based control of cell behaviour has been widely studied and may occur through multiple physical mechanisms-such as ECM micro/nano-topography, elasticity, and mechanical signals that are transmitted from ECM to cells [30]. Substrate nanotopography-ECM interactions play a key role for manipulating cellular behaviour through regulation of ECM presentation (Fig. 1). However, the mechanisms underlying how nanotopographic cues influence cell proliferation and differentiation are not well studied but appear to involve changes in cytoskeletal organization and structure, potentially in response to the geometrical features and their effects on conformation of the ECM.
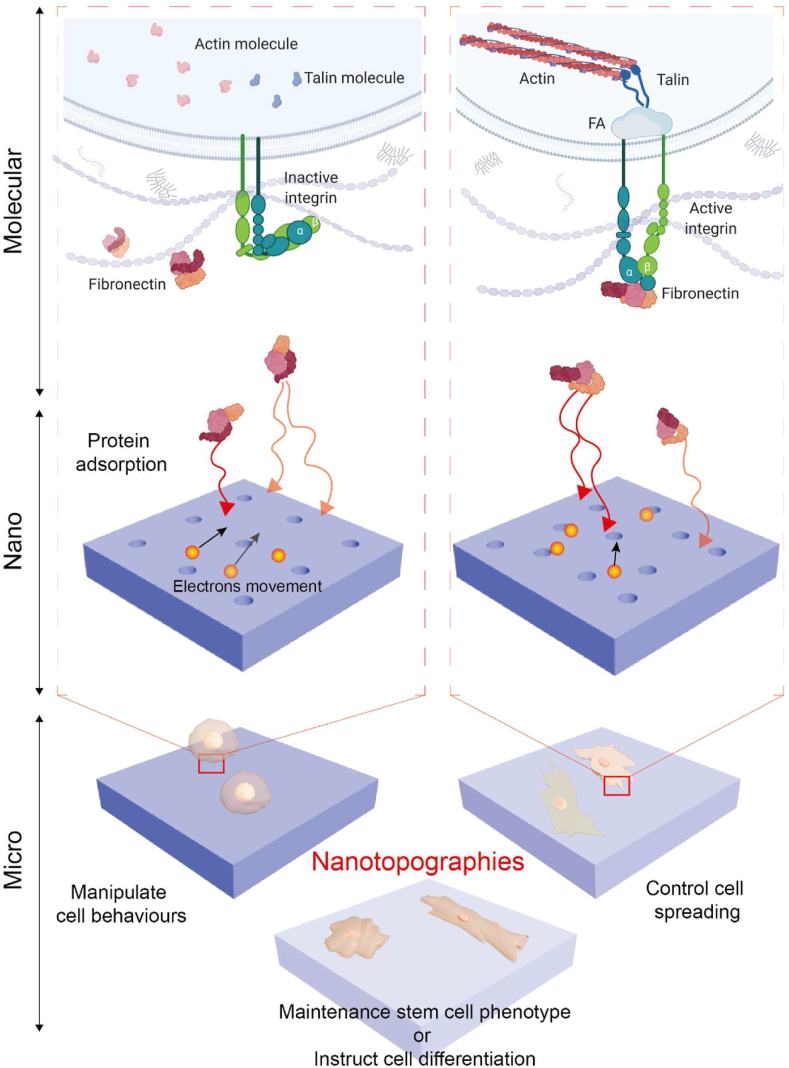
Fig. 1.
Schematic demonstrating influence of nanotopography on cell behaviour through interaction with the extracellular matrix. Cellular behaviours under microscale such as adhesion, spreading and differentiation are influenced by fibronectin interacts with on nanotopography surface electrons and adsorption under nanoscale. Transmembrane receptors like integrins can further interact to fibronectin adsorbed on nanotopography, then behave active or inactive. Upon ligand bonding, the focal adhesions (FAs) is assembled and cytoskeletal filaments are aggregated by actin and talin molecules.
In terms of ECM conformation on biomaterials surfaces, dynamic protein conformation changes have been studied extensively. For example, two conformations can be distinguished for FN in ECM; one is a globular conformation with a diameter of 16–35 nm, the other is a linear conformation, 2 nm in diameter with a flexible length of 120–180 nm [31,32]. It has been shown that the conformation of FN is influenced by material surface properties, e.g. linear conformation was the predominant form on hydrophilic surfaces, and globular conformation was predominant on hydrophobic surfaces [33,34]. This may be due to the higher exposure to negatively charged groups on hydrophilic surfaces, which might interfere with the stabilizing ionic interactions between the two chains in the protein [35]. This demonstrates how subtle differences in substrate properties can lead to significant differences in protein conformation on material surfaces. Factors such as temperature, pH, and ionic strength of the adjacent environment need to be taken into account for the final conformation of ECM [36].
Following ECM adsorption, cells sense the microenvironment through adhesions. Adhesion formation is the first step and plays a fundamental role in subsequent viability, growth and differentiation [37]. The immobilisation of material substrates and ECM can be distinguished into two kinds; chemical bonding functionalisation and physical adsorption [38,39]. Thus, nanotopography acts as an initial physical ‘trigger’ from the material side to initiate downstream molecular reactions through influencing the conformation of ECM, and therefore ligand presentation.
In this review, we will discuss how nanotopography influences the physical adsorption of proteins and how this instructs cell behaviours from interactions at both the nanotopography-ECM interface and ECM-cell interface. We will then discuss the cellular response to various nanotopographies, particularly those that regulate focal adhesions (FAs) formation, cell differentiation and the mechanosensitive Hippo signalling pathway.
2. ECM-nanotopography interactive mechanisms
Cellular sensing of biomaterial features is mediated by the layer of protein adsorbed onto the substrate. Thus, the protein adsorption process plays a fundamental role at the first stage of cell-material interactions [40]. The status of proteins their concentration, conformation, distribution, and strength of interaction, can further influence cell responses [[41], [42], [43], [44]]. During adsorption, the complex composition and structure of proteins causes phenomena such as protein structural rearrangements, cooperative adsorption, overshooting adsorption kinetics, or protein aggregation [45]. For initial contact, soluble matrix proteins like FN, vitronectin (VN), fibrinogen (FG) influence cell response, whereas for long-term contact other ECM proteins dominate, such as collagens (Cols) and laminins (LMs) [46]. FN is a high molecular weight dimeric glycoprotein that is found in a soluble form in blood, other extracellular fluids such as plasma, and in an insoluble form in connective tissues and attached to cell surfaces [26]. FN is able to ligate to cells via integrins through its cell-binding domain (FNШ9-10) containing the arginine-glycine-aspartic (RGD) sequence [47,48]. VN is a glycoprotein with a molecular weight of 75 kDa which also engages the integrin family (αVβ1, αVβ3, αVβ5, αIIbβ3) through the RGD sequence [49]. VN is involved in several cell behaviours: adhesion, migration and integrin-mediated signal transduction [50]. FG is also a large fibrous dimeric glycoprotein with a symmetrical structure [51]. Col is the most prevalent fibrous protein in the matrix, where 28 different types of Col composed of at least 46 distinct polypeptides chains exist [28,52]. LM is a cell adhesion molecule that comprises a family of glycoproteins and features within a thin sheet of ECM that underlie epithelial and endothelial cells and surround muscle cells, schwann cells, and fat cells [53]. These proteins dictate how cells adhere to, and thus interact with, topographical features in their microenvironment. In the following section, we will discuss the factors affecting protein adsorption on biomaterials, then focus on electrostatic interactions between nanotopography and proteins.
2.1. Factors affecting protein adsorption on biomaterials
Based on thermodynamics, the major driving force of protein adsorption is an entropy gain arising from the release of surface adsorbed water molecules and salt ions, and from structural rearrangements inside the protein itself [54]. During the process of adsorption, several factors influence the procedure including external factors like temperature, pH, ionic strength, electrostatic effects and buffer composition [45]. For example, evidence has shown that the amount of adsorbed protein increases as a function of increasing temperature and increasing hydrophobicity [55].
The effect of pH is more complex since the pH determines the electrostatic state of proteins. As the pH equals the isoelectric point (pI) of a protein, the numbers of negative and positive charges are in balance resulting in a net neutral molecule. When pH˂pI, the proteins are positively charged, whereas when pH>pI, the charge of proteins are negative [45]. At the pI point electrostatic protein-protein repulsions are minimized and the maximum adsorption of protein on substrates is achieved around their pI [56]. The concentration of dissolved ions is another parameter affecting protein adsorption processes, which is expressed in terms of ionic strength. This is because ionic strength determines the Debye length correlating with the damping distance of the electric potential of a fixed charge in an electrolyte-increased salt concentration reduces electrostatic repulsion between like-charged materials and decreases electrostatic attraction between oppositely charged materials [57].
Protein adsorption behaviour can be described by several theoretical models, including Langmuir, Freundlich and Brunauer-Emmett-Teller isotherm models [[58], [59], [60]]. Among all models, the Langmuir adsorption model has been widely used for its simplicity. Specifically, there are four basis conditions that an adsorption system must satisfy in order for the Langmuir isotherm model to be applied; (I) All adsorption sites are equivalent, distinguishable and independent (homogeneous adsorption sites). (II) Each adsorption site binds an individual solute molecule. (III) There are no interactions between solutes on the surface that influences their adsorption behaviour. (IV) Adsorption process must represent a dynamically reversible equilibrium. However, due to the complexities of protein adsorption, this leads to difficulties in experimental investigation of the process.
Spreading of proteins can be influenced by cooperative factors, protein internal stability, interaction strength between protein and substrate, and protein-protein interactions. Protein-protein interactions rely on the presence of neighbouring proteins that are affected by the solution concentration. These effects cause saturated adsorption of a protein with monolayer coverage on a substrate, which further changes the spreading and packing of the proteins [61].
To help quantitively analyse the surface protein adsorption, the biological surface adsorption index (BSAI) was developed. The BSAI is a statistics-based model that employs five descriptors to represent the surface energy profile of the substrate: hydrophobicity, hydrogen bonding, polarity, polarizability and lone-pair electrons. Then, by combining with traditional adsorption models, this predictive model can be used in biological applications, for example to predict the safety of nanomaterials [62].
2.2. Electrostatic interactions between nanotopography and ECM
As the BSAI predicts, the protein-surface interactions are not only influenced by the protein's properties, but also affected by the properties of the material, such as surface energy, polarity, charge, and topography [63,64]. Proteins are typically asymmetrical, complex molecules of a few nanometers in size, i.e. the same scale of nanotopgraphical features. The conformation of adsorbed proteins on material surfaces can be traced back to its free energy minimum resulting from attractive Coulomb's force and Van-der-Waals interactions, hydrogen bonds, and the entropy gain of solvent molecules or counter ion release [45]. Due to their complex structure, proteins generally exhibit different affinities dominated by the local composition of their amino acid residues [45]. To investigate the electrical interactions of proteins to a material, it was concluded that the protein surface can be divided into different patches or domains, including hydrophobic, hydrophilic, positively, and negatively charged [65,66]. Obviously, electrostatic-dominated protein adsorption to positively or negatively charged substrates tend to expose oppositely charged regions to surfaces.
Titanium (Ti) is a major biomedical implant-based material, and has been investigated in the context of titanium nanotopography protein interactions. It has been demonstrated that nanotopographical Ti surfaces greatly enhance protein adsorption ability. For example, polished (smooth) and nanotopographical Ti-6Al-7Nb specimens were immersed in albumin- and FN-containing (5 mg/ml) phosphate-buffered saline (PBS), then washed with deionized water. Higher N1s spectra intensity were observed on the nanotopographical Ti-6Al-7Nb specimen measured by X-ray photoelectron spectroscopy (XPS), indicating greater protein adsorption on the Ti-6Al-7Nb nanotopography [67]. Recently, a selection of nanorough TiO2 surfaces was been fabricated with root mean square roughness (Sq) values below 2.7 nm but with different surface morphologies, the proteins myoglobin (MGB), bovine serum albumin (BSA) and thyroglobulin (TGL) adsorption behaviours were investigated in situ [68]. It was observed BSA adsorbed was faster and observed a thicker protein film at saturation, MGB adsorption followed a linear correlation with the surface skewness (Ssk) and the TGL adsorption was mainly influenced by the Ssk, increased Ssk led to faster adsorption and thicker protein film [68]. The proteins adsorption of MGB, BSA and TGL were further investigated on nanorippled TiOx/Ti surfaces, demonstrated that even nanotopographic features with vertical dimensions of less than 2 nm have a surprisingly strong effects on the adsorption and especially the adsorption-induced denaturation of globular proteins [69].
Other biomedical Ti based alloys have also been investigated, for example, nanotopographical porous features of β-type Ti-25Nb-25Zr alloy enhanced albumin and FN adsorption [70]. Moreover, another study showed the density of FN adsorbed on Ti nanotopography was 2–3 times higher than it was on bare Ti; specifically, Ti nanoflakes showed the greatest protein adsorption when compared with pristine Ti; Ti nanowires and Ti nanonests [71]. However, further experimental investigation of the interaction between protein and nanotopography has reported that there is no statistical influence of protein adsorption on titanium with different nanoscale roughness [72]. Titanium films were engineered with similar surface chemistry but different sizes of grains on the surface, leading to different topographical features. Using Atomic force microscopy (AFM) it was shown that surface roughness, even if larger than nanoscale, has little effects on protein adsorption [72]. Additionally, no significant difference was observed in the quantification of FG adsorbing on 53 nm roughness titanium surface (machined), 70 nm roughness (acid etched) and 183 nm roughness (etched and blasted) [73]. However, in contrast, the amount of FN adsorption increased as the titanium surface roughness increased [73]. Other studies have also demonstrated that nanotopography can significantly impact protein adsorption; FG adsorbed on nanotopographical tantalum films was increased 70% with increasing film roughness (from 2.0 nm to 32.9 nm) [74]. Another study concluded that the amount of FG adsorbed on nano-featured colloidal silica surface with 21 nm roughness was significantly lower than on 7 and 14 nm roughness surfaces [75]. The quantification methods in both studies were characterized by quartz crystal microbalance with dissipation (QCM-D). This indicates that even the same protein exhibits different adsorption behaviour on different substrates. The protein-nanotopography interactions strongly rely on the physiochemical properties of the substrate and, the topological features of nanotopography play a key role in protein adsorption.
The ECM captured by the nanotopography is related to the procedure of attractive adsorption which consists of two steps - the direct attachment to the surface of substrates as the first step, and the re-arrangement of the proteins upon adsorption as the second [76]. For instance, FN adsorption on various artificial surfaces depends on several factors of the substrate, such as the physical properties, roughness, wettability and charge [77]. Of these factors, hydrophilicity and charge are thought to be the dominant interactions with FN molecules that affect its conformation [78]. For example, increasing negatively charged M − OH complexes on titanium modulates FN's integrin binding activity probably through a change in conformation [79]. It was also concluded that the binding of a peptide to a titanium-based substrate is strengthened by ionic interactions of charged atoms and polar interactions of neutral atoms [80,81].
Alongside electrostatic-dominated adsorption of proteins to titanium and its alloys, surface oxidation also has an important role. The surface is naturally covered by an oxide layer, such as a TiO2 layer on titanium metal. The oxidation and dissolution of metal cations partially yield to the negative surface potential of the titanium surface [82]. For example, it was shown that the contact between the cell membrane of osteoblasts and the TiO2 layer is established by two steps, firstly the cell membrane makes a non-specific contact because of the electrostatic interactions, then the specific binding takes place [83]. It could be speculated that cells are electrostatically repelled by the negatively charged titanium surface, therefore this indicates some other attractive forces are not present in the system. Recently, an ECM-titanium interactive mechanism was assumed where the negatively charged titanium surface and a negatively charged plasma membrane is mediated by positively charged proteins. A theoretical model by Monte Carlo methodology was created to simulate the distribution and orientation of proteins adjacent to the charged titanium surface. The mediated proteins are featured with a distinctive quadrupolar internal charge distribution on the titanium surface [84].
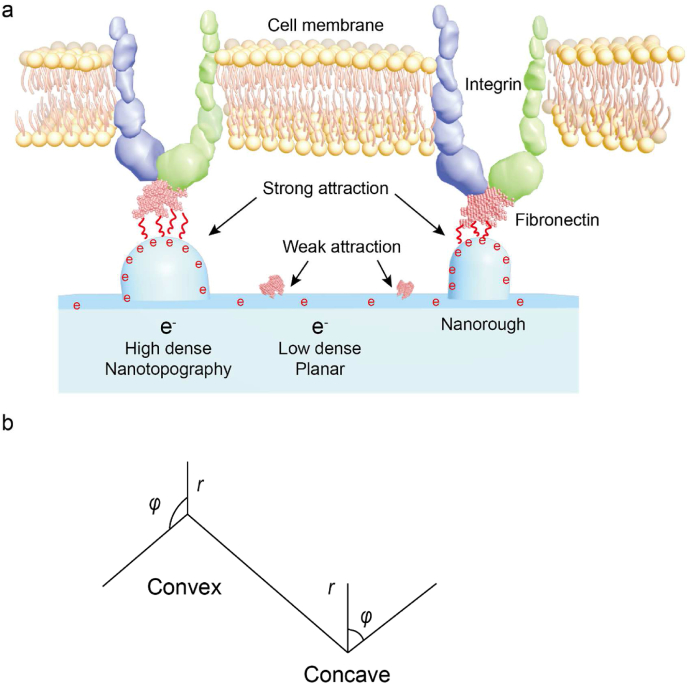
At the nanoscale, it has been proven that nanorough topography creates a surface with an enhanced electric field strength that is strongly attractive (electrostatic attraction, Coulomb's force) to proteins due to the highly curved edge [84] (Fig. 2a.). Fig. 2a illustrates the electric field strength near the highly curved edges of a titanium surface that was estimated in the limit of very sharp edges. Importantly, the cation is also involved as an attractive mediator between FN and titanium and is expected to be more efficient for a high surface charge density [[84], [85], [86]]. Both increased electric field strength and surface charge density at the nanorough area may also promote divalent cation-mediated adsorption of FN to a negatively charged titanium surface [94]. Thus, the charge density of a substrate surface is strongly influenced by the curvature of the nanorough topography. Specifically, based on the Laplace equation, the surface charge density (σ) adjacent to both convex and concave nanotopographies (Fig. 2b.) was calculated at under the equilibrium (boundary) conditions, where the electric potential is constant over the whole conducting metal surface:
| (1) |
| (2) |
Where, σ is the surface charge density of nanorough at convex/concave, r is the distance between FN and convex/concave edge region, is a constant determined from the additional boundary condition, is the permittivity of the free space. While the FN is adsorbing to a convex surface, r→0, leads to the becomes infinitely large and singular. In contrast, the is zero as r→0 [84] (Fig. 2b.). This indicates the surface charge density of convex surfaces would not diverge, instead, it would decrease in magnitude with the increasing curvature radius. Compared with the variation of density on convex surfaces, the charge density would monotonically increase with the increasing curvature radius of the concave edge. Due to the increased surface charge density at the sharp edge, the increased FN accumulation at the nanorough region that can facilitate the adhesion and aggregation of integrin molecules can therefore induce the formation of FAs. The model was then tested in situ using human osteoblasts [84].
Fig. 2.
Fibronectin adsorbed on nanorough regions [[84], [85], [86]]. a. Fibronectin adsorbed on nanorough area which contains higher electric density than a flat area, the nanorough area generate a stronger electric attraction to fibronectin. b. Scheme of Convex and Concave, the electron migration depends on nanotopographical features, such as the curvature; φ.
3. Cellular responses to nanotopographies
It is well established in the recent literature that changes in topographical features of surfaces can significantly influence cell behaviour by altering mechanosensitive responses [[87], [88], [89]]. Since naturally formed titanium oxide film is bioinert with successful biocompatibility, titanium-based alloys have been widely used in orthopaedic reconstruction for various types of bone anchored implants. However, this bioinert film is not integrated into the bone tissue, a lack of bone ingrowth causing Ti implant failures have been documented in clinical case reports [[90], [91], [92], [93], [94]]. Successful osteointegration relies upon acceptance of several key factors: the stable/immobile physical support of an implant under functional loads, apposition of new and re-formed bone in the congruency with the implant without fibrous tissue formation, absence of relative motion between the implant and surrounding bone tissue under loading, and that the components of bone tissue within a thin zone of the implant surface are identified as normal bone and marrow constituents [95]. A critical factor that influences bone ingrowth to the implant are the surface features of the implant; on a micro-scale level, these features regulate the cell responses as they interact with their surrounding microenvironment. Thus, creating nanotopographical features on implants is a feasible method to improve osteointegration of implants, rather than altering the mechanical or chemical components of orthopaedic implants. Among various cell behaviours, the improvement of initial cell adhesion is the earliest challenge, and then of guiding stem cell differentiation. These two cell behaviours play vital roles in the success of early osteointegration, and ultimately, implant longevity [96,97].
Topographical studies on the effects on cell adhesion/spreading, proliferation and differentiation have been extensively investigated [10]. In this section, we will firstly review how the nanotopography manipulates cellular adhesion and spreading behaviour, then we will focus on the differentiation of stem cells and mechanotransductive behaviours triggered by nanotopography.
3.1. Adhesion/spreading behaviour
A variety of approaches have been employed to create different nanotopographies on substrates to explore their influence on the adhesive/spreading behaviour of various cell types, including mesenchymal stem cells (MSCs). For example, Yim et al. observed that a decreased expression of integrin subunits (α2, α6, αV, β2, β3 and β4) occurred in human MSCs (hMSCs) cultured on 350 nm gratings (nanogroove topography) of tissue culture polystyrene (TCPS) and polydimethylsiloxane (PDMS), compared to the planar control [20]. The soft lithography technique was applied on a nanoimprinted poly (methyl methacrylate) (PMMA)-coated Si master moulds to reproduce the nanopattern on PDMS. Nano-embossing pre-patterned PDMS master was used to fabricate nanopatterns onto a TCPS strip or a 35 nm TCPS dish [20]. Here, cellular cytoskeleton exhibited an elongated shape with an aligned arrangement, while a random network arrangement was observed in cells on a control surface [20]. The similar method of soft lithography technique was also applied in another study investigating cellular morphological changes of smooth muscle cells induced by PDMS nanotopography [98]. In this study, the bovine pulmonary artery smooth muscle cells showed significantly elongated and parallel alignment on 350 nm linewidth, 700 nm pitch, and 350 nm depth grooved nanotopography [98]. Indeed, it was shown that rutile TiO2 surfaces promoted better adhesion and a stereoscopic morphology of bone marrow-derived MSCs with a nanorod array topography, compared to a smooth ceramic surface [99]. The TiO2 nanorod array was fabricated by a facile hydrothermal method reported previously [100].
Increasing the roughness of material surfaces can improve the cell adhesion, especially the roughness at the nanoscale. Another study, using PDMS with a variety of different wrinkled topographies, showed that wrinkle dimensions of 7121 nm wavelength and 2561 nm amplitude was optimal for promoting bone marrow-derived MSC adhesion by influencing various factors, such as cytoskeletal arrangement and FAs assembly [101]. This gradient wavelength was prepared by a masked plasma-oxidation procedure with subsequent overoxidation to obtain a SiO2-like surface [101]. It has also been demonstrated that adhesion, spreading and self-renewal of human embryonic stem cells (hECSs) can be regulated by the features of nanorough patterning on silica based glass wafers [102]. It is remarkable that in nanoscale, it has been demonstrated that not only the presence of roughness can promote cell adhesion, but also the arrangement of nanofeatures can change the adhesion and spreading behaviour of cells. It has been shown that repeating nanopit features on PMMA surfaces can lead to a reduction of cell spreading due to the decrease of focal adhesion formation [19]. The nanopits were fabricated by a fast electron-beam lithography (EBL) technique, which can form a defined spot size and each shape is spaced by the beam step size, only by a single exposure [103]. Compared with nanopit features, dot morphology in nanoscale has also attracted attention. Dimers, trimers and extended hexagonals arrangement of nanodots were fabricated by a nanolithographic method, the cell spreading was significantly enhanced on nanodots when at least four liganded sites were spaced within 60 nm or less [104]. Cells are extremely sensitive to nanodot morphologies; indeed, cells have been described as ‘clinging on with their fingertips’ to nanodots that are only 8 nm in height [105].
Cell-specific changes in behaviour has been highlighted in several studies. For example, using Ti-based substrates, it was shown that 70 nm nanoporous surfaces, compared with rough surfaces, induced osteoblast and fibroblast adhesion and alignment along the pores, whereas macrophages remained oval-shaped and sparsely distributed [106]. The TiO2 nanoporous features were formed by electrochemical anodization (EA). Other porous nanotopographies, such as TiO2 nanotubes (NTs), have also been investigated due to their unique nanomorphology that can mimic the natural mineral of bone tissue [24,107]. These nanotubes were also fabricated by the anodization method. It has been demonstrated that nanotubes are able to significantly enhance the proliferation of cells by regulating their adhesion behaviour. Specifically, using a nanotube topography, a significant acceleration in the growth rate of osteoblast cells by as much as 300–400% occurs, compared with pristine titanium surface samples [108]. Osteoblast adhesion behaviour was investigated on ordered and disordered nanopatterns that were functionalized with RGD on polyethylene glycol (PEG). The RGD nanopatterns were fabricated by a novel method based on a Micelle nanolithography technique, the Au nanopatterns were prepared on glass and further constructed by coupling c(-RGDfK-)-thiol ligands via the thiol group, thus each nanoparticle could bind up one integrin [109]. It was observed that when ligand spacing was less than 70 nm, integrins effectively clustered and formed adhesions, whereas integrins did not cluster when ligand spacing was larger than 70 nm. Interestingly, at a global average inter ligand spacing of larger than 70 nm, disordered nanopits displayed some integrin clustering; this may due to a denser local arrangement which was below 70 nm [109]. Recently, RGD functionalized nano gold dots with various spacings of 37, 53, 77, 87 and 124 nm were fabricated on PEG and the smaller RGD spacings induced larger FAs and a distinct, well-organised, cytoskeleton [110].
It has also been shown that self-assembling polystyrene (PS) nanosphere surfaces could enhance spreading behaviour of human adipose-derived MSCs most optimally with nanopit diameters of 300–400 nm between a range of 200–750 nm [111]. Another study generated nanotopography that was optimized to mimic the granularity and porosity of the natural ECM using TiO2 substrates with features on 30 nm thick coatings with an average roughness of 4–5 nm and average surface slope of roughly 8° [112]. This method was reported in a previous study, the TiO2 thin film was produced by supersonic beam deposition of titanium clusters [113]. These nanotopographical features could enhance the spreading behaviour of human MSCs compared to cells on flat control glass surfaces [112]. Also it has been shown that mesangiogenic progenitor cells (MPCs), which are thought to be progenitors to MSCs, showed differences in spreading/adhesive behaviour when cultured on a 400 nm nanograting topography, compared to a flat control, where more protrusive filopodia were observed on nanograting topography suggesting more spreading potential [114]. Table 1 summarises several nanotopographical approaches that influence cell adhesion and spreading.
Table 1.
Nanotopographies affect cell adhesion & spreading behaviours.
| Nanofeatures | Material substrate | Cell type | Functional studies | Ref |
|---|---|---|---|---|
|
Nanopits Diameter-100 nm Depth-100 nm |
Polymethylmethacrylate (PMMA) | Human fibroblasts | Control cell spreading | Dalby M J et al. [19] |
|
Nanodots Dimers, trimers and hexagons arrangement with interdot spacing of 60 nm |
AuPd nanodots | 3T3 mouse fibroblasts | Control cell adhesion receptors arrangement | Schvartzman, M et al. [104] |
|
Nanorods Length-1.5 μm Diameter-100 nm |
TiO2 substrate | Bone marrow-derived MSCs | Osteogenic differentiation | Qiu J et al. [99] |
|
Nano wrinkles Amplitudes-49 to 2561 nm Wavelengths-464 to 7121 nm |
Polydimethylsiloxane (PDMS) | Bone marrow-derived MSCs | Promoting adhesion | Zhou Q et al. [101] |
|
Nanorough Rq=70 nm Rq=150 nm |
Silica based glass | Human embryonic stem cells (hECSs) | Adhesion, spreading and self-renewal | Chen W et al. [102] |
|
Nanogroove 350 nm linewidth and 700 nm pitch 500 nm linewidth and 1 μm pitch |
Tissue culture polystyrene (TCPS) and polydimethylsiloxane (PDMS) | Human mesenchymal stem cell (hMSC) | Induction of focal adhesion and cytoskeleton arrangement | Yim E K F et al. [20] |
|
Nanotubes Outer diameter-100 nm Inner diameter-70 nm Height-250 nm |
TiO2 substrate | MC3T3‐E1 osteoblast cells | Accelerate osteoblasts growth | Oh S et al. [108] |
3.2. Differentiation/maintenance of cell behaviour
Controlling substrate nanotopography is an effective approach for manipulating cell differentiation behaviour (Table 2 and Fig. 3). For example, a study highlighted that by increasing surface roughness of glass using reactive ion etching to generate random nanostructures ranging from 1 to 200 nm, this enhanced osteogenesis of hMSCs and was associated with changes in adhesion, cytoskeletal tension and mechanosensitive transcriptional co-activator with PDZ-binding Motif (TAZ) activation [115]. Using nanopatterned poly (lactic-co-glycolic acid) (PLGA) patches, it was shown that primary osteoblasts displayed enhanced osteogenesis and rat bone regeneration in vivo when cultured on surfaces with 600 nm nanogrooves, compared to flat [116]. Moreover, it has been shown previously that substrates with aligned poly-l-lactic acid (PLLA) nanofibers, compared to random, enhance the osteogenic differentiation of hMSCs through modulation of negative osteogenic regulator miR-125b [117]. It has also been shown that substrates with 120 nm diameter and 100 nm depth disordered nanopits were more effective at enhancing the osteogenesis of human osteoblasts and MSCs than a flat surface [21]. Using PMMA, nanopits of 120 nm in diameter with 100 nm depth were fabricated with different symmetrical arrangements from highly ordered to random distribution. The result has further demonstrated that a controlled disordered arrangement of nanopits generated by the EBL technique can greatly enhance osteogenesis from skeletal stem cells [9]. Interestingly, regulating the ordering level of nanopits can also achieve the maintenance of the MSC phenotype. Absolute square lattice symmetry was also fabricated by EBL on polycaprolactone (PCL) substrates, resulting in a switch from osteogenic stimulation to maintenance of MSC stemness and proliferation, permitting prolonged retention of multipotency for up to eight weeks [5].
Table 2.
Nanotopographies affect cell differentiation behaviour.
| Nanofeatures | Material substrate | Cell type | Functional studies | Ref |
|---|---|---|---|---|
|
Nanorough Rq=1 nm Rq=100 nm Rq=200 nm |
Glass | Human mesenchymal stem cell (hMSC) | Osteogenesis induction | Qian W et al. [115] |
|
Nanopatch Ridge-800 nm Groove-800 nm Height-600 nm |
Poly (lactic-co-glycolic acid) PLGA | Primary osteoblasts | Osteogenesis induction | Lee M S et al. [116] |
|
Nanopits Diameter-120 nm Depth-100 nm Random displacement±50 nm |
Polymethylmethacrylate (PMMA) | Mesenchymal stem cell (MSC) | Osteogenesis induction | Allan C et al. [21] |
|
Ordered/disordered Nanopits Diameter-120 nm Spacing-300 nm Disordered-±50 nm offset Ordered-0 nm offset |
polycaprolactone (PCL) | Mesenchymal stem cell (MSC) | Maintenance of MSC phenotype and multipotency | McMurray R J et al. [5] |
|
Nanopatterns Distancing-30 nm Interdot spacing-106 nm |
TiO2 substrate | Mesenchymal stem cell (MSC) | Osteogenesis induction | Sjöström T et al. [119] |
|
Nanopillar Height-15, 55, 100 nm |
TiO2 substrate | Human mesenchymal stem cell (hMSC) | Bone matrix nodule formation | Sjöström T et al. [120] |
|
Nanoholed Pore sizes-72, 108, 135 amd 195 nm |
Opal film | Rat bone marrow‐derived MSCs (rBMSCs) | Promotes osteogenesis | Xiao Q R et al. [121] |
|
Nanocolloidal Hexagonal lattice spacing-58 nm, 108 nm |
Gold nanoparticle + polyethylene glycol (PEG) | Rat fibroblast | Ligand density regulation | Cavalcanti-Adam E A et al. [123] |
|
Nanopatterns Hexagonal lattice spacing-37, 53, 77, 87 amd 124 nm |
Gold nanoparticle + polyethylene glycol (PEG) | Rat MSCs (rMSCs) | Osteogenic and adipogenic induction | Wang X et al. [110] |
|
Nanopatterns Hexagonal lattice spacing-36, 50, 71, 98, 136 amd 143 nm |
Gold nanoparticle + polyethylene glycol (PEG) | Rat chondrocytes | Maintenance phenotype | Li S et al. [124] |
|
Micro/Nanopatterns Nanospacing 46 and 95 nm and with micropans 35 and 65 μm |
Gold nanoparticle + polyethylene glycol (PEG) | Human mesenchymal stem cell (hMSC) | Independently regulating cell spreading and ligand spacing | Wang X et al. [125] |
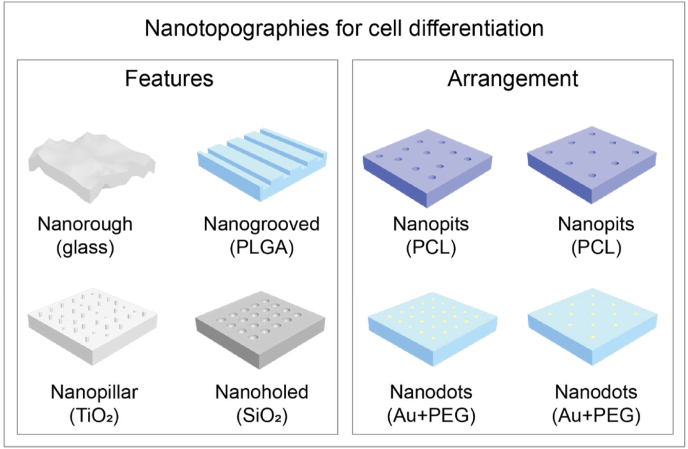
Fig. 3.
Both nanotopographical features type and arrangement can guide cell differentiation. Nanorough glass substrates and PLGA nanopatch have been demonstrated to enhance osteogenesis [115,116]. Osteoinduction and bone matrix nodule formation were proven on TiO2 nanopillar [119,120]. SiO2 nanoholed nanotopography also promoted the osteogenic induction of MSCs [121]. Maintenance of multipotency of stem cells has been strongly influenced by the arrangement of nanopits-ordered/disordered PCL nanopits [5]. Au nanoparticles combined with PEG substrates has been widely used for differentiation guidance by manipulating integrin ligand density [110,[123], [124], [125]].
In terms of stem cells differentiation responses to NTs, researchers have also demonstrated that the behaviour of MSCs is strongly affected by the dimensionality of NTs [118]. On small diameter nanotubes, MSCs display increased cell adhesion and growth with minimal differentiation; this is potentially due to protein aggregate adhesion and distribution configurations induced by the small nanotubes. In contrast, on larger diameter nanotubes, MSCs are forced to elongate and stretch to search for protein aggregates, and as a result, are forced/guided to differentiate specifically into osteoblast cells.
Aside from nanopits and nanotubes, nanodots have also been fabricated by block copolymer techniques. MSCs on 8 nm and 15 nm nanodot patterned surfaces were shown to exhibit an increased number of large osteogenic focal adhesions when compared to polished Ti surfaces [119]. Along with spacing, other features such as height of nanodots influence cell differentiation. Heights of 15 nm, 55 nm and 100 nm nanodots/nanopillars were generated on Ti substrates by anodization; MSCs cultured with nanopillars with a height of 15 nm displayed increased osteogenesis after 21 days [120]. Another study, using nanoholed opal films formed using self-assembly of colloidal SiO2 nanoparticles, showed that a more oriented and stretched nanotopography was favourable for osteogenesis of MSCs, compared to inverse topography [121]. Additionally, it has been shown that gold nanorods, fabricated with similar dimensions to the tobacco mosaic virus, enhanced MSC osteogenesis and chondrogenesis when functionalized to hyaluronic acid hydrogels due to the effects of the highly ordered nanotopographical environment [122]. Differentiation into MSCs from progenitor cells has also been investigated in response to nanotopography where it was shown that MPCs preferentially differentiated into MSCs on a 400 nm nanograting topography than on flat surfaces [114].
In addition to nanotopographical features, ligand density is a leading factor of nanotopographical regulation. Nanocolloidal features have been used to control the number of integrin-adhesive RGD ligands per unit area; cells on 108 nm centre-centre nanopatterns demonstrated a delayed spreading with repeated protrusion-retraction cycles, compared to cells on a 58 nm centre-centre nanopatterned substrate [123]. Gold nanodots used to create these features were ∼8–12 nm in size-integrins are sized between 8 and 12 nm, therefore the gold nanopattern technique is well matched to these single receptor dimensions [126]. Since the size of a nano gold dot is around 10 nm, they are well suited to connect a single integrin to each nano dot. The features of a gold nanopattern creates the possibility of precisely tailoring the dot spacing to regulate the density of integrins. For example, PEG hydrogels with gold dots coated with RGD ligands were prepared to investigate the MSC response. As mentioned in the adhesion section, nano gold dots with spacings of 37, 53, 77, 87 and 124 nm were investigated on these PEG substrates [110]. It was concluded that a small RGD spacing induces strong FAs assembly and distinct cytoskeleton organization, whereas larger spacing of RGD generates weak FAs assembly and an indistinct cytoskeleton. The critical spacing was thus said to be 70 nm, where less spreading of cells was observed on dots with spacing larger than 70 nm. Both osteogenic and adipogenic inductions resulted in greater extents of differentiation on large nanospaced patterns [110]. This method was also shown to improve maintenance of the chondrocyte phenotype when the nano spacing is above 70 nm [124]. Moreover, corresponding effects of micro-nanopatterns with 46 nm and 95 nm spacings were developed to further explore the stem cell response. This technique allows independent investigation of the ligand spacing on cell spreading and regulating differentiation. It was concluded that the small spacing leads to stronger cell tension even in instances of identical cellular size [125]. Thus, the differentiation of cells is influenced by the tension of the cytoskeleton, which is induced by the ligand density which is directly defined by the nanotopography.
Nanopatterning not only manipulates cellular behaviours by shifting features density, but also affects cellular differentiation and multipotency expression through regulated pattern arrangement. Long term maintenance, growth and prolonged retention of MSCs was achieved on ordered square lattice symmetry nanopatterns with dimensions of 120 nm diameter and 300 nm spacing distance that was generated in polycarbonate [5]. In contrast, a surface arrangement of disordered square array with dots displaced randomly up to 50 nm in x and y axes was shown to promote osteogenesis of MSCs. It was speculated that while cell spreading is permitted by the nanotopographical surface, cells will respond to the nanopatterns through regulation of FAs assembly and cytoskeletal tension which leads to further effects on mechanotransductive pathways [9]. A summary scheme of cell differentiation behaviours on substrates such as glass, PLGA, TiO2 and SiO2 with various nanotopographical features are shown in Fig. 3. All these studies illustrate the significant influence of nanotopography on cellular behaviour. However, the interaction between nanotopography-matrix and, matrix-integrin mechanisms have remained elusive, the detailed interactions of these are illustrated in Section 4.
3.3. YAP/TAZ signalling
Yes-associated protein (YAP) and TAZ are downstream regulatory targets of the Hippo signalling pathway. YAP/TAZ translocate to the nucleus and serve as transcriptional co-activators of specific target genes. Nuclear translocation is regulated by large tumour suppressor kinase (LATS)1/2, which phosphorylates YAP/TAZ to allow binding with 14-3-3 protein that causes cytoplasmic sequestration [127,128]. In the absence of YAP/TAZ phosphorylation, a complex is formed with Scalloped (Sd) to permit nuclear translocation where gene expression is facilitated by YAP/TAZ nuclear interactions with DNA-binding partner TEA domain family member (TEAD) [129,130]. Further, YAP/TAZ translocation is highly mechanosensitive to changes in the physical cues of the cellular microenvironment resulting in rapid on-off mechanotransduction [131]. Mechanical inputs regulate cell mechanics and cytoskeletal dynamics that direct YAP/TAZ mechanotransduction to control many key cell responses including proliferation, differentiation, and stem cell maintenance [[132], [133], [134], [135]].
Amongst the many physical cues presented to cells, nanotopography has been shown to significantly influence YAP/TAZ mechanotransduction which regulates gene expression to control various aspects of cell behaviour. Osteogenic differentiation has been widely investigated in terms of YAP/TAZ regulation on different nanotopographies; rough, patterned and fibrous substrates largely support enhanced osteogenesis through nuclear YAP/TAZ activation [136]. For example, it was shown that YAP/TAZ activation and osteogenesis was optimal on hydroxyapatite discs with a surface roughness of 0.77–1.09 μm and a mean distance between peaks of 53.9-39.3 μm [137]. Additionally, YAP activation of MC3T3-E1 preosteoblast cells was enhanced on PDMS micropatterns with a grid topography which coincided with enhanced osteogenesis [138]. Other studies have shown that specific nanopatterns can enhance osteogenesis via TAZ activation, for instance using human MSCs on substrates with 70 nm and 200 nm features [139,140]. Collectively, these studies also suggest that rougher, patterned and fibrous surfaces promote YAP/TAZ-mediated osteogenesis through enhanced cell adhesion and spreading driven by increased integrin clustering and FA formation; this subsequently facilitates increased actin polymerization and cytoskeletal tension via Rho GTPase, focal adhesion kinase (FAK) and mitogen-activated protein kinase (MAPK) signalling [[137], [138], [139], [140]]. Other studies have demonstrated that integrin clustering and FA formation are closely associated with YAP/TAZ-mediated osteogenesis; for instance, in response to cylindrical vs. ribbon-like nanofiber morphologies, enhanced stem cell osteogenesis was facilitated by integrin β1-based FA complex formations [141]. Similarly, using micropatterned substrates that enabled precise control of human MSC spreading and adhesion, larger cell adhesion areas facilitated YAP/TAZ activation and osteogenesis, whereas smaller cell adhesion areas diminished TAP/TAZ activity and favoured adipogenesis [142]. Interestingly, in contrast to previous studies, it was shown in the presence of cell-cell contacts that autophagy-induced YAP degradation permitted nuclear β-catenin translocation to facilitate osteogenic gene expression of MC3T3-E1 preosteoblast cells on titanium nanotopographies [143]. This suggests that the synergistic influence of nanotopograhic guidance and cell-cell contacts can regulate osteogenesis via an alternative mechanism to conventional nuclear YAP/TAZ activation in other environments.
Additionally, neural cell behaviour can be regulated by YAP/TAZ on specific nanofeatures. For instance, nanograting topographies between 200 and 1000 nm were found to enhance neurite alignment of PC12 cells which coincided with increased sub-cellular localization of YAP [144]. Additionally, it has been shown that motor neuron differentiation of human pluripotent stem cells (hPSCs) can be promoted on 200 nm nanorough surfaces via increased nuclear YAP activation [145].
Evidently, YAP/TAZ plays a key role regulating cell behaviour in response to nanotopographical cues; this could be harnessed in tissue engineering and regenerative medicine by directing cell fate towards desired lineages.
4. Mechanotransduction of nanotopographical cues
Nanotopography can provide the physical cues that cell receptors require to organize the cellular cytoskeleton and to propagate mechanical signals towards the nucleus. Direct mechanotransduction relies on ECM being directly coupled with the nucleus through the cytoskeleton, which is postulated to behave like a mechanical tensegrity structure [146]. In contrast, indirect signalling cues via biochemical mechanotransduction, also dominates the cytoskeleton through regulating focal adhesion behaviour and embedded biochemicals such as FAK interacting with extracellular signal related kinase (ERK 1/2 [147]). Thus, alternations in cytoskeleton arrangement, and cell and nucleus morphology can be achieved by manipulating the conformation of matrix. In this procedure, cellular adhesion behaviour plays an initial but vital role.
4.1. Integrin receptors bind to extracellular matrix
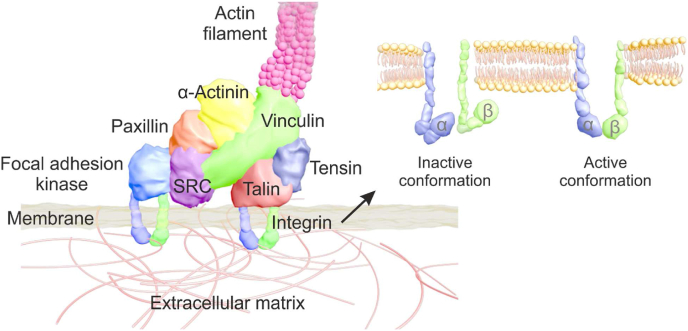
Cell sense the nanotopography during adhesion via thin cell membrane projections, filopodia, that contain integrins. Integrins are essential mechano-sensors when a large fraction of force is transmitted from the ECM to cells [[148], [149], [150]]. Specifically, integrins are transmembrane receptors existing in at least 24 unique combinations of non-covalently interacting α-subunits (18 types) and β-subunits (8 types), containing adhesion complexes and binding to ECM ligands, such as the RGD peptide [149]. Both integrin α-subunits and β-subunits are type I transmembrane proteins which consist of a large number of extracellular domains, a single-pass transmembrane helix, and a short cytoplasmic domain [149]. Binding sites for ECM ligands either comprise epitopes from both subunits or bind to a specific domain of α-subunits. For example, α5β1 and αVβ3 integrins bind to the RGD motif in proteins such as fibronectin and vitronectin [126]. In other cases, the binding site can also reside on specific domains of integrin α-subunits, such as collagen-binding integrins α1β1, α2β1, α10β1 and α11β1 [151]. In a resting cell, integrins exist in a bent, inactive conformation. Then, upon biochemical signals and force activation, these integrins become activated, shifting conformation from ‘bent closed’ (inactive), to ‘extended closed’, and finally to ‘extended open’ (active) [149,152] (Fig. 4). One of the best understood biochemical integrin activation signals is the binding of intracellular adaptor protein talin. Talin is a 270 kDa molecular weight protein that has a key role in the first steps of integrin activation. Upon initial integrin ligation, talin binds to the cytoplasmic tail of the integrin β-subunit, inducing separation of the cytoplasmic domains of integrin α- and β-subunits (opening) and triggers a global conformational change in the extracellular domain [153]. As a result, active integrins have a higher affinity for other ligands and initiate increased adhesion [152].
Fig. 4.
Molecular structure of integrin adhesion complexes (IACs) [159,161] and the inactive/active conformation of integrin [149,152].
The bond between integrins and the ECM is cyclic, rather than permanent, and the lifetime of the bonds will be closely affected by the force, which behaves as a catch bond (or more accurately, a catch-slip bond) [[154], [155], [156]]. The time required to break the bond first increases and then decreases with force. As force keeps on growing, force in individual integrins loads so fast upon binding that integrins become destabilized and disengage before additional integrins can bind [157].
Integrins have no enzymatic activity, but they can recruit and accumulate multiple intracellular proteins at the site of integrin clusters. Such multi-protein structures are termed as integrin adhesion complexes (IACs), which mediate mechanical linkage between the integrin and actin cytoskeleton, and further direct cell spreading and fate [158]. The components of the IACs are highly complex, a large number of proteins have already been demonstrated to be involved in the complex. Recent proteomic analysis on the IACs revealed that over 2000 proteins were identified as IACs components, with a consensus 60 defined as being central to IACs formation [159], encompassing 180 protein-protein interaction nodes [160]. Super-resolution fluorescence microscopy has determined the molecular architecture of IACs down to nanoscale, in which integrins and actin are vertically separated by a 40 nm core region that consists of partially overlapping protein specific layers [161]. Generally, three protein layers assembling the IACs; one is a signalling layer, closest to the plasma membrane, which contains the highly phosphorylated signalling proteins FAK and paxillin. Next, an intermediate force transduction layer, composed of adaptor proteins talin and vinculin, which links the integrin complex to the actomyosin machinery. The third is a distal actin regulatory layer containing zyxin, vasodilator-simulated phosphoprotein (VASP) and α-actinin; these lie up to 60 nm away from the integrin layer [161,162]. The molecular structure of IACs is shown in Fig. 4.
4.2. The bonding between integrins and actin
The formation, disassembly and maturation of IACs is tightly regulated in time and space [163]. Nascent IACs are small and transient, with relatively short lifetimes [164]. This is followed by the formation of focal complexes clustered from nascent IACs. Focal complexes mature into larger, elongated supramolecular complexes called FAs. FAs size is critically important for cell fate. For example, small FAs are featured with ‘dot’ shapes below 1 μm, which are related with cell mobility. Medium sized FAs are in the range of 1–5 μm and matured FAs can reach more than 5 μm, and are associated with stability and tension of cytoskeleton [165]. FAs can themselves further mature into fibrillar adhesions, which support ECM synthesis and remodelling [166]. The adhesion maturation process is closely regulated by the force. The general view is that initial, nascent adhesions form independently of force and then mature in response to force applied either internally by actin structures coupled to integrins at nascent adhesions, or by any of the external factors through the ECM [149,156,167,168]. Once force is applied, adhesions grow and alter their molecular composition. Concomitantly, both adhesions and associated actin fibres align in the direction of force application [149,[169], [170], [171]]. As a result, integrin adhesion maturation induces downstream cell responses involving the recruitment and activation of signalling proteins such as FAK [172], paxillin [173], proto-oncogene tyrosine-protein kinases (SRC) [174] or extracellular signal-related kinases (ERK) [175]. Mature FAs also lead to the enhancement of actin polymerization and the formation of actin stress fibres, which could further connect with mechanotransduction signalling pathways [176].
The adaptor proteins within the force transduction layer of IACs, especially talin and vinculin, have been shown to be involved in mechanosensitive events and are critical for adhesion maturation. It has already been demonstrated that talin tethering spans the whole layer of the IACs structure [161]. The head of talin binds to the cytoplasmic tail of the integrin β-subunit and the rod domain of talin harbours multiple actin-binding and vinculin-binding sites, which mediates the bond between the actin and integrins. As a consequence, integrin–talin–actin axis complexes are formed and serve as mechanical linkages [161] (Fig. 4). Many functional binding sites of talin are mechanosensitive and only become exposed under tensile forces [153]. Once forces are transmitted and sustained from integrins, normally of the order of a few piconewtons, talin shifts to an unfolded conformation and exposes its binding sites, which allows vinculin and actin binding to stabilize talin unfolding; consequently this leads to the strengthening of the adhesion and eventually adhesion maturation [156,177]. It should be noted that the talin unfolding needs time to be completed; talin unfolding is faster when the rate of the force is built up by pulling coming from both sides of the protein (actin filaments and integrins) [178]. Talin unfolding responds to force according to the Bell model [179] as a classical slip bond. That is, when a constant force is applied to a single talin molecule, the time required to unfold decreases exponentially with force [180].
4.3. Molecular clutch model
As aforementioned, integrins are receptors on the cell membrane and their behaviour is significantly influenced by the cellular mechanotransductive pathways which further effects cell behaviours, however the interactive mechanism is still elusive. This interactive mechanism is important for manipulating cells through designing nanotopographies; thus, the integrin receptor has been individually studied. Specifically, several receptors are involved in the interactions between cells and nanotopographical features, and the cellular interactive mechanisms have also been well investigated. For instance, regarding integrin behaviours, the complex responses may be explained by the ‘molecular clutch’ mechanism, where the integrin binding complexes mechanically couple to the force-generating actomyosin system. The adjacent integrins are recruited to respond to force loading until a maximum of integrin recruitment is reached which leads to the collapse of adhesion [88]. Collapse of FAs was observed in cells on a rigid gel (150 kPa) with 100 nm spacing RGD-nanodots; this indicates a general framework for cell sensing of the spatial and physical information at nanoscale. Taken together, the cell mechanosensitive events can be well described as a molecular clutch model, which combines the integrin unbinding catch-bond model and talin unfolding slip-bond model [88,156]. Within this model, the link between actin and an ECM ligand is termed the molecular clutch, which can be bound (engaged) or unbound repeatedly. To engage the clutch, talin unfolding has to occur before integrins unbind from the ECM (this happens with a characteristic biochemical lifetime) [88]. On the other hand, if integrin unbinding (from the ECM) happens before talin unfolding, the clutch cannot engage, and the adhesion is not stabilized [88,178]. Based on this, the clutch model can explain cell response to ECM ligand density and spatial distribution [43]. Increasing density or reducing distance of ECM ligands could increase the number of bound integrins, leading each of them to experience a lower fraction of the force applied either internally by actin structures coupled to integrins at nascent adhesions or by external factors through the ECM [149]. Consequently, the time needed to build up force to unfold talin is high; as a result, integrins unbind from the ECM before talin is unfolded and the clutch does not engage. Reducing density or increased distance of ECM ligands could facilitate the reaching of the threshold of force sensing by integrins, promoting the force-mediated effects. Consequently, integrin adhesions are more prone to grow and induce downstream signalling in response to force loading. When the ligand density reducing or the distance increasing, force is built up faster; this leads to talin unfolding and stabilization by binding to vinculin (i.e., the adhesion grips). The clutch is engaged and stabilized and cell adhesion to the ECM is secured [156]. Of note, if ligands are spaced very far apart (and therefore the force per integrin is extremely high), adhesion growth no longer occurs. That is due to a yet to be characterized mechanism, that high contractile forces dissociate integrin-RGD bonds so fast that integrins become destabilized and disengage before additional integrins can bind [88,104].
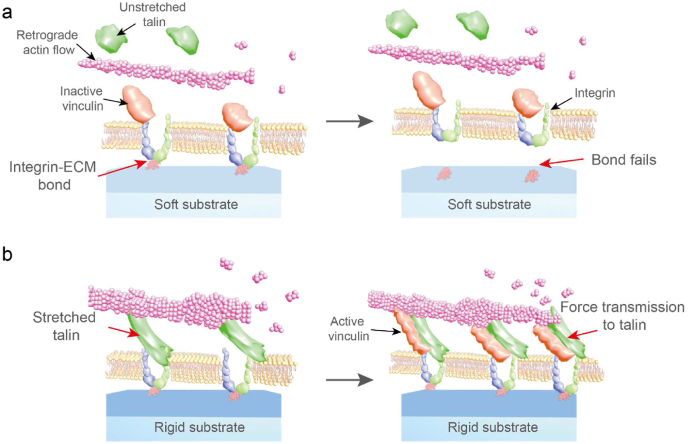
In terms of mathematical modelling of molecular clutch simulation, Roca-Cusachs and colleagues have applied the clutch model to show the agreement of prediction and experimental investigation on a monotonic increase in traction force in response to increasing ECM stiffness [157]. On low-stiffness ECM, the force loading rate on the clutch is slower than the integrin-ECM bond lifetime, and the integrin-ECM bond fails before any force loading can be transmitted to talin (Fig. 5a). In contrast, increasing the rigidity of substrate (>5 kPa) leads to faster clutch loading, when the rigidity threshold is reached (Fig. 5b), the clutch becomes faster than the integrin-ECM lifetime, thereby resulting in force transmitting to talin. Leading to talin unfolding, and binding of vinculin and FAs formation [157,181].
Fig. 5.
The molecular clutch model. a, molecular clutch on soft substrate, clutch is slower than the lifetime of integrin-ECM bond causing bond failure. b, molecular clutch on rigid substrate, clutch is faster than the lifetime of integrin-ECM bond leads to force transmission and unfolding of talin, leading to stabilization of the adhesion [157].
4.4. Direct physical cue: tensegrity structure
Nanotopography also affects cellular mechanotransduction by direct physical cues. The tensegrity theory offers an explanation of how the cellular cytoskeleton could help cells respond as mechanical units [182]. In tensegrity structures, tensional and compressive forces act in unison to maintain the shape of the structure, providing strength and resilience at the same time. This indicates cells are maintained in equilibrium under a balance of forces [183]. When receptors, such as integrins, are pulled by micromanipulation of integrin bound microbeads or micropipettes, the cytoskeletal filaments reorient, nuclei distort and the nucleoli redistribute along the axis of an applied tension field [184,185]. Tensegrity allows a model whereby such forces can be delivered from the ECM to the nucleus (and beyond) through an integrated cytoskeleton. Several models have been established to explain how different orientations based on cytoskeletal stress and strains can convey forces via tensegrity structures. For instance, a simple theoretical model has predicted the dynamics and orientation of cells in both the absence and presence of applied stresses [186]. In this theory, cells are modelled as elastic force dipoles that can change their contractile activity and orientation by reorganizing the FAs and stress fibres in response to external forces. It is noted that this reorganization only occurs if the temporal variation of the force is slower than the time required for FAs and stress fibre reformation [186]. Another substrate strain model explains the relationship between actin cytoskeletal reorganization and substrate deformation applied to cells. Various hypotheses assumed that the normal substrate strain, rather than shear substrate strain, determines the actin cytoskeleton reorganization. Normal substrate strain is transmitted to individual actin filaments, each of the actin filaments has a basal strain energy when the cell adheres to the substrate without stretching, the actin filaments undergo disassembly when their strain energies are decreased to zero or increased to twice that of their basal strain energy. Based on those assumptions, the model predicted that the actin filaments are formed in a direction where their basal strain energy are minimally altered [187], helping explain the difference in cell orientation. In addition, FAs can significantly affect cell orientation. It has been demonstrated that an adhesion cluster is prone to losing its stability under high-frequency loading; this is due to the receptors and ligands having insufficient contact time to form stable bonds because of high speed deformation of the substrate [188]. Furthermore, a cytoskeletal tensegrity planar model of cell reorientation on a substrate under biaxial cyclic stretch was proposed. It was shown that not only the longitudinal stress fibres, but also the lateral actin network also plays a crucial role in cell orientation [189].
5. Conclusion and outlook
The topographical design of biomaterials has been developed since the first studies of cellular contact guidance and has continued to be investigated over the following decades [10,13,20,47,[190], [191], [192]]. Nanotopographical regulation of cell behaviour has been widely employed, however the underlying mechanisms remain elusive. It has been well documented that the nanofeatures manipulate the cell responses through direct physical cues, such as influencing the cytoskeletal structures within in a cell to apply forces upon the nucleus, or indirectly regulating mechanosensitive pathways through integrin receptor linked biochemistry on the cell surface. It is well understood that cells interact with nanotopography through a layer of ECM, and understanding how proteins adsorb onto these substrates is of critical importance to understanding cell responses. However, due to adsorbing characteristics of real time and interfacial features, it is unfeasible to directly measure the interactive mechanisms of protein adsorption. Thus, many of the physical, but fundamental, models have been established based on several hypotheses to explain and predict the adsorption. It has been widely accepted that the dominating interactions can be divided into two: attractive Coulomb's force, electrostatic attraction and Van-der-Waals non-electrostatic interactions [45,54]. It is important to dissect those interactive mechanisms under complex preconditions that can simulate protein adsorption onto nanotopographical surfaces, and predict the conformation of anchored proteins. Those mechanism-focused studies can help to further reveal the cell-nanotopography interactions so that they can be utilized to harness desired cell responses by fabricating appropriate substrates. In terms of nanotopography clinical applications, there are still several barriers that need to be overcome. For example, the infection of orthopaedic implants caused by bacteria is one of the major concerns; however, the bacterial colonization-nanotopography interaction is unclear [193]. It has been reported that transmucosal higher surface roughness/surface free energy facilitates the bacterial biofilm formation; also, the presence of topographical features caused difficulties in sterilisation [194,195]. Sterilisation is also a challenge since it may affect the hydrophobicity and electrical charging of the nanotopographical implant surface [196,197].
In summary, more efforts should be made in nanotopography research, from both fundamental interactive mechanisms and translational medicine perspectives. In this review, we provided a comprehensive discussion on fundamental cell and protein interactions. This review may enlighten readers with designing nanotopographical features to instruct cell behaviours for next generation biomedical implants.
Declaration of competing interest
All authors have given approval to the final version of the manuscript, and have no conflict of interest.
Acknowledgement
We acknowledge support from the Leverhulme Trust through grant RPG-2019-252 and the Engineering and Physical Sciences Research Council (EPSRC) grant EP/P001114/1.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Jiajun Luo, Email: jiajun.luo@glasgow.ac.uk.
Manuel Salmeron-Sanchez, Email: manuel.salmeron-sanchez@glasgow.ac.uk.
References
- 1.Dalby M.J., García A.J., Salmeron-Sanchez M. Receptor control in mesenchymal stem cell engineering. Nat. Rev. Mater. 2018;3 [Google Scholar]
- 2.Benoit D.S., Schwartz M.P., Durney A.R., Anseth K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat. Mater. 2008;7(10):816. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.McMurray R.J., Gadegaard N., Tsimbouri P.M., Burgess K.V., McNamara L.E., Tare R., Murawski K., Kingham E., Oreffo R.O., Dalby M.J. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat. Mater. 2011;10(8):637. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 6.Curran J.M., Chen R., Hunt J.A. The guidance of human mesenchymal stem cell differentiation in vitro by controlled modifications to the cell substrate. Biomaterials. 2006;27(27):4783–4793. doi: 10.1016/j.biomaterials.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Wen J.H., Vincent L.G., Fuhrmann A., Choi Y.S., Hribar K.C., Taylor-Weiner H., Chen S., Engler A.J. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 2014;13(10):979. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trappmann B., Gautrot J.E., Connelly J.T., Strange D.G., Li Y., Oyen M.L., Stuart M.A.C., Boehm H., Li B., Vogel V. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012;11(7):642. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 9.Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D., Oreffo R.O. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 10.Dalby M.J., Gadegaard N., Oreffo R.O. Harnessing nanotopography and integrin–matrix interactions to influence stem cell fate. Nat. Mater. 2014;13(6):558. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 11.Desai T.A. Micro-and nanoscale structures for tissue engineering constructs. Med. Eng. Phys. 2000;22(9):595–606. doi: 10.1016/s1350-4533(00)00087-4. [DOI] [PubMed] [Google Scholar]
- 12.Curtis A., Varde M. Control of cell behavior: topological factors. J. Natl. Cancer Inst. 1964;33(1):15–26. [PubMed] [Google Scholar]
- 13.Clark P., Connolly P., Curtis A., Dow J., Wilkinson C. Cell guidance by ultrafine topography in vitro. J. Cell Sci. 1991;99(1):73–77. doi: 10.1242/jcs.99.1.73. [DOI] [PubMed] [Google Scholar]
- 14.Curtis A. The mechanism of adhesion of cells to glass A study by interference reflection microscopy. J. Cell Biol. 1964;20(2):199–215. doi: 10.1083/jcb.20.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J.Y., Davies J.E. Red blood cell and platelet interactions with titanium implant surfaces. Clin. Oral Implants Res. 2000;11(6):530–539. doi: 10.1034/j.1600-0501.2000.011006530.x. [DOI] [PubMed] [Google Scholar]
- 16.Tan K.S., Qian L., Rosado R., Flood P.M., Cooper L.F. The role of titanium surface topography on J774A. 1 macrophage inflammatory cytokines and nitric oxide production. Biomaterials. 2006;27(30):5170–5177. doi: 10.1016/j.biomaterials.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Linderbäck P., Harmankaya N., Askendal A., Areva S., Lausmaa J., Tengvall P. The effect of heat-or ultra violet ozone-treatment of titanium on complement deposition from human blood plasma. Biomaterials. 2010;31(18):4795–4801. doi: 10.1016/j.biomaterials.2010.02.060. [DOI] [PubMed] [Google Scholar]
- 18.Masaki C., Schneider G.B., Zaharias R., Seabold D., Stanford C. Effects of implant surface microtopography on osteoblast gene expression. Clin. Oral Implants Res. 2005;16(6):650–656. doi: 10.1111/j.1600-0501.2005.01170.x. [DOI] [PubMed] [Google Scholar]
- 19.Dalby M.J., Biggs M.J., Gadegaard N., Kalna G., Wilkinson C.D., Curtis A.S. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell. Biochem. 2007;100(2):326–338. doi: 10.1002/jcb.21058. [DOI] [PubMed] [Google Scholar]
- 20.Yim E.K., Darling E.M., Kulangara K., Guilak F., Leong K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010;31(6):1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allan C., Ker A., Smith C.-A., Tsimbouri P.M., Borsoi J., O'Neill S., Gadegaard N., Dalby M.J., Dominic Meek R. Osteoblast response to disordered nanotopography. J. Tissue Eng. 2018;9 doi: 10.1177/2041731418784098. 2041731418784098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy P., Berger S., Schmuki P. TiO2 nanotubes: synthesis and applications. Angew. Chem. Int. Ed. 2011;50(13):2904–2939. doi: 10.1002/anie.201001374. [DOI] [PubMed] [Google Scholar]
- 23.Luo J., Li B., Ajami S., Ma S., Zhou F., Liu C. Growth of TiO2 nanotube on titanium substrate to enhance its biotribological performance and biocorrosion resistance. J. Bionic Eng. 2019;16(6):1039–1051. [Google Scholar]
- 24.Luo Jiajun, et al. Improving the fretting biocorrosion of Ti6Al4V alloy bone screw by decorating structure optimised TiO2 nanotubes layer. J. Mater. Sci. Technol. 2020;49:47–55. [Google Scholar]
- 25.Hoshiba T., Chen G., Endo C., Maruyama H., Wakui M., Nemoto E., Kawazoe N., Tanaka M. Decellularized extracellular matrix as an in vitro model to study the comprehensive roles of the ECM in stem cell differentiation. Stem Cells Int. 2016. 2016 doi: 10.1155/2016/6397820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankov R., Yamada K.M. Fibronectin at a glance. J. Cell Sci. 2002;115(20):3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 27.Hsiao C.-T., Cheng H.-W., Huang C.-M., Li H.-R., Ou M.-H., Huang J.-R., Khoo K.-H., Yu H.W., Chen Y.-Q., Wang Y.-K. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget. 2017;8(41) doi: 10.18632/oncotarget.19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoulders M.D., Raines R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hytönen V.P., Wehrle-Haller B. Protein conformation as a regulator of cell–matrix adhesion. Phys. Chem. Chem. Phys. 2014;16(14):6342–6357. doi: 10.1039/c3cp54884h. [DOI] [PubMed] [Google Scholar]
- 30.Guilak F., Cohen D.M., Estes B.T., Gimble J.M., Liedtke W., Chen C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehnert M., Gorbahn M., Rosin C., Klein M., Köper I., Al-Nawas B., Knoll W., Veith M. Adsorption and conformation behavior of biotinylated fibronectin on streptavidin-modified TiOX surfaces studied by SPR and AFM. Langmuir. 2011;27(12):7743–7751. doi: 10.1021/la200908h. [DOI] [PubMed] [Google Scholar]
- 32.Lin H., Lal R., Clegg D.O. Imaging and mapping heparin-binding sites on single fibronectin molecules with atomic force microscopy. Biochemistry. 2000;39(12):3192–3196. doi: 10.1021/bi991624o. [DOI] [PubMed] [Google Scholar]
- 33.Keselowsky B.G., Collard D.M., García A.J. Surface chemistry modulates fibronectin conformation and directs integrin binding and specificity to control cell adhesion. J. Biomed. Mater. Res. Part A: An Official Journal of The Society for Biomaterials, The Japanese Society for Biomaterials, and The Australian Society for Biomaterials and the Korean Society for Biomaterials. 2003;66(2):247–259. doi: 10.1002/jbm.a.10537. [DOI] [PubMed] [Google Scholar]
- 34.García A.J., Vega M.a.D., Boettiger D. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell. 1999;10(3):785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergkvist M., Carlsson J., Oscarsson S. Surface‐dependent conformations of human plasma fibronectin adsorbed to silica, mica, and hydrophobic surfaces, studied with use of Atomic Force Microscopy. J. Biomed. Mater. Res. Part A: An Official Journal of The Society for Biomaterials, The Japanese Society for Biomaterials, and The Australian Society for Biomaterials and the Korean Society for Biomaterials. 2003;64(2):349–356. doi: 10.1002/jbm.a.10423. [DOI] [PubMed] [Google Scholar]
- 36.Sousa S., Brás M.M., Moradas-Ferreira P., Barbosa M. Dynamics of fibronectin adsorption on TiO2 surfaces. Langmuir. 2007;23(13):7046–7054. doi: 10.1021/la062956e. [DOI] [PubMed] [Google Scholar]
- 37.Gumbiner B.M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84(3):345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 38.Wilson C.J., Clegg R.E., Leavesley D.I., Pearcy M.J. Mediation of biomaterial–cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11(1–2):1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 39.Hersel U., Dahmen C., Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 40.Grinnell F. Focal adhesion sites and the removal of substratum-bound fibronectin. J. Cell Biol. 1986;103(6):2697–2706. doi: 10.1083/jcb.103.6.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia A.J. Springer; 2006. Interfaces to Control Cell-Biomaterial Adhesive Interactions, Polymers for Regenerative Medicine; pp. 171–190. [Google Scholar]
- 42.Salmerón-Sánchez M., Altankov G. Cell-protein-material interaction in tissue engineering. Tissue Eng. 2010:77–103. [Google Scholar]
- 43.Anselme K., Ponche A., Bigerelle M. Relative influence of surface topography and surface chemistry on cell response to bone implant materials. Part 2: biological aspects. Proc. IME H J. Eng. Med. 2010;224(12):1487–1507. doi: 10.1243/09544119JEIM901. [DOI] [PubMed] [Google Scholar]
- 44.Ballet T., Boulange L., Brechet Y., Bruckert F., Weidenhaupt M. Protein conformational changes induced by adsorption onto material surfaces: an important issue for biomedical applications of material science, Bulletin of the Polish Academy of Sciences. Tech. Sci. 2010;58(2):303–315. [Google Scholar]
- 45.Rabe M., Verdes D., Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011;162(1–2):87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Rico P., Cantini M., Altankov G., Salmerón-Sánchez M. 2015. Matrix–protein Interactions with Synthetic Surfaces; pp. 91–146. [Google Scholar]
- 47.González-García C., Sousa S.R., Moratal D., Rico P., Salmerón-Sánchez M. Effect of nanoscale topography on fibronectin adsorption, focal adhesion size and matrix organisation. Colloids Surf. B Biointerfaces. 2010;77(2):181–190. doi: 10.1016/j.colsurfb.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Dolatshahi-Pirouz A., Jensen T., Kraft D.C., Foss M., Kingshott P., Hansen J.L., Larsen A.N., Chevallier J., Besenbacher F. Fibronectin adsorption, cell adhesion, and proliferation on nanostructured tantalum surfaces. ACS Nano. 2010;4(5):2874–2882. doi: 10.1021/nn9017872. [DOI] [PubMed] [Google Scholar]
- 49.Madsen C.D., Sidenius N. The interaction between urokinase receptor and vitronectin in cell adhesion and signalling. Eur. J. Cell Biol. 2008;87(8–9):617–629. doi: 10.1016/j.ejcb.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Webster T.J., Schadler L.S., Siegel R.W., Bizios R. Mechanisms of enhanced osteoblast adhesion on nanophase alumina involve vitronectin. Tissue Eng. 2001;7(3):291–301. doi: 10.1089/10763270152044152. [DOI] [PubMed] [Google Scholar]
- 51.Mosesson M. Fibrinogen and fibrin structure and functions. J. Thromb. Haemostasis. 2005;3(8):1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 52.Brinckmann J. Springer; 2005. Collagens at a Glance, Collagen; pp. 1–6. [Google Scholar]
- 53.Durbeej M. Laminins, Cell and tissue research. 2010;339(1):259. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- 54.Norde W. Macromolecular Symposia, Wiley Online Library; 1996. Driving Forces for Protein Adsorption at Solid Surfaces; pp. 5–18. [Google Scholar]
- 55.Sigal G.B., Mrksich M., Whitesides G.M. Effect of surface wettability on the adsorption of proteins and detergents. J. Am. Chem. Soc. 1998;120(14):3464–3473. [Google Scholar]
- 56.Demanèche S., Chapel J.-P., Monrozier L.J., Quiquampoix H. Dissimilar pH-dependent adsorption features of bovine serum albumin and α-chymotrypsin on mica probed by AFM. Colloids Surf. B Biointerfaces. 2009;70(2):226–231. doi: 10.1016/j.colsurfb.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 57.Jones K.L., O'Melia C.R. Protein and humic acid adsorption onto hydrophilic membrane surfaces: effects of pH and ionic strength. J. Membr. Sci. 2000;165(1):31–46. [Google Scholar]
- 58.Langmuir I. The constitution and fundamental properties of solids and liquids. Part I. Solids. J. Am. Chem. Soc. 1916;38(11):2221–2295. [Google Scholar]
- 59.Freundlich H. Over the adsorption in solution. Phys. Chem. 1906;57(385471):1100–1107. [Google Scholar]
- 60.Brunauer S., Emmett P.H., Teller E. Adsorption of gases in multimolecular layers. J. Am. Chem. Soc. 1938;60(2):309–319. [Google Scholar]
- 61.Wei Y., Thyparambil A.A., Latour R.A. Quantification of the influence of protein-protein interactions on adsorbed protein structure and bioactivity. Colloids Surf. B Biointerfaces. 2013;110:363–371. doi: 10.1016/j.colsurfb.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia X.R., Monteiro-Riviere N.A., Mathur S., Song X., Xiao L., Oldenberg S.J., Fadeel B., Riviere J.E. Mapping the surface adsorption forces of nanomaterials in biological systems. ACS Nano. 2011;5(11):9074–9081. doi: 10.1021/nn203303c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsapikouni T.S., Missirlis Y.F. Protein–material interactions: from micro-to-nano scale. Mater. Sci. Eng., B. 2008;152(1–3):2–7. [Google Scholar]
- 64.Pasche S., Vörös J., Griesser H.J., Spencer N.D., Textor M. Effects of ionic strength and surface charge on protein adsorption at PEGylated surfaces. J. Phys. Chem. B. 2005;109(37):17545–17552. doi: 10.1021/jp050431+. [DOI] [PubMed] [Google Scholar]
- 65.Leermakers F., Ballauff M., Borisov O. On the mechanism of uptake of globular proteins by polyelectrolyte brushes: a two-gradient self-consistent field analysis. Langmuir. 2007;23(7):3937–3946. doi: 10.1021/la0632777. [DOI] [PubMed] [Google Scholar]
- 66.Wittemann A., Ballauff M. Interaction of proteins with linear polyelectrolytes and spherical polyelectrolyte brushes in aqueous solution. Phys. Chem. Chem. Phys. 2006;8(45):5269–5275. doi: 10.1039/b609879g. [DOI] [PubMed] [Google Scholar]
- 67.Huang H.-H., Wu C.-P., Sun Y.-S., Yang W.-E., Lee T.-H. Surface nanotopography of an anodized Ti–6Al–7Nb alloy enhances cell growth. J. Alloys Compd. 2014;615:S648–S654. [Google Scholar]
- 68.Yang Y., Knust S., Schwiderek S., Qin Q., Yun Q., Grundmeier G., Keller A. Protein adsorption at nanorough titanium oxide surfaces: the importance of surface statistical parameters beyond surface roughness. Nanomaterials. 2021;11(2):357. doi: 10.3390/nano11020357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Y., Yu M., Böke F., Qin Q., Hübner R., Knust S., Schwiderek S., Grundmeier G., Fischer H., Keller A. Effect of nanoscale surface topography on the adsorption of globular proteins. Appl. Surf. Sci. 2021;535 [Google Scholar]
- 70.Huang H.-H., Wu C.-P., Sun Y.-S., Yang W.-E., Lin M.-C., Lee T.-H. Surface nanoporosity of β-type Ti–25Nb–25Zr alloy for the enhancement of protein adsorption and cell response. Surf. Coating. Technol. 2014;259:206–212. [Google Scholar]
- 71.Li K., Liu S., Hu T., Razanau I., Wu X., Ao H., Huang L., Xie Y., Zheng X. Optimized nanointerface engineering of micro/nanostructured titanium implants to enhance cell–nanotopography interactions and osseointegration. ACS Biomater. Sci. Eng. 2020;6(2):969–983. doi: 10.1021/acsbiomaterials.9b01717. [DOI] [PubMed] [Google Scholar]
- 72.Cai K., Bossert J., Jandt K.D. Does the nanometre scale topography of titanium influence protein adsorption and cell proliferation? Colloids Surf. B Biointerfaces. 2006;49(2):136–144. doi: 10.1016/j.colsurfb.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 73.Sela M.N., Badihi L., Rosen G., Steinberg D., Kohavi D. Adsorption of human plasma proteins to modified titanium surfaces. Clin. Oral Implants Res. 2007;18(5):630–638. doi: 10.1111/j.1600-0501.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- 74.Rechendorff K., Hovgaard M.B., Foss M., Zhdanov V., Besenbacher F. Enhancement of protein adsorption induced by surface roughness. Langmuir. 2006;22(26):10885–10888. doi: 10.1021/la0621923. [DOI] [PubMed] [Google Scholar]
- 75.Lord M., Cousins B., Doherty P., Whitelock J., Simmons A., Williams R., Milthorpe B. The effect of silica nanoparticulate coatings on serum protein adsorption and cellular response. Biomaterials. 2006;27(28):4856–4862. doi: 10.1016/j.biomaterials.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 76.Giacomelli C.E., Esplandiú M.J., Ortiz P.I., Avena M.J., De Pauli C.P. Ellipsometric study of bovine serum albumin adsorbed onto Ti/TiO2 electrodes. J. Colloid Interface Sci. 1999;218(2):404–411. doi: 10.1006/jcis.1999.6434. [DOI] [PubMed] [Google Scholar]
- 77.Baujard-Lamotte L., Noinville S., Goubard F., Marque P., Pauthe E. Kinetics of conformational changes of fibronectin adsorbed onto model surfaces. Colloids Surf. B Biointerfaces. 2008;63(1):129–137. doi: 10.1016/j.colsurfb.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 78.Campillo-Fernández A.J., Unger R.E., Peters K., Halstenberg S., Santos M., Sánchez M.S., Dueñas J.M.M., Pradas M.M., Ribelles J.L.G., Kirkpatrick C.J. Analysis of the biological response of endothelial and fibroblast cells cultured on synthetic scaffolds with various hydrophilic/hydrophobic ratios: influence of fibronectin adsorption and conformation. Tissue Eng. 2009;15(6):1331–1341. doi: 10.1089/ten.tea.2008.0146. [DOI] [PubMed] [Google Scholar]
- 79.Rapuano B.E., MacDonald D.E. Surface oxide net charge of a titanium alloy: modulation of fibronectin-activated attachment and spreading of osteogenic cells. Colloids Surf. B Biointerfaces. 2011;82(1):95–103. doi: 10.1016/j.colsurfb.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carravetta V., Monti S. Peptide− TiO2 surface interaction in solution by ab initio and molecular dynamics simulations. J. Phys. Chem. B. 2006;110(12):6160–6169. doi: 10.1021/jp056760j. [DOI] [PubMed] [Google Scholar]
- 81.Monti S., Alderighi M., Duce C., Solaro R., Tiné M.R. Adsorption of ionic peptides on inorganic supports. J. Phys. Chem. C. 2009;113(6):2433–2442. [Google Scholar]
- 82.Butt H.-J., Graf K., Kappl M. John Wiley & Sons; 2013. Physics and Chemistry of Interfaces. [Google Scholar]
- 83.Monsees T.K., Barth K., Tippelt S., Heidel K., Gorbunov A., Pompe W., Funk R.H.W. Effects of different titanium alloys and nanosize surface patterning on adhesion, differentiation, and orientation of osteoblast-like cells. Cells Tissues Organs. 2005;180(2):81–95. doi: 10.1159/000086749. [DOI] [PubMed] [Google Scholar]
- 84.Gongadze E., Kabaso D., Bauer S., Slivnik T., Schmuki P., van Rienen U., Iglic A. Adhesion of osteoblasts to a nanorough titanium implant surface. Int. J. Nanomed. 2011;6:1801–1816. doi: 10.2147/IJN.S21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kabaso D., Gongadze E., Perutková Š., Matschegewski C., Kralj-Iglič V., Beck U., van Rienen U., Iglič A. Mechanics and electrostatics of the interactions between osteoblasts and titanium surface. Comput. Methods Biomech. Biomed. Eng. 2011;14(5):469–482. doi: 10.1080/10255842.2010.534986. [DOI] [PubMed] [Google Scholar]
- 86.Heath M.D., Henderson B., Perkin S. Ion-specific effects on the interaction between fibronectin and negatively charged mica surfaces. Langmuir. 2010;26(8):5304–5308. doi: 10.1021/la100678n. [DOI] [PubMed] [Google Scholar]
- 87.Yang Y., Wang K., Gu X., Leong K.W. Biophysical regulation of cell behavior—cross talk between substrate stiffness and nanotopography. Engineering. 2017;3(1):36–54. doi: 10.1016/J.ENG.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oria R., Wiegand T., Escribano J., Elosegui-Artola A., Uriarte J.J., Moreno-Pulido C., Platzman I., Delcanale P., Albertazzi L., Navajas D. Force loading explains spatial sensing of ligands by cells. Nature. 2017;552(7684):219–224. doi: 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- 89.Fu J., Wang Y.-K., Yang M.T., Desai R.A., Yu X., Liu Z., Chen C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7(9):733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geetha M., Singh A.K., Asokamani R., Gogia A.K. Ti based biomaterials, the ultimate choice for orthopaedic implants - a review. Prog. Mater. Sci. 2009;54(3):397–425. [Google Scholar]
- 91.Bozic K.J., Kurtz S.M., Lau E., Ong K., Chiu V., Vail T.P., Rubash H.E., Berry D.J. The epidemiology of revision total knee arthroplasty in the United States. Clin. Orthop. Relat. Res. 2010;468(1):45–51. doi: 10.1007/s11999-009-0945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bozic K.J., Kurtz S.M., Lau E., Ong K., Vail T.P., Berry D.J. The epidemiology of revision total hip arthroplasty in the United States. J Bone Joint Surg Am. 2009;91A(1):128–133. doi: 10.2106/JBJS.H.00155. [DOI] [PubMed] [Google Scholar]
- 93.Sundfeldt M., Carlsson L.V., Johansson C.B., Thomsen P., Gretzer C. Aseptic loosening, not only a question of wear: a review of different theories. Acta Orthop. 2006;77(2):177–197. doi: 10.1080/17453670610045902. [DOI] [PubMed] [Google Scholar]
- 94.Raphel J., Holodniy M., Goodman S.B., Heilshorn S.C. Multifunctional coatings to simultaneously promote osseointegration and prevent infection of orthopaedic implants. Biomaterials. 2016;84:301–314. doi: 10.1016/j.biomaterials.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah F.A., Thomsen P., Palmquist A. Osseointegration and current interpretations of the bone-implant interface. Acta Biomater. 2019;84:1–15. doi: 10.1016/j.actbio.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 96.Chen S., Guo Y., Liu R., Wu S., Fang J., Huang B., Li Z., Chen Z., Chen Z. Tuning surface properties of bone biomaterials to manipulate osteoblastic cell adhesion and the signaling pathways for the enhancement of early osseointegration. Colloids Surf. B Biointerfaces. 2018;164:58–69. doi: 10.1016/j.colsurfb.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 97.Su E., Justin D., Pratt C., Sarin V., Nguyen V., Oh S., Jin S. Effects of titanium nanotubes on the osseointegration, cell differentiation, mineralisation and antibacterial properties of orthopaedic implant surfaces. Bone Joint J. 2018;100(1_Supple_A):9–16. doi: 10.1302/0301-620X.100B1.BJJ-2017-0551.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yim E.K., Reano R.M., Pang S.W., Yee A.F., Chen C.S., Leong K.W. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26(26):5405–5413. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu J., Li J., Wang S., Ma B., Zhang S., Guo W., Zhang X., Tang W., Sang Y., Liu H. TiO2 nanorod array constructed nanotopography for regulation of mesenchymal stem cells fate and the realization of location‐committed stem cell differentiation. Small. 2016;12(13):1770–1778. doi: 10.1002/smll.201503946. [DOI] [PubMed] [Google Scholar]
- 100.Liu B., Aydil E.S. Growth of oriented single-crystalline rutile TiO2 nanorods on transparent conducting substrates for dye-sensitized solar cells. J. Am. Chem. Soc. 2009;131(11):3985–3990. doi: 10.1021/ja8078972. [DOI] [PubMed] [Google Scholar]
- 101.Zhou Q., Castañeda Ocampo O., Guimarães C.F., Kühn P.T., van Kooten T.G., van Rijn P.J.A.a.m. interfaces, Screening platform for cell contact guidance based on inorganic biomaterial micro/nanotopographical gradients. ACS Appl. Mater. Interface. 2017;9(37):31433–31445. doi: 10.1021/acsami.7b08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen W., Villa-Diaz L.G., Sun Y., Weng S., Kim J.K., Lam R.H., Han L., Fan R., Krebsbach P.H., Fu J. Nanotopography influences adhesion, spreading, and self-renewal of human embryonic stem cells. ACS Nano. 2012;6(5):4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gadegaard N., Thoms S., Macintyre D., Mcghee K., Gallagher J., Casey B., Wilkinson C. Arrays of nano-dots for cellular engineering. Microelectron. Eng. 2003;67:162–168. [Google Scholar]
- 104.Schvartzman M., Palma M., Sable J., Abramson J., Hu X., Sheetz M.P., Wind S.J. Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 2011;11(3):1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McNamara L.E., Sjöström T., Seunarine K., Meek R.D., Su B., Dalby M.J. Investigation of the limits of nanoscale filopodial interactions. J. Tissue Eng. 2014;5 doi: 10.1177/2041731414536177. 2041731414536177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gulati K., Moon H.-J., Li T., Kumar P.S., Ivanovski S. Titania nanopores with dual micro-/nano-topography for selective cellular bioactivity. Mater. Sci. Eng. C. 2018;91:624–630. doi: 10.1016/j.msec.2018.05.075. [DOI] [PubMed] [Google Scholar]
- 107.Roy P., Berger S., Schmuki P. TiO2 nanotubes: synthesis and applications. Angew. Chem. Int. Ed. 2011;50(13):2904–2939. doi: 10.1002/anie.201001374. [DOI] [PubMed] [Google Scholar]
- 108.Oh S., Daraio C., Chen L.H., Pisanic T.R., Finones R.R., Jin S. Significantly accelerated osteoblast cell growth on aligned TiO2 nanotubes. J. Biomed. Mater. Res. 2006;78A(1):97–103. doi: 10.1002/jbm.a.30722. [DOI] [PubMed] [Google Scholar]
- 109.Huang J., Grater S.V., Corbellini F., Rinck S., Bock E., Kemkemer R., Kessler H., Ding J., Spatz J.P. Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 2009;9(3):1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X., Yan C., Ye K., He Y., Li Z., Ding J. Effect of RGD nanospacing on differentiation of stem cells. Biomaterials. 2013;34(12):2865–2874. doi: 10.1016/j.biomaterials.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 111.Zhao C., Song X., Lu X. Directional osteo-differentiation effect of hADSCs on nanotopographical self-assembled polystyrene nanopit surfaces. Int. J. Nanomed. 2020;15:3281. doi: 10.2147/IJN.S240300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petecchia L., Usai C., Vassalli M., Gavazzo P. Biophysical characterization of nanostructured TiO2 as a good substrate for hBM-MSC adhesion, growth and differentiation. Exp. Cell Res. 2017;358(2):111–119. doi: 10.1016/j.yexcr.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 113.Carbone R., Giorgetti L., Zanardi A., Marangi I., Chierici E., Bongiorno G., Fiorentini F., Faretta M., Piseri P., Pelicci P.G. Retroviral microarray-based platform on nanostructured TiO2 for functional genomics and drug discovery. Biomaterials. 2007;28(13):2244–2253. doi: 10.1016/j.biomaterials.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 114.Antonini S., Montali M., Jacchetti E., Meucci S., Parchi P.D., Barachini S., Panvini F.M., Pacini S., Petrini I., Cecchini M. Nanotopography induced human bone marrow mesangiogenic progenitor cells (MPCs) to mesenchymal stromal cells (MSCs) transition. Front. Cell Dev. Biol. 2016;4:144. doi: 10.3389/fcell.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qian W., Gong L., Cui X., Zhang Z., Bajpai A., Liu C., Castillo A.B., Teo J.C., Chen W. Nanotopographic regulation of human mesenchymal stem cell osteogenesis. ACS Appl. Mater. Interfaces. 2017;9(48):41794–41806. doi: 10.1021/acsami.7b16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee M.S., Lee D.H., Jeon J., Oh S.H., Yang H.S. Topographically defined, biodegradable nanopatterned patches to regulate cell fate and acceleration of bone regeneration. ACS Appl. Mater. Interfaces. 2018;10(45):38780–38790. doi: 10.1021/acsami.8b14745. [DOI] [PubMed] [Google Scholar]
- 117.Izadpanahi M., Seyedjafari E., Arefian E., Hamta A., Hosseinzadeh S., Kehtari M., Soleimani M. Nanotopographical cues of electrospun PLLA efficiently modulate non-coding RNA network to osteogenic differentiation of mesenchymal stem cells during BMP signaling pathway. Mater. Sci. Eng. C. 2018;93:686–703. doi: 10.1016/j.msec.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 118.Oh S., Daraio C., Chen L.H., Pisanic T.R., Finones R.R., Jin S. Significantly accelerated osteoblast cell growth on aligned TiO2 nanotubes. J. Biomed. Mater. Res. Part A: An Official Journal of The Society for Biomaterials, The Japanese Society for Biomaterials, and The Australian Society for Biomaterials and the Korean Society for Biomaterials. 2006;78(1):97–103. doi: 10.1002/jbm.a.30722. [DOI] [PubMed] [Google Scholar]
- 119.Sjöström T., McNamara L.E., Meek R.D., Dalby M.J., Su B. 2D and 3D nanopatterning of titanium for enhancing osteoinduction of stem cells at implant surfaces. Adv. Healthcare Mater. 2013;2(9):1285–1293. doi: 10.1002/adhm.201200353. [DOI] [PubMed] [Google Scholar]
- 120.Sjöström T., Dalby M.J., Hart A., Tare R., Oreffo R.O., Su B. Fabrication of pillar-like titania nanostructures on titanium and their interactions with human skeletal stem cells. Acta Biomater. 2009;5(5):1433–1441. doi: 10.1016/j.actbio.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 121.Xiao Q.R., Zhang N., Wang X., Man X.Y., Yang K., Lü L.X., Huang N.P. Oriented surface nanotopography promotes the osteogenesis of mesenchymal stem cells. Adv. Mater. Interfac. 2017;4(3) [Google Scholar]
- 122.Metavarayuth K., Maturavongsadit P., Chen X., Sitasuwan P., Lu L., Su J., Wang Q. Nanotopographical cues mediate osteogenesis of stem cells on virus substrates through BMP-2 intermediate. Nano Lett. 2019;19(12):8372–8380. doi: 10.1021/acs.nanolett.9b02001. [DOI] [PubMed] [Google Scholar]
- 123.Cavalcanti-Adam E.A., Volberg T., Micoulet A., Kessler H., Geiger B., Spatz J.P. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92(8):2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li S., Wang X., Cao B., Ye K., Li Z., Ding J. Effects of nanoscale spatial arrangement of arginine–glycine–aspartate peptides on dedifferentiation of chondrocytes. Nano Lett. 2015;15(11):7755–7765. doi: 10.1021/acs.nanolett.5b04043. [DOI] [PubMed] [Google Scholar]
- 125.Wang X., Li S., Yan C., Liu P., Ding J. Fabrication of RGD micro/nanopattern and corresponding study of stem cell differentiation. Biomaterials. 2015;15(3):1457–1467. doi: 10.1021/nl5049862. [DOI] [PubMed] [Google Scholar]
- 126.Xiong J.-P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 127.Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L.C., Yaffe M.B. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19(24):6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z.C., Guan K.L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vassilev A., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15(10):1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu S., Liu Y., Zheng Y., Dong J., Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14(3):388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 131.Piccolo S., Dupont S., Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 2014;94(4):1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 132.Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaalouk D.E., Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10(1):63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mammoto A., Ingber D.E. Cytoskeletal control of growth and cell fate switching. Curr. Opin. Cell Biol. 2009;21(6):864–870. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 135.Wozniak M.A., Chen C.S. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10(1):34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heng B.C., Zhang X., Aubel D., Bai Y., Li X., Wei Y., Fussenegger M., Deng X. Role of YAP/TAZ in cell lineage fate determination and related signaling pathways. Front. Cell Dev. Biol. 2020;8:735. doi: 10.3389/fcell.2020.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang W., Han W., He W., Li J., Wang J., Feng H., Qian Y. Surface topography of hydroxyapatite promotes osteogenic differentiation of human bone marrow mesenchymal stem cells. Mater Sci Eng C Mater Biol Appl. 2016;60:45–53. doi: 10.1016/j.msec.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Y., Gong H., Sun Y., Huang Y., Fan Y. Enhanced osteogenic differentiation of MC3T3-E1 cells on grid-topographic surface and evidence for involvement of YAP mediator. J. Biomed. Mater. Res. 2016;104(5):1143–1152. doi: 10.1002/jbm.a.35648. [DOI] [PubMed] [Google Scholar]
- 139.Hwang J.H., Lee D.H., Byun M.R., Kim A.R., Kim K.M., Park J.I., Oh H.T., Hwang E.S., Lee K.B., Hong J.H. Nanotopological plate stimulates osteogenic differentiation through TAZ activation. Sci. Rep. 2017;7(1):3632. doi: 10.1038/s41598-017-03815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qian W., Gong L., Cui X., Zhang Z., Bajpai A., Liu C., Castillo A.B., Teo J.C.M., Chen W. Nanotopographic regulation of human mesenchymal stem cell osteogenesis. ACS Appl. Mater. Interfaces. 2017;9(48):41794–41806. doi: 10.1021/acsami.7b16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Arslan E., Hatip Koc M., Uysal O., Dikecoglu B., Topal A.E., Garifullin R., Ozkan A.D., Dana A., Hermida-Merino D., Castelletto V., Edwards-Gayle C., Baday S., Hamley I., Tekinay A.B., Guler M.O. Supramolecular peptide nanofiber morphology affects mechanotransduction of stem cells. Biomacromolecules. 2017;18(10):3114–3130. doi: 10.1021/acs.biomac.7b00773. [DOI] [PubMed] [Google Scholar]
- 142.Wang X., Hu X., Dulinska-Molak I., Kawazoe N., Yang Y., Chen G. Discriminating the independent influence of cell adhesion and spreading area on stem cell fate determination using micropatterned surfaces. Sci. Rep. 2016;6 doi: 10.1038/srep28708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li L., Yang S., Xu L., Li Y., Fu Y., Zhang H., Song J. Nanotopography on titanium promotes osteogenesis via autophagy-mediated signaling between YAP and beta-catenin. Acta Biomater. 2019;96:674–685. doi: 10.1016/j.actbio.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 144.Tonazzini I., Masciullo C., Savi E., Sonato A., Romanato F., Cecchini M. Neuronal contact guidance and YAP signaling on ultra-small nanogratings. Sci. Rep. 2020;10(1):3742. doi: 10.1038/s41598-020-60745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen W., Han S., Qian W., Weng S., Yang H., Sun Y., Villa-Diaz L.G., Krebsbach P.H., Fu J. Nanotopography regulates motor neuron differentiation of human pluripotent stem cells. Nanoscale. 2018;10(7):3556–3565. doi: 10.1039/c7nr05430k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ingber D.E. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 2003;116(7):1157–1173. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 147.Tsimbouri P.M., McMurray R.J., Burgess K.V., Alakpa E.V., Reynolds P.M., Murawski K., Kingham E., Oreffo R.O., Gadegaard N., Dalby M.J. Using nanotopography and metabolomics to identify biochemical effectors of multipotency. ACS Nano. 2012;6(11):10239–10249. doi: 10.1021/nn304046m. [DOI] [PubMed] [Google Scholar]
- 148.Geiger B., Spatz J., Bershadsky A. Environmental sensing by cells through focal adhesions. Nat. Rev. Mol. Cell Biol. 2008;23 doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 149.Kechagia J.Z., Ivaska J., Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 2019:1. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- 150.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 151.Emsley J., Knight C.G., Farndale R.W., Barnes M.J., Liddington R.C. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101(1):47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 152.Shattil S.J., Kim C., Ginsberg M.H. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 2010;11(4):288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sun Z., Costell M., Fässler R. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 2019;21(1):25–31. doi: 10.1038/s41556-018-0234-9. [DOI] [PubMed] [Google Scholar]
- 154.Kong F., García A.J., Mould A.P., Humphries M.J., Zhu C. Demonstration of catch bonds between an integrin and its ligand. JCB (J. Cell Biol.) 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen Y., Lee H., Tong H., Schwartz M., Zhu C. Force regulated conformational change of integrin αVβ3. Matrix Biol. 2017;60:70–85. doi: 10.1016/j.matbio.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Elosegui-Artola A., Oria R., Chen Y., Kosmalska A., Pérez-González C., Castro N., Zhu C., Trepat X., Roca-Cusachs P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 2016;18(5):540–548. doi: 10.1038/ncb3336. [DOI] [PubMed] [Google Scholar]
- 157.Elosegui-Artola A., Trepat X., Roca-Cusachs P. Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 2018;28(5):356–367. doi: 10.1016/j.tcb.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 158.Horton E.R., Astudillo P., Humphries M.J., Humphries J.D. Mechanosensitivity of integrin adhesion complexes: role of the consensus adhesome. Exp. Cell Res. 2016;343(1):7–13. doi: 10.1016/j.yexcr.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 159.Horton E.R., Byron A., Askari J.A., Ng D.H., Millon-Frémillon A., Robertson J., Koper E.J., Paul N.R., Warwood S., Knight D. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 2015;17(12):1577–1587. doi: 10.1038/ncb3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zaidel-Bar R., Geiger B. The switchable integrin adhesome. J. Cell Sci. 2010;123(9):1385–1388. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., Waterman C.M. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu J., Wang Y., Goh W.I., Goh H., Baird M.A., Ruehland S., Teo S., Bate N., Critchley D.R., Davidson M.W. Talin determines the nanoscale architecture of focal adhesions. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112(35):E4864–E4873. doi: 10.1073/pnas.1512025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vicente-Manzanares M., Horwitz A.R. Adhesion dynamics at a glance. J. Cell Sci. 2011;124(23):3923–3927. doi: 10.1242/jcs.095653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116(22):4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 165.Biggs M.J., Richards R.G., Gadegaard N., Wilkinson C.D., Oreffo R.O., Dalby M.J. The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells. Biomaterials. 2009;30(28):5094–5103. doi: 10.1016/j.biomaterials.2009.05.049. [DOI] [PubMed] [Google Scholar]
- 166.Pankov R., Cukierman E., Katz B.-Z., Matsumoto K., Lin D.C., Lin S., Hahn C., Yamada K.M. Integrin dynamics and matrix assembly tensin-dependent translocation of α5β1 integrins promotes early fibronectin fibrillogenesis. JCB (J. Cell Biol.) 2000;148(5):1075–1090. doi: 10.1083/jcb.148.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Galbraith C.G., Yamada K.M., Sheetz M.P. The relationship between force and focal complex development. J. Cell Biol. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Elosegui-Artola A., Bazellières E., Allen M.D., Andreu I., Oria R., Sunyer R., Gomm J.J., Marshall J.F., Jones J.L., Trepat X. Rigidity sensing and adaptation through regulation of integrin types. Nat. Mater. 2014;13(6):631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brockman J.M., Blanchard A.T., Pui-Yan V., Derricotte W.D., Zhang Y., Fay M.E., Lam W.A., Evangelista F.A., Mattheyses A.L., Salaita K. Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods. 2018;15(2):115–118. doi: 10.1038/nmeth.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Swaminathan V., Kalappurakkal J.M., Mehta S.B., Nordenfelt P., Moore T.I., Koga N., Baker D.A., Oldenbourg R., Tani T., Mayor S. Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114(40):10648–10653. doi: 10.1073/pnas.1701136114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nordenfelt P., Moore T.I., Mehta S.B., Kalappurakkal J.M., Swaminathan V., Koga N., Lambert T.J., Baker D., Waters J.C., Oldenbourg R. Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 2017;8(1):1–16. doi: 10.1038/s41467-017-01848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pasapera A.M., Schneider I.C., Rericha E., Schlaepfer D.D., Waterman C.M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. JCB (J. Cell Biol.) 2010;188(6):877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lee R.T., Lee Y.C. Affinity enhancement by multivalent lectin–carbohydrate interaction. Glycoconj. J. 2000;17(7–9):543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 174.Guilluy C., Swaminathan V., Garcia-Mata R., O'Brien E.T., Superfine R., Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 2011;13(6):722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lai C.-F., Chaudhary L., Fausto A., Halstead L.R., Ory D.S., Avioli L.V., Cheng S.-L. Erk is essential for growth, differentiation, integrin expression, and cell function in human osteoblastic cells. J. Biol. Chem. 2001;276(17):14443–14450. doi: 10.1074/jbc.M010021200. [DOI] [PubMed] [Google Scholar]
- 176.Kirby T.J., Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 2018;20(4):373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang X., Jiang G., Cai Y., Monkley S.J., Critchley D.R., Sheetz M.P. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 2008;10(9):1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cantini M., Donnelly H., Dalby M.J., Salmeron‐Sanchez M. The plot thickens: the emerging role of matrix viscosity in cell mechanotransduction. Adv. Healthcare Mater. 2019 doi: 10.1002/adhm.201901259. [DOI] [PubMed] [Google Scholar]
- 179.Likodimos V., Stergiopoulos T., Falaras P., Kunze J., Schmuki P. Phase composition, size, orientation, and antenna effects of self-assembled anodized titania nanotube arrays: a polarized micro-Raman investigation. J. Phys. Chem. C. 2008;112(33):12687–12696. [Google Scholar]
- 180.Yao M., Goult B.T., Chen H., Cong P., Sheetz M.P., Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 2014;4(1):1–7. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Swaminathan V., Waterman C.M. The molecular clutch model for mechanotransduction evolves. Nat. Cell Biol. 2016;18(5):459–461. doi: 10.1038/ncb3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang N., Naruse K., Stamenović D., Fredberg J.J., Mijailovich S.M., Tolić-Nørrelykke I.M., Polte T., Mannix R., Ingber D.E. Proc. Natl. Acad. Sci. Cell. 2001;98(14):7765–7770. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ingber D.E. The riddle of morphogenesis: a question of solution chemistry or molecular cell engineering? Cell. 1993;75(7):1249–1252. doi: 10.1016/0092-8674(93)90612-t. [DOI] [PubMed] [Google Scholar]
- 184.Maniotis A.J., Chen C.S., Ingber D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. Unit. States Am. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hu S., Chen J., Butler J.P., Wang N. Prestress mediates force propagation into the nucleus. Biochem. Biophys. Res. Commun. 2005;329(2):423–428. doi: 10.1016/j.bbrc.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 186.De R., Zemel A., Safran S.A. Dynamics of cell orientation. Nat. Phys. 2007;3(9):655–659. [Google Scholar]
- 187.Wang J.H.-C. Substrate deformation determines actin cytoskeleton reorganization: a mathematical modeling and experimental study. J. Theor. Biol. 2000;202(1):33–41. doi: 10.1006/jtbi.1999.1035. [DOI] [PubMed] [Google Scholar]
- 188.Kong D., Ji B., Dai L. Stability of adhesion clusters and cell reorientation under lateral cyclic tension. Biophys. J. 2008;95(8):4034–4044. doi: 10.1529/biophysj.108.131342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xu G.-K., Li B., Feng X.-Q., Gao H. A tensegrity model of cell reorientation on cyclically stretched substrates. Biophys. J. 2016;111(7):1478–1486. doi: 10.1016/j.bpj.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mendonça G., Mendonça D.B., Aragao F.J., Cooper L.F. Advancing dental implant surface technology–from micron-to nanotopography. Biomaterials. 2008;29(28):3822–3835. doi: 10.1016/j.biomaterials.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 191.Ngandu Mpoyi E., Cantini M., Reynolds P.M., Gadegaard N., Dalby M.J., Salmerón-Sánchez M. Protein adsorption as a key mediator in the nanotopographical control of cell behavior. ACS Nano. 2016;10(7):6638–6647. doi: 10.1021/acsnano.6b01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lee L.C., Gadegaard N., De Andrés M.C., Turner L.-A., Burgess K.V., Yarwood S.J., Wells J., Salmeron-Sanchez M., Meek D., Oreffo R.O. Nanotopography controls cell cycle changes involved with skeletal stem cell self-renewal and multipotency. Biomaterials. 2017;116:10–20. doi: 10.1016/j.biomaterials.2016.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mas‐Moruno C., Su B., Dalby M.J. Multifunctional coatings and nanotopographies: toward cell instructive and antibacterial implants. Adv. Healthcare Mater. 2019;8(1) doi: 10.1002/adhm.201801103. [DOI] [PubMed] [Google Scholar]
- 194.Teughels W., Van Assche N., Sliepen I., Quirynen M. Effect of material characteristics and/or surface topography on biofilm development. Clin. Oral Implants Res. 2006;17(S2):68–81. doi: 10.1111/j.1600-0501.2006.01353.x. [DOI] [PubMed] [Google Scholar]
- 195.Badihi Hauslich L., Sela M.N., Steinberg D., Rosen G., Kohavi D. The adhesion of oral bacteria to modified titanium surfaces: role of plasma proteins and electrostatic forces. Clin. Oral Implants Res. 2013;24:49–56. doi: 10.1111/j.1600-0501.2011.02364.x. [DOI] [PubMed] [Google Scholar]
- 196.de Fátima Balderrama Í., Cardoso M.V., Stuani V.T., Oliveira R.C., Matos A.A., Greghi S.L.A., Sant'Ana A.C.P. Residual decontamination chemical agents negatively affect adhesion and proliferation of osteoblast-like cells on implant surface. Int. J. Implant Dent. 2020;6(1):1–9. doi: 10.1186/s40729-020-00278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.de Fátima Balderrama Í., de Toledo Stuani V., Cardoso M.V., Oliveira R.C., Lopes M.M.R., Greghi S.L.A. The influence of implant surface roughness on decontamination by antimicrobial photodynamic therapy and chemical agents: a preliminary study in vitro. Photodiagnosis Photodyn. Ther. 2021;33 doi: 10.1016/j.pdpdt.2020.102105. [DOI] [PubMed] [Google Scholar]